Abstract
Bacteriophage lambda infection of Escherichia coli can result in distinct cell fate outcomes. For example, some cells lyse whereas others survive as lysogens. A quantitative biophysical model of lambda infection supports the hypothesis that spontaneous differences in the timing of individual molecular events during lambda infection leads to variation in the selection of cell fates. Building from this analysis, the lambda lysis–lysogeny decision now serves as a paradigm for how intrinsic molecular noise can influence cellular behavior, drive developmental processes, and produce population heterogeneity. Here, we report experimental evidence that warrants reconsidering this framework. By using cell fractioning, plating, and single-cell fluorescent microscopy, we find that physical differences among cells present before infection bias lambda developmental outcomes. Specifically, variation in cell volume at the time of infection can be used to help predict cell fate: a ≈2-fold increase in cell volume results in a 4- to 5-fold decrease in the probability of lysogeny. Other cell fate decisions now thought to be stochastic might also be determined by pre-existing variation.
Keywords: deterministic, extrinsic variation, lysis–lysogeny, stochastic
One of the best-studied natural biological systems for exploring cell fate commitment is bacteriophage lambda. Lambda-infected cells typically become lytic or lysogenic. Cells that become lytic produce and release progeny phage into the environment following lysis of the host cell. Alternatively, if infected cells become lysogenic, the phage genome integrates into the bacterial chromosome and the resulting prophage is passively replicated within the surviving cell and its offspring (1–3).
In studying the lambda lysis–lysogeny “decision,” a valuable starting observation is that, across many conditions, genetically identical cells grown in the same environment and each infected with a single lambda particle select different cell fates: Some cells lyse whereas other cells become lysogens (4–6). This variability in cell fate is unlikely to be due to genetic variation within the phage population or to an artifact of the experimental methods used to grow infected cells or quantify developmental outcome (Figs. S1 and S2) (4, 5). Given this observation, how do genetically identical cells infected with the same number of phage particles give rise to distinct cell fates?
In considering this question, cell fate selection during lambda infection has emerged as a paradigm for how biochemical “noise” might account for differences in developmental outcomes (7–10). In particular, Arkin and colleagues (7) used a detailed stochastic chemical kinetics model of lambda infection to analyze whether the lysis–lysogeny decision might be driven by spontaneous differences in the timing of individual biochemical reaction events. In the Arkin model, initially identical newly infected cells are expected to spontaneously accumulate quantitative differences in the abundances of key regulatory molecules, which then propagate through the lysis–lysogeny “decision circuitry” and result in distinct cell fates (Fig. 1A).
Fig. 1.
Alternative models for cell fate selection in a population of genetically identical cells. (A) Variation during infection (for example, spontaneous chemical kinetic noise) leads to qualitative differences in cell fate. (B) Variation in the physical state of individual cells before infection predetermines cell fate.
The Arkin model is important for at least 2 reasons. First, the model revitalized the study of how lambda-infected cells produce distinct cell fates, recognizing that the existing detailed descriptions of the individual molecular components of phage lambda are not sufficient to explain such system-level behavior. Second, Arkin and colleagues (7) mapped the process of cell fate selection onto an explicit model of intracellular physics, which forces a recognition of the fact that many systems in biology are composed of components whose cellular abundances are far below levels for which continuous approximations of chemical kinetics are valid (11) and for which precision of behavior cannot be expected to emerge via the bulk averaging of many individual reaction events (12).
The Arkin model also makes a strong claim—that cell fate differences in lambda-infected Escherichia coli are due to spontaneous biochemical noise during infection (Fig. 1A). However, to our knowledge, no experiments have been carried out to test this hypothesis. Thus, we considered and tested an alternative model for lambda infection in which cell–cell variability that exists before infection determines individual cell fates (Fig. 1B) (5, 13). In this alternative model, lambda does not use intrinsic variation (14) to produce multiple cell fates; rather, the phage-infected cell responds to extrinsic cell–cell variation that is present before the start of infection.
To identify candidate predictors of cell fate we considered past work showing that the frequency of lysogeny increases with the average number of infecting phage per cell (5, 15). To explain this observation, Kourilsky proposed that lysogeny requires the concentrations of one or more phage-encoded factors to exceed a critical threshold (16, 17). The number of pro-lysogeny factors produced in an infected cell may be greater when the levels of their corresponding genes are high, either because many phage are simultaneously infecting the same cell (15) or because the infected cell is small (this work). For cells infected with the same number of phage we might therefore predict that, at least early during infection, smaller cells will have an increased concentration of pro-lysogeny gene products and may tend to become lysogens; larger cells will have a lower concentration of pro-lysogeny gene products and might favor lytic growth.
