Abstract
Mitochondrial fission in mammals is mediated by at least two proteins, DLP1/Drp1 and hFis1. DLP1 mediates the scission of mitochondrial membranes through GTP hydrolysis, and hFis1 is a putative DLP1 receptor anchored at the mitochondrial outer membrane by a C-terminal single transmembrane domain. The cytosolic domain of hFis1 contains six α-helices (α1-α6) out of which α2-α5 form two tetratricopeptide repeat (TPR) folds. In this study, by using chimeric constructs, we demonstrated that the cytosolic domain contains the necessary information for hFis1 function during mitochondrial fission. By using transient expression of different mutant forms of the hFis1 protein, we found that hFis1 self-interaction plays an important role in mitochondrial fission. Our results show that deletion of the α1 helix greatly increased the formation of dimeric and oligomeric forms of hFis1, indicating that α1 helix functions as a negative regulator of the hFis1 self-interaction. Further mutational approaches revealed that a tyrosine residue in the α5 helix and the linker between α3 and α4 helices participate in hFis1 oligomerization. Mutations causing oligomerization defect greatly reduced the ability to induce not only mitochondrial fragmentation by full-length hFis1 but also the formation of swollen ball-shaped mitochondria caused by α1-deleted hFis1. Our data suggest that oligomerization of hFis1 in the mitochondrial outer membrane plays a role in mitochondrial fission, potentially through participating in fission factor recruitment.
Keywords: mitochondria, mitochondrial fission, hFis1, DLP1, Drp1, oligomerization
Introduction
Mitochondria in cultured mammalian cells commonly form reticular networks comprised of filamentous tubules. These mitochondria are dynamic, displaying continuous movement and shape changes [1-3]. The main processes that determine mitochondrial morphology are fission and fusion of mitochondrial tubules, which occur in a balanced frequency to maintain the normal morphology [4]. Aberration in the fission/fusion balance causes terminally deformed mitochondria that are often associated with pathological conditions [5-10], suggesting that the proper maintenance of mitochondrial morphology is important for normal cell function.
While multiple proteins are implicated in mitochondrial fission [6, 11-16], the most studied mitochondrial fission process in mammalian cells is the one mediated by two proteins: a dynamin-like large GTPase, DLP1/Drp1, and a small helix-rich mitochondrial outer membrane protein, hFis1. DLP1 has the capacity to deform spherical liposomes into tubules of a defined dimension in vitro [17], consistent with its presumed activity of membrane squeezing and severing during mitochondrial fission. The 17-kD mitochondrial protein hFis1 is thought to be a receptor for the recruitment of cytosolic DLP1 to the mitochondrial surface during fission [13, 18]. The C-terminal single transmembrane domain anchors the hFis1 molecule in the mitochondrial outer membrane, leaving the bulk of the molecule exposed to the cytosol. Structural studies of the N-terminal cytosolic domain of hFis1 have revealed the presence of six α helices (α1-α6) that contain two tetratricopeptide repeat (TPR)-like folds, TPR1 and TPR2 [19, 20]. While TPR1 and TPR2 are formed by α2-α3 and α4-α5, respectively, the α1 helix has been suggested to function as a regulatory component [19], presumably by negatively controlling the fission factor binding at the TPR [18].
Despite the well-defined structural features of the Fis1 protein, how hFis1 functions in mitochondrial fission is not fully understood. Our previous studies indicated that hFis1 transiently interacts with DLP1 and that the hFis1 α1-helix has a pivotal role in regulating the hFis1-DLP1 interaction [13] [18]. In addition, mutations in the hFis1 TPR helices cause a dominant-negative effect when overexpressed, suggesting that hFis1 interacts with each other for correct fission of mitochondrial tubules [18]. In the current study, using chimeric, mutational, and crosslinking approaches, we identified regions of hFis1 that are important for its function. Our study indicated that these regions participate in hFis1 oligomer formation that is necessary for hFis1-mediated morphological changes of mitochondria. These results provide insight for the mitochondrial fission mechanisms mediated by hFis1.
Materials and Methods
Cell culture
The cell lines BHK-21 (ATCC CCL-10) and Clone 9 (ATCC CRL-1439) were used for all experiments. Cells were cultured in Ham's F-12K medium (Clone 9) or in DMEM (Dulbecco's modified eagle's medium; BHK-21), supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin under 5% CO2 at 37°C. Clone 9 cells stably expressing green fluorescent protein (GFP) in the mitochondrial matrix [13] were maintained in 200 μg/mL G418.
Plasmid construction and transfection
Deletion and point mutants and chimeras were constructed in the pcDNA3 mammalian expression vector (Invitrogen) using the standard PCR technique. The mutations and correct sequences were verified through DNA sequencing. For cell transfection, plasmids were purified using the Maxiprep Plasmid purification system (Quiagen). Cells were plated in either 35-mm or 100-mm tissue culture dishes. Transfections were performed using LipofectAMINE® (Invitrogen) per manufacturer's instructions. Transfected cells were allowed to recover for 16 to 24 hours before processing for whole cells crosslinking, immunofluorescence, or mitochondrial isolation.
Whole cell crosslinking
Whole cell crosslinking was done in 35-mm culture dishes. BHK-21 cells transfected with the appropriate plasmids were rinsed once with MEM Buffer (100 mM MES, pH 6.6, 1 mM EGTA, 0.5 mM MgCl2) and crosslinked by incubating in 1 mM EDC (1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride; Pierce BioTech. Inc.) in MEM at room temperature for 1 hour with gentle rocking. Crosslinking was stopped by adding ethanolamine (50 mM final) and incubating for 15 minutes. After rinsing, the cells were collected in 1× SDS PAGE sample buffer. Crosslinked samples were analyzed through Tris/Tricine gel electrophoresis and immunoblot analyses using anti-Myc antibody (Clone 9E10, Sigma). The Image J program (Wayne Rasband, NIH) was used for quantification of the hFis1 band densities from immunoblotting.
Isolation of mitochondria and crosslinking
BHK-21 cells were plated in 100-mm tissue culture plates and transfected with the DNA constructs. Cells were rinsed in Isolation Buffer (IB: 10 mM Hepes, pH 7.2, 1 mM EDTA, 320 mM Sucrose) and collected in 1mL of cold isolation buffer per plate. Cells were centrifuged at 700 ×g and resuspended in IB containing protease inhibitor. Cells were homogenized in Dounce homogenizer by 20 strokes, and the homogenate was spun down at 700 ×g and the supernatant collected. The remaining pellet was homogenized again and the supernatant was collected and pooled with the one from the previous step. The pooled supernatant was spun at 17,000 ×g and the brown pellet was collected as the mitochondrial fraction and resuspended in Isolation buffer. For crosslinking of the isolated mitochondria, the mitochondrial fraction was further diluted in MEM buffer and crosslinked with 1 mM EDC for 15 minutes at room temperature. Samples were prepared for gel electrophoresis in SDS-PAGE sample buffer and analyzed as describe above.
Indirect immunofluorescence
Clone 9 cells or Clone 9 cells carrying GFP in the mitochondrial matrix were used for immunofluorescence. Cells were plated on coverslips housed in 35-mm Petri dishes and transfected with an appropriate plasmid. At 16-24 hour post transfection, cells were fixed with 3% paraformaldehyde (Electron Microscopy Sciences) and permeabilized with 0.2% Triton X-100. Samples were then incubated in blocking buffer containing 5% horse serum for 1 hour at 37°C. Mouse monoclonal anti-c-Myc (Clone 9E10: Sigma) and rabbit polyclonal anti-DLP1 (REF) antibodies were used for primary antibodies. For secondary antibodies, Alexa 488- or 594-conjugated anti-mouse or rabbit antibodies (Molecular Probes Inc.) were used. After appropriate rinsings, coverslips were mounted in ProLong antifade reagent (Molecular Probes Inc.) on glass slides and cells were viewed with an Olympus IX71 epifluorescence microscope. Fluorescence images were acquired with an Evolution QEi camera (Mediacybernetics, Inc.) driven by IPLab imaging software (Scanalytics, Inc.). Acquired images were adjusted using Adobe Photoshop (Adobe Systems Inc.) software.
Results
The N-terminal cytosolic domain of hFis1 contains information for the hFis1-specific function in mitochondrial morphology
To identify functionally relevant domains of hFis1, we employed a chimeric approach and tested the functional importance of the cytosolic and the transmembrane/C-terminal tail (TM/C) domains. We constructed Myc epitope-tagged chimeras between the two proteins hFis1 and OMP25 (Fig. 1A). Similar to hFis1, OMP25 is a small protein (145 amino acids) anchored on the mitochondrial outer membrane through a C-terminal single transmembrane region [21]. We tested the functionality of the chimeric proteins by examining intracellular locations and mitochondrial morphologies in cells transiently expressing individual chimeras. It has been shown that overexpression of full-length hFis1 causes fragmentation of mitochondrial tubules [13, 15, 18] (Fig. 2A). We found that the chimera in which the TM/C of hFis1 was substituted with that from OMP25 (Fis/OMP) was correctly localized to mitochondria and induced mitochondrial fragmentation when overexpressed (Fig. 2B, B′). This fragmented mitochondrial phenotype is indistinguishable from the one induced by the overexpression of full-length hFis1, indicating that the mitochondrial fission-related function of hFis1 resides within the cytosolic domain of hFis1.
