Abstract
Gene annotation underpins genome science. Most often protein coding sequence is inferred from the genome based on transcript evidence and computational predictions. While generally correct, gene models suffer from errors in reading frame, exon border definition, and exon identification. To ascertain the error rate of Arabidopsis thaliana gene models, we isolated proteins from a sample of Arabidopsis tissues and determined the amino acid sequences of 144,079 distinct peptides by tandem mass spectrometry. The peptides corresponded to 1 or more of 3 different translations of the genome: a 6-frame translation, an exon splice-graph, and the currently annotated proteome. The majority of the peptides (126,055) resided in existing gene models (12,769 confirmed proteins), comprising 40% of annotated genes. Surprisingly, 18,024 novel peptides were found that do not correspond to annotated genes. Using the gene finding program AUGUSTUS and 5,426 novel peptides that occurred in clusters, we discovered 778 new protein-coding genes and refined the annotation of an additional 695 gene models. The remaining 13,449 novel peptides provide high quality annotation (>99% correct) for thousands of additional genes. Our observation that 18,024 of 144,079 peptides did not match current gene models suggests that 13% of the Arabidopsis proteome was incomplete due to approximately equal numbers of missing and incorrect gene models.
Keywords: annotation, genomics, proteomics
A fundamental goal of genome projects is to generate a protein-coding catalog. Much of modern biological research depends on a complete and accurate proteome. Extensive proteomic catalogs have been developed through the integration of gene prediction algorithms, cDNA sequences, and comparative genomics (1, 2). As emerging research is incorporated into annotation pipelines and manual curation efforts, gene models continue to improve. High throughput gene annotation pipelines use a variety of information sources, and benefit most significantly when new data contains information that is orthogonal to the kinds currently available (3).
Recent advances in chemistry and algorithms for peptide mass spectrometry have enabled the production of large proteomics datasets with broad coverage of the proteome (4–6). Proteo-genomics (using proteomic information to annotate the genome) complements nucleotide-based annotation in that it unambiguously determines reading frame, translation start and stop sites, splice boundaries, and the validity of short ORFs. By combining DNA-based annotation with proteogenomics, an accurate and more complete protein-coding catalog can be obtained (6–10). With its clear potential for improving genome annotation, proteogenomics could be integrated with genome projects.
A recent publication by Baerenfaller et al. (4) demonstrated the ability of extensive resampling to provide good coverage of the Arabidopsis proteome. From 1,354 LC runs the authors identified 86,456 distinct peptides covering 13,029 proteins. In addition to providing an organ specific proteome catalog, they demonstrated the ability of proteomics to refine plant genome annotation by presenting evidence for 57 new gene models, including 7 from intergenic regions not suspected to contain genes.
We reported a proteogenomic study of humans that described an exon splice graph that enabled efficient searches of potential coding sequences, including peptides that span splice junctions (6). We reasoned that we could extend the observations of Baerenfaller et al. deeper into the unmapped proteome by building an exon splice graph of Arabidopsis and obtaining a novel set of peptides. We used two strategies to obtain novel peptides. First we used a nested 3D LC strategy to obtain much greater peptide separation permitting a deeper sampling of the proteome. This is reflected by our yield of 144,079 distinct peptides from only 45 LC runs, with a false-discovery rate <1%. Second we used TiO2 to enrich for phosphopeptides. Phosphorylated proteins are less abundant and are mostly missing from profiles of whole proteomes. Considering only cases in which we observed 2 or more previously non-annotated peptides mapping within 1 kb of each other, we discovered 1,473 new or revised genes; a model was generated for each using the gene finder AUGUSTUS (11). Two hundred eighty genes were previously unrecognized, 498 were previously annotated as pseudogenes, and 695 were revisions of known genes that were annotated in the wrong reading frame, with missing exons, or with incomplete exons. Extrapolating from our sample we estimate that 13% of Arabidopsis protein-coding genes were either not yet identified or they contained significant errors in their exon definitions. We have remedied ≈40% of these deficiencies.
Results
Coverage.