Results
We designed experiments to test the hypothesis that variation in cell volume present before infection controls the fate of infected cells. To control the number of infecting phage per cell (a quantity known as the “multiplicity of infection,” or MOI) in our experiments, we infected cells at a low average phage-to-cell ratio to ensure that virtually all infected cells were infected at an MOI = 1 (18). Practically, wild-type cells grown to early exponential phase and infected with a single phage typically follow the lytic pathway exclusively (15, 19). To raise the frequency of lysogeny to measurable levels, we starved the cells by growing them to stationary phase, a growth phase reported to give balanced fractions of both lytic events and lysogens (5, 6, 15). To produce cell fractions with different mean cell volumes (MCVs) from a naturally asynchronous population of stationary phase cells, we first adapted and optimized methods of counterflow centrifugal elutriation (SI Text, Fig. S3, and Tables S1 and S2). We collected cell fractions with MCVs between 0.82 and 1.33 μm3, giving a MCV ratio of ≈1.6 between the largest and smallest fractions (Fig. 2 and Fig. S4).
Fig. 2.
Genetically identical cells can be separated into many fractions. Asynchronous cultures of stationary phase E. coli MG1655 were sorted by using counterflow centrifugal elutriation (see Experimental Procedures). (A–C) The distribution of cell volumes obtained by microscopy (n > 500) and electronic (n > 10,000) measurements are shown for representative small (A) and large (B) fractions, as well as for the starting asynchronous population (C). (See Fig. S4 for additional fractions.) CoV, coefficient of variation. (D–F) Representative phase contrast pictures are shown to the right of the corresponding volume distributions. (Scale bars: 5 μm.)
We infected each cell fraction and quantified cell fate outcomes by using macroscopic plating assays. Across the ≈1.6-fold range of MCV fractions, we observed a ≈3-fold difference in the percentage lysogeny (Fig. 3). For example, 66.8 ± 2.4% SEM (n = 3; MCV ± SEM = 0.83 ± 0.01 μm3) of the cells from the smallest MCV fractions underwent lysogeny, whereas only 21.6 ± 2.4% (n = 3; MCV = 1.30 ± 0.02 μm3) of the cells from the largest MCV fractions formed lysogens. For comparison, 44.2 ± 4.0% (n = 3; MCV = 1.07 ± 0.01 μm3) of the cells taken directly from the starting asynchronous cell populations formed lysogens. The correlation coefficient between fraction MCV and frequency of lysogeny is high (n = 21, r2 = 0.95, P < 0.001), with smaller cells being ≈3-fold (P = 0.002) more likely to form lysogens following infection (Fig. S5). We also independently observed corresponding differences in the fraction of lytic cells, with smaller cells being ≈3-fold less likely than larger cells to lyse (P = 0.002; Fig. S5). Across all fractions, the sum of our 2 independent measurements of lytic cells and lysogens was consistent with a third independent measure of the total number of infected cells (Fig. 3, orange data points); we used this internal control as a first-order test to check that all lysogens and lytic events were detected. Additional control experiments indicated that our results are unlikely due to artifacts of the elutriation procedure or to differences in cell viability, kanamycin sensitivity, spontaneous resistance to kanamycin, or cell division before plating (SI Text and Fig. S6).
Fig. 3.
Variation in developmental outcome is a function of physical variation that exists before infection. Independent measurements of the fraction of lytic events and lysogeny as a function of the mean cell volume for different cell fractions. Developmental outcome is expressed as a percentage of an independent measure of the total number of infected cells for each fraction. Circles, triangles, and squares depict fractions collected from 3 independent experiments. The 3 starting asynchronous (unfractionated) cultures are also shown († near data points). Error bars indicate the Poisson standard error of the mean from plating. Dashed lines indicate linear regression lines for percentage lysis and lysogeny and the sum of lytic events and lysogens.
No cell fraction obtained by using the elutriation and plating method produced 100% lysogens or 100% lytic events (Fig. 3). However, each elutriated fraction was comprised of cells spanning a range of volumes (Fig. 2 and Fig. S4). Thus, we were unsure if the occurrence of both lysis and lysogeny in each fraction might be due to stochastic processes intrinsic to infection within individual cells or to remaining preexisting extrinsic differences in cell volume (or another variable) that were beyond the segregation capabilities of our elutriation method. Given the observed sensitivity to preexisting differences in cell volume, we were particularly curious whether volume at the time of infection might entirely account for the eventual fate of infected cells. To quickly consider this possibility we used a single parameter abstract “all-or-none” model to posit that any cells smaller than a “critical volume” might produce a lysogen, whereas any larger cell would undergo lysis. Our analysis (Fig. S7) suggested that the cell fraction data are not obviously inconsistent with an all-or-none deterministic model for lambda infection (Fig. 1B) in which cells smaller than a putative critical volume (≈1 μm3) will become lysogens.
