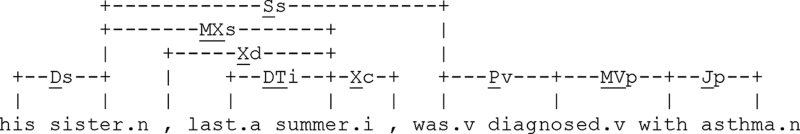Abstract
Objectives
The authors study two approaches to assertion classification. One of these approaches, Extended NegEx (ENegEx), extends the rule-based NegEx algorithm to cover alter-association assertions; the other, Statistical Assertion Classifier (StAC), presents a machine learning solution to assertion classification.
Design
For each mention of each medical problem, both approaches determine whether the problem, as asserted by the context of that mention, is present, absent, or uncertain in the patient, or associated with someone other than the patient. The authors use these two systems to (1) extend negation and uncertainty extraction to recognition of alter-association assertions, (2) determine the contribution of lexical and syntactic context to assertion classification, and (3) test if a machine learning approach to assertion classification can be as generally applicable and useful as its rule-based counterparts.
Measurements
The authors evaluated assertion classification approaches with precision, recall, and F-measure.
Results
The ENegEx algorithm is a general algorithm that can be directly applied to new corpora. Despite being based on machine learning, StAC can also be applied out-of-the-box to new corpora and achieve similar generality.
Conclusion
The StAC models that are developed on discharge summaries can be successfully applied to radiology reports. These models benefit the most from words found in the ± 4 word window of the target and can outperform ENegEx.
Introduction
The narrative in patient records contains information about the medical problems of patients. Given the medical problems mentioned in a record, for each mention of each medical problem, assertion classification aims to determine whether the problem is present (as stated by a positive assertion), absent (as stated by a negative assertion), or uncertain in the patient (as stated by an uncertain assertion), or is associated with someone other than the patient (as stated by an alter-association assertion).
Related Work
Extraction of key concepts from narrative medical records requires studies of medical language. Medical language processing systems enable studies of patient data repositories (e.g., for developing decision support systems 1 and for automatic diagnosis) 1,2 by extracting information from narrative patient records. Determining the nature of the assertion made on each mention of each medical problem is a step towards interpreting medical narratives. 1 To this end, there have been some efforts in the literature.
Fiszman et al. 1,2 developed the SymText system for encoding information from chest x-ray reports. SymText processes each sentence in a document independently, parses text syntactically, and fills semantic templates either with words extracted from the text or with broader concepts derived from these words. Bayesian networks applied to these templates can interpret one sentence at a time, e.g., can determine based on a single sentence the probably that a disease is present in the patient. Fiszman et al. used SymText's output with a rule-based system for determining whether a concept was present in a record. Identifying a single mention of the concept (or a term related to the concept) as present or possible was sufficient to qualify the concept as present in the record. They found that SymText's performance on this task “was similar to that of physicians” 1 ; its performance was better than that of keyword search systems when these systems considered a concept to be present unless it was accompanied by an explicit negation.
Friedman et al.'s MedLEE 3 uses domain-specific vocabulary and semantic grammar to process medical record narratives. It identifies the concepts in a report, maps the concepts to semantic categories, and maps the semantic categories to semantic structures. The resulting semantic representation captures information on status, location, and certainty of each mention of each concept. Hripcsak et al. 4 processed these semantic representations through Medical Logic Modules that could determine whether each mention of a disease was indicated as present or absent. They studied mentions of six diseases and found that the performance in determining presence of these diseases was not significantly different from that of physicians, but was significantly better than that of a keyword-based system that used negation phrases to identify absence. 4
For determining positive and negative assertions, Chapman's NegEx 5 studies candidate diseases and findings identified by the Unified Medical Language System (UMLS), and employs dictionaries of pre- and post-UMLS phrases that are indicative of negation. NegEx uses heuristics to limit the scope of indicative phrases, and identifies negative assertions with 78% recall (sensitivity) and 84% precision (positive predictive value) on 1,235 findings and diseases found in 1,000 sentences taken from discharge summaries. 5 Informal evaluations of NegEx report 78% recall and 86% precision on uncertain assertions. 6 Application of NegEx to identify the experiencer of a medical problem by ConText achieves 50% recall and 100% precision on a corpus containing 8 instances (out of 1,620) of alter-association assertions. 7
Mutalik et al.'s Negfinder 8 employs techniques and tools used for creating programming language compilers, makes use of a lexical scanner that is based on regular expressions, and runs a parser based on a restricted context-free grammar. The NegFinder finds negated concepts in discharge summaries and surgical notes with 95.7% recall and 91.8% specificity when evaluated on 1,869 concepts found in 10 medical documents from a variety of specialties.
