SUMMARY OF RECENT ADVANCES
Developmental patterning events involve cell fate specification and maintenance processes in diverse, multicellular organisms. The simple arrangement of tissue layers in the Arabidopsis thaliana root provides a highly tractable system for the study of these processes. This review highlights recent work addressing the patterning of root tissues focusing on the factors involved and their complex regulation. In the last two years studies of root patterning have indicated that chromatin remodeling, protein movement, transcriptional networks, and an auxin gradient all contribute to the complexity inherent in developmental patterning events within the root. As a result, future research advances in this field will require tissue specific information at both the single gene and global level.
INTRODUCTION
Over the past two decades studies of primary root patterning in Arabidopsis thaliana initiated using classical genetic approaches were extended to the molecular level by the advent of molecular biology techniques, and then were propelled into the genomics era with the sequencing of the Arabidopsis genome. These approaches have led to substantial insights into cell fate specification and the positioning of the stem cell niche within the root. The simplicity and transparency of the Arabidopsis are the keys that have unlocked these discoveries. The Arabidopsis primary root is composed of concentric rings of tissue layers along the radial axis and morphologically distinguishable developmental zones along the longitudinal axis.
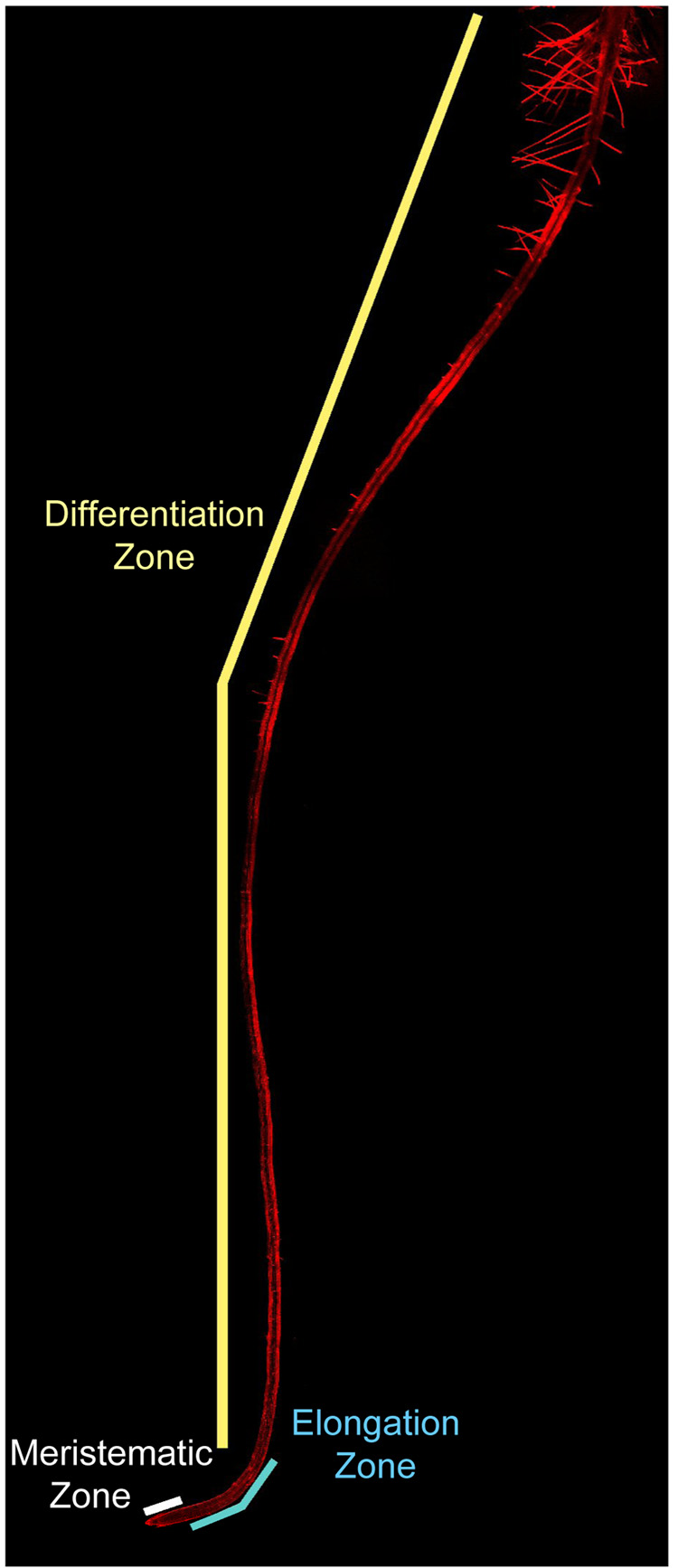
Specifically, from the root tip to the root-stem junction exist distinct zones that are visible as regions of small, actively dividing cells (called meristematic cells), elongated cells, and terminally differentiated cells marked by the root hairs of the epidermis (Figure 1, Figure 3). As such, the position of each cell along this axis can be used to infer its developmental age.
