Abstract
Objective
To develop methods for building corpus-specific sense inventories of abbreviations occurring in clinical documents.
Design
A corpus of internal medicine admission notes was collected and instances of each clinical abbreviation in the corpus were clustered to different sense clusters. One instance from each cluster was manually annotated to generate a final list of senses. Two clustering-based methods (Expectation Maximization—EM and Farthest First—FF) and one random sampling method for sense detection were evaluated using a set of 12 clinical abbreviations.
Measurements
The clustering-based sense detection methods were evaluated using a set of clinical abbreviations that were manually sense annotated. “Sense Completeness” and “Annotation Cost” were used to measure the performance of different methods. Clustering error rates were also reported for different clustering algorithms.
Results
A clustering-based semi-automated method was developed to build corpus-specific sense inventories for abbreviations in hospital admission notes. Evaluation demonstrated that this method could largely reduce manual annotation cost and increase the completeness of sense inventories when compared with a manual annotation method using random samples.
Conclusion
The authors developed an effective clustering-based method for building corpus-specific sense inventories for abbreviations in a clinical corpus. To the best of the authors knowledge, this is the first time clustering technologies have been used to help building sense inventories of abbreviations in clinical text. The results demonstrated that the clustering-based method performed better than the manual annotation method using random samples for the task of building sense inventories of clinical abbreviations.
Introduction
Electronic medical records (EMR) are increasingly used in healthcare. Clinical computerized applications, such as decision support systems, have shown their value for improving healthcare quality, by using patient information from available clinical records. 1,2 However those types of applications require coded information, which are not always available. Natural language processing (NLP) 3–5 has received great attention because it provides an automated way to unlock information from clinical narrative text.
The first step for a medical NLP system is to accurately recognize and identify biomedical entities. A common problem for most types of clinical notes is the frequent use of abbreviations, 6,7 which are often ambiguous (one abbreviation can refer to several different meanings) 8 and specialized for a particular subdomain of medicine. In this paper, abbreviations refer to both acronyms (e.g., “dm”—“diabetes mellitus”) and shortened forms of words (e.g., “ca”—“carcinoma”). The possible senses of an abbreviation refer to the possible corresponding full terms of the abbreviation (e.g., “diabetes mellitus”). Several studies have focused on developing methods to automatically disambiguate clinical abbreviations, and some abbreviation disambiguation methods 9,10 based on supervised machine learning were reported as achieving high performance. All those disambiguation methods assume the existence of a set of predetermined senses for each abbreviation from either existing knowledge sources or expert knowledge. However, studies 11 showed that the coverage of sense inventories generated from existing knowledge sources (e.g., UMLS) were not sufficient for specific clinical corpora, such as admission notes. Therefore, it is essential to develop corpus-based methods that are able to detect specific senses for any abbreviation in a given corpus.
In this paper, we describe a semi-automated method for building sense inventories of abbreviations from a clinical corpus. Instances of an abbreviation are collected from the corpus and clustered to different sense clusters. Then, one instance from each cluster is shown to an expert who manually annotates it to get a final list of senses. To the best of our knowledge, this is the first time that machine learning techniques have been applied to a clinical corpus to build sense inventories of clinical abbreviations.
Background
The easiest way to build sense inventories of abbreviations would be to use existing knowledge sources, such as the UMLS, which contains senses for many clinical abbreviations. However, there are two issues associated with using the UMLS. One is low coverage—some abbreviations and their senses that occur in a clinical corpus are not covered by any existing knowledge sources, such as the abbreviation “2/2” (“secondary to”). A study 11 showed that sense inventories generated from the UMLS only covered about 35% of senses of abbreviations in hospital admission notes at NYPH (New York Presbyterian Hospital). The other problem is the noisiness of the senses covered by the UMLS. Since the UMLS covers a broader domain of biomedicine than the clinical domain, some senses of abbreviations never appear in a particular clinical corpus. For example, only four different senses for “pt:” “patient,” “physical therapy,” “posterior tibial,” and “prothrombin time assay,” occur in the corpus of general medicine inpatient admission notes at NYPH. However the UMLS knowledge source has more than 10 senses for “pt”, some of which are very unlikely to appear in inpatient admission notes corpus (e.g., “Platinum”).