Aronow et al.'s NegExpander 9 finds negation phrases through rules applied to part-of-speech tagged radiology reports, studies conjunctions that split negations, and expands negation phrases across conjunctions to make explicit the negation of individual concepts. The NegExpander gives 93% precision on radiology reports.
Elkin et al. 10 employ a rule base to mark positive, negative, and uncertain assertions on text which is preprocessed into its tokens and parsed. They achieve 97.2% recall and 91.2% precision on the assignment of negations.
The above-mentioned systems employ contextual features of various complexity with algorithms and tools of various complexity. We extend their studies on negation and uncertainty extraction to recognition of alter-association. We expect that the context immediately surrounding a medical problem holds valuable information regarding the assertion made on that medical problem. Although language allows extensive variation in the expression of assertions, we hypothesize that a significant portion of assertions are marked with clear contextual characteristics. While testing this hypothesis, we explore the significance of one form of syntactic information in assertion classification.
In general, rule-based approaches to assertion classification can be applied out-of-the-box to new corpora. On the other hand, supervised learning approaches are usually retrained for use on new corpora. This can make rule-based approaches more desirable over supervised learning approaches, even if the choice of a rule-based over a supervised learning approach trades off some performance for convenience. Ideally, the choice of an assertion classifier for a task would not trade off performance for convenience; the assertion classifier used would be convenient to apply and would outperform the alternatives. We hypothesize that supervised learning approaches hold some potential for achieving these two goals simultaneously. We check the feasibility of building a statistical assertion classifier that can be used out-of-the-box and that can maintain a performance advantage over its rule-based counterparts.
Our end product is a statistical assertion classifier, StAC, that can automatically capture the contextual clues for negative, uncertain, and alter-association assertions. The StAC approach makes use of lexical and syntactic context in conjunction with Support Vector Machines 11 (SVMs). We evaluate StAC on discharge summaries and on radiology reports. We compare StAC with Extended NegEx (ENegEx), our implementation of the NegEx algorithm extended to capture alter-association in addition to positive, negative, and uncertain assertions. We employ ENegEx as a representative rule-based assertion classifier. We show that ENegEx can give good results on our corpora. We also show that StAC need only use the words that appear in ± 4 word window of the target problem (i.e., the problem to be classified with an assertion type) to recognize most of the assertions in the same corpora. The models captured by StAC are most useful when they are specific to each corpus. However, the models built on one corpus can also identify assertions on a new corpus. As a result, StAC can be applied to new corpora out-of-the-box, in the same manner as ENegEx, and demonstrates potential for performance gain over this rule-based counterpart.
Data
We studied assertion classification on two corpora of discharge summaries and one corpus of radiology reports. The studies of these corpora were approved by the relevant Institutional Review Boards.
Beth Israel Deaconess Medical Center (BIDMC) Corpus
The BIDMC corpus consisted of 48 deidentified discharge summaries, consisting of a total of 5,166 sentences and including 2,125 medical problem mentions, from various departments in the BIDMC.
Challenge Corpus
The Challenge corpus consisted of 142 deidentified discharge summaries, consisting of 15,042 sentences and including 8,279 medical problem mentions, from various departments of hospitals in Partners Health Care.
Computational Medicine Center (CMC) Corpus
The CMC corpus consisted of 1,954 deidentified radiology reports, consisting of 6,406 sentences and including 6,325 medical problem mentions, for the 2007 CMC challenge 12 of the University of Cincinnati.
We used the BIDMC corpus for development; we used the Challenge and CMC corpora for evaluation.
Annotations
Assertion classification, as tackled in this paper, assumes that mentions of medical problems in clinical records have already been identified, and aims to determine whether each mentioned medical problem is present, absent, or uncertain in the patient, or associated with someone other than the patient. Therefore, before studying assertions, we annotated our corpora in two ways: we identified the medical problems in them (the summary numbers that resulted from this annotation are in the descriptions of the corpora above) and we determined the assertion class of each identified medical problem.