Figure 1.
The Arabidopsis primary root (6 days old) as viewed by laser confocal microscopy under 10X magnification. Three distinct zones mark the longitudinal axis of the root, namely, the meristematic (white bar), elongation (blue bar), and differentiation (yellow bar) zones. A close up of the meristematic and elongation zones is shown in Figure 3. Cell walls are visible in red from propidium iodide staining.
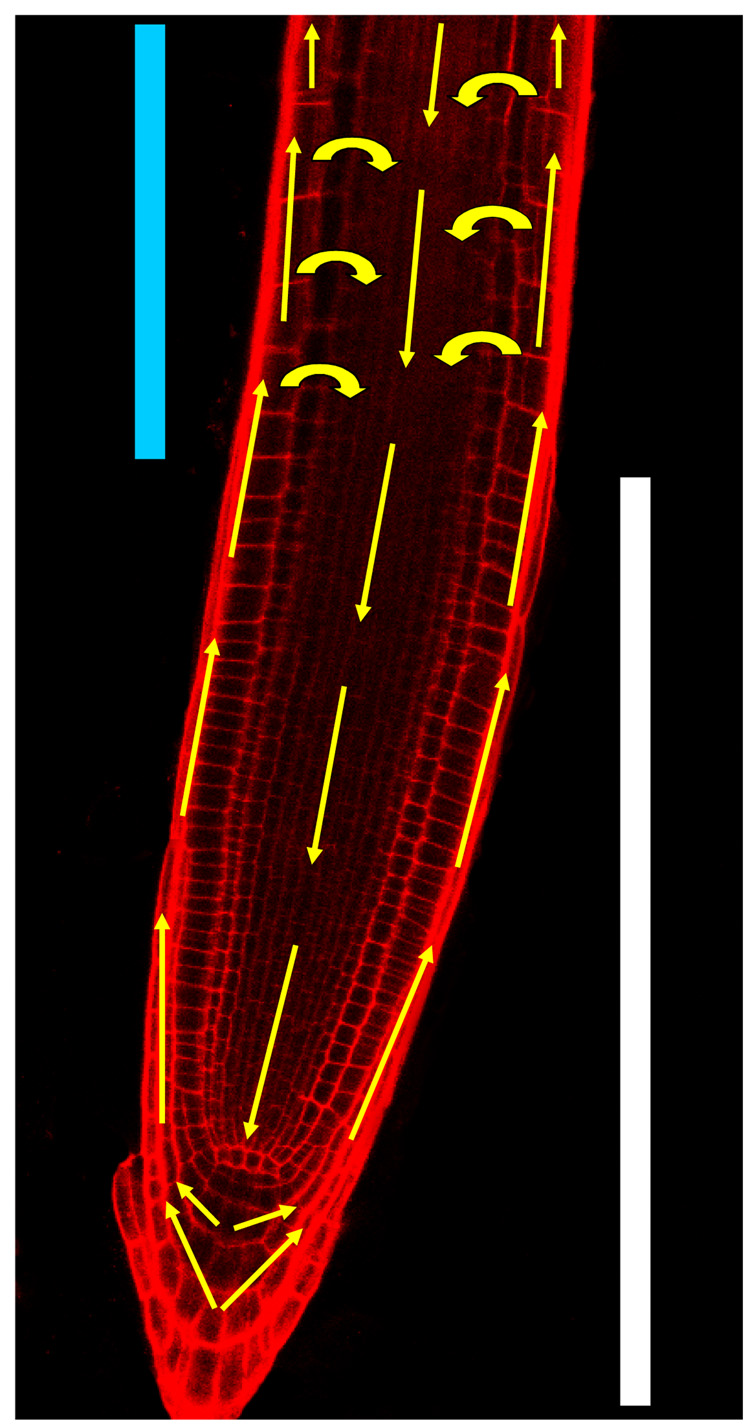
Figure 3.
Simplified schematic of the efflux routes of auxin resembling an inverted fountain in the Arabidopsis primary root (7 days old) as viewed by laser confocal microscopy under 25X magnification. Auxin is thought to be transported from the shoot downward to the root tip via PIN carriers in the central cell layers of the root. Once auxin reaches the tip it is believed to be directed outward to the outer cell layers by different PIN carriers. It is then thought to be transported upward toward the shoot by still other PIN carriers. Some of this auxin may also be recycled from the outer root layers back into the inner ones to provide a ‘battery’ like mechanism that maintains auxin levels within the root tip. The meristematic (white bar) and elongation (light blue bar) zones are shown. Cell walls are visible in red from propidium iodide staining.