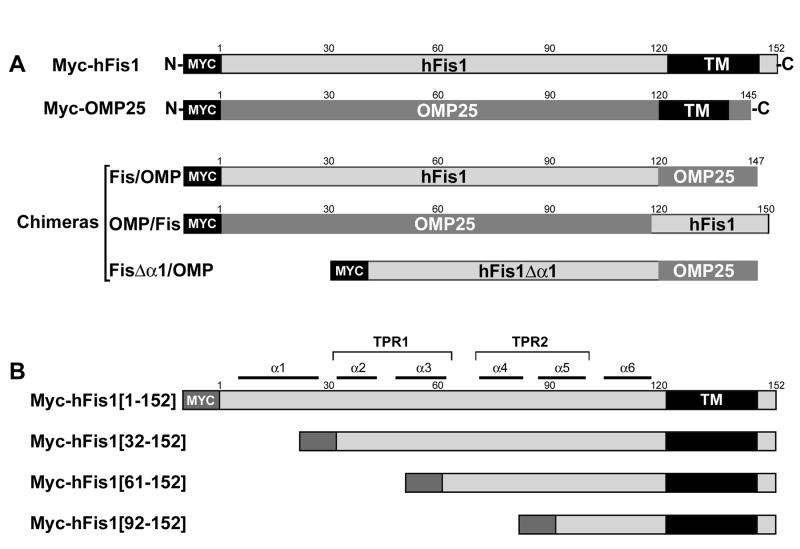
Figure 1. Chimeric and deletion constructs of hFis1 and OMP25.
(A) Chimeric constructs between hFis1 and OMP25. The N-terminal cytosolic domain of hFis1 (aa 1-120) and the C-terminal transmembrane/tail region of OMP25 (aa 119-145) were fused to make the Fis/OMP. The converse chimera OMP/Fis was made of the cytosolic domain of OMP (aa 1-118) and the C-terminal region of hFis1 (aa 121-152). In FisΔα1/OMP, the α1 helix (aa 1-31) was deleted from the Fis/OMP chimera. (B) hFis1 full-length and N-terminal deletion constructs. Myc-hFis1[1-152] is the full length molecule. N-terminal 31, 60, and 91 amino acids were deleted in Myc-hFis1[32-152], Myc-hFis1[61-152], and Myc-hFis1[92-152], respectively. All constructs were tagged with the Myc-epitope at the N-terminus.
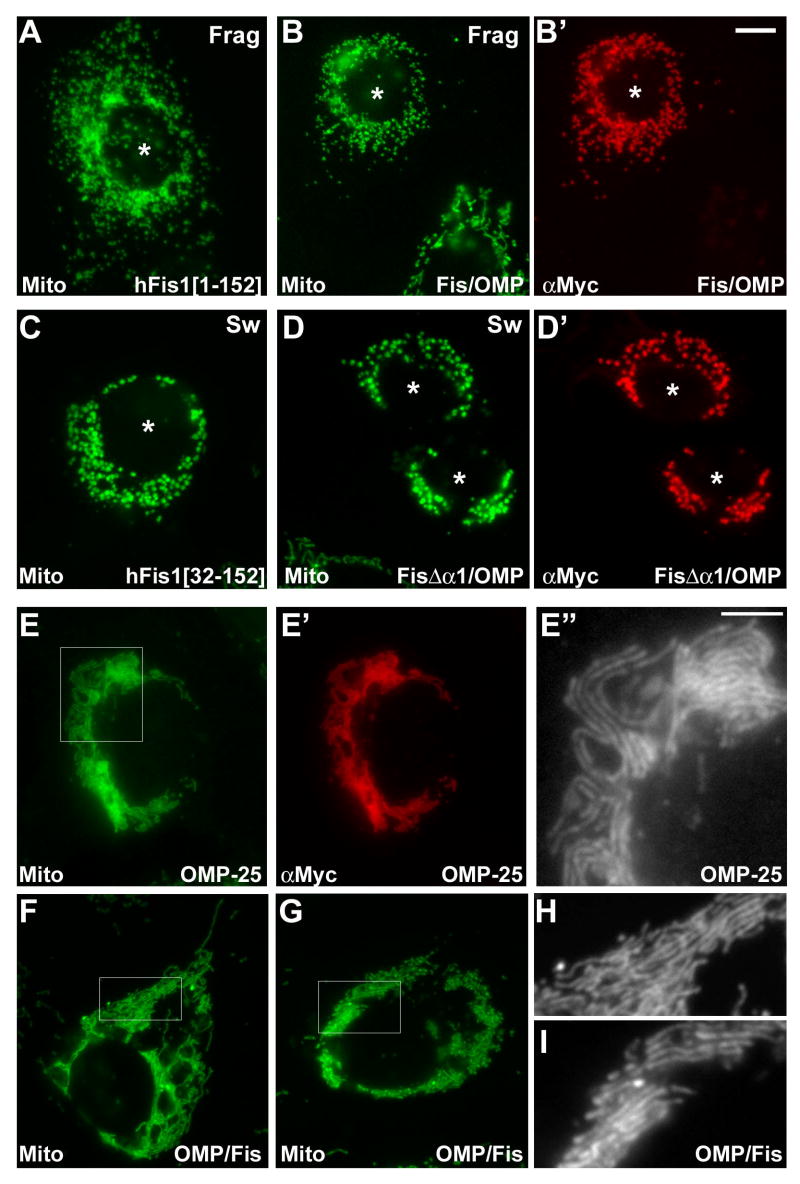
Figure 2. The N-terminal cytosolic domain of hFis1 contains the necessary information for hFis1-specific function.
Chimeric proteins between hFis1 and OMP-25 were transfected into Clone 9 cells expressing GFP in mitochondria and immunostained with anti-Myc antibody. Fis/OMP that has the hFis1 cytosolic domain and the OMP-25 TM/C is localized to mitochondria (B′) and induces mitochondrial fragmentation (B), which is the same functional characteristics as the wild type hFis1 (A). (C) Overexpression of α1-deleted hFis1 (Myc-hFis1[32-152]) induces the formation of swollen ball-shape mitochondria. The chimera that has the α1 deletion in the Fis/OMP (FisΔα1/OMP) causes the same swollen mitochondrial morphology (D′) and correctly distributes to the mitochondria (D). Asterisks in A-D′ denote transfected cells. Myc-OMP-25 is localized to mitochondria (E′) and induces perinuclear clustering of mitochondria (E). Higher magnification image show that mitochondrial tubules are aligned and stacked (E″). (F, G, H, I) The chimera that has the OMP-25 cytosolic domain and hFis1 TM/C induces the same mitochondrial phenotype as the wild type Myc-OMP-25. Panels E″, H, and I are enlarged images of the boxed regions in E, F, and G, respectively. The scale bar in the panel E″ (5 μm) represents the magnification of the images E″, H, and I, whereas the one in B′ (10 μm) is for the rest of the images.
We and others have shown that the overexpression of α1-deleted hFis1 induces the formation of swollen ball-shaped mitochondria (Fig. 2C) [18, 22]. Deletion of the α1 helix from the full-length chimera (FisΔα1/OMP) distributed to mitochondria and induced the formation of ball-shaped mitochondria (Fig. 2D, D′), identical to the morphology induced by overexpression of hFis1[32-152] (Fig. 2C). Cell counting for the different mitochondrial morphologies indicated no significant difference between hFis1 and Fis/OMP or between hFis1Δα1 and FisΔα1/OMP (supplemental figure 1). These results suggest that the hFis1 TM/C region is dispensable for the hFis1-specific function. In support of this notion, the converse chimera that has N-terminal OMP25 and the TM/C from hFis1 (OMP/Fis) showed perinuclear clustering of mitochondria, the same phenotype that was reported in cells overexpressing wild type OMP25 [21]. Interestingly, we found that mitochondria are uniquely organized in OMP25 overexpressing cells. Higher magnification images revealed that mitochondrial tubules appeared to interact laterally displaying stacks of mitochondrial tubules aligned in a same direction (Fig. 2E, E″). Although the nature of this morphology is unknown, the same mitochondrial phenotype was observed with the chimera OMP/Fis (Fig. 2F-I), suggesting that this chimera preserves the OMP25-specific function. Taken together, these data indicate that transmembrane and tail regions of hFis1 and OMP25 are interchangeable without affecting their respective functions in mitochondrial morphology and that the C-terminus of hFis1 (aa 121-152) can be substituted by the one from OMP25 (aa 119-145) for proper mitochondrial targeting and membrane anchoring. Importantly, these results demonstrate that the N-terminal domain of hFis1 exposed to the cytosol contains the necessary information for the fission activity of hFis1.