We next developed single-cell fluorescent microscopy methods to directly examine whether differences in cell volume (or correlated variables) predetermine an all-or-none cell fate process and to confirm that our observed predetermination of cell fate outcomes was not specific to a particular culture condition, genetic background, or experimental approach. To this end, we developed a lambda strain that combines a constitutive GFP reporter cassette integrated into the b region of the lambda genome, a region that is not known to encode functions associated with the cell fate selection process (20). We also wanted to prevent progeny phage released from cells that lysed early from reinfecting neighboring infected cells, thereby possibly disrupting their cell fate selection process by increasing the MOI. We therefore introduced an amber mutation in gene A (Aam19), which encodes an essential phage DNA packaging enzyme not involved in the cell fate selection process (SI Text) (21). Bacterial strains that do not suppress the amber mutation, such as those used in our experiments, can only release empty (uninfectious) lambda particles. Note that infected cells can still lyse because the phage-encoded holin-endolysin system for host cell lysis (genes R and S) remains intact.
The resulting phage (λ Aam19 b::GFP cI857) produces bright fluorescence upon infection, allowing easy detection of infected cells. In a typical experiment we infected cells at low multiplicity (average phage-to-cell ratio of ≈0.03), recorded time-lapse movies via automated fluorescence and brightfield microscopy, and determined the starting cell size of infected cells by direct manual measurement of brightfield images. We established cell fates as described in Table 1. Briefly, we scored an infection as a lytic event when a GFP-expressing cell or both of its daughters lysed. We did not observe any instance of lysis in cells that did not previously express GFP. An infected cell was presumed to be a lysogen when it survived the infection, producing 2 daughters that also divided. Stable lysogeny requires integration of lambda DNA into the host chromosome (19). Although our fluorescent reporter system did not allow us to directly determine whether integration took place, the overall cell fate selection statistics from our microscopy experiments were equivalent to those obtained by using our elutriation and plating method (see next paragraph). Finally, in a minority of cases, we observed an infected cell giving rise to 2 daughters: one that lysed and one that survived and divided. Such events were scored as “mixed fate.”
Table 1.
Cell fate selection statistics determined by using time-lapse microscopy
| Cell fates |
||||||
|---|---|---|---|---|---|---|
| Infected cell | Daughters of infected cell |
Classification | Example movies | Percentage of total (counts) |
||
| Daughter A | Daughter B | Wild-type E. coli grown to stationary phase | hflK−E. coli mutant grown to exponential phase | |||
| Lyses | n/a | n/a | Lytic event | S1, S2 | 55.6 (175) | 85.1 (382) |
| Divides | Lyses | Lyses | Lytic event | S4, S5 | 0.3 (1) | 1.3 (6) |
| Divides | Lyses | Divides | Mixed fate | S3, S6, S7 | 5.1 (16) | 1.6 (7) |
| Divides | Divides | Divides | Lysogeny | S1 | 39.0 (123) | 12.0 (54) |
| Total | 100 (315) | 100 (449) | ||||
Bacterial cultures were infected at low API (phage-to-cell, ≈1:30) with λ Aam19 b::GFP cI857. The developmental outcome of infected (GFP-expressing) cells and, if applicable, their daughter cells, was monitored by time-lapse microscopy. The classification column outlines our mapping of developmental outcomes (lysis or division) of infected cells to overall cell fate outcomes. Cell fate statistics were collected for 2 conditions with different cell strains, growth media and growth phase at time of harvest. Example movies for the different combinations of cell fate outcomes are provided (Movies S1–S7). In some cases, the fate of an infected cell or one or both of its daughters could not be determined either because (i) the cell moved out of the field of view or became obscured by neighboring cells or (ii) the cell did not divide or lyse before the end of the time-lapse movie. Such ambiguous cases were not included in this table or in any analyses.