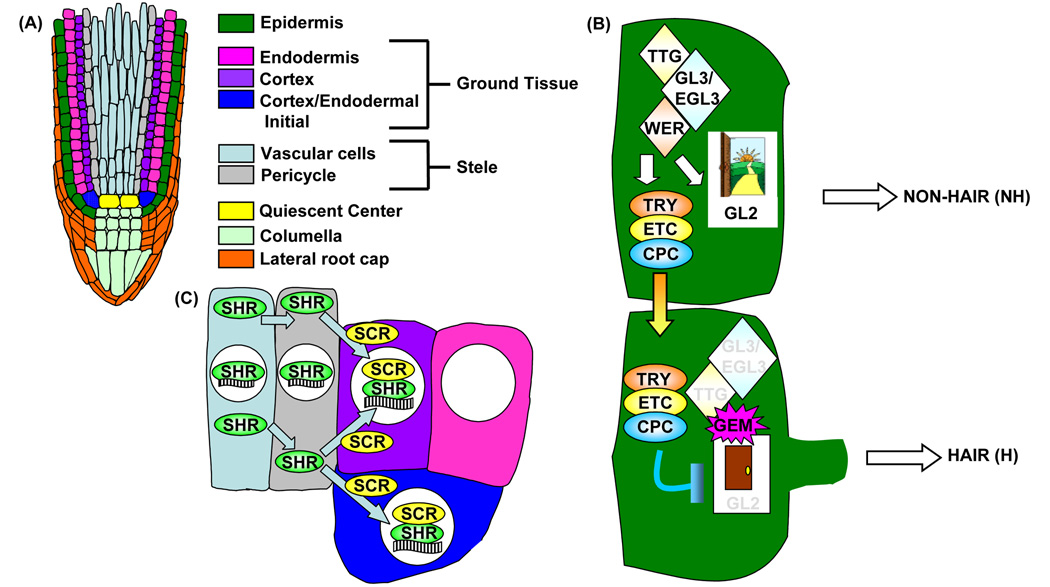
The root has an outer layer of epidermis and an inner core of vascular tissue that is spatially separated by a layer of ground tissue (Figure 2A). This elegant radial organization is derived from asymmetric cell divisions of stem cell initials and the daughter cells they produce. For instance, the ground tissue is generated from asymmetric division of the cortex-endodermal initial cell (CEI) to renew itself and produce a daughter cell (CED) that subsequently divides to generate the endodermal and cortex cell lineages [1•].
Figure 2.
The simple structure of the Arabidopsis primary root is specified by complex regulatory mechanisms. (A) Schematic drawing of a longitudinal slice of the primary root tip. (B) Non-hair epidermal cell fate is specified by high levels of GL2. GL2 is activated by TTG/GL3/EGL3/WER transcriptional complexes that also activate TRY, ETC, and CPC. CPC (and presumably TRY and ETC) moves to neighboring epidermal cells to repress GL2 expression, resulting in hair cell fate. GEM chromatin modifications regulate the ‘open’ or ‘closed’ configuration of the GL2 promoter. (C) SHR moves from the stele into adjacent cells where it is sequestered by SCR into the nucleus. Nuclear SHR-SCR complexes activate SCR expression, specifying the endodermis. White circles in (C) denote the nucleus, waved black/white-striped rectangle represents the SCR promoter DNA. Throughout the figure, cell colors indicate the associated cell-type shown in (A) and colored arrows depict protein movement.
A small population of cells that rarely divides, called the quiescent center (QC) is surrounded by undifferentiated stem cells, such as the CEI, from which the different tissue layers arise. Analogous to animal systems, the QC and stem cells of the Arabidopsis root, together termed the plant stem cell niche, possess the ability to renew themselves and are essentially ageless unlike the daughter cells they produce [2, 3]. Since plants are immobile, this pattern is not achieved by cell migration as it is in many animals, but rather specified by positional information exchanged between cells [1•].
In the last two years, microarray profiling of these cell populations comprising the QC, root cell layers and longitudinal zones, termed the ‘root map’, has resulted in abundant gene expression information in both space and time with resolution unprecedented in any other multicellular organism [4•]. Modeling of the localization and directional activity of the PINFORMED1 (PIN) proteins that transport auxin has unveiled an auxin gradient robust to perturbations in auxin concentrations [5••]. Remarkably, the PLETHORA (PLT) transcription factors are expressed in graded patterns resembling this auxin gradient and when mutated, shift the boundaries between zones along the longitudinal axis [6•].
In contrast, along the radial axis the GRAS family transcription factor SHORT ROOT (SHR) has been demonstrated to specify a single layer of endodermis within the ground tissue via its movement from the central vascular tissue into neighboring cells where its interaction with a transcription factor of the same family, SCARECROW (SCR), leads to its sequestration into the nucleus. Once in the nucleus, SHR is restricted from moving to outer cell layers and specifies this single layer of endodermis by regulating a number of transcription factors, including SCR which in turn positively regulates itself [7••]. This is just one example of a tightly-controlled, complex regulatory process embedded in the beguilingly simple patterning of the root. A direct link was also recently demonstrated between chromatin remodeling of the upstream region of the HDZIP transcription factor GLABRA2 (GL2) by the GL2 modulator (GEM) and epidermal patterning [8••]. This review focuses on these landmarks in root patterning research in relation to other findings over the last two years in a framework of the root’s tissue layers and their underlying complexity at the molecular level.