Identifying Medical Problems
For our purposes, medical problems refer to the diseases and symptoms of the patient. Diseases include the UMLS semantic types pathological function, disease or syndrome, mental or behavioral dysfunction, cell or molecular dysfunction, virus, neoplastic process, anatomic abnormality, injury or poisoning, congenital abnormality, and acquired abnormality. 13 Symptoms correspond to UMLS's signs or symptoms. Using this mapping, two undergraduate computer science students independently marked the medical problems in the BIDMC corpus. 13,14 Two other undergraduate computer science students independently marked the medical problems in the challenge corpus. This required two months of full time effort from each annotator. Given time and resource constraints, the medical problems in the CMC corpus were tagged using MetaMap. 15 Given the possible errors of MetaMap on this task, 13 the output of MetaMap was manually corrected and finalized by a nurse librarian and by a graduate student. The use of MetaMap for marking medical problems cut the annotation time per annotator by approximately 75%.
Determining Assertion Classes
Given the patient medical problems, we defined four classes of assertions:
• Positive assertions state that the problem, marked in square brackets, is/was present in the patient. e.g., “She had [airway stenosis].”
• Negative assertions state that the problem is absent in the patient. e.g., “Patient denies [headache].”
• Uncertain assertions state that the patient may have the problem. e.g., “… was thought possibly to be a [neoplasm].”
• Alter-association assertions state that the problem is not associated with the patient. e.g., “Sick contact positive for family member with [cough].” We do not differentiate between present, absent, or uncertain alter-association assertions.
While positive, negative, and uncertain assertions are often studied in negation and uncertainty extraction, alter-association assertions are usually not studied as a part of this task. We believe that alter-association assertions make sense in the context of more general assertion classification as they indicate whether the medical problem directly or indirectly affects the patient. We therefore include this assertion class in our studies.
Using the above assertion class definitions, for each occurrence of each problem in each corpus, one nurse-librarian and one information studies graduate student marked its assertion class. Initial agreement between the annotators as measured by kappa (K) 16 was 0.93 on the BIDMC corpus, 0.8 on the challenge corpus, and 0.92 on the CMC corpus. In general, K ≥ 0.8 is considered “almost perfect agreement”. 16 The annotators discussed and resolved their disagreements, providing us with the gold standard (see ▶).
Table 1.
Table 1 Instances and Percentages of Medical Problems in Each Assertion Class
| Assertion Class | Number in BIDMC | Number in Challenge | Number in CMC |
|---|---|---|---|
| Positive | 1,537 (72%) | 6,702 (81%) | 4,761 (75%) |
| Negative | 398 (19%) | 1,249 (15%) | 811 (13%) |
| Uncertain | 169 (8%) | 259 (3%) | 742 (12%) |
| Alter-Association | 21 (1%) | 69 (1%) | 11 (0%) |
| Total | 2,125 (100%) | 8,279 (100%) | 6,325 (100%) |
BIDMC = Beth Israel Deaconess Medical Center; CMC = Computational Medicine Center.
Methods
Given the medical problems mentioned in a clinical record, both ENegEx and StAC classify the assertion made on each medical problem by processing the records one sentence at a time and one medical problem at a time. They treat each occurrence of each medical problem independently of all others.
Extended NegEx (ENegEx)
In the absence of direct access to NegEx in time for this study, we implemented our own version of this program using the algorithm and the pre- and post-UMLS indicative phrases of NegEx 6 . We extended NegEx to alter-association assertions by studying the BIDMC corpus. We added to NegEx dictionaries consisting of:
1 Preceding alter-association phrases: that precede a problem and imply that it is associated with someone other than the patient, e.g., cousin, sister, and brother.
2 Succeeding alter-association phrases: that succeed a problem and imply that it is associated with someone other than the patient.
The resulting number of alter-association indicative phrases was 14. These indicative phrases were a superset of ConText's 7 dictionaries. We applied the NegEx algorithm 6 with the extended set of indicative phrases to our data, and called this system ENegEx.