We performed single cell experiments under 2 sets of conditions: (i) by using the same bacterial strain and culture conditions as were used in the elutriation and plating experiments [wild-type E. coli grown to stationary phase in tryptone broth (TB)] and (ii) by using a high frequency of lysogeny mutant (hflK−) grown in a defined M9-based medium and infected during exponential phase growth. Under the stationary phase culture conditions, the percentage lysogeny from our microscopy experiments (41.1%) closely matched that obtained by using our elutriation and plating method (44.2 ± 4.0% SEM). The majority of infected cells that divided produced lysogens (123 of 140). A minority of infected, dividing cells (16 of 140) produced mixed fate events.
Under the exponential culture conditions, most infected cells lysed (382 of 449), with the majority of the remaining cells producing lysogens (54 of 67). Fig. 4Upper presents a time-lapse of 2 infected cells: One cell produces a lytic response whereas the other cell survives the infection as a presumed lysogen. We again observed a small number of infected, dividing cells whose offspring obtained distinct cell fates (7 of 449; and see Fig. 4 Lower). In both stationary and exponential phase cultures, a few infected cells divided, producing daughters that both lysed. Complete cell fate selection statistics and representative time-lapse fluorescent microscopy movies for all qualitatively distinct combinations of cell fate outcomes are provided (Table 1 and Movies S1–S7).
Fig. 4.
Cell fate selection in individual cells. Images from time-lapse movies of exponentially growing JW4132 (hflK−) cells infected at a phage-to-cell ratio of ≈1:30 with a lambda strain carrying a fluorescent reporter expression cassette. Numbers indicate the number of minutes at which the images were taken after the start of each movie. (Upper) Two neighboring cells are infected, each presumably at a multiplicity of one, and both cells turn green over time. The upper-right infected cell is larger and near the end of its cell division cycle, as indicated by the presence of a membrane invagination (developing septum) at mid-cell. This cell lyses between 132 and 177 min. The lower-left infected cell, which is presumably the product of a recent cell division event and is relatively small, continues to progress through a typical cell division cycle. The infected cell was presumably lysogenized by lambda as it survives the infection, producing viable progeny which also divide between 461 and 520 min. (Lower) An infected cell produces daughter cells with apparently distinct fates: the rightmost daughter lyses between 527 and 559 min, whereas the leftmost daughter divides ≈372 min, producing progeny which themselves divide ≈714 min.
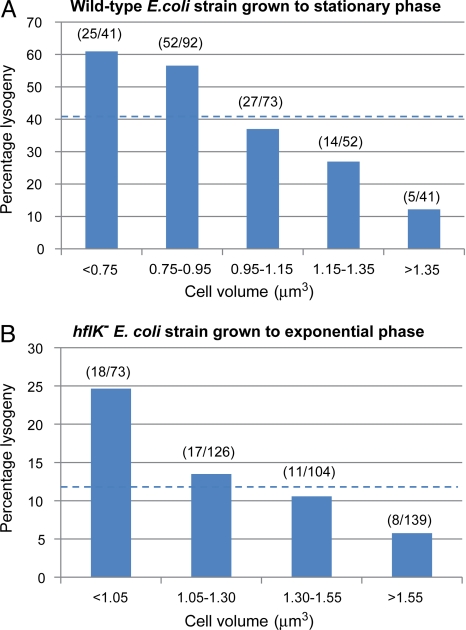
We estimated the initial size of each infected cell by measuring cell length immediately after infection (SI Text). We combined the cell size and cell fate data to determine to what extent the size of individual cells predetermines cell fates. Consistent with our elutriation-based results, we observed that in individual stationary phase cells the probability of lysogeny is greatest in the smallest cells (P = 0.61) and decreases with increasing cell size (P = 0.12) (Fig. 5A). Similarly, in individual exponential phase cells the probability of lysogeny is greatest in the smallest cells (P = 0.25) and decreases with increasing size (P = 0.06) (Fig. 5B). In both cases, although volume was strongly correlated with cell fate, we did not find a critical volume below which all cells became lysogens and above which all cells underwent lysis (Fig. 5 and Fig. S8).
Fig. 5.
Extrinsic variation helps determine developmental outcome of single infected cells. Probability of lysogeny as a function of the volume of individual MG1655 (hflK+) grown to stationary phase (A) or JW4132 (hflK−) grown to exponential phase (B). The numbers in parentheses represent the number of lysogens over the total number of infected cells within each volume bin (excluding “mixed fate” cells). The dashed lines correspond to the average percentage lysogeny in each condition.