To achieve broad coverage of the proteome, we acquired 21 million mass spectra from protein extracts of 4 Arabidopsis organs (leaf, root, flower, and silique) and a cell culture (MM2d). In addition, phosphopeptides were enriched from MM2d proteins. Inspect was used to search spectra against 3 reference databases: TAIR7; a 6 frame translation of the genome; and an exon splice-graph that compactly encodes putative splicing events (6) (See Fig. 1 for an overview of the method). The data were filtered to a 1% cumulative false-discovery rate (FDR) at the spectrum level (Fig. S1). We required at least two peptides per protein for identification, so our 1% spectrum-level FDR provided an empirical, protein-level FDR of 0.6%.
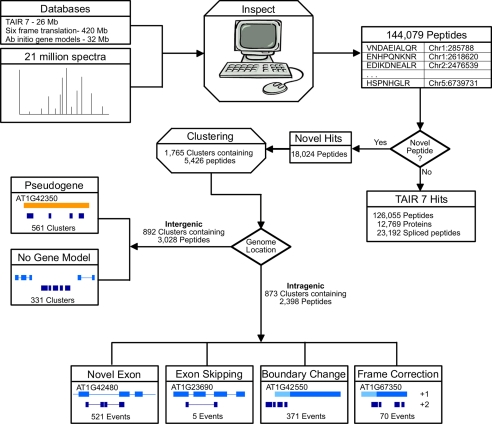
Fig. 1.
Work flow. All mass spectra are compared with three databases using Inspect. Spectra are filtered to a 1% false discovery rate and grouped into peptides. Novel peptides are separated from those that appear in TAIR7 and clustered. It is important to note that only a subset of the novel peptides appear in a peptide cluster. Novel peptide clusters are then segregated based on genome location. Those that overlap a current gene model (intragenic) are further classified by how they refine the model. Peptides that do not overlap a gene model (intergenic) are classified by whether they overlap a pseudogene. The peptide clusters, along with evidence from cDNA and current gene annotations, are given to the gene predictor AUGUSTUS to produce new gene models. Not all peptides in the peptide clusters are included in the final AUGUSTUS models.
A total of 144,079 distinct peptides were mapped to at least 1 of our 3 Arabidopsis protein databases. Most (126,055 peptides) resided in TAIR7 gene models (12,769 proteins confirmed). We mapped 18,024 peptides not present in the TAIR7 annotation including 4,018 peptides (22%) that were derived from mRNA splicing. Of these, 16,348 peptides mapped to single loci (i.e., “uniquely-located”) in the genome, whereas the rest were shared between 2 or more related proteins. The 6-frame translation and the spliced-exon databases contributed equally to the discovery of novel peptides and their contributions had little overlap; only 5% were found in both databases. This indicates that both types of database should be used for proteogenomic studies because they provide complementary novelty. Every reported peptide can be uploaded as a track in TAIR8. These files are available at http://peptide.ucsd.edu. The AUGUSTUS model building was restricted to nuclear genes and they encompass 2,873 novel peptides. These models can be accessed through Table S1 and from http://peptide.ucsd.edu.
Novel Genes.
Using the protein identification standard of 2 peptides per protein, we focused on 1,765 novel peptide clusters containing 5,426 novel peptides, 4,575 of which are uniquely located (see SI Materials and Methods). An additional 6,361 novel peptides were observed outside of clusters but with a unique genomic location and a local FDR <0.05. These were not analyzed in detail. We classified novel peptide clusters according to their position relative to annotated protein coding models. We defined intragenic clusters as those falling within the boundaries of a known protein coding gene and intergenic clusters as those falling in the intergenic space (i.e., these indicate novel genes). Some of the novel clusters overlapped loci that had been annotated as non-coding pseudogenes (31% of the peptides or 1,420 peptides derived from 561 clusters) or genes that had not been recognized at all by gene finding programs or annotators (20% of the peptides or 905 peptides derived from 331 clusters).
With our novel intergenic peptides, we defined 778 new genes consisting of 930 transcripts using the gene finder AUGUSTUS. Evidence from peptides plus EST alignments, and genomic conservation with rice, poplar, and Medicago, were given as “hints” to AUGUSTUS, which derives gene models that are in agreement with the hints and that have high likelihood in an ab initio probabilistic gene structure model. Resulting gene models include alternative splice variants, if suggested by the evidence. Of the 778 novel genes, 55 have EST and homology support, in addition to peptides; 455 genes have support by the peptides and ESTs; and 70 genes are supported by the peptides and homology only. The remaining 198 genes have no other support than the peptides. As an independent validation of our discoveries, 52 of the 778 loci have now been incorporated in the newest Arabidopsis genome release (TAIR8).