hFis1 forms oligomers in an α1-helix-dependent manner
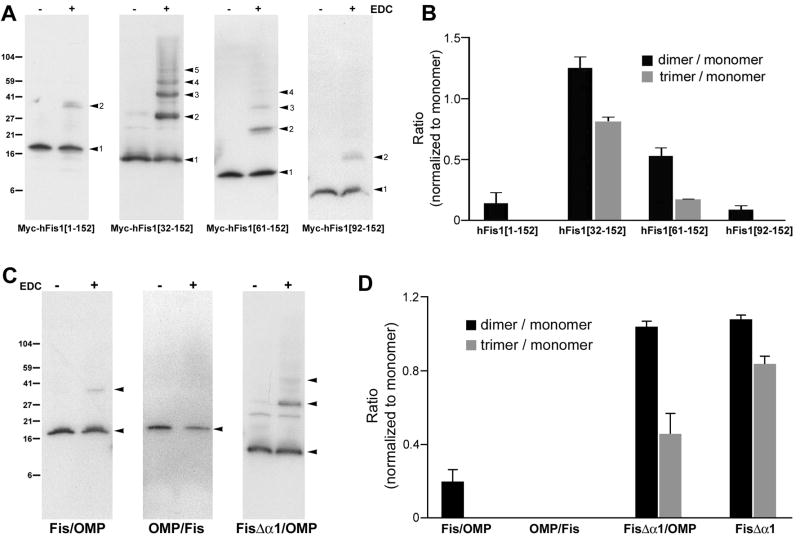
It has been shown that mutations in the hFis1 TPR helices cause a dominant-negative effect when overexpressed [18]. One possibility for this dominant-negative phenotype is that the mutant protein binds to endogenous hFis1 to inhibit the fission process, suggesting that hFis1 self-interaction may be necessary for mitochondrial fission. The results from the chimeric proteins demonstrated that the functional information of hFis1 resides within the N-terminal cytosolic domain containing the TPR motifs. It is possible that this functional information may include self-interaction of hFis1. Therefore, we carried out whole cell crosslinking using the zero-length crosslinker EDC in order to capture the state of hFis1 molecules in the cell. Transient transfection of Myc-tagged full-length hFis1 (Myc-hFis1[1-152]) (Fig. 1B) was performed, followed by EDC crosslinking on whole cells. Upon immunoblotting of crosslinked cell lysate with anti-Myc antibody, we detected a small amount of a dimeric band (Fig. 3A), suggesting that hFis1 self-interacts to form dimers. A doublet band was occasionally observed at the dimer position, suggesting that endogenous hFis1 might also be crosslinked to the transfected Myc-tagged protein. To test the contribution of the hFis1 cytosolic domain to the self-interaction, we performed the crosslinking experiments with cells transfected with N-terminal deletion mutants (Fig. 1B). Surprisingly, we found that deletion of the α1 helix (Myc-hFis1[32-152]) greatly increased the dimer formation (Fig. 3A). In addition, we observed a discrete ladder pattern of hFis1 with single molecule-size increments, indicative of homo-oligomer formation of the α1-deleted hFis1. We were able to detect up to pentameric hFis1[32-152]. Formation of hFis1 oligomers along with the greatly increased dimer formation upon α1 deletion suggests that the α1 helix functions as a negative element for hFis1 self-interaction. The hFis1 mutant missing the α1-α3 (Myc-hFis1[61-152]) showed an oligomer pattern similar to the Myc-hFis1[32-152], but in a reduced extent. Experiments with cells expressing the hFis1 mutant lacking a majority of its N-terminal sequence (deletion of the helices α1-α4 and part of α5: Myc-hFis1[92-152]) displayed no oligomer formation with a small amount of occasional dimer. These results suggest that the region responsible for hFis1 oligomer formation may reside downstream of the first TPR motif, and possibly includes the second TPR motif of hFis1. For the quantitative measure of the hFis1 homo-oligomerization, densitometric analyses were performed in which dimer and trimer band intensities were normalized against the monomer intensity within the crosslinked hFis1 protein. Although the expression levels of different hFis1 constructs often vary in transient transfection, this quantification method allows correct assessment of the oligomerization efficiency of given hFis1 proteins regardless of their varying expression levels. As shown in Fig. 3B, the α1 deletion greatly increased dimeric and trimeric forms and further deletions decreased them.
Figure 3. hFis1 forms oligomers in an α1-helix dependent manner and the oligomerization occurs within the cytosolic domain.
(A) EDC crosslinking of cells transiently expressing different Myc-tagged hFis1 constructs. Cell lysates were separated by SDS-gel electrophoresis and hFis1 proteins were detected by anti-Myc immunoblotting. Myc-hFis1[32-152] and Myc-hFis1[61-152] show oligomeric complexes whereas Myc-hFis1[1-152] and Myc-hFis1[92-152] showed only a small amount of dimer. (B) Densitometric analyses of the hFis1 oligomerization. Monomeric, dimeric, and trimeric bands were subjected to densitometry, and densities of dimer and trimer were normalized to that of the monomeric band. Results from three different experiments were analyzed. Error bars represent SEM. (C) EDC crosslinking of cells expressing the chimeric constructs. OMP/Fis shows no complex formation whereas a small amount of dimer is seen in Fis/OMP. The α1-deleted chimera FisΔα1/OMP shows an increased dimer and trimer formation. (D) Densitometric analyses for oligomerization of chimeras. The same method described in (B) was used.
Self-interaction of hFis1 occurs within the N-terminal cytosolic domain
To test whether the hFis1 TM/C is necessary for the hFis1 self-interaction, we used hFis1/OMP25 chimeric constructs for crosslinking experiments. We found that cells transfected with the Fis/OMP chimera in which the TM/C of hFis1 was substituted with that from OMP25 showed a small amount of dimeric band (Fig. 3C, D), same as the full-length hFis1. In addition, the α1-deleted chimera (FisΔα1/OMP) increased the levels of dimeric and oligomeric bands similar to Myc-hFis1[32-152] (Fig. 3C, D). These results indicate that the hFis1 TM/C region is not required for the oligomer formation. Accordingly, the converse OMP/Fis chimera showed no dimeric and oligomeric bands (Fig. 3C, D), indicating that dimerization and oligomerization by self-interaction are specific features of the hFis1 cytosolic domain. Together with morphological data shown in Fig. 2, these results suggest that the hFis1 self-interaction within the cytosolic domain participates in the hFis1-mediated change of mitochondrial morphology.
hFis1 oligomerization requires membrane anchoring
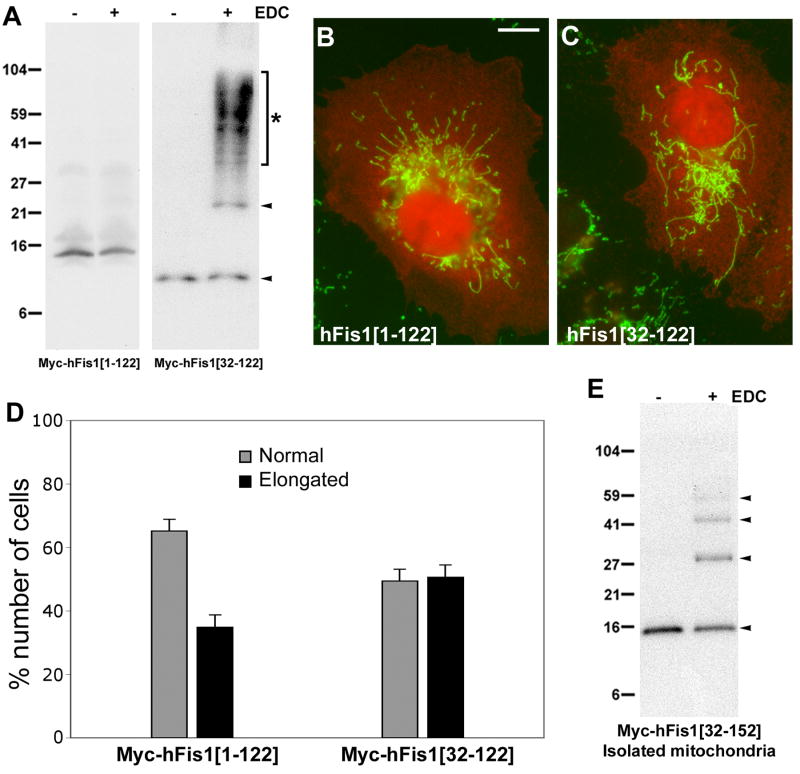
To further test whether the N-terminal cytosolic domain of hFis1 is sufficient to form oligomers, we performed crosslinking experiments using cells transfected with constructs in which the TM/C region is deleted. We previously reported that the deletion of this region causes diffuse cytosolic distribution of the protein due to the loss of mitochondrial membrane anchoring [13]. The ΔTM/C construct that has the intact N-terminal region (Myc-hFis1[1-122]) showed no detectable oligomeric band, consistent with the notion that the α1 helix is an inhibitory element for hFis1 oligomerization (Fig. 4A). Immunostaining showed that Myc-hFis1[1-122] was localized to the cytosol and failed to induce mitochondrial fragmentation (Fig. 4B). Upon deletion of the α1 helix along with the TM/C (Myc-hFis1[32-122]), we observed a drastic increase of crosslinking. However, unlike the membrane-anchored α1 deletion that showed a discrete oligomeric ladder pattern upon crosslinking, this cytosolically localized mutant displayed a large smear between 30 and 104 kD in addition to the dimeric band (Fig. 4A). The absence of discrete bands upon crosslinking indicates that this mutant protein can bind to a multitude of proteins in the cytosol, presumably due to the exposed TPR region from α1-deletion. Despite the α1 deletion, Myc-hFis1[32-122] was unable to induce swollen ball-shaped mitochondria without mitochondrial localization (Fig. 4C). Cells overexpressing Myc-hFis1[1-122] or Myc-hFis1[32-122] contained tubular mitochondria but portions of transfected cells (approximately 35 and 50%, respectively) showed elongated and entangled mitochondria, indicative of blocked fission (Fig. 4C, D).