From the outside in: Epidermal patterning in the root
Specification of hair (H) versus non-hair (NH) cell fate in the epidermal tissue layer is known to involve an intricate network of transcription factors. Specifying NH cells are the GLABRA2 (GL), GLABRA3 (GL3), ENHANCER OF GLABRA3 (EGL3), TRANSPARENT TEST GLABRA (TTG), and WEREWOLF (WER) transcription factors, while the CAPRICE (CPC), TRIPTYCHON (TRY), and ENHANCER of TRIPTYCHON and CAPRICE (ETC) transcription factors specify H cells [9]. The exact model of NH versus H cell fate is an active subject of controversy in this field. However, most models include results from previous studies suggesting a TTG/GL3/EGL3/WER transcriptional complex binds to the GL2 promoter to repress root hair cell fate, while the same complex simultaneously induces CPC expression in NH cells [9]. CPC then moves into neighboring epidermal cells to repress GL2 expression, resulting in H cell specification [9].
A new player to this model, TRANSPARENT TESTA GLABRA2 (TTG2) was implicated in epidermal cell specification from experiments using transgenic plants expressing a chimeric protein of TTG2 fused to the EAR repression domain (TTG2:SRDX) [10]. In these transgenic roots, GL2 expression, as detected using a GL2:GUS promoter-reporter fusion construct, was repressed resulting in ectopic root hair formation. Further analysis of reporter lines in the TTG2:SRDX background showed that TTG2 itself, GL2, and CPC expression was reduced, while TTG1, WER, GL3, and EGL3 expression was relatively unchanged [10]. These data imply that in NH cells, TTG2 can activate expression of itself, GL2, and CPC independently of a TTG1/WER/GL3/EGL3 transcriptional complex. Using various promoter-deletion constructs and one-hybrid analysis they also showed that WER most likely binds to a specific MYB regulatory element in the TTG2 promoter [10]. Thus while TTG2 can activate GL2 independently, it does not do so until it is activated by the TTG1/WER/GL3/EGL3 complex. This suggests a new step exists in the regulatory cascade of epidermal patterning involving this WRKY family transcription factor in NH cells. However, as ttg2 does not exhibit epidermal patterning defects in the root, it is unclear how critical this new step is in the cascade. This is also true of the newly proposed TRY lateral inhibition feedback loop [11]. The try mutants have a normal epidermal cell type pattern, even though the author’s expression studies of cpc try and gl2 mutants indicate that TRY, like CPC, is part of a regulatory loop in which GL2 promotes TRY expression and then TRY represses GL2 expression in cells located in the H position [11]. Nevertheless, in the future these two loops may prove important in the timing and/or levels of protein movement and transcriptional complex action.
Elegant complementation and gel-shift experiments with chimeric proteins of the R3 MYB regions of WER and CPC revealed that the CPC R3 region cannot functionally substitute for the WER R3 region in H cell differentiation. They also showed that the CPC chimera protein containing the WER R3 motif binds to the GL2 promoter, but not the WER chimera protein containing the CPC R3 [12•]. These data support a model of competition between WER and CPC in transcriptional complexes regulating GL2 expression in epidermal patterning [12•]. The recent identification of yet another CPC gene in Arabidopsis [13] further illustrates the point that the number of transcription factor complexes and combinatorial and competitive interactions is dauntingly large. Mathematical and computational models will be needed to fully understand their dynamics and a comprehensive model of all the factors involved. To test these models, experimental techniques will need to be developed and used to visualize transcription factor complex dynamics and biochemically purify complexes from H or NH cells in planta.
These studies suggest that a delicate balance exists between the different transcription factor complexes converging on promoters of GL2, CPC, and perhaps others, to specify hair and non-hair cells in the appropriate position within the epidermis (Figure 2B). A recent report of the GL2 expression modulator (GEM), a protein identified as interacting with the Arabidopsis homologue of the eukaryotic licensing factor for DNA replication (CDT) [8••], supports a new layer of complexity in the specification of H and NH epidermal cells [8••, 14]. Arabidopsis plants over-expressing GEM1 have reduced GL2 messenger RNA levels that correlate with the increased root hair density of these plants. The authors tested three possibilities for the GEM effect on GL2: (1) Direct binding of the GEM protein to the GL2 promoter, (2) GEM regulates expression of GL2 transcriptional regulators, and (3) GEM is recruited specifically to the GL2 promoter via protein-protein interaction with the TTG-GL3-EGL3-WER/CPC transcriptional machinery. Their results eliminated the former two possibilities and showed by a combination of yeast two- and three-hybrid experiments and pull-down assays that the third possibility was most likely from observed interactions between GEM and TTG1 [8••]. Not stopping there, the authors tested the histone modification status of GL2 and CPC promoters by ChIP experiments. They found that both promoters contained histone H3K9acK14AC acetylation and H3K9 methylation in gem-1 plants that was absent in GEM over-expression plants, suggesting a repressive role for GEM in GL2 and CPC expression by histone modification [8••]. This is exciting because GEM provides a direct connection between the TTG transcriptional complex and chromatin modification of GL2. It will be interesting to learn if this is a tightly controlled switch of cell fate as previously proposed for the role of chromatin remodeling in root epidermal specification [15•]. It is also possible that chromatin remodeling provides a graded response mechanism corresponding to the amount of histone modification. In this context, it will be interesting to see if the degree of chromatin modification correlates with the developmental competence of epidermal cells along the root’s longitudinal axis and/or the duration and intensity of exposure to environmental stresses in the modulation of root epidermal cell specification.