An alternative method is to build sense inventories of abbreviations directly from a corpus. In the biomedical literature, abbreviations usually occur together with their long forms (definitions) at least once in the document, typically with the patterns of “long form (short form)” or “short form (long form),” e.g., “coronary artery bypass graft (CABG)” or “CABG (coronary artery bypass graft)” although other patterns also occur but less frequently. Various approaches have been developed to map those abbreviation-definition patterns, which were then applied to Medline abstracts to build databases 12–14 that contain abbreviations and their possible senses. However, those methods are not applicable to a clinical corpus, because abbreviations in most clinical reports, such as admission notes, usually do not occur along with their expanded long forms.
One possible method for discovering the senses is to manually annotate occurrences of abbreviations that are randomly selected from a clinical corpus. However, to achieve an acceptable degree of sense completeness, it would require a large manual effort and would not be cost-effective. Therefore, automated methods that can help reduce the manual effort will be very useful for building sense inventories. Word sense discrimination methods, 15–17 which automatically cluster senses within a text corpus, should be an effective way to help select samples. Similar sampling methods are also used for different tasks. For example, Liu 18 reported a clustering method and used it to reduce annotation cost for supervised machine learning methods for word sense disambiguation (WSD). Other sampling methods, such as active learning, 19 have been developed to reduce the annotation cost of supervised machine learning methods by selecting the most informative samples.
We hypothesized that a clustering-based method for building sense inventories would detect more senses, while reducing the load of work of annotation when compared with manual annotation methods based on using random samples.
Methods
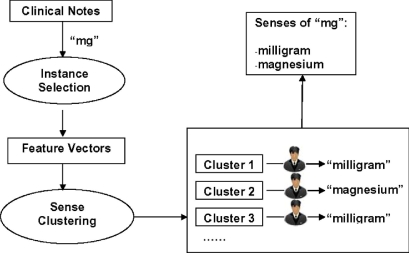
Some clinical notes, such as discharge summaries and admission notes in NYPH, consist of multiple sections, such as “chief complaint” and “history of present illness.” Each section contains multiple sentences of free text, where abbreviations occur. For each occurrence of every ambiguous abbreviation in this study, we collect surrounding words of the abbreviation within a window size and the section name where the abbreviation occurs. We call this set of information an “instance” of the particular abbreviation. ▶ shows an overview of the semi-automated sense generation method for the abbreviation “mg.” To build the sense inventory of the abbreviation “mg,” several instances of “mg” are collected from clinical notes. Then, information from surrounding words and section names is used to form feature vectors for clustering. Instances of the abbreviation “mg” are clustered into different sense clusters using different clustering algorithms. Next, one instance from each sense cluster is selected and shown to an expert who determines the appropriate sense. The final sense inventory is then formed by combining the annotated senses from different clusters. In this paper, we studied two clustering methods, and evaluated performance by comparing manual annotation methods with random sampling methods.
Figure 1.
An overview of a clustering-based method for building sense inventory of clinical abbreviations. To build the sense inventory of an abbreviation “mg”, a number of instances of “mg” are collected from a corpus of clinical notes. Then, information from surrounding words and section names are used to form feature vectors for clustering. Instances of the abbreviation “mg” are clustered into different sense clusters using different clustering algorithms. Next, one instance from each sense cluster is selected and shown to an expert who determines the appropriate sense. The final sense inventory is then formed by combining the annotated senses from the different clusters.