Discussion
Our results show that the fate of individual lambda-infected cells is strongly correlated with preexisting variation in host volume. For example, a ≈2-fold increase in the size of stationary phase cells led to a ≈5-fold decrease (61% to 12.2%) in frequency of lysogeny (Fig. 5A). We initially observed that lambda was sensitive to host volume by using elutriation as a method for segregating larger and smaller cells, and macroscopic plate tests (plaques and colonies) for determining cell fate (Fig. 3). Our initial finding was confirmed at the single-cell level by using time-lapse fluorescent microscopy. We observed that the frequency of lysogeny decreased with increased cell size in 2 conditions, which differed from one another with respect to strain genotypes, culture medium, and growth phase (Fig. 5). We also found no correlation between cell volume and the activity of a representative endogenous stationary phase promoter, as expected if the sensitivity to cell volume is not specific to stationary phase (Fig. S9). Sensitivity to preexisting variation in host volume may thus be a general feature of the lysis–lysogeny regulatory network of phage lambda.
Our findings challenge the view that differences in lambda cell fate outcomes are solely driven by intrinsic noise during infection. The lambda regulatory network is not only composed of phage-encoded components; rather, it operates within a bacterial environment that can also exhibit variation. Future biophysical models of lambda cell fate selection might do well to represent such variation.
The correlation between the size and fate of lambda-infected cells is particularly strong considering the limitations of the methods we used to observe and analyze the lysis–lysogeny decision. For instance, the size measurements of individual cells (Fig. 5) were made manually, considered individual cell lengths only, and represented the size of cells at the start of imaging rather than at some critical time window during infection when the lysis–lysogeny decision is being determined.
However, although host size before infection is a strong predictor of cell fate, it appears that the lambda cell fate selection process may still be sensitive, in part, to other types of variation, including intrinsic variation during infection. For example, Fig. 5 shows that there is no critical volume above or below which all infected cells produce an identical developmental response. In addition, a small number of infected cells (1.6–5.1%) divide and give rise to 2 daughters who obtain distinct cell fates: one that undergoes lysis and another that continues to grow and divide. If cell volume immediately before infection completely determined cell fate, we would expect the developmental response of daughter cells to be identical. Thus, mixed fate outcomes might represent an ideal case of intrinsic noise in gene expression or in partitioning of molecules (protein, DNA, and mRNA) between daughter cells during cell division. Alternatively, mixed fate outcomes may still represent a deterministic process; for instance, the “age” of the poles (and other variables) of the 2 daughter cells might differ and affect cell fate selection following division of the originally infected cell (22). It will be exciting to explore further whether the lysis–lysogeny decision is absolutely determined by cell–cell variation present before infection (Fig. 1B) or whether lambda might “play dice” to some degree or on rare occasions (Fig. 1A).
More generally, the sensitivity of lambda infection to preexisting physical variation highlights an opportunity to study how evolution shapes or delimits the molecular mechanisms underlying the behavior of biological systems. One possibility is that cell–cell variation in volume reflects physiological differences that lambda must sample to make optimal developmental decisions at the single cell level. For example, the lytic response may be preferred in infected cells capable of producing a large burst of progeny phage. Because larger cells presumably contain more material (amino acids, nucleotides, etc.) than smaller cells, such cells may produce greater bursts of phage particles. Alternatively, the response to preexisting variation in host size may be an artifact of circuitry selected to be responsive to other, more dominant factors. For example, lambda's regulatory network may be tuned to respond to the MOI by measuring the concentration (rather than the absolute number per se) of phage infecting a given cell. If true, lambda would not be able to distinguish between variations in phage concentration due to differences in MOI or in host volume.
There are many known molecular mechanisms that could point toward a model for explaining how preexisting variation in cell volume biases infected cells toward lysis or lysogeny. As discussed earlier, smaller and larger cells, infected with the same number of phage, might produce different concentrations of phage-encoded pro-lysogeny gene products early during infection. Examples of such pro-lysogeny genes are cII and cIII, whose products are thought to be critical regulators of the lysis–lysogeny decision (1, 3, 23). Alternatively, small and large cells may be in different physiological states, perhaps because of their presumably distinct positions within the cell cycle. As a result, variation in volume may correlate with differences in the amount, localization or activity of host-encoded regulators of lambda development such as HflB/FtsH or RNase III (1, 3, 23–26).