To discover homology with the novel genes, we excised the surrounding nucleotide sequence and searched against the nonredundant database of proteins (National Center for Biotechnology Information nr version 03/26/08) (Table S2). For 539 of the loci, the underlying sequence revealed a close homolog (e value <1e-10), providing additional validation, and functional assignments for the new genes. Although many of the novel genes we discover are homologous to genes of unknown function, we highlight a novel gene involved in photosynthesis. Our predicted protein, supported by 13 novel and uniquely located peptides, aligns with proteins targeted to the chloroplast thylakoid lumen (e value 1 × 10−75). It also contains the PsbP pfam domain characteristic of photosystem II (Fig. 2). A second novel locus containing 4 uniquely located peptides on chromosome 4 shows strong similarity (e value 1 × 10−85) with a heat-shock protein (AT4G12770).
Fig. 2.
Novel gene supported discovered by proteogenomics. (A) A cluster of 13 uniquely located peptides that do not overlap a current gene model (Chr3). The prediction track shows the single exon gene model produced by AUGUSTUS. (B) The predicted sequence shows strong homology to a Thylakoid lumen family protein (sp|P82658|TL19_ARATH). It also shows strong similarity to proteins in both grapevine (emb|CAO40861.1 a hypothetical gene) and rice (Os08g0504500 a cDNA derived gene).
We also note several interesting structural features of the intergenic clusters. First, a significant fraction (64%) of intergenic clusters overlap annotated pseudogenes or transposons. An example of a translated pseudogene is at locus AT2G15040, ATRLP18: Receptor like protein 18, which has high homology to disease resistance proteins in both Arabidopsis and other plants. We identify 5 peptides, 3 of which are uniquely located at this locus, confirming translation. It is presumed that pseudogenes do not produce proteins, but transposons (which like pseudogenes are not typically included in the proteome) can contain active protein-coding genes. We find evidence in transposons of translated proteins that are unrelated to transposon activity. For example, we identified 3 peptides within the locus AT4G07947 (Fig. 3A). Although annotated as a pseudogene in TAIR7 it has been reclassified as a transposable element gene in TAIR8. The genomic region containing these peptides has high similarity to the ubiquitin-like protease (Ulp1) family in Arabidopsis (Fig. 3B), suggesting this may be a gene traveling as “cargo” with the transposable element (12).
Fig. 3.
Peptides overlapping a predicted transposable element gene. (A) 5 peptides, 4 unique, overlap locus AT4G07947, which is annotated as a transposable element gene. (B) Sequence alignment to an Arabidopsis Ulp1 (ubiquitin like protease) showing strong conservation (56% identity, e value 0.0). Observed peptides highlighted.
Since the release of TAIR7, there have been several community annotation efforts, including publication of short ORFs (13). We compared the novel peptides for overlap with this set. Hanada et al. (13) reported that 7,442 non-annotated small ORFs in Arabidopsis are transcribed. Our peptides confirm the translation of 155 of these predicted ORFs. An additional 85 ORFs overlap at least 1 of our novel peptides, but the peptides indicate that the frame of the ORF may be incorrect.
Refined Gene Models.
In addition to the novel genes, we discovered peptide clusters overlapping annotated gene models, suggesting refinement of the existing annotation, e.g., a new exon, exon boundary change, exon skipping, or modified translation boundaries. The refinement events can be classified according to their type, location, and the transcript being modified (Table S3). A majority (521) of the events are novel exons, of which 314 are located within introns and 207 are in untranslated regions (UTR) of TAIR7 gene models. Of the 314 instances of novel coding sequence predicted between 2 exons of a gene, 26 are observed in the same frame as both adjacent exons and may indicate a single exon, a portion of which may be spliced out in some isoforms. Exon boundary changes were also prevalent, with typical instances including 5′ extension of the first exon and alternative donor/acceptor splice sites. We find evidence for 180 instances of exon extension, and 191 instances of exon shortening. In 5 transcripts, peptide evidence supports an exon skipping event. Some intragenic loci indicate gene extension beyond the borders of the annotated gene model; 323 of these gene extension events were discovered. Using AUGUSTUS to refine existing models with the new peptide evidence, we predict 964 new or altered transcripts in 695 genes.