Figure 4. Membrane anchoring of hFis1 is necessary for oligomerization.
(A) Crosslinking of Myc-hFis1[1-122] and Myc-hFis1[32-122]. The lack of the C-terminal transmembrane and tail domain in these constructs results in the loss of membrane anchoring. No crosslinked bands were detected with Myc-hFis1[1-122] whereas a high molecular weight smear between the 27 and 104 kD markers (asterisk) was seen upon crosslinking of cells expressing Myc-hFis1[32-122]. (B, C) Clone 9 cells harboring GFP in mitochondria were transfected with Myc-hFis1[1-122] (B) and Myc-hFis1[32-122] (C), and immunofluorescence was performed using anti-Myc antibodies. Both mutants showed diffuse cytosolic distribution (red fluorescence). Mitochondrial morphologies were tubular with both mutants, indicating the loss of the ability to induce fragmentation and swelling. Mitochondria in many transfected cells were elongated and entangled (C), indicating blocked fission. Scale bar: 10 μm. (D) Cell counting for mitochondrial morphology shows that 35 and 50% of cells transfected with Myc-hFis1[1-122] and Myc-hFis1[32-122], respectively, contain elongated mitochondria. (E) Crosslinking of the mitochondrial fraction isolated from cells expressing Myc-hFis1[32-152]. Mitochondria were isolated by differential centrifugation. Isolated mitochondria were resuspended in the crosslinking buffer and subjected to the EDC crosslinking. Dimeric, trimeric, and tetrameric bands were detected, indicating the presence of oligomeric forms of hFis1.
Inability of Myc-hFis1[32-122] to form oligomers even without the α1-helix suggests that the correct anchoring of hFis1 in the membrane is necessary for hFis1 oligomerization. Without anchoring on the mitochondrial membrane, randomly oriented hFis1 in the cytosol may not be able to take a preferred binding configuration for oligomerization. To further support this notion, we isolated the mitochondrial fraction from cells transfected with Myc-hFis1[32-152] and performed crosslinking. As shown in Fig. 4E, EDC crosslinking of isolated mitochondria showed the characteristic oligomeric ladder pattern of hFis1 bands. These results indicate that hFis1 membrane anchoring is necessary for oligomerization, presumably by providing the correct orientation of hFis1 molecules on the membrane.
The α3-α4 linker and the α5 helix contribute to the hFis1 oligomerization
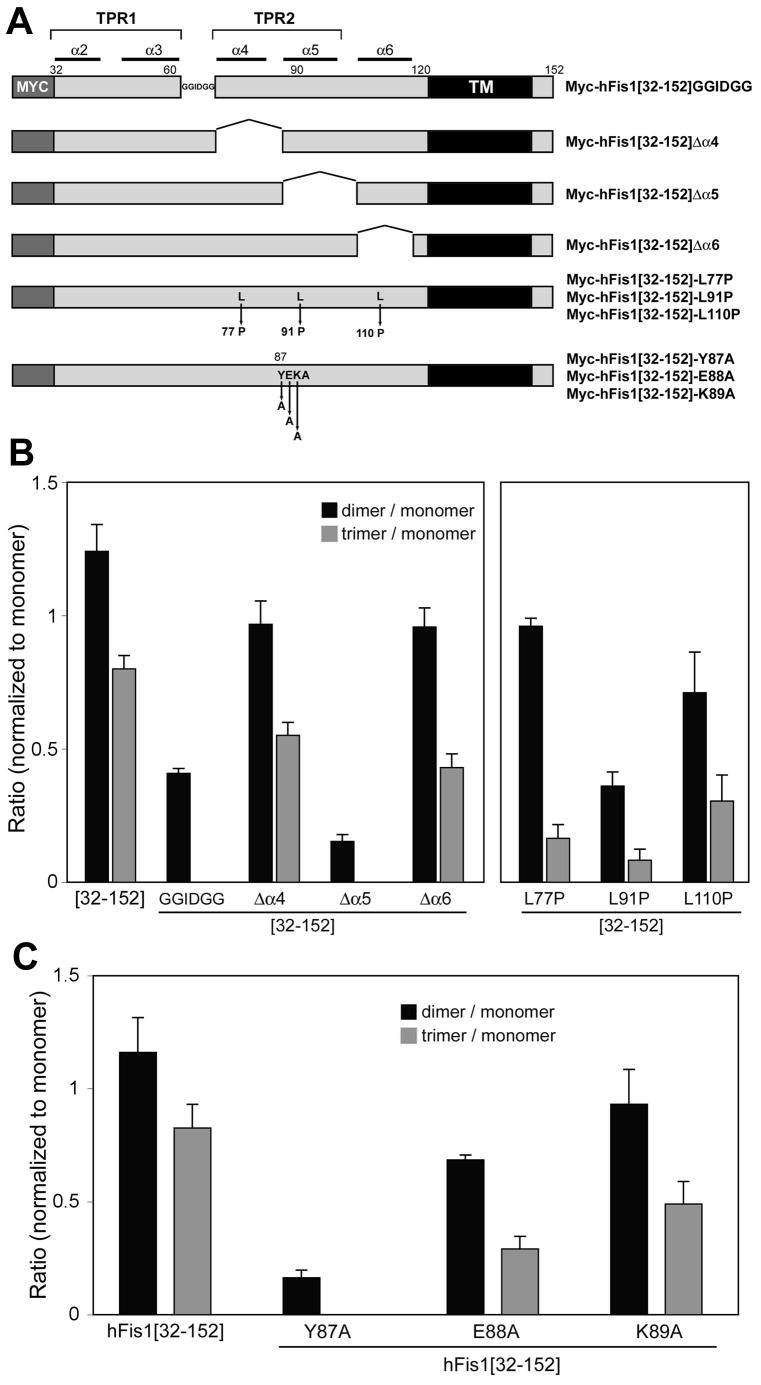
Our data suggest that the amino acids [62-91] including the second TPR fold (α4-α5) may contain the hFis1-oligomerizing motif (Fig. 3). To determine the hFis1 regions participating in the oligomer formation, we made three additional deletion constructs. We deleted amino acids [64-70] (KGSKEEQ) and replaced them with GGIDGG to test the region linking α3 and α4 helices (Myc-hFis1[32-152]GGIDGG, Fig. 5A). For testing α4 and α5 helices, amino acids [71-86] and [87-104], respectively, were deleted (Myc-hFis1[32-152]Δα4 and Myc-hFis1[32-152]Δα5, Fig. 5A). These mutant constructs were made in the Myc-hFis1[32-152] background to test the efficiency of oligomer formation. The EDC crosslinking of cells expressing Myc-hFis1[32-152]GGIDGG or Myc-hFis1[32-152]Δα5 showed a significant decrease in oligomer formation whereas the α4 deletion (Myc-hFis1[32-152]Δα4) did not affect the oligomerization. Approximately 3 and 8 fold decreases were observed in dimer formation with Myc-hFis1[32-152]GGIDGG and Myc-hFis1[32-152]Δα5, respectively, compared to that with Myc-hFis1[32-152] (Fig. 5B). No trimer or higher order oligomers were detected with these two mutants. Additional crosslinking of cells expressing the α6 deletion mutant showed an oligomer formation similar to that of the α4 deletion mutant. In support of these results, the helix-breaking mutation in α5 (Myc-hFis1[32-152]-L91P) caused a reduction in oligomerization whereas the α4 and α6 mutants (Myc-hFis1[32-152]-L77P and L110P) showed no significant effect on oligomerization (Fig. 5B), indicating that the correct conformation of the α5 helix is important for hFis1 oligomerization. These experimental results suggest that the region linking α3 and α4 helices as well as the α5 helix contribute to hFis1 oligomer formation.
Figure 5. Identification of hFis1 regions participating in oligomerization.
(A) Internal deletion and point mutants used for crosslinking experiments. In Myc-hFis1[32-152]GGIDGG, the loop sequence KGSKEEQ that links the α3 and α4 helices was substituted with GGIDGG. Amino acids [71-86], [87-104], and [105-118] were deleted in Myc-hFis1[32-152]Δα4, Myc-hFis1[32-152]Δα5, Myc-hFis1[32-152]Δα6, respectively. Helix-breaking L-to-P mutations (L77P, L91P, and L110P) were made in α4, α5, and α6 helices. The three amino acids in the α5-helix were mutated to alanine (Y87A, E88A, and K89A). (B, C) Densitometric analyses of hFis1 oligomeric bands for crosslinking experiments. Dimeric and trimeric band densities were normalized against the monomer density. Results from three different experiments were analyzed. Error bars represent SEM. Myc-hFis1[32-152]GGIDGG and Myc-hFis1[32-152]Δα5 showed a marked decrease in oligomerization whereas Δα4 and Δα6 formed dimers and trimers to the extent comparable to Myc-hFis1[32-152] (B). The α5 helix-breaking mutation (L91P) also decreased oligomerization, but the ones in the α4 and α6 did not (B). No trimer formation and very little dimer were observed with Myc-hFis1[32-152]-Y87A (C), indicating that the tyrosine residue in the α5 helix plays a role in hFis1 oligomerization.
