Abstract
Modularity is known to be one of the most relevant characteristics of biological systems and appears to be present at multiple scales. Given its adaptive potential, it is often assumed to be the target of selective pressures. Under such interpretation, selection would be actively favouring the formation of modular structures, which would specialize in different functions. Here we show that, within the context of cellular networks, no such selection pressure is needed to obtain modularity. Instead, the intrinsic dynamics of network growth by duplication and diversification is able to generate it for free and explain the statistical features exhibited by small subgraphs. The implications for the evolution and evolvability of both biological and technological systems are discussed.
Keywords: complex networks, modularity, evolvability, tinkering, network biology
1. Introduction
Biological and technological systems both exhibit a common pattern of modular organization. A modular system is formed by quasi-independent parts that not only are tightly integrated within themselves but also exhibit a certain degree of interdependency among themselves (Schlosser & Wagner 2004). Modularity is considered to be a prerequisite for the adaptation of complex organisms and their evolvability (Raff 1996; Gerhart & Kirschner 1997; Calabretta et al. 2000).
Modularity is particularly obvious in cellular networks (Ravasz et al. 2002), where it can be detected at the topological level. These networks include the webs of interactions among proteins, genes, enzymes and metabolites or signalling molecules. It has been argued that modularity is likely to have been selectively favoured by evolution. In that case, explaining its origins would require a functional view of biological networks (Hartwell et al. 1999).
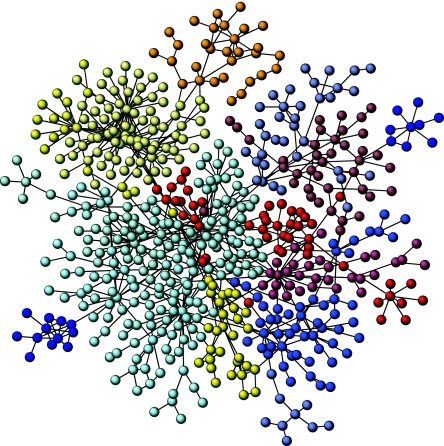
Within the context of network theory (Bornholdt & Schuster 2003; Dorogovtsev & Mendes 2003; Boccaletti et al. 2006; Koonin et al. 2006), the given system is represented as a graph Ω=(V, E) composed of a set of N nodes (say proteins), V={vi}, and a set of links, eij∈E, indicating if a connection exists between nodes vi and vj. An example is shown in figure 1: the human protein interaction network. Here we can see a few proteins having a large number of links (the hubs) surrounded by many proteins having just a few connections. This type of heterogeneous networks is very common and is characterized by a probability distribution P(k) of having a node with k links, which falls off as a power law with a cut-off, i.e.
| (1.1) |
where k0 is a constant and 2<γ<3 denotes the scaling exponent (typically close to γ∼2.5). The cut-off kc is a characteristic degree indicating the presence of a maximum number of links. The hubs tend to have important roles (Albert et al. 2000), particularly when looking at regulatory elements such as transcription factors (Rodriguez-Caso et al. 2005), where the most connected nodes are often proto-oncogenes or tumour suppressor genes and their failure typically involves some proliferative disorder.
Figure 1.
A modular network is illustrated by means of the human proteome (data obtained from the DIP database: http://dip-doe-mbi.ucla.edu). Nodes are proteins and links indicate their physical (protein–protein) interaction. A standard algorithm for identifying topological modules has been used (see text), and it effectively detects several well-defined groups of tightly related proteins. Modules are indicated by different colours.