Regardless of the molecular mechanism through which the lambda cell fate selection process responds to extrinsic variation, in considering stochastic cellular behavior more generally it is interesting to note the extent to which developmental decisions are deterministic. Several hundred reports have discussed the importance of stochastic variation in gene expression and other cellular processes (e.g., refs. 27–29). A few cell fate decision processes—for example, the transient differentiation of Bacillus subtilis into a state of competence (30–32) or stem cell commitment and differentiation (33)—are thought to be (partly) stochastic. However, it is unclear whether stochastic variation during development actually has an impact on the outcome of many cell fate decisions. For comparison, the patterns of programmed death and terminal differentiation during embryonic development of Caenorhabditis elegans are nearly invariant from one individual to the next (34). Understanding how and the extent to which natural biological systems tolerate, buffer, or correct for spontaneous molecular variation during development to produce deterministic behavior deserves more attention (10, 35–39).
Experimental Procedures
A more detailed version of the procedures is provided in SI Text.
Preparation of Stationary and Exponential Phase Cells.
To prepare stationary phase cells, we grew E. coli MG1655 to near saturation in TB at 37 °C (Fig. S2a). To prepare exponentially growing cells, we grew the hflK− strain JW4132 in M9GlyM (M9 supplemented with 0.4% glycerol, 0.2% maltose, 2 mM MgSO4, and 0.1 mM CaCl2) + 1 mM IPTG (to induce GFP expression) to early exponential phase (OD600 ≈ 0.07).
Segregation of Cells by Using Counterflow Centrifugal Elutriation.
We size-fractionated cells according to differences in terminal sedimentation velocity by using 2 JE5.0 counterflow centrifugal elutriation systems, each equipped with a single, 40-mL chamber (Beckman Coulter). One elutriation system was used to obtain the fraction of smallest cells; we collected all other fractions by using the second system. In both cases we followed custom protocols (SI Text) and performed the fractionation at 4 °C to prevent further cell growth.
Electronic and Microscopy Measurements of Cell Size from Elutriated Fractions.
We measured the volumes of individual cells in each fraction microscopically and electronically. For microscopy measurements, we manually measured the long axis of >500 cells per fraction under 60× dark medium (DM) phase contrast optics and 1.5× intermediate magnification or by using 100× DM phase contrast optics (Nikon; Hamamatsu). Measurements of lengths were converted into estimates of cell volume by using methods described in SI Text. To obtain electronic volume measurements, we first stained cells with 2 μM of the nucleic acid dye Syto-9 (Invitrogen). We streamed cells into a 25-μm flow chamber in a NPE Cell Quanta Hg/488 system (NPE Systems), triggering on fluorescence (excitation: 488 nm laser; emission: 525/30 nm) and recording electronic volume signals according to the Coulter Counter principle (40, 41). We calibrated our electronic volume measurements by spiking cell suspensions with 1.51-μm fluorescent polystyrene beads (Bangs Labs). Electronic volume and fluorescence were recorded for >10,000 cells and 5,000 beads per fraction. Volumes obtained electronically or by microscopy were usually similar (Fig. 2 and Figs. S4 and S10); for simplicity, we therefore only reported volumes determined by phase contrast microscopy (unless noted otherwise).
Quantification of Developmental Outcomes by Using Macroscopic Plate Tests.
We typically added phage (λ cI857 bor::kanR) at an average phage-to-cell ratio of 0.005. After adsorption, we removed unadsorbed phages by centrifugation and incubated cells at 30 °C for 45 min in liquid medium. We selected for lysogens by spreading cells on agar supplemented with 20 μg/ml kanamycin sulfate. We incubated agar plates at 30 °C and scored lysogens by counting the number of colony-forming units. We measured lytic events by plating on TB agar, incubating at 30 °C and counting plaque-forming units (PFU). We also measured the total number of infected cells by preparing plates as if to count lytic events, with the exception that the plates were incubated at 42 °C, a nonpermissive temperature for allele cI857; we scored infected cells as PFU.
Construction of λ Aam19 b::GFP cI857.
We constructed λ Aam19 b::GFP cI857 by first recombineering a GFP cassette into wild-type lambda by using published methods (42). The GFP cassette contains GFP variant GFPmut3b (43) as well as a kanamycin marker to select recombinants. The 2,662-nt cassette was inserted into the b region of lambda (20), replacing sequence between lambda coordinates 20430–22278, producing a phage only slightly longer (813 nt or 1.7%) than wild-type λ. We confirmed the presence of the cassette by fluorescence microscopy and by sequencing. We next engineered the Aam19 (C→T at lambda coordinate 1917, ref. 45) and cI857 (C→T at lambda coordinate 37742; ref. 2) mutations onto λ b::GFP by using published methods of recombineering using single-stranded oligonucleotides (42, 44). We identified recombinants by plaque morphology and confirmed the presence of the mutations by sequencing.
Single-Cell Time-Lapse Microscopy.