Data Set
The Columbia University Medical Center (CUMC) Clinical Data Repository (CDR) is a central pool of clinical data, which includes narrative data consisting of different types of clinical reports. In 2004, CUMC implemented a new Physician Data Entry system called eNote, 20 which allows physicians to directly key in various types of notes, such as hospital admission notes, progress notes and discharge summaries. Now that these notes are entered into the computer directly by providers, they contain more abbreviations than those that are dictated and transcribed. For this study, we collected all the inpatient admission notes from the Hospitalist Service that were entered via the eNote system during 2004–2006, amounting to 16,949 notes. Using a decision-tree based method as described in a previous study, 11 a total of 19,965 abbreviations were detected from the admission notes corpus. Among them, only 977 abbreviations occurred more than 100 times. However, those 977 abbreviations accounted for 94.6% of the total occurrences of all abbreviations. In addition, some abbreviations are more relevant to us than others because of other applications we are working on. For example, abbreviations with a disease sense and abbreviations that are ambiguous are more interesting to us because they will be used for subsequent studies concerning detection of correlations between diseases and other clinical entities, and for developing disambiguation methods. Therefore, we focused our research on ambiguous abbreviations with at least one disease sense based on the UMLS. We linked the 977 abbreviations to the UMLS and found 171 abbreviations that were ambiguous and had at least one disease sense according to the UMLS. Due to the cost of manual annotation only 12 of the most frequent ambiguous abbreviations in the corpus were selected from the 171 abbreviations and used for this study: ca, cc, cm, dm, hd, lad, le, mi, mg, pe, pt, ra. Those 12 abbreviations accounted for about 40% of total occurrences of all 171 abbreviations.
For each abbreviation, 1,000 instances were randomly selected and clustered using different clustering algorithms. To evaluate the performance of the clustering algorithms, a domain expert manually annotated the senses for 200 instances that were randomly selected from the above 1,000 instances for each abbreviation. Any instance whose sense could not be determined by the domain expert was removed from the 200 annotated data sets. For example, the abbreviation “ca” had one instance whose sense could not be determined by the domain expert. Therefore, that instance was removed from the set of 200 instances of “ca”.
▶ lists the 12 abbreviations and their senses obtained from the 200 annotated instances. The relative frequency of each sense is also listed in the third column. According to the 200 annotated instances, 11 of the 12 abbreviations were ambiguous (having more than one sense), and only the abbreviation “mi” had one sense myocardial infarction.
Table 1.
Table 1 Twelve Abbreviations and Their Senses
| ABB Review | Sense | Sense Frequency |
|---|---|---|
| ca | Calcium | 0.477 |
| Carcinoma | 0.518 | |
| Carbohydrate antigen | 0.005 | |
| cc | Cubic centimeter | 0.355 |
| Chief complaint | 0.635 | |
| Clubbing, cyanosis | 0.010 | |
| cm | Cardiomyopathy | 0.071 |
| Costal margin | 0.015 | |
| Cardiac monitoring | 0.005 | |
| Centimeter | 0.828 | |
| Cardiomegaly | 0.081 | |
| dm | Diabetes mellitus | 0.990 |
| dematomyositis | 0.005 | |
| Diastolic murmur | 0.005 | |
| hd | High dose | 0.010 |
| Hors decubitus at bedtime | 0.005 | |
| Hemodynamic | 0.080 | |
| Heart disease | 0.005 | |
| Hodgkin's Disease | 0.005 | |
| Huntington's Disease | 0.010 | |
| Hospital day | 0.005 | |
| Hemodialysis | 0.880 | |
| lad | Left atrial dilation | 0.005 |
| Lymphadenopathy | 0.715 | |
| Anterior descending branch of left coronary artery | 0.140 | |
| Left axis deviation | 0.140 | |
| le | Leukocyte esterase | 0.185 |
| Lower extremity | 0.815 | |
| mi | Myocardial infarction | 1.000 |
| mg | Magnesium | 0.055 |
| Milligram | 0.945 | |
| pe | Physical examination | 0.325 |
| Pulmonary embolism | 0.675 | |
| pt | Posterior tibial | 0.005 |
| Patient | 0.905 | |
| Physical therapy | 0.035 | |
| Prothrombin time assay | 0.055 | |
| ra | Right atrium | 0.030 |
| Room air | 0.900 | |
| Rheumatoid arthritis | 0.070 |
Clustering Methods
Each occurrence of an abbreviation is treated as an instance for clustering. Three types of features were used to form the feature vector of an instance for clustering an abbreviation: (1) stemmed words within a window size of 5 of the target abbreviation; (2) positional information and stemmed words within a window size of 5 of the target abbreviation; (3) section name of the admission note where the abbreviation occurs. All the features were weighted using Pointwise Mutual Information (PMI) (see Eq 1) between the feature word and the target abbreviation. Since mutual information is known to be biased towards infrequent words, the above mutual information value is multiplied by a discounting factor 17 as described in Eq 2. The modified Pointwise Mutual Information could be viewed as a measure of association between a feature word and the target abbreviation. Important feature words will have higher PMI values, as well as higher weights in the feature vectors.