It is difficult to determine whether a new transcript predicted by AUGUSTUS is a refinement of a gene model or an additional isoform of an alternatively spliced gene. To better distinguish between these two cases, we compared each new transcript and the TAIR7 transcripts to all available cDNA evidence. For 122 genes, EST evidence, in addition to the peptide evidence, was found in support of the new transcript; no ESTs were found in support of the TAIR transcript. For an additional 130 genes, EST evidence was found to support both the new transcript and the TAIR7 gene models, suggesting that the peptides are produced by a newly discovered isoform.
To provide additional support to our gene refinements, we excised the predicted amino acid sequence surrounding a novel cluster and searched for homology to the nonredundant database of proteins (National Center for Biotechnology Information nr version 03/26/08) (Table S2). For 348 loci, we found a close homolog (e value <1 × 10−10). Several genes that have been extensively studied are included among the refined gene models. For example, we found an additional 200-aa exon in the 5′ UTR of MAPK phosphatase (AT3G55270; Fig. S2). Also, we identified 8 peptides corresponding to 4 missing or mispredicted exons at locus AT1G79920 (heat shock protein 70). The new sequence completes the canonical HSP70 pFam domain. A final example is the gene PMI1 (AT1G42550), which, when mutated, results in impaired plastid movement and localization (14). We found 6 peptides upstream of the annotated start codon, providing at least 130 aa of additional sequence.
We identified 70 cases in which the annotated reading frame is different from the observed peptides (Table S4). Assignment of reading frame is particularly difficult for nucleotide-based genome annotation (e.g., cDNA). However, proteomic evidence unambiguously defines the frame of translation. The 70 frame corrections are supported by multiple peptides and extensive homology to other proteins (see SI Materials and Methods). We will use two proteins to illustrate: first, a whole gene frame correction; and second, a partial gene correction. Locus AT3G22240 is a 51-aa protein with no discernable homologs. Four of our peptides indicate translation in different frame than has been annotated. Translation in the new reading frame yields a protein with high sequence identity to PCC1, pathogen and clock controlled protein. The second example is AT1G63500, a protein kinase, which has 4 novel peptides in the annotated 5′ UTR. These peptides point to a large expansion of the gene and a misprediction of the current first exon (Fig. 4).
Fig. 4.
Refined Gene Model. TAIR locus AT1G63500 encodes a protein kinase. (A) Four novel peptides map within the 5′ UTR and the first exon. (B) Zoom of the region shows that the current first exon (frame 3) is out of frame with the peptides (frame 2). (C) Sequence alignment with Arabidopsis and grapevine proteins supports translation in the frame supported by peptides (observed peptides highlighted in alignment).
In addition to the peptides that are described above, 3,534 uniquely located singleton peptides with high confidence (lFDR <0.05) overlap genes and indicate refinement events. These peptides (see Table S5) likely indicate corrections to gene models and are a starting point for further investigation.
Similarly, 2,827 singleton peptides (also uniquely located and with lFDR <0.05) are found in intergenic regions. Some of the peptides may be mis-annotations, however, subsequent work has indicated that many are correct: 665 peptides are contained in ORFs with strong sequence similarity to known proteins (BLAST e value <1 × 10−10). Spectral counts are also an indication of strength of an annotation; 291 peptides have higher spectral counts than 50% of all peptides identified in this study. The intergenic peptides indicate novel coding regions that may have produced a single detectible peptide for several reasons including protein composition or protein length.
Validated Gene Models.
In addition to discovering new protein-coding loci, we identified 126,055 distinct peptides (1.72 million amino acids) that confirmed annotated gene models for 12,769 proteins (40% of the TAIR7 genes). Our claims of coverage are conservative. We count only proteins covered by at least two peptides, one of which must uniquely map to the designated locus. A total of 11,801 peptides were lone supporters of proteins or shared peptides, and therefore were not counted toward the confirmed proteins. Of the sequenced peptides, 87% map to a unique genomic location, unambiguously identifying 10,692 proteins. In addition, we observed proteins from highly homologous gene groups that could not be attributed to a single locus (see SI Materials and Methods). The Arabidopsis genome has high rates of tandem and segmental duplication and many loci contain multiple gene predictions that differ only in the non-translated regions (15). We observed peptides from 913 groups of indistinguishable proteins (2,077 proteins), bringing the total confirmed gene models to 12,769.
Splicing.