At its smallest scale, modules are defined by means of subgraphs involving three or four elements (Wolf & Arkin 2003). These subgraphs have received considerable attention in relation to the so-called network motifs (Milo et al. 2002, 2004). Roughly speaking, motifs are patterns of interconnections occurring in complex networks at numbers that are significantly higher than those in randomized networks. The analysis of their statistical distribution reveals that each class of natural and artificial networks seems to display a common pattern of motif abundances. The statistical pattern is thus interpreted as functionally meaningful. Under this view, motif abundances—as well as modularity—would be a consequence of selective forces. Is that the case? Recently, a model for the evolution of modularity and network motifs has been suggested, based on a genetic programming approach (Lipson et al. 2002; Kashtan & Alon 2005). The model evolves electronic circuits under an environment that changes itself in a modular manner. However, the view of network substructures as resulting from pure selection or optimization has been questioned in a number of studies (Guimerà et al. 2004; Solé et al. 2002a; Banzhaf & Kuo 2004; Mazurie et al. 2005; Rice et al. 2005; Valverde & Solé 2005; Kuo et al. 2006; Solé & Valverde 2006), suggesting that the abundance of motifs does not necessarily reflect functional advantages.
In this paper we show that an alternative explanation for modularity exists, associated with the inevitable constraints imposed by the rules driving the growth of cellular networks. Specifically, since biological entities typically evolve by tinkering (Jacob 1976; Solé et al. 2002a), widely reusing, combining and reconnecting available parts, some patterns of network organization will be essentially inevitable. As a consequence, both subgraph patterns and modular features would be largely a by-product of the network generative rules.
2. Growing networks by duplication
The approach taken here is inspired by a physics view of biological complexity (Albert & Barabási 2002), namely searching for generic mechanisms responsible for global patterns. The question being addressed here is how much of the modular organization of complex networks might result from just the rules of network growth by tinkering (Wagner 2003). The view here is thus topological, with no direct link to functional traits. In this context, we will restrict ourselves to a graph-theoretical description of protein–protein interactions, as previously followed by several authors (Wuchty 2001; Kim et al. 2002; Solé et al. 2002b; Pastor-Satorras et al. 2003; Vazquez et al. 2003; Colizza et al. 2005; Goh et al. 2005; Ispolatov et al. 2005; Foster et al. 2006). These models involve some type of duplication–divergence (DD) growth dynamics. This approach considers single-gene duplication events as the leading mechanism of genome growth (Ohno 1970). This is of course an approximation to the real complexities associated with genome growth dynamics. Although single-gene duplication is considered to be the driving force behind the evolution of complex organisms (Ohno 1970), several scales of duplication need to be considered, including whole-genome duplications (Maere et al. 2005).
After gene duplication has taken place, rapid divergence occurs and many redundant genes become silenced (i.e. become pseudogenes). Changes in wiring are associated with the emergence of novelty and new functionalities (Patthy 1999). In our work presented here we consider only such simple approximation based on single-gene events. We will use one of the simplest DD models of protein network evolution (Vazquez et al. 2003), which involves the following set of rules, to be applied a given number of times, until N nodes are present. Assuming that we have a graph of size n, we iterate the following rules.
Duplication. Choose a node vi∈V at random and duplicate it, thus generating a new node vn+1.
Link deletion. The new node shares a set of neighbouring nodes {vj} with its predecessor. For each common pair of common links, i.e. ei,j and en+1,j, we choose one of them and delete it with probability δ. This rule thus removes (probabilistically) redundant relations among proteins.
Link addition. A link is added among nodes vi and vn+1 with probability α. This is a small number and allows new functionalities to emerge by linking the twin proteins.
This model has been solved analytically and it has been shown that it exhibits a phase transition at a given deletion rate. This can be shown by constructing a dynamical equation for the average degree Kn of the simulated protein network after n nodes have been introduced. It can be shown that Kn evolves following the discrete system (Vazquez et al. 2003)
| (2.1) |
where we can see that the number of proteins n is also a time scale. Using the continuous approximation Kn+1−Kn∼dKn/dn and assuming that n is large, we have a differential equation
| (2.2) |
By solving it, we obtain the time evolution of the average degree
| (2.3) |
It is easy to check that a steady degree K* is achieved for δ>δc=1/2, namely
| (2.4) |
whereas for δ<δc, link removal is too slow and the average connectivity is high. There is thus a critical deletion rate δc=1/2 separating a strongly connected proteome from a sparse one (which would also include many small subgraphs). Since real protein maps are known to be rather sparse (with average connectivities around 〈k〉∼3–5), we should expect to find appropriate removal rates at values δ>δc. Actually, it has been shown (Vazquez et al. 2003) that for α=0.1 and δ=0.7, the model is consistent with several properties observed in the yeast proteome (such as scale-free topology, small-world behaviour, graph correlations and robustness against node deletion). By using appropriate measures, we will show that both modules and non-random distributions of subgraphs are an expected by-product of network growth by duplication.