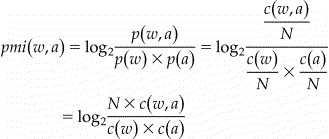 |
Equation 1: Definition of pointwise mutual information between a context word w and an abbreviation a, where p w denotes the probability of word w occurring in a corpus with N words, circa(w) denotes the count of occurrences of word w, and circa(w, a) denotes the count of occurrences where word w and a occur together within the specified window size, or at specified positions for positional features.
Equation 2: Discounting Factor for Pointwise Mutual Information.
Two clustering algorithms were studied using the above generated feature vectors. The first one is the standard Expectation-Maximization (EM) algorithm, 21 which provides a way to estimate the values of the hidden parameters of a model and has been widely used in clustering analysis. The second one is the Farthest-First (FF) Traversal Algorithm by Hochbaum and Shmoys, 22 which is an approximation algorithm for hierarchical clustering. Both of the clustering algorithms were implemented by the Weka Machine Learning Package 3.4, 23 which was used in this study. Two different numbers of clusters (K) were used for both algorithms: 10 and 20. Therefore, we have a total of four different clustering-based methods, notated as “EM_10,” “EM_20,” “FF_10,” and “FF_20.”
Cluster Sense Annotation
After clustering, instances of an abbreviation were grouped into different clusters, which ideally should correspond to the different senses of the abbreviation. To determine the sense of a cluster, an instance that was closest to the centroid of the cluster was selected and shown to a domain expert for annotation. The annotated sense for the selected instance was assigned as the sense of the cluster. The final sense inventory of an abbreviation was determined by combining senses determined by the expert from all clusters.
An annotation tool was also developed to facilitate the annotation process. This tool highlights the occurrence of the target abbreviation and lists previous annotated senses of the same abbreviation to facilitate annotation and to help make it more consistent.
Evaluation
The overall goal of the clustering-based sense generation system is to determine a list of possible senses of an abbreviation in a given corpus, while minimizing the manual annotation effort. We define two measurements to evaluate the performance of the current system toward that goal. One is called “Sense Completeness”, which is the ratio between the number of senses detected via the clustering-based method and the number of senses detected via manual annotation using 200 randomly selected instances for each abbreviation. The other is “Annotation Cost”, which measures the ratio between the number of annotated instances for the clustering-based method and the total number of annotated instances by the manual annotation method (200). For example, the abbreviation “cm” had 5 different senses based on the 200 annotated instances, and the EM-10 method detected 3 senses based on annotation of 10 instances. Then, the “Sense Completeness” for the EM-10 method will be 3/5 = 0.6 and the “Annotation Cost” will be 10/200 = 5% for the abbreviation “cm.” Methods that obtain senses by randomly selecting different numbers of instances from the 200 manually annotated instances were also evaluated together with the clustering-based methods. We randomly picked 10, 20, 40, and 100 annotated instances and evaluated their performance in terms of “Sense Completeness” and “Annotation Cost.” Then we repeated the above evaluation 50 times for each random selection method and reported the mean and standard deviations of the “Sense Completeness”.
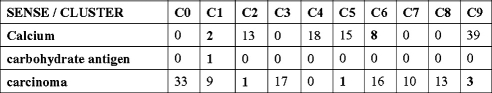
We focused on evaluating the completeness of senses because the goal of this study was to detect all possible senses of an abbreviation in the corpus. It is also interesting to evaluate the performance of the clustering methods for the task of sense discrimination. We developed a method to automatically link the clusters to their senses based on the manually sense-annotated data sets. The clustering results and the manually annotated results based on the 200 instances were compared with each other. A result matrix was formed and used to determine which cluster should be assigned to which sense. ▶ shows an example of a result matrix for the abbreviation “ca”. Clusters 0–9 were from the EM-10 clustering method. For each cluster, the number of instances for each sense of “ca” was counted based on the manually annotated data. A cluster c is assigned to a particular sense s if it satisfies the following condition: among all the manually annotated elements in the cluster c, more than 50% of the elements have the sense s according to the manual annotation. If a cluster does not have 50% of elements with the same sense, it will not map to any sense. For the example in ▶, cluster C0 would be assigned to sense “carcinoma”, and C1 would be assigned to “calcium.” By that definition, multiple clusters could map to one sense. After clusters are mapped to senses, incorrectly clustered instances could be counted and error rates of the sense clustering methods could be reported. We also used a majority sense based method, which always takes the majority sense as the correct sense, as a baseline for error rate calculation.