Because deletions of internal helices are likely to change the overall folding and structure of hFis1, which potentially causes nonspecific effects, we turned to point mutations. In our deletion experiments, Myc-hFis1[92-152] (Δ1-91) and the α5 deletion (Δ87-104) showed a great reduction in oligomerization whereas the α4 deletion (Δ71-86) had little effect, suggesting that the first five amino acids of the α5 helix (87-91: YEKAL) may contain important information for the hFis1 oligomer formation. Therefore, we made single point mutations at the Y87, E88, and K89 positions by changing to alanine residues in the Myc-hFis1[32-152] background (Fig. 5A). Upon crosslinking of cells transfected with these mutants, Myc-hFis1[32-152]-Y87A displayed a drastic reduction in oligomerization, showing a small amount of dimer and no trimer or higher order oligomers (Fig. 5C). However, Myc-hFis1[32-152]-E88A and Myc-hFis1[32-152]-K89A still showed significant dimer and trimer formation. These data indicate that, along with the correct conformation of α5 as shown with L91P mutation, the specific tyrosine residue (Y87) in the α5 helix plays an important role in hFis1 oligomerization.
Oligomerization-defective hFis1 loses the ability to induce ball-shaped mitochondria
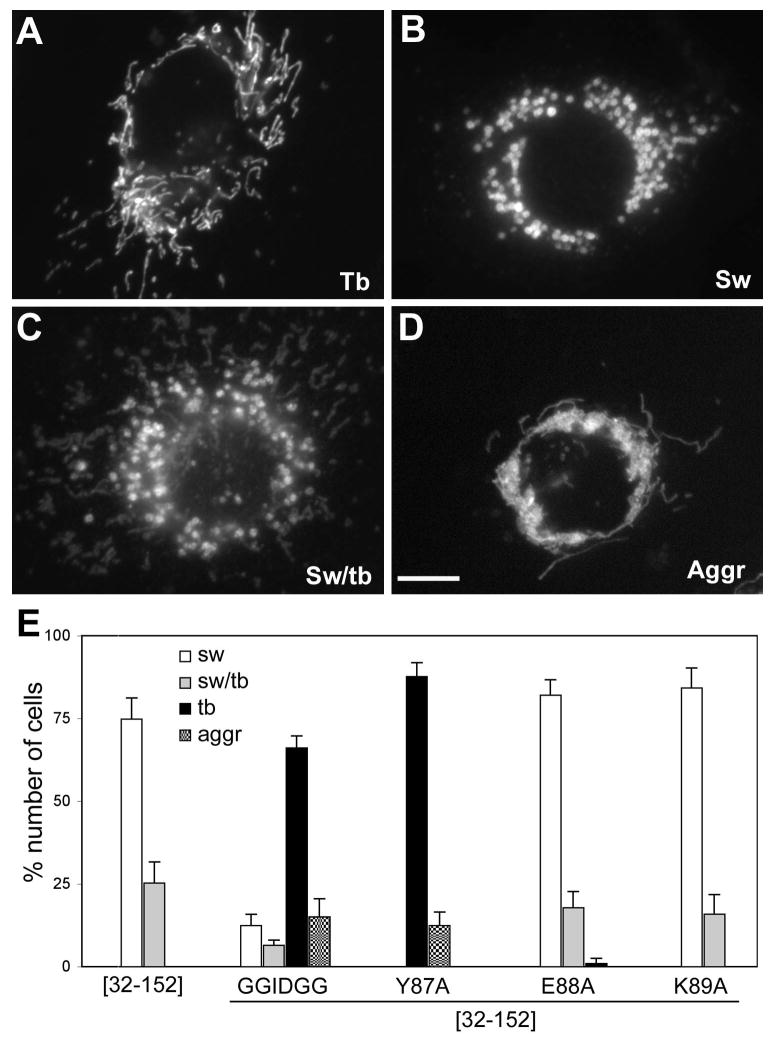
Overexpression of the α1-deleted hFis1 induces swollen ball-shaped mitochondria (Fig. 2C) [18, 22]. In the current study, we found that the α1 deletion also increases the hFis1 oligomer formation, suggesting that increased oligomerization of hFis1 could contribute to the swollen mitochondrial phenotype. To test this, we examined the effects of the oligomerization-defective mutations on the formation of swollen mitochondria. Approximately 75% of the cells overexpressing the α1-deleted hFis1 (Myc-hFis1[32-152]) contained swollen mitochondria and the rest showed a mixture of swollen and tubular mitochondria (Fig. 6B, C and E). Transfection of the empty vector DNA did not have effect on mitochondrial morphology, showing normal tubular mitochondria in more than 95% of transfected cells, similar to untransfected cells. The point mutants that did not affect the oligomerization (Myc-hFis1[32-152]-E88A and Myc-hFis1[32-152]-K89A) still induced the swollen phenotype to an extent similar to the Myc-hFis1[32-152] upon overexpression (Fig. 6E). However, we observed that overexpression of Myc-hFis1[32-152]GGIDGG or Myc-hFis1[32-152]-Y87A that has greatly reduced oligomerization abolished the formation of swollen ball-shaped mitochondria. Cell counting for different mitochondrial morphologies indicate that very few cells transfected with oligomerization-defective mutants contained swollen mitochondria. Less than 20% of cells expressing Myc-hFis1[32-152]GGIDGG showed either swollen or a mixture of swollen and tubular mitochondrial morphology and more than 65% displayed the normal tubular mitochondrial phenotype (Fig. 6A and E). The abolition of mitochondrial swelling was more pronounced in cells expressing Myc-hFis1[32-152]-Y87A, showing no swollen mitochondria in transfected cells (Fig. 6E). In addition, a small portion of cells transfected with Myc-hFis1[32-152]GGIDGG or Myc-hFis1[32-152]-Y87A displayed an aggregated and collapsed mitochondrial phenotype (Fig. 6D and E). These morphological data suggest that the oligomeric form of hFis1 stabilized by the α1 deletion may cause the formation of ball-shaped mitochondria.
Figure 6. Mitochondrial swelling is abolished in cells expressing oligomerization-defective mutants.
Clone 9 cells harboring GFP in the mitochondrial matrix were transfected with different constructs, and mitochondrial morphologies were evaluated. (A, B, C, D) Different mitochondrial morphologies: normal tubular morphology (A, Tb), swollen ball-shape mitochondria (B, Sw), The mixture of swollen and tubular mitochondria (C, Sw/tb), and aggregated and collapsed mitochondria (D, Aggr). Scale bar: 10μm. (E) Cell counting for different mitochondrial phenotypes. More than 300 transfected cells were examined from three separate experiments. Most of the cells overexpressing Myc-hFis1[32-152] contained swollen mitochondria or a mixture of swollen and tubular mitochondria. These phenotypes were mostly absent in cells transfected with the oligomerization defective mutants Myc-hFis1[32-152]GGIDGG and Myc-hFis1[32-152]-Y87A, but not with E88A and K89A mutants. The normal tubular mitochondrial morphology was predominant in cells transfected with the oligomerization-defective mutants. Some cells showed the aggregated and entangled mitochondrial phenotype. Error bars represent SEM.