3. Modularity from tinkering
3.1 Modules
We will first analyse the emergence of modular patterns in the previously described model. In order to provide a quantitative measure, we will use a specific algorithm1 of community detection (Clauset et al. 2004; see also Newman 2006; Reichardt & Bornholdt 2006). The method considers a decomposition of the graph Ω (figure 2) into a set Γμ of subgraphs Ci∈Γμ defining a partition . Obviously, many possible Cr-partitions are possible. Using the adjacency matrix of the graph, A=(aij), and assuming a given partition, the fraction of edges that fall within subsets of will be given by
| (3.1) |
where δ(a, b)=1 if a=b and zero otherwise. Using , we can also write
| (3.2) |
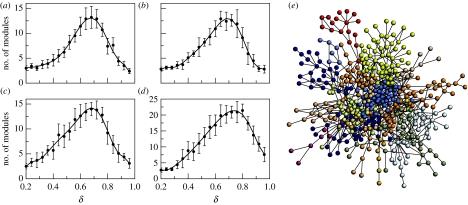
Figure 2.
Modularity in tinkered model networks. Here we show (a–d) the average number of modules (μ) of networks generated using the DD model. Here μ is computed on the largest connected component for each δ and averaged over 50 replicas. We used four different α values ((a) α=0.005, (b) α=0.01, (c) α=0.10 and (d) α=0.50) and a maximal network size N=1000. A well-defined maximum is observed at δ*≈0.7 in all cases. The network shown in (e) is an example of the modular graphs obtained at δ*.
In order to define an appropriate modularity index, the previous measure needs to be compared with the expectation from a randomly wired graph with identical number of nodes and links. Let us indicate as ki the degree of vi, which is obtained from the adjacency matrix as .
The expected probability of having a link connecting two arbitrary nodes vi and vj will be simply kikj/2m, and thus we can defined modularity Q in terms of the average difference between the observed and the expected value of f, namely
| (3.3) |
which is properly normalized between 0 (random network) and 1 (a single module is present).
The modularity of a network will be defined as the maximum as evaluated by the search algorithm (Clauset et al. 2004). The size of the best partition μ defines the (potential) number of modules. Here we apply this measure to the largest connected component of the network generated by the algorithm described in §2. For each δ, and a fixed α value, we generate 50 simulated networks, each one starting from a small graph of four fully connected elements and ending once a graph with N=103 nodes is obtained. Four different values of α have been used. The results are shown in figure 2, where a one-hump curve μ(δ) is obtained in all four cases. A maximum is reached for δ*≈0.7, which is actually the deletion rate that gave best-fit statistics compared with yeast proteome data (Vazquez et al. 2003).
The origins of the maximum can be understood in terms of two basic conflicting components associated with the growth rules. At low δ values, the system is highly connected, being all node members of the giant component and having a large degree. As a consequence, we should not expect to observe a large number of modules. On the other hand, as δ increases, the network becomes more and more sparse and for δ>δc, it starts to get fragmented. The largest component in this domain will be formed by highly heterogeneous groups of loosely linked subgraphs. Close to the transition δc, many elements belong to the largest connected component, but the number of modules is not large since they share many links. With increasing δ, it is more likely to find groups of connected nodes that still share few links. But further increasing δ implies breaking of Ω into many small subgraphs, which will display a low modularity.
3.2 Subgraph census
A second level of analysis can be performed by considering the frequency of subgraphs of a given size, also known as subgraph census (see Wasserman & Faust 1994 and references therein). Here we have studied the census of n=4 subgraphs, since it provides a reasonable number of different structures. More importantly, most examples whose functional relevance has been described in detail fall within this class (Milo et al. 2002, 2004; Klemm & Bornholdt 2005; Valverde & Solé 2005).