Figure 2.
The clustering result matrix of EM-10 for the abbreviation “ca.” The clustering results and the manually annotated results on the 200 instances were compared with each other. A result matrix was formed and used to determine which cluster should be assigned to which sense. A cluster c will be assigned to a particular sense s if the cluster c has more than 50% of elements that have the sense s according to the manual annotation. For this example, C0, C1, C3, C6, C7 and C8 map to sense “carcinoma;” C2, C4, C5, and C9 map to sense “calcium;” none of the clusters map to sense “carbohydrate antigen.” Out of a total of 199 instances, there were 16 incorrectly clustered instances (in bold) based on the above cluster-sense mappings. Therefore, the error rate for this example is 16/199 = 8.04%.
Results
▶ shows the results of the random sampling methods for building sense inventories of the 12 abbreviations, when different numbers of random samples were selected (10, 20, 40, 100, and 200). The average values of “Sense Completeness” and “Annotation Cost” across the 12 abbreviations are reported. Standard deviations of “Sense Completeness” measurements for random sampling methods are shown in parenthesis. As expected, when the number of selected samples increased, which also indicated a higher “Annotation Cost,” the “Sense Completeness” increased as well.
Table 2.
Table 2 Results of the Random Sampling Method for Building Sense Inventories of Clinical Abbreviations
| Method | Sense Completeness (Average of 12 Abbreviations) | Annotation Cost (Average of 12 Abbreviation) |
|---|---|---|
| Random-200 | 1.0 | 100% |
| Random-100 | 0.89 (0.031) | 50% |
| Random-40 | 0.79 (0.025) | 20% |
| Random-20 | 0.72 (0.031) | 10% |
| Random-10 | 0.64 (0.038) | 5% |
▶ shows the results of clustering-based methods for building sense inventories of the 12 abbreviations. When the “Annotation Cost” was fixed (e.g., 10%), the “Sense Completeness” of both clustering-based methods (EM-20-0.79 and FF-20-0.76) were better than that of the Random method (Random-20-0.72). The EM clustering method seemed to be better than the FF method, though statistical significance was not assessed. When EM-10 was compared with the Random-40 method, which had similar “Sense Completeness” values to those of EM-10, the reduction of “Annotation Cost” (from 20 to 10%) was large.
Table 3.
Table 3 Results of Clustering-based Methods for Building Sense Inventories of Clinical Abbreviations
| Method | Sense Completeness (Average of 12 Abbreviations) | Annotation Cost (Average of 12 Abbreviation) |
|---|---|---|
| EM-20 | 0.79 | 10% |
| FF-20 | 0.76 | 10% |
| EM-10 | 0.67 | 5% |
| FF-10 | 0.65 | 5% |
As described in the Methods section, we automatically mapped clusters to senses and calculated the error rates of clustering methods. ▶ shows the results of the averaged error rates across the 12 abbreviations for different clustering methods. The majority sense method had the highest average error rate of 23.9%, and the EM-20 method reached the lowest error rate of 6.31%.
Table 4.
Table 4 Error Rates of Sense Clustering Algorithms
| Method | Error Rate (Average of 12 Abbreviations) |
|---|---|
| EM-20 | 6.31% |
| FF-20 | 8.18% |
| EM-10 | 7.94% |
| FF-10 | 11.06% |
| Majority sense | 23.9% |
We also recorded the time for sense annotation. The result showed that the sense annotation speed was about 10-seconds per instance when the expert used the annotation tool. This annotation speed is very reasonable and it indicates it is a feasible task if we want to annotate all of the 977 frequent abbreviations from the admission notes corpus.