It is difficult to estimate the true extent of alternative splicing, given that the alternative splice forms are often not as highly expressed, and might not be sampled. However, our deep proteogenomic sampling revealed a total of 47 genes in which multiple splice forms were observed (see Figure S3 for observed splice types). We estimate (see SI Materials and Methods) that with high probability, the number of genes with alternative splice forms is between 6,718 and 8,983. This is considerably higher than the number of alternatively spliced genes in TAIR7 (3,799) and the number recently predicted by cDNA and ESTs (4,707 at the transcript level) (16).
Discussion
In tandem mass spectrometry, a peptide (from an enzymatic digestion of a protein mixture) is fragmented, usually through collisions. While the physics of the fragmentation is incompletely understood, the fragmentation pattern is consistent, and the collection of fragments (the spectrum) can be used to “fingerprint” the peptide. Recent advances in mass resolution and the availability of software tools to analyze spectra make mass spectrometry the tool of choice for proteomics. Nevertheless, technological limitations create many challenges for the approach.
First, the sampled peptides are biased toward the more abundant proteins in the cell. To comprehensively sample the proteome, a diversity of samples must be assayed (see SI Materials and Methods). Second, incomplete fragmentation patterns and spectral noise “smudge” the fingerprint and introduce errors in peptide identification. Additionally, identification is typically based upon looking up a database of known peptides to pick the most likely candidate. If the true peptide is not in the database, it will not be identified. Finally, posttranslational modifications change the mass and pattern of the fragments, making identification harder. Our study addresses each of these issues. Broad sampling of the proteome was achieved through assaying multiple plant organs and phosphopeptide enriched peptides. We address identification error rates through the introduction of a local false discovery rate. The genome is explicitly and thoroughly queried for potential protein coding sequences. Finally, we use a phosphopeptide spectra specific algorithm for sensitive and efficient annotation of phosphorylated peptides.
The database search tool we used, Inspect, contributed significantly to our ability to extensively annotate spectra. Inspect's Bayesian scoring function is more sensitive than that of SEQUEST, annotating more spectra at a given false-discovery rate. The exon splice-graph database allowed us to detect peptides that span splice boundaries. Our experimental techniques enabled a sampling of the phosphoproteome, which typically contains low abundance proteins. We used 3D LC, which provides much greater resolution and renders unnecessary the extensive resampling that is typical of LC ESI MS/MS experiments. To illustrate, we identified 67% more total peptides using only 3% as many LC runs compared with a study based on resampling (4). We used our novel peptides and an automated gene prediction pipeline to derive 1,473 new and revised gene models. The technical advances reported here dramatically reduce the time and cost required to obtain deep proteome coverage.
Historically, the proteomic and genomic communities have operated independently, with the genomic community in charge of annotation efforts. The predicted proteome is then passed over to the proteomics community for validation, and identification of posttranslational events. We assert that much is to be gained by joining forces, and incorporating proteomic evidence upfront into the genomics pipelines. Proteogenomics provides an orthogonal data source to predict gene models, with levels of sensitivity that are complementary to cDNA sequencing. By investing in proteogenomics to complement more traditional cDNA and EST data at the onset of genome annotation, a more complete and accurate proteome can be achieved even in the early releases. Here, we provide proteomic evidence for 778 new genes and refine 695 current gene models, using the reference annotation from TAIR7. Recently, TAIR has release the next revision of the genome/proteome, TAIR8. Only a small number of our novel peptides (3%) appear in the TAIR8 release indicating that the proteogenomic approach is complementary to computer-based annotation.
Materials and Methods
Sample Chemistry.
In total 21,170,989 MS/MS spectra were collected from 45 LC-MS/MS profiles of Arabidopsis organ samples (leaf, root, flower, and silique) and MM2d cells. Frozen organs were ground in 50 mL of cold (−20 °C) methanol containing 0.2 mM Na3VO4, incubated, and then spun down at 4,000 × g for 5 min. Two more methanol washes were followed by 3 acetone washes. Sample pellets were dried and proteins were extracted by dissolving in 1 mL of 0.2% RapiGest (Waters) with 0.2 mM Na3VO4. Contaminants were spun down and discarded. Cysteines were reduced and alkylated and then proteins were digested with trypsin. Peptides were separated using 2D-LC and charged with electrospray ionization. Spectra were acquired using LTQ linear ion trap tandem mass spectrometers. The data associated with this manuscript may be downloaded from Tranche (http://tranche.proteomecommons.org) using the following hash: eTyqbeZEgF7KOZNqcE0OAbFGAmrIzV1xKx4OCC0CJN9A1MwZmuP2drhEsT + 7XohMx8FM8wtckHv7, mqSnWHLhVuGmrsYAAAAAAASfeg==. The Tranche hash can also be used to verify that files have not changed since publication.