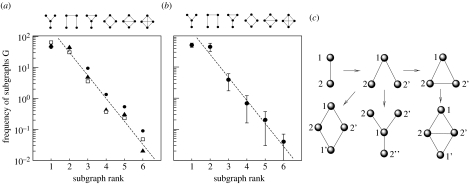
The results are shown in figure 3a,b, where we can compare the observed patterns of subgraph abundances in real and simulated networks, respectively. For the real datasets, we have used the human interactome, the subset of transcription factors and the yeast proteome. The plots display the per cent of subgraphs found in each network against the subgraph rank r. After a plateau, the frequency of subgraphs rapidly decays as an exponential function, i.e.
| (3.4) |
with η≈1.8. Such pattern is also found in the distribution of subgraphs obtained from the DD model. Using the δ* value that gives the maximum number of modules, we also obtain an exponential decay, with ηDD=1.71±0.20, consistently with the real datasets.
Figure 3.
(a) Subgraph census for three different protein networks: human interactome (HI, filled triangles); yeast proteome (YP, filled circles); and the subset of human transcription factors (HTF, open squares). The exponential fit for r≥2 gives ηHI=1.81±0.20, ηYP=1.79±0.21 and ηHTF=1.58±0.21. In (b), the corresponding graph census is shown from runs of the DD model, averaged over 50 replicas. The exponent here is ηDD=1.71±0.20. The data for the protein networks were obtained from the DIP database: http://dip-doe-mbi.ucla.edu and from Rodriguez-Caso et al. (2005). In (c), some examples of the transitions between different subgraphs (using DD rules) are shown.
The common pattern shared by both real and simulated graphs is consistent with a rule-driven mechanism of network evolution. In this context, it is interesting to see that the different subgraphs are easily connected through DD events (figure 3c).
4. Discussion
What drives the emergence of modularity in evolution? Is it a function-driven mechanism or instead the by-product of more fundamental, dynamical rules, as it has been suggested in other contexts (Kauffman 1993)? Although it is true that modularity is an essential feature of biological structures, our analysis suggests that it might be a by-product of the multiplicative nature of duplication–rewiring mechanisms. Such a tinkering process (Jacob 1976; Solé et al. 2002a,b) inevitably leads to fluctuations in network structure due to its multiplicative nature: the rich gets richer and the graph will be organized around hubs. Moreover, the local amplification of subgraph abundances obtained from the DD process is also responsible for the decay observed in N(r).
The presence of an optimal level of modularity at a given δ value is an important result of our study with potential implications for evolution by selection, at least at some levels. Selection might have been present at the level of link deletion. By removing the right amount of links, a large connected graph can be obtained, which will be both heterogeneous (and thus robust against random node deletion) and modular. In this context, although our results do not rule out the role of selection and functional adaptation in explaining modularity, they suggest that strong constraints are imposed by the rules of network growth. Thus, topological patterns (including heterogeneity and modular organization) would be an emergent property of evolutionary tinkering. Evolvability might have strongly benefited from such features, since heterogeneity and modularity immediately favour robustness and specialization, respectively. Further work should explore how these results can be extended to a more detailed level of description of network evolution.
Acknowledgments
We thank the members of the Complex Systems Lab for their useful discussions. This work has been supported by a grant MCyT FIS2004-05422, the European Union within the Sixth Framework Programme under contracts FP6-001907 (DELIS), the James S. McDonnell Foundation and Santa Fe Institute.
Footnotes
All these approaches are heuristics. It has been shown that modularity maximization is an NP-complete problem (Brandes et al. 2006) and thus there is in general no optimal partition.