Discussion
Our results showed that a semi-automated method based on clustering could substantially reduce manual annotation effort and increase sense completeness, when compared with manual annotation methods using random samples.
This method was applied to internal medicine admission notes, but it is a generalizable method and could be applied to other similar clinical text with sections. For clinical notes without sections, this method is still applicable by using local context features alone, but the performance needs to be investigated further.
For clustering, we only used local contextual features that occur within a window of 5 of the target abbreviation. We used a small window size of 5 because admission notes usually contain short sentences. In an initial study, a large window size of 10 was also tested but no explicit improvement was observed.
We did not use global features from the entire document because the Assumption “one sense per discourse” 24 does not seem to be valid in clinical notes. Based on our observations, one abbreviation could appear in the same clinical note with more than one meaning. Several abbreviations, such as “ca”, “cm”, “mg”, and “pt”, were observed to have different meanings within one clinical document. For example, for the abbreviation “pt,” one admission note had the two different senses “patient” and “physical therapy” when “pt” was mentioned more than once. However, we also expect some global features will improve the performance of word sense discrimination systems. For example, if one of the senses of an abbreviation is a “disease” sense and that disease is highly associated with particular drugs, then using the words in the medication section of the note as features should improve the performance of the clustering methods. However, this study is only based on the Assumption “one sense per collocation” 25 (local context determine the sense of a term), not “one sense per discourse.” 24
One of the limitations of this study is that we only reported results of clustering-based methods from 1,000 randomly selected instances of each abbreviation. Ideally it would be better if we could repeat the evaluation multiple times for both clustering-based methods and the random sampling method using randomly selected samples. In that way, statistical analysis could be performed to determine if there is significant difference among different methods. That would require a very large manual annotation effort. We plan to increase the sample sizes for clustering and perform more evaluations on multiple sample sets.
We also analyzed errors where a sense was not found using the clustering-based methods. We noticed that most of the senses that were not detected were rare senses. The current clustering algorithms we use are not especially designed to detect rare senses. In the future, a new clustering algorithm for word sense discrimination with a focus on detecting rare sense is needed, and it should further improve the performance of clustering-based methods for building sense inventories. In this study, we used a set of the most frequent abbreviations, which are more likely to have a majority sense and a few rare senses. This could make the task more difficult because there might be more senses that are rare in the set of 12 most frequent abbreviations.
In this study, we used two clustering algorithms: EM and FF, which both require a predetermined number of clusters. Because of the cost of manual annotation, we studied clustering-based methods using 10 and 20 clusters only. In general, increasing the number of clusters will increase the “Sense Completeness.” However, it also requires higher “Annotation Cost,” which is not very practical. Therefore, the optimal goal is to reach the maximum “Sense Completeness” while maintaining the minimum number of clusters. In the future, larger number of clusters will be used and the correlation between the gained improvement of the clustering-based methods and the number of clusters will be studied as well. Furthermore, we will also investigate other clustering algorithms that can automatically determine the number of clusters.
Because some senses of clinical abbreviations are unknown to any of the biomedical knowledge sources, it is difficult to automate the mapping from clusters to their corresponding senses and therefore some manual annotation is still needed. In the future, we plan on developing methods to map sense clusters to known senses in existing knowledge sources in order to further reduce the effort required for manual annotation.
Conclusions
We developed an effective clustering-based, semi-automated method for building corpus-specific sense inventories for abbreviations in admission notes from a general medicine service. Instances of an abbreviation are clustered to different sense clusters and one instance from each cluster is manually annotated and then the senses from the different clusters are combined to obtain a final list of senses. Evaluation on a set of clinical abbreviations that were manually sense annotated showed that this method reduced manual annotation cost by half, and increased the completeness of sense inventories, when compared with manual annotation using random samples. In the future, we will develop methods that will increase the coverage of rare senses while maintaining a reduced cost of manual annotation.
Footnotes
Dr. Xu is currently with the Department of Biomedical Informatics, Venderbilt University, Nashville, TN.
Supported by grants LM007659, LM008635 and K22LM008805 from the NLM.