These studies emphasize the importance of future study of transcriptional complex dynamics, protein movement, and histone modification as aspects of regulation in root epidermal patterning. Next steps in the field will also include connecting information from these studies with that from studies regarding root hair outgrowth/shape and Ca++ signaling [16], auxin regulation of root hair positioning within a root hair cell [17•], and the role of signaling in transcriptional control of epidermal patterning [18].
Ground tissue patterning in the root
In the ground tissue patterning field, the buzz was all about the long sought after elucidation of the downstream targets of the GRAS family transcription factor SHORTROOT and its mode of action. The longitudinal cell division of the cortex/endodermal initial daughter cell (CED) does not occur in shr mutants and due to this only a single layer with cortex features is present in these mutants [19–20].
Meta-analysis of the results from microarray experiments of shr-2, an inducible SHR line in the shr-2 background, and cells sorted for SHR:GFP expression identified 8 direct targets of SHR [21•]. Four of these were confirmed in vivo by ChIP-qPCR including the predicted target, the GRAS transcription factor SCARECROW (SCR), and two closely related C2H2 zinc finger genes NUTCRACKER (NUC) and MAGPIE (MGP) [21•]. The recently identified JACKDAW (JKD) is also a member of this subfamily [22]. SCR expression is abated in jkd mutant roots and yeast-two hybrid and onion bombardment Bimolecular Fluorescent Complementation (BiFC) assays suggest JKD interacts with itself as well as with SCR and SHR [21]. Taken together, the studies in [21•] and [22] imply a regulatory network exists between the GRAS family transcription factors and this C2H2 zinc finger subfamily. It will be interesting to see if the protein interactions found in yeast and onion cells can be demonstrated in Arabidopsis and how the different complexes formed by these families act to effect transcriptional changes. Based on the sizes of these families, one can imagine the interactions may be as entangled and challenging to study as those of the proteins involved in epidermal patterning mentioned above.
SHR action has been a hot topic since the vascular tissue (stele)-specific gene expression of SHR was first reported, seemingly at odds with the loss of endodermis observed in the ground tissue of shr-2 mutants [19]. The SHR protein was later shown to move from the stele into the adjacent ground tissue, where it became nuclear localized [23–24]. However, while there were hints that this movement was limited by SCR [24], the mechanism for it remained unknown. Confocal microscopy of RNAi lines knocking down SCR (SCRi) to different levels and expressing SHR-GFP and pSCR::GFP led to insights into this mechanism [7••]. SCRi lines displayed supernumerary layers inversely correlating with SCR transcript levels and both pSCR::GFP and SHR-GFP were expressed in these layers, SHR-GFP primarily being localized to the nucleus. SHR-GFP was present in both daughter cells of the CEI and after each additional division the level of SHR-GFP was reduced [7••]. These results, taken together with the authors’ ChIP-qPCR data and demonstrated in vivo protein-protein interaction between SHR and SCR [7••], strongly support their hypothesis that SCR restricts SHR movement by sequestering it into the nucleus to create a SHR/SCR-dependent positive feedback loop for SCR transcription specifying endodermis (Figure 2C).
Studying the dynamics of SHR/SCR is an obvious next step. Another question is: are there other proteins that facilitate SHR movement?
In summary, this ground tissue patterning research points to exciting downstream transcriptional targets that now can be linked to later differentiation processes and an enlightening mechanism for how cell fate can be specified by a combination of protein movement, interaction, and transcription control. So far, the SCR/SHR pathway does not seem to involve the plant hormone auxin. This is intriguing given that auxin seems to be implicated in most developmental processes of the root, including QC and stele function as discussed in the next sections.
Stele patterning in the root
Little is known about the factors involved in the specification of cell types in the stele [25]. The role of auxin is prominent in this tissue and the action of PINFORMED (PIN) auxin transporters affects vascular patterning in the stele [26]. This is perhaps due to the comparatively complex number and locations of different cell types within this tissue. Genes expressed in protophloem have recently been identified from enhancer trap screens [27], including the previously characterized transcription factor BREVIS RADIX (BRX) that mediates feedback between brassinosteroids and the plant hormone auxin [28•,29–30]. Regulation of the bilateral symmetry within the stele was recently shown to be eliminated in lonesome highway roots [31]. As this gene encodes a protein with similarity to bHLH transcription factors, it will be interesting to know if it interacts with SHR and/or BRX to control stele patterning as well as to know of the nature of any overlap between the targets of these transcription factors.