We ran ENegEx on the BIDMC corpus (see ▶), manually checked its output, and reestablished that its algorithm complied with the specifications of NegEx. We double-checked that the low recall on uncertain assertions was due to a weakness in NegEx's dictionaries, which for uncertain assertions consisted solely of morphological and syntactic variants of the phrase “rule out.” 6 We verified that our newly introduced alter-association indicative phrases were complete in their coverage of the alter-association assertions in the BIDMC corpus.
Table 2.
Table 2 ENegEx on BIDMC Corpus
| Assertion Class | EnegEx |
||
|---|---|---|---|
| Precision | Recall | F-Measure | |
| Positive | 0.88 | 0.99 | 0.93 |
| Negative | 0.97 | 0.84 | 0.90 |
| Uncertain | 1.00 | 0.08 | 0.15 |
| Alter-Association | 1.00 | 1.00 | 1.00 |
BIDMC = Beth Israel Deaconess Medical Center.
Statistical Assertion Classifier (StAC)
To test the hypothesis that contextual features capture the information necessary for assertion classification, and to explore the contribution of one form of syntactic information to this task, we built StAC. The StAC applies SVMs to a binary feature vector. We define a feature as a characteristic that can have a multitude of values, e.g., for a person, “eye color” is a feature with several possible values, e.g., green. For each feature of StAC, the binary feature vector lists all possible values of that feature in the corpus as its columns, and for each target medical problem to be classified (row), it sets the columns observed to 1, leaving the rest at zero. 14 If the target has no value for a feature, then all columns representing this feature will be set to zero.
We armed StAC with a variety of contextual features, which included some simple lexical information and some more complex syntactic information. For each target, StAC uses features extracted from the sentence containing the target. Upon request, the code for extracting these features will be made available for research purposes.
Lexical context features of StAC include:
• ± 4 word window, i.e., words that appear within a ± 4 word window of the target. Given the target at the nth position in the sentence, the ± 4 word window captures the words found in the (n-1)th, (n-2)th, (n-3)th, (n-4)th, (n+1)th, (n+2)th, (n+3)th, and (n+4)th positions in the sentence. Our knowledge representation treats each of the above positions as an individual feature, lists all possible values for each feature, and identifies the value of the feature in the context of each target by setting only that value to one. For some targets, one or more of the features can have no values specified, e.g., the third word of the sentence will have all possible values of the (n-4)th position set to zero.
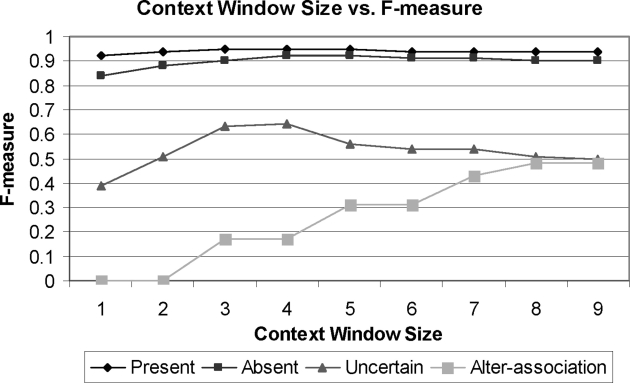
The ± 4 word window subsumes ± 1, ± 2, and ± 3 word windows so that any strings captured by these smaller windows are also captured by the larger window of ± 4. The focus on a ± 4 word window was determined by cross-validation on the BIDMC corpus. ▶ shows the F-measures of StAC when run only with various ± n word window features and indicates that windows greater than ± 4 can hurt performance on three of the assertion classes.
• Section headings, i.e., whether the target appears in a section whose heading contains the word “Family”, e.g., family history. This feature is represented by a single column which is set to one only if the target appears in a section whose title contains the word “Family”.
Figure 1.
Context window size (± n) vs. F-measure on each assertion class on BIDMC corpus.
Syntactic context features include:
• Verbs preceding and succeeding the target, e.g., verb showed preceding a problem suggests that the problem is present, verb cured after a problem suggests that the problem is absent. We treat the verb preceding and the verb succeeding the target as two separate features, each with numerous possible values.