References
- 1.Bates DW, Cohen M, Leape LL, et al. Reducing the frequency of errors in medicine using information technology J Am Med Inform Assoc 2001;8:299-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teich JM, Wrinn MM. Clinical decision support systems come of age MD Comput 2000;17(1):43-46[PubMed]. [PubMed] [Google Scholar]
- 3.Haug PJ, Christensen L, Gundersen M, et al. A natural language parsing system for encoding admitting diagnoses Proc AMIA Annu Fall Symp 1997:814-818. [PMC free article] [PubMed]
- 4.Aronson AR. Effective mapping of biomedical text to the UMLS matathesaurus: The MetaMap program Proc AMIA Symp 2001:17-21. [PMC free article] [PubMed]
- 5.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural language text processor for clinical radiology J Am Med Inform Assoc 1994;1:161-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long WJ. Parsing free text nursing notes AMIA Annu Symp Proc 2003:917. [PMC free article] [PubMed]
- 7.Stetson PD, Johnson SB, Scotch M, Hripcsak G. The sublanguage of cross-coverage Proc AMIA Symp 2002:742-746. [PMC free article] [PubMed]
- 8.Liu H, Lussier YA, Friedman C. A study of abbreviations in the UMLS Proc AMIA Symp 2001:393-397. [PMC free article] [PubMed]
- 9.Liu H, Teller V, Friedman C. A multi-aspect comparison study of supervised word sense disambiguation J Am Med Inform Assoc 2004;11:320-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pakhomov S. Semi-supervised maximum entropy based approach to acronym and abbreviation normalization in medical texts. Proceedings of the 40th Annual Meeting of the Association of Computational Linguistics (ACL), Philadelphia, 2002, pp 160–167.
- 11.Xu H, Stetson PD, Friedman C. A study of abbreviations in clinical notes AMIA Annu Symp Proc 2007:821-825. [PMC free article] [PubMed]
- 12.Chang JT, Schutze H, Altman RB. Creating an online dictionary of abbreviations from Medline J Am Med Inform Assoc 2001;9:612-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adar E. SaRAD: A simple and robust abbreviation dictionary Bioinformatics 2004;20:527-533. [DOI] [PubMed] [Google Scholar]
- 14.Zhou W, Torvik VI, Smalheiser NR. Adam: Another database of abbreviations in Medline Bioinformatics 2006;22:2813-2818. [DOI] [PubMed] [Google Scholar]
- 15.Pedersen T, Bruce R. Distinguishing word senses in untaggged text. Proceedings of the second Conference on Empirical Methods in Natural Language Processing 1997, pp 197–207.
- 16.Schutze H. Automatic word sense discrimination Comput Linguist 1998;24–1:97-123. [Google Scholar]
- 17.Pantel P, Lin D. Discovering word sense from text. Proceedings of the Eighth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 2002, pp 613–9.
- 18.Liu H. Corpus-based ambiguity Resolution of biomedical terms using knowledge bases and machine learning. Ph.D. dissertation, 2002.
- 19.Thompson C, Califf ME, Mooney R. Active learning for natural language parsing and information extraction Proceedings of the Sixteenth International Conference on Machine Learning 1999:406-414.
- 20.Stetson PD, Keselman A, Rappaport D, et al. Electronic discharge summaries AMIA Annu Symp Proc 2005:1121. [PMC free article] [PubMed]
- 21.Dempster A, Laird N, Rubin D. Maximum likelihood from incomplete data via the EM algorithm J Royal Stat Soc 1977;39:1-38. [Google Scholar]
- 22.Hochbaum DS, Shmoys DB. A best possible heuristic for the k-center problem Math Oper Res 1985;10(2):180-184. [Google Scholar]
- 23.Witten I, Frank E. Data Mining: Practical Machine Learning Tools and Techniques2nd edn. San Francisco, CA: Morgan Kaufmann Publishers; 2005.
- 24.Gale WA, Church KW, Yarowsky D. One sense per discourse. Proceedings of the Workshop on Speech and Natural Language, HLTC-ACL, 1992, pp 233–7.
- 25.Yarowsky D. One sense per collocation. Proceedings of the Workshop on Speech and Natural Language, HLTC-ACL, 1993, pp 266–71.