References
- Albert R, Barabási A.-L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002;74:47–97. doi: 10.1103/RevModPhys.74.47. [DOI] [Google Scholar]
- Albert R, Jeong H, Barabási A.-L. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- Banzhaf W, Kuo P.D. Network motifs in natural and artificial transcriptional regulatory networks. J. Biol. Phys. Chem. 2004;4:85–92. [Google Scholar]
- Boccaletti S, Latora V, Moreno Y, Chavez M, Hwang D.U. Complex networks: structure and dynamics. Phys. Rep. 2006;424:175–308. doi: 10.1016/j.physrep.2005.10.009. [DOI] [Google Scholar]
- Bornholdt S, Schuster H.G. Wiley; Berlin, Germany: 2003. Handbook of graphs and networks. [Google Scholar]
- Brandes, U., Delling, D., Gaertler, M., Goerke, R., Hoefer, M., Nikoloski, Z. & Wagner, D. 2006 Maximizing modularity is hard. arXiv:physics/0608255.
- Calabretta R, Nolfi S, Parisi D, Wagner G.P. Duplication of modules facilitates the evolution of functional specialization. Artif. Life. 2000;6:69–84. doi: 10.1162/106454600568320. [DOI] [PubMed] [Google Scholar]
- Clauset A, Newman M.E.J, Moore C. Finding community structure in very large networks. Phys. Rev. E. 2004;70:066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- Colizza V, Flammini A, Maritan A, Vespignani A. Characterization and modeling of protein–protein interaction networks. Physica A. 2005;352:1–27. doi: 10.1016/j.physa.2004.12.030. [DOI] [Google Scholar]
- Dorogovtsev S.N, Mendes J.F.F. Oxford University Press; New York, NY: 2003. Evolution of networks: from biological nets to the Internet and WWW. [Google Scholar]
- Foster D.V, Kauffman S.A, Socolar E.S. Network growth models and genetic regulatory networks. Phys. Rev. E. 2006;73:031912. doi: 10.1103/PhysRevE.73.031912. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Kirschner M. Blackwell Science; Malden, MA: 1997. Cells, embryos, and evolution. [Google Scholar]
- Goh K.-I, Kahng B, Kim D. Evolution of the protein interaction network of budding yeast: role of the protein family compatibility constraint. J. Korean Phys. Soc. 2005;46:551–555. [Google Scholar]
- Guimerà R, Sales-Pardo M, Amaral L. Modularity from fluctuations in random graphs and complex networks. Phys. Rev. E. 2004;66:056120. doi: 10.1103/PhysRevE.66.026704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H, Hopfield J.J, Leibler S, Murray A.W. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- Ispolatov I, Krapivsky P.L, Yuryev A. Duplication-divergence model of protein interaction network. Phys. Rev. E. 2005;71:061911. doi: 10.1103/PhysRevE.71.061911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F. Evolution as tinkering. Science. 1976;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- Kashtan N, Alon U. Spontaneous evolution of modularity and network motifs. Proc. Natl Acad. Sci. USA. 2005;102:13 773–13 778. doi: 10.1073/pnas.0503610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman S.A. Oxford University Press; New York, NY: 1993. The origins of order. [Google Scholar]
- Kim J, Krapivsky P.L, Kahng B, Redner S. Infinite-order percolation and giant fluctuations in a protein interaction network. Phys. Rev. E. 2002;66:055101. doi: 10.1103/PhysRevE.66.055101. [DOI] [PubMed] [Google Scholar]
- Klemm K, Bornholdt S. Topology of biological networks and reliability of information processing. Proc. Natl Acad. Sci. USA. 2005;102:18 414–18 419. doi: 10.1073/pnas.0509132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V, Wolf Y.I, Karev G.P, editors. Power laws, scale-free networks and genome biology. Springer; New York, NY: 2006. [Google Scholar]