• ± 2 link window, i.e., syntactic links within a ± 2 link window of the target (and of the verbs preceding and succeeding the target) and the words they link to the target (and to the verbs preceding and succeeding the target). We extract the links and the words they link to from the output of the Link Grammar Parser17 (LGP). We use a version of LGP whose lexicon has been extended to improve coverage on medical corpora.18 Even in the absence of a fully-correct parse for each sentence, this parser provides useful parses for phrases.13,14
The choice of ± 2 link window over windows of any other size was based on cross-validation on the BIDMC corpus. Given a target (or a verb) at the nth position in the sentence, its ± 2 link window is represented by the (n-1)th, (n-2)th, (n+1)th, and (n+2)th links and the words to which they link. e.g., for asthma in for asthma“His sister, last summer, was diagnosed with asthma”, the −2 link window is given by the set {(Jp, with),(MVp, diagnosed)} where MVp links verbs to their prepositional phrases and Jp connects prepositions to their objects (see ▶). Our knowledge representation treats each of (n-1)th, (n-2)th, (n+1)th, and (n+2)th links and each of their words as an individual feature with its own set of possible values. For some targets, one or more of the link and word positions can have no values assigned, i.e., all possible values of that feature are set to zero. When absent among words within short range lexical window, the ± 2 link window features clarify the modifier–noun relationships and help eliminate false positives of lexical context that would result from mere lexical proximity. When present within long range lexical window, the ± 2 link window features can capture the long distance dependencies and help eliminate false negatives that would be missed by our lexical context. For example, the connection between sister and asthma would be missed by the lexical context but is captured by the {(Pv, was),(Ss, sister)} links of the verb diagnosed (see Sibanda 14 ).
Figure 2.
Sample link grammar parse.
The StAC employs SVMs with a linear kernel. The choice of SVMs over other classifiers is motivated by their ability to robustly handle large feature sets and by their ability to often find globally optimum solutions. 19 In our case, the number of features in the set is on the order of thousands. We use the multiclass SVM implementation of LIBSVM. 20
We evaluate StAC using single train–test cycles and using cross-validation. For both train–test cycles and cross-validation, we create the binary feature vector from only the development data used for each round. As a result, the feature values of the targets that appear only in the validation and test sets of that round do not appear in the feature vector, i.e., the feature vector is not overfit to the validation and test sets. The performance of StAC on the BIDMC corpus is given in ▶.
Table 3.
Table 3 StAC Cross-Validated on the BIDMC Corpus
| Assertion Class | StAC |
||
|---|---|---|---|
| Precision | Recall | F-Measure | |
| Positive | 0.93 | 0.97 | 0.95 |
| Negative | 0.95 | 0.93 | 0.94 |
| Uncertain | 0.70 | 0.56 | 0.63 |
| Alter-Association | 1.00 | 0.81 | 0.89 |
BIDMC = Beth Israel Deaconess Medical Center.
Evaluation Methods
We evaluate system performances in terms of precision (P), recall (R), and F-measure (F). Precision (positive predictive value) measures the proportion of predictions in a class that were correct. Recall (sensitivity) measures the proportion of true class instances that were correctly identified. F-measure is the harmonic mean of precision and recall. We test significance of the differences in performances of the two systems using the Z test on two proportions. This test considers the size of the sample in a class to make a judgment of significance on the difference of performances (proportions) in that class. 21,22 We present precision, recall, and F-measure values for all of our experiments; however, we base our observations on the F-measure which provides a single convenient number for comparing systems.
Evaluation, Results, and Discussion
For evaluation, we ran ENegEx on the challenge and CMC corpora. ▶ shows that ENegEx is strongest in recognizing positive and negative assertions, weakest in recognizing uncertain and alter-association assertions. Although the performance of ENegEx can be improved by tuning it to the corpora on which it is to be run, even in the absence of such tuning, ENegEx maintains itself as a simple algorithm that can recognize positive from negative assertions. Most of ENegEx's mistakes come from scope and from incomplete dictionaries. For example, in “She is an obese white female in no acute distress with a hoarse voice,” ENegEx finds both acute distress and hoarse voice are within the scope of the indicative phrase no. In “… frozen section analysis revealed this to be adenocarcinoma, metastatic disease from the colon most likely,” ENegEx misses the subtle uncertainty expressed by most likely.
Table 4.