Patterning the root’s stem cell niche
Auxin response and transport are central to recent work on the patterning of the root’s stem cell niche. A maximum of auxin response visualized by reporter genes up-regulated by auxin corresponds to the position of the QC [32], suggesting the QC’s position is defined by this maximum. Protonated auxin can move into cells by passive diffusion, while the PIN auxin transporters facilitate the movement of negatively ionized auxin out of cells because auxin is a weak acid and the extracellular pH is lower than cytoplasmic pH in root cells [33]. ‘Inverted fountain models’ have been proposed based on the asymmetric distribution of PIN proteins within single root cells to describe the direction and magnitude of auxin fluxes (Figure 3) [34].
Based largely on these models combined with experimental information about the spatial localization of the PIN family members in the root, and accounting for simple diffusion of auxin, a recent mathematical model correctly predicted the position of this auxin maximum, but also proposed an auxin gradient in the root [5••]. Although direct quantification of auxin levels in individual cells would definitively prove the existence of this gradient, this has not yet been achievable in plants. Moreover, while the authors base their model largely upon PIN localization, they do not describe simulations of auxin gradients expected in pin mutants. Despite this, the model has an impressive ability to simulate a variety of perturbations to the auxin maximum that correctly match experimental observations, including laser ablation of the QC, high levels of auxin applied to the root, and amazingly, decapitation of the plant removing the root’s main source of auxin [5••]. Decapitated plants were able to survive for 10–30 days depending upon the amount of ‘reflux’, which depended upon PIN localization to the lateral face of cells to direct auxin flux back into the downward-directed flow within the vascular tissue [5••]. In this sense, the root possesses an auxin ‘battery’ that holds charge, but slowly loses it at a rate proportional to inefficient reflux.
Remarkably, the PLETHORA (PLT) transcription factors are expressed in a pattern resembling this gradient [6•, 35]. Various plt double and triple mutants have reductions in PIN expression [6•, 35], suggesting a connection between PLTs and an auxin gradient involving PINs. A direct relationship between PLTs and PINs has not yet been demonstrated, but one may not expect there to be one, because QC ablation experiments suggest there is a significant lag time between the appearance of the QC auxin maxima and PIN protein localization [36•].
A host of transcription factors involved in QC specification and maintenance may function during this lag time, including WUSCHEL-RELATED HOMEOBOX 5 (WOX5), SHR, and SCR, whose expression have already been shown to appear during it [36•]. Since SHR, SCR, and WOX5 have roles in the QC [19–20, 23, 37–40], they represent attractive candidates for connecting PLTs and PIN derived gradients. Knowledge of the PLT transcriptional targets will be pertinent to future evaluations of their role in the auxin gradient, but information about PLT upsteam regulators will be as well. Interestingly, double mutants in the recently identified OBERON1 and OBERON2 plant homeodomain finger proteins were shown to lack PLT1, WOX5, and SCR expression, suggesting the nuclear-localized OBE1 and OBE2 may be upstream regulators in QC identity and specification [41].
Although this information about PLT pathways will be valuable, knowledge of PIN localization, regulation, and dynamics is just as beneficial to understanding the nature of an auxin gradient guiding root patterning and growth. It is thus worthwhile for researchers to consider how transcriptional information impacts and is integrated with other proteins modulating PIN, such as P-glycoproteins, AUXIN RESPONSE FACTORS, VACUOLAR PROTEIN SORTING 29, protein phosphatases 2A/ROOT CURLING ON NPA, and PINOID kinases [42–49].
CONCLUSION
Study of the patterning mechanisms establishing and maintaining the patterning of the Arabidopsis root began over two decades ago. In the last two years, it has become increasingly clear that the deceivingly simple structure of this organ is specified and maintained by many, potentially redundant, factors. Recent work has uncovered complex layers of regulation controlling these root patterning factors, such as chromatin remodeling, protein movement, transcriptional complexes, and an auxin gradient. An additional regulator that was not mentioned here is the potential role of small RNAs in root patterning [50]. Genome-wide studies of protein abundance and interactions, small RNAs, and histone modification at the resolution level achieved in [4•] should be of considerable use in future research dissecting this complexity.
ACKNOWLEDGEMENTS
Funding for work on radial patterning was from NIH (GM043778), MCB (0618304), and P50 (1P50-GM081883) grants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1. Benfey PN, Scheres B. Root development. Curr Biol. 2000;10:R813–R815. doi: 10.1016/s0960-9822(00)00814-9. This is considered by many as the basic primer of root development.
- 2.Jiang K, Feldman LJ. Regulation of root apical meristem development. Annu Rev Cell Dev Biol. 2005;21:485–509. doi: 10.1146/annurev.cellbio.21.122303.114753. [DOI] [PubMed] [Google Scholar]
- 3.Scheres B. Stem-cell niches: nursery rhymes across kingdoms. Nat Rev Mol Cell Biol. 2007;8:345–354. doi: 10.1038/nrm2164. [DOI] [PubMed] [Google Scholar]
- 4. Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. This is an excellent resource for gene expression in both space and time within the root for your gene(s) of interest. Dominant expression patterns were identified in both space and time using clustering methods. Interestingly, potentially oscillating gene expression patterns were also uncovered.