Oligomerization-defective mutations abolish the increased DLP1-mitochondria association mediated by the α1-deleted hFis1
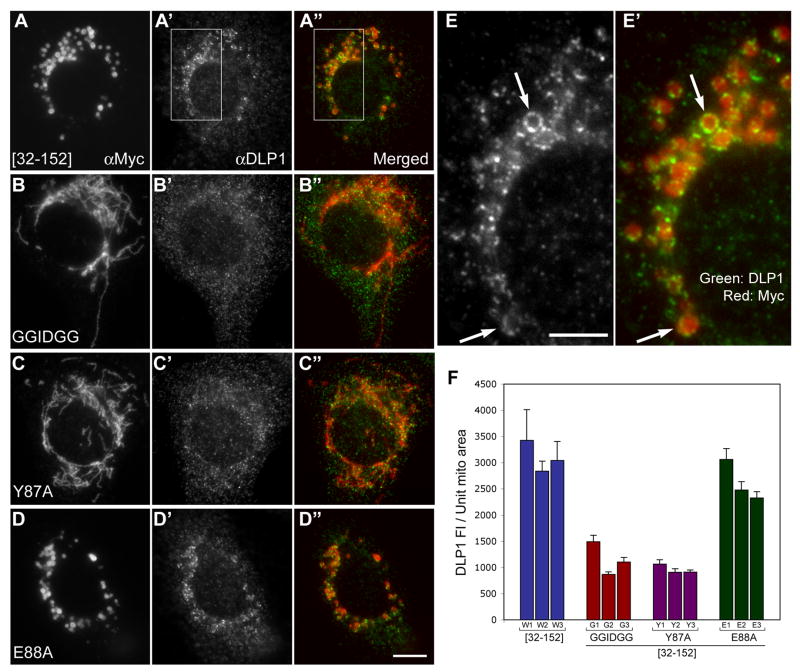
It has been shown that, at any given moment, only a small population of DLP1 distributes to mitochondrial tubules, displaying a punctate pattern often at the mitochondrial tips and constrictions [11, 12]. Our previous study showed an increased association of DLP1 with the α1-deleted hFis1 at the surface of the swollen mitochondria, suggesting a negative regulatory role of the hFis1 α1 in the DLP1 recruitment to mitochondria [18]. Because we observed increased oligomerization of hFis1 by the same α1 deletion, it is possible that hFis1 oligomerization plays a role in DLP1 recruitment to mitochondria. To test this possibility, we performed immunofluorescence for DLP1 in cells overexpressing α1-deleted forms of oligomerization-defective mutants. Because these hFis1 mutant constructs were Myc-tagged and localized to the mitochondrial surface, the DLP1 association with mitochondria was assessed by DLP1 and Myc double labeling. As shown in Fig. 7, DLP1 puncta associate with the swollen mitochondria formed by overexpressed Myc-hFis1[32-152]. DLP1 appeared to form coalesced patches in many mitochondria as well as to coat the entire circumference of swollen mitochondria in some cases (Fig. 7E, E′). However, we found that less DLP1 puncta were associated with mitochondrial tubules in cells overexpressing Myc-hFis1[32-152]GGIDGG or Myc-hFis1[32-152]-Y87A, indicating a reduced binding of DLP1 to these mutant hFis1 on the mitochondrial surface. On the other hand, the Myc-hFis1[32-152]-E88A mutant shows a DLP1 staining pattern around the swollen mitochondria similar to Myc-hFis1[32-152]. For quantitative analyses, we measured the fluorescence intensity of DLP1 associated with a mitochondrion and normalized against the area of the same mitochondrion to calculate the amount of mitochondria-associated DLP1 per unit mitochondrial area. We found a significant reduction of DLP1 associated with mitochondria in cells overexpressing the oligomerization-defective mutants (Fig. 7F). Fluorescence intensity of mitochondria-associated DLP1 was reduced in these mutants by approximately three fold. On the other hand, a similar level of DLP1 was found associated with mitochondria in cells overexpressing the Myc-hFis1[32-152]-E88A mutant, compared to the control Myc-hFis1[32-152]. These results indicate that the oligomerization-defective mutations abolish the increased DLP1-mitochondria association mediated by the α1-deleted hFis1, suggesting that hFis1 oligomerization may play a role in DLP1 recruitment to the mitochondrial membrane.
Figure 7. Oligomerization-defective mutations abolish the ability of the α1-deleted hFis1 to increase the DLP1 association to the mitochondria.
Indirect immunofluorescence was carried out for DLP1 and the Myc epitope in cells transfected with Myc-hFis1[32-152] (A, A', A″, E. E′), Myc-hFis1[32-152]-GGIDGG (B, B', B″), Myc-hFis1[32-152]-Y87A (C, C', C″), and Myc-hFis1[32-152]-E88A (D, D', D″). (A, A', A″) Myc-hFis1[32-152] is localized to the swollen mitochondria (A) brighter DLP1 puncta are concentrated on and around the ball-shape mitochondria (A′). DLP1 forms patches and often coats the entire circumference of swollen mitochondria (arrows in E, E′). (B, B', B″, C, C', C″) No significant DLP1 association is apparent with mitochondrial tubules stained for Myc-hFis1[32-152]-GGIDGG (B, B', B″) and Myc-hFis1[32-152]-Y87A (C, C', C″). (D, D', D″) Myc-hFis1[32-152]-E88A mutant show the DLP1 staining pattern around the swollen mitochondria (D', D″) similar to the Myc-hFis1[32-152]. (E, E′) Enlarged images of boxed regions in A′ and A″, respectively. The scale bar in the panel E (5 μm) represents the magnification of the images E and E′ whereas the one in D″ (10 μm) is for the rest of the images. (F) Quantification of DLP1 association with mitochondria. 5 to 11 isolated, discernable mitochondria were selected from each cell and the mitochondrial area was measured. Total DLP1 fluorescence associated with a given mitochondrion was measured and normalized against the area of the same mitochondrion to calculate the amount of mitochondria-associated DLP1 per unit mitochondrial area. The graph shows the measurements of three individual cells from Myc-hFis1[32-152] (W1-W3), Myc-hFis1[32-152]-GGIDGG (G1-G3), Myc-hFis1[32-152]-Y87A (Y1-Y3), and Myc-hFis1[32-152]-E88A (E1-E3). An approximately three-fold reduction of mitochondria-associated DLP1 fluorescence was observed in oligomerization-defective mutants compared to Myc-hFis1[32-152] and Myc-hFis1[32-152]-E88A. Error bars represent SEM.
hFis1 oligomerization is necessary for mitochondrial fission
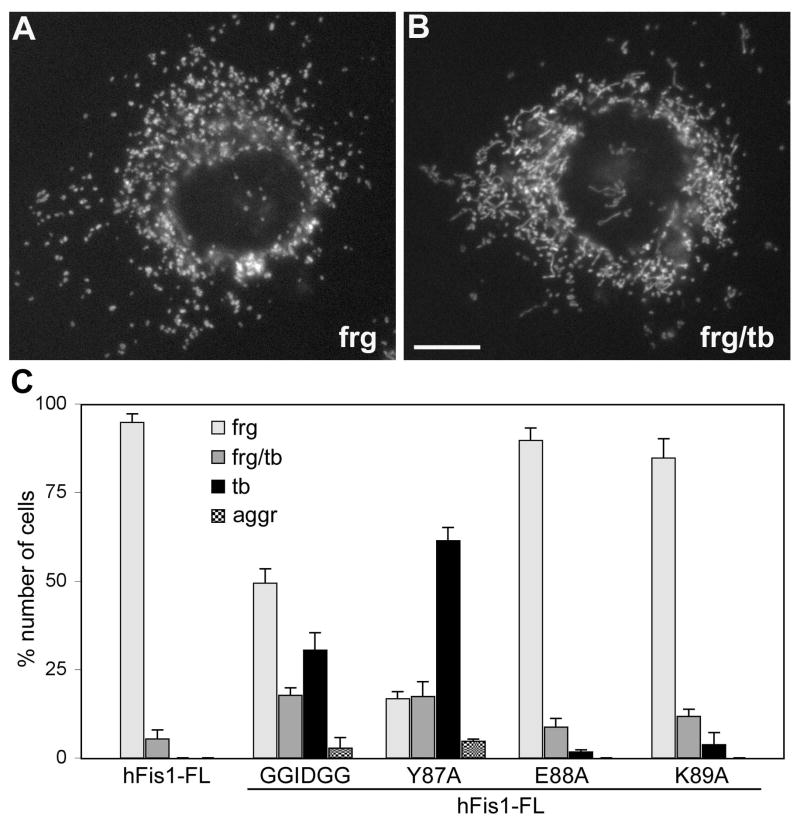
Increased number of hFis1 molecules in the mitochondrial surface by overexpression increases the fission frequency, inducing mitochondrial fragmentation (Fig. 2A) [13, 15, 18]. Therefore, we used hFis1 overexpression-induced mitochondrial fragmentation as a functional readout to test whether hFis1 oligomerization is necessary for mitochondrial fission. To this end, we made the oligomerization-defective mutations in the hFis1 full-length background, and examined their effects on mitochondrial morphology. Overexpression of wild type hFis1 causes mitochondrial fragmentation in a majority of the transfected cells (Fig. 8A). The fragmented mitochondrial phenotype was also observed upon overexpression of the mutants Myc-hFis1-E88A or Myc-hFis1-K89A, indicating these mutations have no significant effect on hFis1 function (Fig. 8C). However, Myc-hFis1-Y87A and Myc-hFis1-GGIDGG greatly reduced the ability to induce mitochondrial fragmentation upon overexpression. Cells containing fragmented mitochondria comprise less than 20% of the cells transfected with the Y87A mutant and more than 60% of them showed tubular mitochondria (Fig. 8C). Another 17% of transfected cells showed a mixture of fragmented and tubular mitochondria. A small population (<5%) showed an aggregated mitochondrial phenotype, the same phenotype shown in Fig. 6D. The GGIDGG mutation was less effective than the Y87A in preventing mitochondrial fragmentation. Approximately 50% and 30% of the cells overexpressing Myc-hFis1-GGIDGG contained fragmented and tubular mitochondria, respectively (Fig. 8C). These results indicate that both mutants have a reduced ability to promote mitochondrial fission upon overexpression, which is presumably caused by the decreased self-interaction. These data demonstrate that Y87 and the α3-α4 linker are necessary for mitochondrial fission, suggesting that hFis1 oligomerization plays an important role in hFis1-mediated mitochondrial fission.
Figure 8. Oligomerization-defective mutants have a diminished ability to induce mitochondrial fragmentation.
Mitochondrial morphologies were examined in cells transfected with different hFis1 constructs. Overexpression of full-length Myc-tagged wild type hFis1 caused the fragmented mitochondrial phenotype (A, frag). Some cells contained a mixture of fragmented and tubular mitochondria (B, frg/tb). Scale bar: 10μm. (C) Cell counting of more than 300 cells transfected with each construct was performed in three experiments. Decreased numbers of cells showed fragmented mitochondria among cells transfected with the oligomerization-defective mutants, GGIDGG and Y87A. Myc-hFis1-E88A and K89A had no significant effect on mitochondrial fragmentation upon overexpression. Error bars represent SEM.