Table 4 ENegEx on the Challenge and CMC Corpora
| Class | Challenge |
CMC |
||||
|---|---|---|---|---|---|---|
| p | R | F | p | R | F | |
| Positive | 0.92 | 0.99 | 0.95 | 0.83 | 0.99 | 0.90 |
| Negative | 0.93 | 0.74 | 0.83 | 0.93 | 0.74 | 0.82 |
| Uncertain | 0.96 | 0.08 | 0.16 | 1.00 | 0.00 | 0.01 |
| Alter-Association | 0.71 | 0.81 | 0.76 | 0.50 | 0.07 | 0.13 |
CMC = Computational Medicine Center.
We evaluate StAC in two different ways:
• Cross-validation experiments: We developed the assertion classification approach of StAC, with its specific methods and features, on the BIDMC corpus. Would the methods and features of StAC be as useful on other corpora? To answer this question, we cross-validated StAC on the challenge and CMC corpora. Cross-validation developed and validated models on each corpus separately.
• Generality experiments: ENegEx can be applied to new data sets as is and would give reasonable results. Could StAC be similarly applicable to new corpora? To answer this question, we trained StAC on the BIDMC corpus and we ran it, without retraining or cross-validating, on the challenge and CMC corpora. While cross-validating StAC on a corpus tests the generality of StAC's approach on that corpus, running StAC on a corpus as trained on another corpus checks whether the model obtained from one corpus helps assertion classification in the other corpus.
In both of the above experiments, we use ENegEx (▶) as a benchmark. We use * to mark the performances of StAC that are significantly different from the corresponding performance of ENegEx at α = 0.05. Bold marks performances of StAC that are equal to or greater than the corresponding performance of ENegEx.
Cross-validation Experiments
▶ shows that StAC's approach to extracting key contextual clues for recognizing assertion classes generalize to all of our corpora. The StAC approach applies the methods and features identified on the BIDMC corpus to the challenge and CMC corpora. It extracts specific contextual clues from each corpus within the limits of these methods and features, and gives good results.
Table 5.
Table 5 Cross-Validation of StAc on the Challenge and CMC Corpora
| Class | Challenge |
CMC |
||||
|---|---|---|---|---|---|---|
| p | R | F | p | R | F | |
| Positive | 0.96* | 0.97* | 0.97* | 0.97* | 0.98* | 0.98* |
| Negative | 0.91 | 0.88* | 0.90* | 0.95 | 0.95* | 0.95* |
| Uncertain | 0.65* | 0.53* | 0.58* | 0.90* | 0.88* | 0.89* |
| Alter-Association | 0.93* | 0.81 | 0.87 | 1.00* | 0.21 | 0.35 |
CMC = Computational Medicine Center.
Generality Experiments
In general, rule-based approaches like ENegEx can be applied out-of-the-box to new corpora. To test whether StAC, based on supervised learning, can be used out-of-the-box in a manner analogous to ENegEx, we trained StAC on the BIDMC corpus and tested the resulting model as is on the challenge and CMC corpora. We found that with the models trained on the BIDMC corpus, StAC could outperform ENegEx, when both systems are run (compare ▶ and ▶) on the challenge and CMC corpora. The performance gain of StAC is more pronounced on the F-measures from the CMC corpus.
Table 6.
Table 6 StAC Trained on BIDMC and Run on Challenge and CMC Corpora
| Class | Challenge |
CMC |
||||
|---|---|---|---|---|---|---|
| p | R | F | p | R | F | |
| Positive | 0.96* | 0.93* | 0.94* | 0.89* | 0.98* | 0.93* |
| Negative | 0.82* | 0.89* | 0.85 | 0.90* | 0.82* | 0.86* |
| Uncertain | 0.31* | 0.50* | 0.38* | 0.87* | 0.45* | 0.60* |
| Alter-Association | 0.90* | 0.75 | 0.82 | 0* | 0 | 0 |
BIDMC = Beth Israel Deaconess Medical Center; CMC = Computational Medicine Center.
Naturally, StAC gives its best results when it is cross-validated because cross-validation allows it to tune its context to each corpus (compare ▶ and ▶). However, even in the absence of cross-validation, the information pertinent to classifying assertions on one corpus aids classification of assertions in another corpus.