Both stationary and exponential phase cells were infected with λ Aam19 b::GFP cI857 at a phage-to-cell ratio of ≈1:30. After adsorption at 4 °C, we removed unadsorbed phage by centrifugation and spread cells on TB (stationary phase cells) or M9GlyM (exponential phase cells) agarose (2%) pads supplemented with 1 mM IPTG to allow expression of the GFP cassette. We incubated the cells in a microscope heating chamber set to 30 °C, taking phase contrast and GFP fluorescence images (75 ms) every ≈15 min at 60–100× magnification. We identified infected cells by using GFP fluorescence produced by our specialized lambda strain. We manually measured the lengths of individual infected cells by using the first frame of our timelapse movies. We converted length measurements into volume measurements (SI Text).
Supplementary Material
Acknowledgments.
We thank Donald Court for suggesting cell volume as a potential marker of cell fate; Lynn Thomason (Gene Regulation and Chromosome Biology Laboratory, Center for Cancer Research, National Cancer Institute-Frederick, Frederick, MD) for the generous gift of phage strains (wild-type λ, λ cI857 bor::kanR and λ cI26) and technical expertise; members of laboratories of Stephen P. Bell and G. R. Fink (Massachusetts Institute of Technology, Cambridge, MA) for use of their elutriators; Mingjie Li for the measurements of cell size from the time-lapse movies and for experiments with the reporters of stationary phase; Felix Moser for help with the construction of λ Aam19 b::GFP cI857 and the reporters of stationary phase; and I. J. Molineux, T. Sauer, A. P. Arkin, R. Ward, A. Grossman, R. Brent, S. Adya, G. Grigoryan, and past members of the Endy Lab for encouraging discussions. E. coli strains MG1655 (CGSC #7740) and JW4132 (CGSC #10975) were from the Coli Genetic Stock Center (New Haven, CT). This work was supported by U.S. National Institute of General Medical Sciences Grant 5R01GM076147-03. F.S.-P. was supported in part by a postgraduate scholarship from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808831105/DCSupplemental.
References
- 1.Herskowitz I, Hagen D. The lysis–lysogeny decision of phage lambda: Explicit programming and responsiveness. Annu Rev Genet. 1980;14:399–445. doi: 10.1146/annurev.ge.14.120180.002151. [DOI] [PubMed] [Google Scholar]
- 2.Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Lambda II. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1983. [Google Scholar]
- 3.Ptashne M. A Genetic Switch: Phage Lambda Revisited. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2004. [Google Scholar]
- 4.Lieb M. Studies on lysogenization in Escherichia coli. Cold Spring Harb Symp Quant Biol. 1953;18:71–73. doi: 10.1101/sqb.1953.018.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Lieb M. The establishment of lysogenicity in Escherichia coli. J Bacteriol. 1953;65:642–651. doi: 10.1128/jb.65.6.642-651.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobiler O, Koby S, Teff D, Court D, Oppenheim AB. The phage lambda CII transcriptional activator carries a C-terminal domain signaling for rapid proteolysis. Proc Natl Acad Sci USA. 2002;99:14964–14969. doi: 10.1073/pnas.222172499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arkin A, Ross J, McAdams HH. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1998;149:1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thattai M, van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA. 2001;98:8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell. 2005;122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Maheshri N, O'Shea EK. Living with noisy genes: How cells function reliably with inherent variability in gene expression. Annu Rev Biophys Biomol Struct. 2007;36:413–434. doi: 10.1146/annurev.biophys.36.040306.132705. [DOI] [PubMed] [Google Scholar]
- 11.Gillespie DT. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J Phys Chem. 1976;22:403–434. [Google Scholar]
- 12.Schrödinger E. What Is Life? The Physical Aspect of the Living Cell. Cambridge, UK: Cambridge Univ Press; 1945. [Google Scholar]
- 13.Kourilsky P, Gros D. Lysogenization by bacteriophage lambda IV inhibition of phage DNA synthesis by the products of genes cII and cIII. Biochimie. 1976;58:1321–1327. doi: 10.1016/s0300-9084(77)80015-1. [DOI] [PubMed] [Google Scholar]
- 14.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kourilsky P. Lysogenization by bacteriophage lambda. I. Multiple infection and the lysogenic response. Mol Gen Genet. 1973;122:183–195. doi: 10.1007/BF00435190. [DOI] [PubMed] [Google Scholar]