- 5. Grieneisen VA, Xu J, Marée AF, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. A strong model of biological patterning reproduces not only steady state patterns, but also patterns resulting from alterations in model components and environmental conditions. The authors describe a model that does just this by using known localization patterns of PIN proteins and modeling diffusion and PIN-facilitated auxin transport in and across cells within a virtual root lattice of appropriately sized and shaped cells. Simulations of the model produce the known auxin maximum present in the plant stem cell niche as well as the ‘battery’ power of this auxin maximum (the ability to maintain the maximum in the absence of shoot-derived auxin).
- 6. Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. The authors draw connections between a putative auxin gradient, PLETHORA expression and function, and root patterning using plt single, double, and triple mutants. In an interesting experiment, they use ink toner to mark the boundary of the root elongation zone before induction of 35S-PLT2-GR to show overexpression of PLT both inhibits cell expansion within the elongation zone and sustains cell division of only the cells that are still cycling.
- 7. Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. The authors’ ChIP-qPCR, protein pull-down, and gene dosage experiments unveil a new and dynamic mechanism of cell fate specification of the endodermis by SHR and SCR protein-protein interaction, localization, and a positive feedback loop regulating SCR transcription.
- 8. Caro E, Castellano MM, Gutierrez C. A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature. 2007;447:213–217. doi: 10.1038/nature05763. The authors report the discovery of a protein (GEM) modulating the expression of GLABRA2 (GL2), a transcription factor that specifies epidermal cell fate, via histone modifications. Pull-down and ChIP assays in this work support a GEM interaction with TRANSPARENT TESTA GLABRA1 to modify histones affecting CAPRICE and GL2 expression and their control of epidermal patterning.
- 9.Ishida T, Kurata T, Okada K, Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol. 2008 doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- 10.Ishida T, Hattori S, Sano R, Inoue K, Shirano Y, Hayashi H, Shibata D, Sato S, Kato T, Tabata S, et al. Arabidiopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell. 2007;19:2531–2543. doi: 10.1105/tpc.107.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J. Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev Biol. 2007;311:566–578. doi: 10.1016/j.ydbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 12. Tominaga R, Iwata M, Okada K, Wada T. Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell. 2007;19:2264–2677. doi: 10.1105/tpc.106.045732. Elegant domain swapping experiments performed by the authors elucidate the functional nature of the MYB R3 domain of the MYB family proteins WEREWOLF and CAPRICE in cell fate determination of the epidermis.
- 13.Tominaga R, Iwata M, Sano R, Inoue K, Okada K, Wada T. Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering in addition to trichome and root formation. Development. 2008;135:1335–1345. doi: 10.1242/dev.017947. [DOI] [PubMed] [Google Scholar]
- 14.Wildwater M, The I, van den Heuvel S. Coordination of cell proliferation and differentiation: finding a GEM in the root? Dev Cell. 2006;12:841–842. doi: 10.1016/j.devcel.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 15. Costa S, Shaw P. Chromatin organization and cell fate switch respond to positional information in Arabidopsis. Nature. 439:493–496. doi: 10.1038/nature04269. The authors conduct interesting three-dimensional fluorescence in situ hybridization (3D FISH) studies in various mutant backgrounds. Using a GL2 bacterial artificial chromosome (BAC) they find that chromatin at this locus is ‘open’ in NH cell and ‘closed’ in H cells.
- 16.Takeda S, Gapper C, Kaya H, Bell E, Kuchitsu K, Dolan L. Local positive feedback regulation determine cell shape in root hair cells. Science. 2008;319:1241–1244. doi: 10.1126/science.1152505. [DOI] [PubMed] [Google Scholar]
- 17. Fischer U, Ikeda Y, Ljung K, Serralbo O, Singh M, Heidstra R, Palme K, Scheres B, Grebe M. Vectorial information for Arabidopsis planar polarity is mediated by combined AUX1, EIN2, and GNOM activity. Curr Biol. 2006;16:2143–2149. doi: 10.1016/j.cub.2006.08.091. The authors investigate the relationship between the planar polarity of root hairs and auxin, ethylene, and vesicle trafficking. Although they are not the focus of the article, experiments applying beads with 1-NAA to the outer face of an epidermal cell to study the planar positioning of a root hair along a single epidermal cell are worth checking out.
- 18.Kwak SH, Schiefelbein J. The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Dev Biol. 2007;302:118–131. doi: 10.1016/j.ydbio.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development. 1993;119:57–70. doi: 10.1242/dev.119.Supplement.57. [DOI] [PubMed] [Google Scholar]
- 20.Helariutta Y, Fukari H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- 21. Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4:e143. doi: 10.1371/journal.pbio.0040143. For those interested in addressing developmental questions using microarray analyses, this is a recommended read. The authors describe a meta-analysis of three different microarray experiments that identify robust transcriptional targets.