Feature Evaluation
To understand the source of the strength of StAC, we cross-validated it with each of its features separately. ▶ and ▶, where italics mark the best F-measures, show that the words in the ± 4 word window are the most informative features of StAC on all of our corpora, indicating that the nature of the assertions made about a problem is mostly captured by the lexical context of the problem. Only for determining the alter-association assertions on the discharge summaries does lexical context drastically benefit from Section Headings. The ± 2 link window is the second most informative feature for StAC. The ± 2 link window features contribute to lexical features by correcting false positives that occur when a negation indicator such as no appears within the ± 4 word window but does not in fact modify a disease, e.g., “no intervention due to cardiovascular disease” where the ± 2 link window clarifies that no modifies intervention and not cardiovascular disease. Their value in our experiments is only limited by the number of such examples in our corpora.
Table 7.
Table 7 F-Measures of StAC When Run on BIDMC Corpus with Subsets of Features Best F-Measures in Italics
| Corpus | Feature | Positive | Negative | Uncertain | Alter-Ass'n. | |
|---|---|---|---|---|---|---|
| BIDMC | Lexical Context | ±4 Word Window | 0.95 | 0.92 | 0.64 | 0.17 |
| Section headings | 0.84 | 0 | 0 | 0.92 | ||
| ±4 word window + section headings | 0.96 | 0.93 | 0.64 | 0.92 | ||
| Syntactic Context | ±2 link window | 0.90 | 0.76 | 0.59 | 0.08 | |
| Verbs | 0.85 | 0.37 | 0.17 | 0 | ||
| ±2 link window + verbs | 0.91 | 0.79 | 0.51 | 0.08 |
BIDMC = Beth Israel Deaconess Medical Center.
Table 8.
Table 8 F-Measure of StAC When Run on Challenge and CMC Corpora with Subsets of Features Best in Italics
| Corpus | Feature | Positive | Negative | Uncertain | Alter-Ass'n. | |
|---|---|---|---|---|---|---|
| Challenge | Lexical Context | ±4 word window | 0.97 | 0.90 | 0.58 | 0.49 |
| Section headings | 0.90 | 0 | 0 | 0.82 | ||
| ±4 word window + section headings | 0.97 | 0.90 | 0.58 | 0.86 | ||
| Syntactic Context | ±2 link window | 0.94 | 0.70 | 0.49 | 0.47 | |
| Verbs | 0.90 | 0.21 | 0.05 | 0.38 | ||
| ±2 link window + verbs | 0.94 | 0.72 | 0.48 | 0.52 | ||
| CMC | Lexical Context | ±4 word window | 0.98 | 0.95 | 0.89 | 0.42 |
| Section headings | 0.85 | 0 | 0 | 0 | ||
| ±4 word window + section headings | 0.98 | 0.95 | 0.89 | 0.42 | ||
| Syntactic Context | ±2 link window | 0.92 | 0.61 | 0.73 | 0.13 | |
| Verbs | 0.88 | 0.21 | 0.53 | 0 | ||
| ±2 link window + verbs | 0.92 | 0.62 | 0.75 | 0.25 |
CMC = Computational Medicine Center.
Limitations
Despite correctly classifying most of the assertions, StAC makes several recurring mistakes. For example, it misinterprets the scope of some phrases: in “No JVP, 2+ swelling, no pain”, JVP and pain appear to be absent, while swelling is present. However, the lack of a consistent indicative context prevents StAC from recognizing this information.
The results in ▶ show that StAC can obtain much of the contextual information necessary for assertion classification on all of our corpora just from the BIDMC corpus. Our choice of the BIDMC corpus for development was guided by its decent size and by its genre, which had previously been used for assertion classification. 5 If trained on a corpus that was weaker in its representation of information pertinent to assertion classes, both in terms of the number of examples of each assertion class and in terms of capturing the variety of contexts indicating the various assertion classes, the results presented for StAC and its generalizability could change (as would the results and generality of ENegEx if developed under the same conditions). The results on the alter-association class support this claim: this class could benefit from further studies on corpora that may be richer in their examples for it.