- 16.Kourilsky P. Lysogenization by bacteriophage lambda. II. Identification of genes involved in the multiplicity dependent processes. Biochimie. 1974;56:1511–1516. [PubMed] [Google Scholar]
- 17.Kourilsky P, Knapp A. Lysogenization by bacteriophage lambda. III. Multiplicity dependent phenomena occurring upon infection by lambda. Biochimie. 1974;56:1517–1523. [PubMed] [Google Scholar]
- 18.Delbruck M. The burst size distribution in the growth of bacterial viruses (bacteriophages) J Bacteriol. 1945;50:131–135. doi: 10.1128/JB.50.2.131-135.1945. [DOI] [PubMed] [Google Scholar]
- 19.Oppenheim AB, Kobiler O, Stavans J, Court DL, Adhya S. Switches in bacteriophage lambda development. Annu Rev Genet. 2005;39:409–429. doi: 10.1146/annurev.genet.39.073003.113656. [DOI] [PubMed] [Google Scholar]
- 20.Court D, Oppenheim AB. In: Lambda II. Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1983. pp. 251–277. [Google Scholar]
- 21.Feiss M, Becker A. In: Lambda II. Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1983. pp. 305–330. [Google Scholar]
- 22.Nystrom T. A bacterial kind of aging. PLoS Genet. 2007;3:e224. doi: 10.1371/journal.pgen.0030224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobiler O, Rokney A, Oppenheim AB. Phage lambda CIII: A protease inhibitor regulating the lysis-lysogeny decision. PLoS ONE. 2007;2:e363. doi: 10.1371/journal.pone.0000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson HR, Yu D, Peters HK, III, Zhou JG, Court DL. The global regulator RNase III modulates translation repression by the transcription elongation factor N. EMBO J. 2002;21:4154–4161. doi: 10.1093/emboj/cdf395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wehrl W, Niederweis M, Schumann W. The FtsH protein accumulates at the septum of Bacillus subtilis during cell division and sporulation. J Bacteriol. 2000;182:3870–3873. doi: 10.1128/jb.182.13.3870-3873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobiler O, Oppenheim AB, Herman C. Recruitment of host ATP-dependent proteases by bacteriophage lambda. J Struct Biol. 2004;146:72–78. doi: 10.1016/j.jsb.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 27.Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colman-Lerner A, et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- 29.Sigal A, et al. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- 30.Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 31.Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–1719. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 33.Enver T, Heyworth CM, Dexter TM. Do stem cells play dice? Blood. 1998;92:348–351. and discussion (1998) 92:352. [PubMed] [Google Scholar]
- 34.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 35.Von Neumann J. In: Automata Studies. Shannon CE, McCarthy J, Ashby WR, editors. Princeton: Princeton Univ Press; 1956. pp. 43–98. [Google Scholar]
- 36.Hopfield JJ. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao CV, Wolf DM, Arkin AP. Control, exploitation and tolerance of intracellular noise. Nature. 2002;420:231–237. doi: 10.1038/nature01258. [DOI] [PubMed] [Google Scholar]
- 38.Houchmandzadeh B, Wieschaus E, Leibler S. Establishment of developmental precision and proportions in the early Drosophila embryo. Nature. 2002;415:798–802. doi: 10.1038/415798a. [DOI] [PubMed] [Google Scholar]
- 39.Arias AM, Hayward P. Filtering transcriptional noise during development: Concepts and mechanisms. Nat Rev Genet. 2006;7:34–44. doi: 10.1038/nrg1750. [DOI] [PubMed] [Google Scholar]
- 40.Coulter WH, Hogg WR, Moran JP, Claps WA. Particle analyzing device. US patent 3. 1966;259:942. [Google Scholar]
- 41.Kubitschek HE. Electronic counting and sizing of bacteria. Nature. 1958;182:234–235. doi: 10.1038/182234a0. [DOI] [PubMed] [Google Scholar]
- 42.Thomason L, et al. Recombineering: Genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol. 2007 doi: 10.1002/0471142727.mb0116s78. Chap 1:Unit 1.16. [DOI] [PubMed] [Google Scholar]
- 43.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 44.Oppenheim AB, Rattray AJ, Bubunenko M, Thomason LC, Court DL. In vivo recombineering of bacteriophage lambda by PCR fragments and single-strand oligonucleotides. Virology. 2004;319:185–189. doi: 10.1016/j.virol.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 45.Davidson AR, Gold M. Mutations abolishing the endonuclease activity of bacteriophage λ terminase lie in two distinct regions of the A gene, one of which may encode a “leucine zipper” DNA-binding domain. Virology. 1992;189:21–30. doi: 10.1016/0042-6822(92)90677-h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.