- 22.Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 21:2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- 24.Sena G, Jung JW, Benfey PN. A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development. 2004;131:2817–2826. doi: 10.1242/dev.01144. [DOI] [PubMed] [Google Scholar]
- 25.Helariutta Y. Cell signalling during vascular morphogenesis. Biochem Soc Trans. 2007;35:152–155.24. doi: 10.1042/BST0350152. [DOI] [PubMed] [Google Scholar]
- 26.Vieten A, Sauer M, Brewer PB, Friml J. Molecular and cellular aspect of auxin-transport-mediated development. Trends Plant Sci. 2007;12:160–168. doi: 10.1016/j.tplants.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 27.Bauby H, Divol F, Truernit E, Grandjean O, Palauqui JC. Protophloem differentiation in early Arabidopsis thaliana development. Plant Cell Physiol. 2007;48:97–109. doi: 10.1093/pcp/pcl045. [DOI] [PubMed] [Google Scholar]
- 28. Mouchel CF, Osmont KS, Hardtke CS. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature. 2006;443:458–461. doi: 10.1038/nature05130. The authors use a combination of microarray, expression, and phenotypic analyses to show that brx root defects are due to a loss of expression of a rate-limiting enzyme in brassinosteroid synthesis. Intriguingly, auxin induces BRX expression AND brx mutants have impaired expression of auxin-response genes, suggesting feedback between brassinosteroid levels and auxin response.
- 29.Mouchel CF, Briggs GC, Hardtke CS. Characterization of the plant-specific BREVIS RADIX gene family reveals limited genetic redundancy despite high sequence conservation. Plant Physiol. 2006;140:1306–1316. doi: 10.1104/pp.105.075382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mouchel CF, Briggs GC, Hardtke CS. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev. 2004;18:700–714. doi: 10.1101/gad.1187704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohashi-Ito K, Bergmann DC. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development. 2007;134:2959–2968. doi: 10.1242/dev.006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malmay J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. An auxin-dependent distal organizer or pattern and polarity in Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- 33.Blakeslee JJ, Peer WA, Murphy AS. Auxin transport. Curr Opin Plan Biol. 2005;8:494–500. doi: 10.1016/j.pbi.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 34.Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 35.Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 36. Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B. A molecular framework for plant regeneration. Science. 2006;20:525–536. doi: 10.1126/science.1121790. The authors employ confocal microscopy in conjunction with laser ablation to study the re-appearance of the quiescent center using fluorescent reporters of QC identity. The ‘new’ QC is marked by these reporters in cells positioned several tiers above the location of the former QC. The reason and mechanism for the upward shift of the QC toward vascular tissue, rather than a downward shift, remains a mystery.
- 37.Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 38.Heidstra R, Welch D, Scheres B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 18:1964–1969. doi: 10.1101/gad.305504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131:657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- 40.Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- 41.Saiga S, Furumizu C, Yokoyama R, Kurata T, Sato S, Kato T, Tabata S, Suzuki M, Komeda Y. The Arabidopsis OBERON1 and OBERON2 GENES encode plant homeodomain finger proteins are required for apical meristem maintenance. Development. 2008;135:1751–1759. doi: 10.1242/dev.014993. [DOI] [PubMed] [Google Scholar]
- 42.Boutté Y, Ikeda Y, Grebe M. Mechanisms of auxin-dependent cell and tissue polarity. Curr Opin Plant Biol. 10:616–623. doi: 10.1016/j.pbi.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 43.Bandyopadhyay A, Blakeslee JJ, Lee OR, Mrave J, Sauer M, Titapiwatanaku B, Makam SN, Bouchard, Geisler M, Martinoia E, et al. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Biochem Soc Trans. 2007;35:137–141. doi: 10.1042/BST0350137. [DOI] [PubMed] [Google Scholar]
- 44.Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, et al. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell. 2007;19:131–147. doi: 10.1105/tpc.106.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parry G, Estelle M. Auxin receptors: a new role for F-box proteins. Curr Opin Cell Biol. 2006;18:152–156. doi: 10.1016/j.ceb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Sauer M, Balla J, Luschnig C, Wisniewska J, Reinöhl V, Friml J, Benková E. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 2006;20:2902–2011. doi: 10.1101/gad.390806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaillais Y, Santambrogio M, Rozier F, Fobis-Loisy I, Miège C, Gaude T. The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell. 2007;130:1057–1070. doi: 10.1016/j.cell.2007.08.040. [DOI] [PubMed] [Google Scholar]
- 48.Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 49.Blakeslee JJ, Zhou HW, Heath JT, Skottke KR, Barrios JA, Liu SY, Delong A. Specificity of RCN1-mediated protein phosphatase 2A in meristem organization and stress response in roots. Plant Physiol. 2008;146:539–553. doi: 10.1104/pp.107.112995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA. 2006;103:6055–6060. doi: 10.1073/pnas.0510607103. [DOI] [PMC free article] [PubMed] [Google Scholar]