Conclusions
We presented StAC and used it in exploring the contribution of various contextual features to assertion classification. Using ENegEx as a benchmark, we showed that StAC can capture assertion classes on discharge summaries and radiology reports by making use of the information contained in the immediate context of target problems. The information contained in the words found in the ± 4 word window of target goes a long way towards this goal. More importantly, information obtained from one corpus can help assertion classification on other corpora.
Footnotes
Supported in part by the National Institutes of Health through research grants 1 RO 1 EB001659 and U54LM008748. IRB approval has been granted for the studies presented in this manuscript. The authors thank Peter Szolovits, Ted Pedersen, Ira Goldstein, and the anonymous reviewers of AMIA and JAMIA for their insightful comments and constructive feedback.
References
- 1.Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug P. Automatic detection of acute bacterial pneumonia from chest x-ray reports J Am Med Inform Assoc 2000;7(6):593-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiszman M, Chapman WW, Evans SR, Haug P. Automatic identification of pneumonia related concepts on chest x-ray reports AMIA Annu Symp Proc 1999:67-71. [PMC free article] [PubMed]
- 3.Friedman C, Alderson PO, Austin JHM, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology J Am Med Inform Assoc 1994;1:161-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hripcsak G, Friedman C, Alderson PO, et al. Unlocking clinical data from narrative reports: A study of natural language processing Ann Intern Med 1995;122(9):681-688. [DOI] [PubMed] [Google Scholar]
- 5.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries J Biomed Inform 2001;34(5):301-310. [DOI] [PubMed] [Google Scholar]
- 6. NegEx version 2: A simple algorithm for identifying pertinent negatives in textual medical recordshttp://www.dbmi.pitt.edu/chapman/NegEx.html 2001. accessed Jul 28, 2008.
- 7.Chapman WW, Chu D, Dowling JN. ConText: An algorithm for identifying contextual features from clinical text. BioNLP 2007: Biological, translational, and clinical language processing. Prague 2007:81-88.
- 8.Mutalik PG, Deshpande A, Nadkarni PM. Use of general-purpose negation detection to augment concept indexing of medical documents: A quantitative study using the UMLS J Am Med Inform Assoc 2001;8(6):598-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aronow D, Feng F, Croft WB. Ad hoc classification of radiology reports J Am Med Inform Assoc 1999;6(5):393-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkin PL, Brown SH, Bauer BA, et al. A controlled trial of automated classification of negation from clinical notes BMC Med inform Decis Mak 2005;5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortes C, Vapnik V. Support-vector networks Machine Learn 1995;20(3):273-297. [Google Scholar]
- 12.Pestian JP, Brew C, Matykiewicz P, et al. A shared task involving multi-label classification of clinical free text. BioNLP 2007: Biological, translational, and clinical language processing. Prague 2007:97-104.
- 13.Sibanda T, He T, Szolovits P, Uzuner Ö. Syntactically-informed semantic category recognizer for discharge summaries AMIA Annu Symp Proc 2006:714-718. [PMC free article] [PubMed]
- 14.Sibanda T. Was the patient cured? Understanding semantic categories and their relationships in patient records. Master's Thesis, MIT. June 2006.
- 15.Aronson A. Effective mapping of biomedical text to the UMLS metathesaurus: The Metamap program AMIA Annu Symp Proc 2001:17-21. [PMC free article] [PubMed]
- 16. What is kappa?http://www.musc.edu/dc/icrebm/kappa.html 2001. accessed: Jul 28, 2008.
- 17.Sleator D, Temperley D. Parsing English with a Link Grammar. Technical Report CMU-CS-91-196. Carnegie Mellon University; 1991.
- 18.Szolovits P. Adding a medical lexicon to an English parser AMIA Annu Symp Proc 2003:639-643. [PMC free article] [PubMed]
- 19.Burges CJC. A tutorial on support vector machines for pattern recognition Data Min Knowl Discov 1998;2(2):121-167. [Google Scholar]
- 20.Chang C, Lin C. LIBSVM: A Library for Support Vector Machines Department of Computer Science and Information Engineering. Taipei, Taiwan: National Taiwan University; 2001.
- 21.Osborn CE. Statistical Applications for Health Information Management2nd edn. Boston: Jones & Bartlett Publishing; 2005.
- 22. Z test for two proportionshttp://www.dimensionresearch.com/resources/calculators/ztest.html 2005. accessed Jun 19, 2008.