Discussion
Although mitochondrial fission mediated by DLP1 and hFis1 has been under investigation for some time, the details of the mitochondrial fission process is still elusive. Because DLP1 is a dynamin-like GTPase, it is presumed to act as a mitochondrial scission protein, but the definitive action of DLP1 during mitochondrial fission has not been demonstrated. Studies in yeast provided information that Fis1p (hFis1 homologue) acts as a fission factor receptor [23-25] and that Mdv1p or Caf4p as a linker protein interacting with both Dnm1p (DLP1 homologue) and Fis1p [26-28]. Interestingly, studies with both yeast and mammalian cells indicated that functions of these mitochondrial fission proteins appear to be shared in the division process of peroxisomes [29-33], suggesting that metabolic cross-talk between the two organelles might have contributed to the sharing of the fission machinery [34].
Structural studies of hFis1 and Fis1p produced important information regarding how this molecule might interact with other fission proteins [19, 20, 35, 36]. Our previous studies indicate that hFis1 interacts with a fission complex containing DLP1 through the hFis1 TPR in a transient manner, and this interaction is regulated by the α1 helix of hFis1 [18]. In the present study, using chimera, mutation, and crosslinking approaches, we identified hFis1 self-interaction as a potential mechanism for the hFis1-mediated mitochondrial fission.
Morphological data obtained using chimeric proteins between hFis1 and OMP25 showed that the cytosolic domain of hFis1 contains information necessary for inducing mitochondrial fragmentation or swelling (Fig. 2). In addition, results from chimera experiments suggest that the transmembrane and short C-terminal sequences of hFis1 and OMP25 were functionally interchangeable in providing mitochondrial localization. However, a recent report indicates that the hFis1 C-terminal sequence may contain an additional function in inducing apoptosis independent of mitochondrial fission [22]. Overexpression of hFis1 induces mitochondrial fragmentation and leads to apoptosis [14, 18]. However, a conservative mutation at the C-terminal lysine148 to arginine abolishes the hFis1-induced apoptosis although it still induces mitochondrial fragmentation, suggesting that two processes, mitochondrial fission and apoptosis, mediated by hFis1 are separate events that require the cytosolic domain and the C-terminal sequence, respectively [22]. While testing the apoptosis-inducing activity in chimera experiments is beyond the scope of this study, our results confirmed that the hFis1 function necessary for mitochondrial fission is confined within the cytosolic domain (Fig. 2).
The zero-length crosslinker EDC forms a crosslinking intermediate with a very short half-life before covalently linking a carboxyl group to a primary amine. Therefore, EDC would preferably crosslink between molecules whose interaction interface is close to each other and relatively stable, which renders a higher specificity of crosslinking between interacting molecules. Indeed, we were able to detect the specific oligomerization of α1-deleted hFis1 by EDC crosslinking. We found that deletion of the α1 helix greatly increased the oligomer formation. Given that we detected oligomers of the α1-deleted hFis1 using EDC that preferentially crosslinks more stably interacting molecules, we predict that the α1 helix may play a role in destabilizing the hFis1 oligomer. Because no oligomer formation was detected in the presence of α1, it is possible that the functional oligomer formation of hFis1 in cells is a transient event.
Based on the EDC crosslinking of overexpressed full-length hFis1 molecule, endogenous hFis1 is not expected to show the oligomer pattern due to the presence of the α1 helix that has a negative effect on oligomer formation. Our crosslinking experiments for cellular endogenous hFis1 protein showed a small amount of the potential dimeric band (supplementary figure 2), suggesting that endogenous hFis1 behaves similarly to the transfected full-length hFis1 in self-interaction.
Crosslinked α1-deleted hFis1 showed one molecule-size increments, indicating that hFis1 forms homo-oligomers (Fig. 3). While we detected up to the pentamer-sized oligomer in our crosslinking, it is possible that the actual hFis1 oligomer could be larger. Although a previous study with rat Fis1 suggested that it forms a ∼200 kD complex [37], it is not clear whether the complex corresponds to the homo-oligomeric form of Fis1. Native gel electrophoresis and immunoprecipitation were used to identify the rat Fis1 complex [37], which did not discriminate Fis1 homo-oligomers from other interactions. Based on the potentially unstable nature of the oligomeric form of the full-length hFis1 as suggested in our study, it is possible that the 200-kD rat Fis1 complex detected after the detergent extraction represents a more stable complex containing Fis1 and other proteins rather than the Fis1 homo-oligomer. In that study, the self-interaction of rat Fis1 detected by immunoprecipitation has been shown to require the intact transmembrane region, partly contradicting to our result. Although the reason for this discrepancy is currently unclear, our experimental data using hFis1/OMP25 chimeric constructs clearly demonstrate that the cytosolic domain of hFis1 contains all the necessary information for not only mitochondrial fission but also oligomerization of hFis1 (Figs. 2 and 3). Additional analyses using deletion and point mutations suggest that the α3-α4 linker and the α5 helix contribute to the hFis1 oligomer formation (Fig. 5).
It is interesting that the crosslinking of hFis1[32-122] produces a high molecular weight smear (Fig. 4). This observation indicates that the exposure of the TPR in the cytosol allows the binding of a multitude of proteins. The lack of the hFis1 oligomeric ladder in the hFis1[32-122] crosslinking indicates that hFis1 membrane anchoring is necessary for its oligomerization. In addition, this observation also suggests that the correct membrane localization of hFis1 may render a specificity of protein-protein interactions involving hFis1 during mitochondrial fission. Morphological observations indicate that overexpression of mutants lacking TM/C induced a fission-defective mitochondrial phenotype. It is possible that the overexpressed hFis1 mutants sequester hFis1-binding proteins in the cytosol, preventing the normal interaction with hFis1 in mitochondria for fission. One of these proteins was identified to be cyclin B2 by mass spectrometric analyses (M. N. Serasinghe and Y. Yoon, unpublished). Cdk1/cyclin B has been shown to phosphorylate the fission protein DLP1 in mitotic cells to fragment mitochondria [38]. hFis1 on the mitochondrial surface may provide a scaffold for cdk1/cyclin B and DLP1 during mitotic fission of mitochondria. The absence of high molecular weight complex formation with hFis1[1-122] suggests that the access to the binding site (TPR) may be physically hindered by the sequence within the first 31 amino acids of hFis1. The N-terminal sequence of yeast Fis1p has been shown to bind to the TPR region [35]. A recent report indicated that the deletion of this N-terminal “arm” greatly increases the Dnm1p binding to the Fis1p TPR region [25], supporting our findings that the hFis1 α1 helix negatively regulates fission factor binding, potentially by modulating hFis1 oligomerization.
Overexpression of hFis1 increases the mitochondrial fission frequency and causes mitochondrial fragmentation via DLP1 activity [13, 15, 18]. Our morphological data showed that the oligomerization-defective mutants have a greatly reduced ability to induce mitochondrial fragmentation when overexpressed, suggesting that hFis1 oligomerization plays a role in mitochondrial fission (Fig. 8). The same mutations in the α1-deleted hFis1 almost completely abolished the ability to form swollen ball-shaped mitochondria (Fig. 6). It has been suggested that the formation of ball-shaped mitochondria is induced by increased DLP1 interaction with the α1-deleted hFis1 at the mitochondrial surface [18]. It is possible that the oligomeric form of hFis1 may bind to a DLP1-containing complex. Indeed, our data indicate that oligomerization-defective mutants lose the ability to increase the DLP1 association to mitochondria when the α1-helix is deleted (Fig. 7). Perhaps, transient assembly and disassembly of hFis1 oligomers on the mitochondrial surface recruits and releases the DLP1-containing complex to the fission site through a manner regulated by hFis1 α1-helix [18].
According to the structure solved through X-ray crystallography and NMR [19, 20], the two potential interacting regions that we identified, the α3-α4 linker and Y87, lie on opposite sides of the α4-helix. Because these two regions are not spatially close to each other within a single hFis1 molecule, it is likely that these two regions are involved in an inter-molecular interaction rather than an intramolecular interaction. Dimeric structures have been observed in the protein crystal of the hFis1 cytosolic domain [19]. This dimer appears to be formed by interaction of α1 helix of one molecule with the concave surface formed by α2, α4, and α6 helices of the other molecule. While this interaction suggests the ability of the α1 helix to interact with downstream helices either intra- or inter-molecularly, the participation of the α3-α4 linker and Y87 in this particular dimer formation is currently unclear. We speculate that the α3-α4 linker and Y87 would be in close proximity between adjacent hFis1 molecules anchored in the membrane with the same orientation, allowing lateral interactions that lead to the oligomerization. One attractive scenario in this type of interaction is that, due to the relatively fixed positions of these two sites within the rigid helix bundle, the interaction of the two sites would occur with a tilted angle. Thus, a string of interactions of multiple hFis1 on the mitochondrial tubule would form a helical ring-like structure instead of a closed ring. Furthermore, because of the presence of the transmembrane anchor, the hFis1 helical assembly with a fixed angle may generate a membrane tension that may produce a force to constrict the membrane tubules. Dynamins are presumed to form helical ring-like structures with a defined dimension for membrane scission, which may fit to the membrane constriction. Perhaps, hFis1 oligomerization may provide a site for DLP1 action, serving as a template for DLP helical ring assembly and also possibly mediating the initial constriction of the mitochondrial tubule.