- Kuo P.D, Banzhaf W, Leier A. Network topology and the evolution of dynamics in an artificial regulatory network model created by whole genome duplication and divergence. Biosystems. 2006;85:177–200. doi: 10.1016/j.biosystems.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Lipson H, Pollack J.B, Suh N.P. On the origin of modular variation. Evolution. 2002;56:1549–1556. doi: 10.1111/j.0014-3820.2002.tb01466.x. [DOI] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl Acad. Sci. USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurie A, Bottani S, Vergassola M. An evolutionary and functional assessment of regulatory network motifs. Genome Biol. 2005;6:R35. doi: 10.1186/gb-2005-6-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Milo R, Itzkovitz S, Kashtan N, Levitt R, Shen-Orr S, Ayzenshtat I, Sheffer M, Alon U. Superfamilies of designed and evolved networks. Science. 2004;303:1538–1542. doi: 10.1126/science.1089167. [DOI] [PubMed] [Google Scholar]
- Newman M.E.J. Modularity and community structure in networks. Proc. Natl Acad. Sci. USA. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Springer; Berlin, Germany: 1970. Evolution by gene duplication. [Google Scholar]
- Pastor-Satorras R, Smith E, Solé R.V. Evolving protein interaction networks from gene duplication. J. Theor. Biol. 2003;222:199–210. doi: 10.1016/S0022-5193(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Patthy L.Protein evolution1999Blackwell; Oxford, UK [Google Scholar]
- Raff R.A. Chicago University Press; Chicago, IL: 1996. The shape of life. [Google Scholar]
- Ravasz E, Somera S.L, Mongru D.A, Oltvai Z.N, Barabási A.-L. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- Reichardt J, Bornholdt S. Statistical mechanics of community detection. Phys. Rev. E. 2006;74:01640. doi: 10.1103/PhysRevE.74.016110. [DOI] [PubMed] [Google Scholar]
- Rice J.J, Kershenbaum A, Stolovitzky G. Lasting impressions: motifs in protein–protein maps may provide footprints of evolutionary events. Proc. Natl Acad. Sci. USA. 2005;102:3173–3174. doi: 10.1073/pnas.0500130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Caso C, Medina M.A, Solé R.V. Topology, tinkering and evolution of the human transcription factor network. FEBS J. 2005;272:6423–6434. doi: 10.1111/j.1742-4658.2005.05041.x. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Wagner G.P. Chicago University Press; Chicago, IL: 2004. Modularity in development and evolution. [Google Scholar]
- Solé R.V, Valverde S. Are network motifs the spandrels of cellular complexity? Trends Ecol. Evol. 2006;21:419–422. doi: 10.1016/j.tree.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Solé R.V, Ferrer i Cancho R, Montoya J.M, Valverde S. Selection, tinkering, and emergence in complex networks. Complexity. 2002a;8:20–33. doi: 10.1002/cplx.10055. [DOI] [Google Scholar]
- Solé R.V, Pastor-Satorras R, Smith E, Kepler T.S. A model of large-scale proteome evolution. Adv. Complex Syst. 2002b;5:43–54. doi: 10.1142/S021952590200047X. [DOI] [Google Scholar]
- Valverde S, Solé R.V. Network motifs in computational networks: a case study in software architecture. Phys. Rev. E. 2005;72:026107. doi: 10.1103/PhysRevE.72.026107. [DOI] [PubMed] [Google Scholar]
- Vazquez A, Flammini A, Maritan A, Vespignani A. Modeling of protein interaction networks. ComPlexUs. 2003;1:38–44. doi: 10.1159/000067642. [DOI] [Google Scholar]
- Wagner A. Does natural selection mold molecular networks? Science STKE. 2003:41–43. doi: 10.1126/stke.2003.202.pe41. [DOI] [PubMed] [Google Scholar]
- Wasserman S, Faust K. Cambridge University Press; Cambridge, UK: 1994. Social network analysis. [Google Scholar]
- Wolf D.M, Arkin A.P. Motifs, modules and games in bacteria. Curr. Opin. Microbiol. 2003;6:125–134. doi: 10.1016/S1369-5274(03)00033-X. [DOI] [PubMed] [Google Scholar]
- Wuchty S. Scale-free behavior in protein domain networks. Mol. Biol. Evol. 2001;18:1694–1702. doi: 10.1093/oxfordjournals.molbev.a003957. [DOI] [PubMed] [Google Scholar]