Database Construction and Use.
For gene model confirmation, we used the TAIR7 release of the Arabidopsis proteome (www.arabidopsis.org). For proteomic discovery, we constructed a 6-frame translation of the Arabidopsis genome and a spliced-exon graph containing ab initio gene predictions from AUGUSTUS. For the MS/MS searches, all three databases were combined with decoy sequences formed by shuffling each target sequence and then searched using Inspect. All results are filtered to 1% spectrum-level false discovery rate using the decoy database strategy. We report proteins with two or more peptides, and at least 1 uniquely mapped peptide. For proteins groups that have exactly identical coding sequences we report the group of proteins, because they share all peptides and do not have any uniquely mapped peptides. Our 1% spectrum-level FDR translated to an empirical 0.6% protein-level FDR. The source code for Inspect is available at our lab website, http://peptide.ucsd.edu.
For further details, please refer to SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Anand Patel for his efforts in homology searches and alignment visualization. This work was supported by National Science Foundation IGERT Plant Systems Biology Training Grant DGE-0504645 (to S.H.P.); National Science Foundation Grant IBN 0619411 (to Z.S. and S.P.B.); National Institutes of Health Grants R01-RR16522 and 1P41RR024851-01 (V.B. and N.E.C.); and Deutsche Forschungsgemeinschaft Grant STA 1009/4-1 (M.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The spectra reported in this article have been deposited in the Tranche database, http://tranche.proteomecommons.org (hash eTyqbeZEgF7KOZNqcE0OAbFGAmrIzV1xKx4OCC0CJN9A1MwZmuP2drhEsT+7XohMx8FM8wtckHv7, mqSnWHLhVuGmrsYAAAAAAASfeg==).
This article contains supporting information online at www.pnas.org/cgi/content/full/0811066106/DCSupplemental.
References
- 1.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 2.Lin MF, et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 2007;17:1823–1836. doi: 10.1101/gr.6679507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brent MR. Steady progress and recent breakthroughs in the accuracy of automated genome annotation. Nat Rev Genet. 2008;9:62–73. doi: 10.1038/nrg2220. [DOI] [PubMed] [Google Scholar]
- 4.Baerenfaller K, et al. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science. 2008;320:938–941. doi: 10.1126/science.1157956. [DOI] [PubMed] [Google Scholar]
- 5.Brunner E, et al. A high-quality catalog of the Drosophila melanogaster proteome. Nat Biotechnol. 2007;25:576–583. doi: 10.1038/nbt1300. [DOI] [PubMed] [Google Scholar]
- 6.Tanner S, et al. Improving gene annotation using peptide mass spectrometry. Genome Res. 2007;17:231–239. doi: 10.1101/gr.5646507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savidor A, et al. Expressed peptide tags: An additional layer of data for genome annotation. J Proteome Res. 2006;5:3048–3058. doi: 10.1021/pr060134x. [DOI] [PubMed] [Google Scholar]
- 8.Gupta N, et al. Whole proteome analysis of post-translational modifications: Applications of mass-spectrometry for proteogenomic annotation. Genome Res. 2007;17:1362–1377. doi: 10.1101/gr.6427907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fermin D, et al. Novel gene and gene model detection using a whole genome open reading frame analysis in proteomics. Genome Biol. 2006;7:R35. doi: 10.1186/gb-2006-7-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desiere F, et al. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 2005;6:R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanke M, et al. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–9. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang B, Bao Z, Zhang X, Eddy SR, Wessler SR. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:163–167. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 13.Hanada K, Zhang X, Borevitz JO, Li WH, Shiu SH. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res. 2007;17:632–640. doi: 10.1101/gr.5836207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeBlasio SL, Leusse DL, Hangarter RP. A plant-specific protein essential for blue-light-induced chloroplast movements. Plant Physiol. 2005;139:101–114. doi: 10.1104/pp.105.061887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B, Brendel V. Genomewide comparative analysis of alternative splicing in plants. PNAS. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.