In this study, we have shown that hFis1 can self-interact to form homo-oligomeric complexes, which is negatively regulated by the α1-helix. Two potential interaction sites were identified in the second half of the TPR-like folds. Our data suggest that oligomerization of hFis1 in the mitochondrial outer membrane plays an important role in mitochondrial fission, possibly by mediating DLP1 recruitment.
Supplementary Material
Supplementary figure 1. Quantification of mitochondrial morphologies in cells transfected with hFis1/OMP25 chimeric proteins. 200-300 cells transfected with each construct were counted for morphology assessment. (A) Fis/OMP chimera in which the C-terminal region of hFis1 is substituted with that of OMP25 caused the fragmented mitochondrial phenotype, similar to the one seen in cells overexpressing full-length hFis1. (B) Similar to cells overexpressing α1-deleted hFis1 (hFis1Δα1), swollen mitochondria are prevalent in cells overexpressing the α1-deleted form of the Fis/OMP chimera (FisΔα1/OMP). Frag: cells containing fragmented mitochondria, Frag/Tb: cells containing both fragmented and tubular mitochondria, Tb: cells containing tubular mitochondria, Sw: cells containing swollen mitochondria, Sw/Tb: cells containing both swollen and tubular mitochondria. Error bars represent SEM.
Supplementary figure 2. Detection of potential dimer of endogenous hFis1. BHK 21 cells were crosslinked with 0.2 and 1.0 mM EDC and crosslinked proteins were subjected to immunoblotting using anti-hFis1 antibodies. Similar to overexpressed full-length hFis1, a small amount of potential dimer was observed upon the EDC crosslinking. Because this dimer-sized band was also faintly visible in the uncrosslinked sample, it is possible that it is a non-specific band or a stable SDS-resistant hFis1 dimer.
Acknowledgments
This study was supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (DK061991 and DK073858).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yoon Y. Regulation of mitochondrial dynamics: Another process modulated by Ca2+ signal? Sci STKE. 2005;2005:pe18. doi: 10.1126/stke.2802005pe18. [DOI] [PubMed] [Google Scholar]
- 2.Yoon Y. Sharpening the scissors: mitochondrial fission with aid. Cell Biochem Biophys. 2004;41:193–205. doi: 10.1385/CBB:41:2:193. [DOI] [PubMed] [Google Scholar]
- 3.Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int Rev Cytol. 1990;122:1–63. doi: 10.1016/s0074-7696(08)61205-x. [DOI] [PubMed] [Google Scholar]
- 4.Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, Bhattacharya SS, Wissinger B. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- 6.Niemann A, Ruegg M, La Padula V, Schenone A, Suter U. Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot-Marie-Tooth disease. J Cell Biol. 2005;170:1067–1078. doi: 10.1083/jcb.200507087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schroder JM, Vance JM. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 8.Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel CP. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 9.Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 10.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Pitts KR, Yoon Y, Krueger EW, McNiven MA. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol Biol Cell. 1999;10:4403–4417. doi: 10.1091/mbc.10.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 15.Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J Cell Sci. 2004;117:1201–1210. doi: 10.1242/jcs.01058. [DOI] [PubMed] [Google Scholar]
- 16.Tondera D, Czauderna F, Paulick KRS, Kaufmann J, Santel A. The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J Cell Sci. 2005;118:3049–3059. doi: 10.1242/jcs.02415. [DOI] [PubMed] [Google Scholar]
- 17.Yoon Y, Pitts KR, McNiven MA. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol Biol Cell. 2001;12:2894–2905. doi: 10.1091/mbc.12.9.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu T, Fox RJ, Burwell LS, Yoon Y. Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1. J Cell Sci. 2005;118:4141–4151. doi: 10.1242/jcs.02537. [DOI] [PubMed] [Google Scholar]
- 19.Dohm JA, Lee SJ, Hardwick JM, Hill RB, Gittis AG. Cytosolic domain of the human mitochondrial fission protein Fis1 adopts a TPR fold. Proteins. 2004;54:153–156. doi: 10.1002/prot.10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki M, Jeong SY, Karbowski M, Youle RJ, Tjandra N. The solution structure of human mitochondria fission protein Fis1 reveals a novel TPR-like helix bundle. J Mol Biol. 2003;334:445–458. doi: 10.1016/j.jmb.2003.09.064. [DOI] [PubMed] [Google Scholar]
- 21.Nemoto Y, De Camilli P. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 1999;18:2991–3006. doi: 10.1093/emboj/18.11.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alirol E, James D, Huber D, Marchetto A, Vergani L, Martinou JC, Scorrano L. The Mitochondrial Fission Protein hFis1 Requires the Endoplasmic Reticulum Gateway to Induce Apoptosis. Mol Biol Cell. 2006;17:4593–4605. doi: 10.1091/mbc.E06-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated Mitochondrial Fission Is a Multi-step Process Requiring the Novel Integral Membrane Component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karren MA, Coonrod EM, Anderson TK, Shaw JM. The role of Fis1p-Mdv1p interactions in mitochondrial fission complex assembly. J Cell Biol. 2005;171:291–301. doi: 10.1083/jcb.200506158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells RC, Picton LK, Williams SC, Tan FJ, Hill RB. Direct binding of the dynamin-like GTPase, Dnm1, to mitochondrial dynamics protein Fis1 is negatively regulated by the Fis1 N-terminal arm. J Biol Chem. 2007;282:33769–33775. doi: 10.1074/jbc.M700807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tieu Q, Okreglak V, Naylor K, Nunnari J. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J Cell Biol. 2002;158:445–452. doi: 10.1083/jcb.200205031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerveny KL, Jensen RE. The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol Biol Cell. 2003;14:4126–4139. doi: 10.1091/mbc.E03-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffin EE, Graumann J, Chan DC. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J Cell Biol. 2005;170:237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motley AM, Hettema EH. Yeast peroxisomes multiply by growth and division. J Cell Biol. 2007;178:399–410. doi: 10.1083/jcb.200702167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motley AM, Ward GP, Hettema EH. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J Cell Sci. 2008;121:1633–40. doi: 10.1242/jcs.026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2005;16:5077–86. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Gould SJ. The dynamin-like GTPase DLP1 is essential for peroxisome dvision and is recruited to peroxisomes in part by PEX11. J Biol Chem. 2003;4:4. doi: 10.1074/jbc.M212031200. [DOI] [PubMed] [Google Scholar]
- 33.Koch A, Thiemann M, Grabenbauer M, Yoon Y, McNiven MA, Schrader M. Dynamin-like protein 1 is involved in peroxisomal fission. J Biol Chem. 2003;278:8597–8605. doi: 10.1074/jbc.M211761200. [DOI] [PubMed] [Google Scholar]
- 34.Schrader M, Yoon Y. Mitochondria and peroxisomes: are the ‘big brother’ and the ‘little sister’ closer than assumed? Bioessays. 2007;29:1105–14. doi: 10.1002/bies.20659. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki M, Neutzner A, Tjandra N, Youle RJ. Novel structure of the N terminus in yeast Fis1 correlates with a specialized function in mitochondrial fission. J Biol Chem. 2005;280:21444–21452. doi: 10.1074/jbc.M414092200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Chan DC. Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc Natl Acad Sci U S A. 2007;104:18526–18530. doi: 10.1073/pnas.0706441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jofuku A, Ishihara N, Mihara K. Analysis of functional domains of rat mitochondrial Fis1, the mitochondrial fission-stimulating protein. Biochem Biophys Res Commun. 2005;333:650–659. doi: 10.1016/j.bbrc.2005.05.154. [DOI] [PubMed] [Google Scholar]
- 38.Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Quantification of mitochondrial morphologies in cells transfected with hFis1/OMP25 chimeric proteins. 200-300 cells transfected with each construct were counted for morphology assessment. (A) Fis/OMP chimera in which the C-terminal region of hFis1 is substituted with that of OMP25 caused the fragmented mitochondrial phenotype, similar to the one seen in cells overexpressing full-length hFis1. (B) Similar to cells overexpressing α1-deleted hFis1 (hFis1Δα1), swollen mitochondria are prevalent in cells overexpressing the α1-deleted form of the Fis/OMP chimera (FisΔα1/OMP). Frag: cells containing fragmented mitochondria, Frag/Tb: cells containing both fragmented and tubular mitochondria, Tb: cells containing tubular mitochondria, Sw: cells containing swollen mitochondria, Sw/Tb: cells containing both swollen and tubular mitochondria. Error bars represent SEM.
Supplementary figure 2. Detection of potential dimer of endogenous hFis1. BHK 21 cells were crosslinked with 0.2 and 1.0 mM EDC and crosslinked proteins were subjected to immunoblotting using anti-hFis1 antibodies. Similar to overexpressed full-length hFis1, a small amount of potential dimer was observed upon the EDC crosslinking. Because this dimer-sized band was also faintly visible in the uncrosslinked sample, it is possible that it is a non-specific band or a stable SDS-resistant hFis1 dimer.