Abstract
Genomic islands (GEI) comprise a recently recognized large family of potentially mobile DNA elements and play an important role in the rapid differentiation and adaptation of bacteria. Most importantly, GEIs have been implicated in the acquisition of virulence factors, antibiotic resistances or toxic compound metabolism. Despite detailed information on coding capacities of GEIs, little is known about the regulatory decisions in individual cells controlling GEI transfer. Here, we show how self-transfer of ICEclc, a GEI in Pseudomonas knackmussii B13 is controlled by a series of stochastic processes, the result of which is that only a few percent of cells in a population will excise ICEclc and launch transfer. Stochastic processes have been implicated before in producing bistable phenotypic transitions, such as sporulation and competence development, but never before in horizontal gene transfer (HGT). Bistability is instigated during stationary phase at the level of expression of an activator protein InrR that lays encoded on ICEclc, and then faithfully propagated to a bistable expression of the IntB13 integrase, the enzyme responsible for excision and integration of the ICEclc. Our results demonstrate how GEI of a very widespread family are likely to control their transfer rates. Furthermore, they help to explain why HGT is typically confined to few members within a population of cells. The finding that, despite apparent stochasticity, HGT rates can be modulated by external environmental conditions provides an explanation as to why selective conditions can promote DNA exchange.
Keywords: integrase, integrative and conjugative element, regulation, 3-chlorobenzoate, clc element
Horizontal gene transfer (HGT), the exchange of DNA between species other than by vertical descent, is of major importance for prokaryotic evolution (1, 2). Various mechanisms play a role in DNA exchange, most commonly known as conjugation, transformation and phage transduction (3). Most pertinently, HGT is responsible for the rapid adaptation of bacteria to acquire new virulence factors and antibiotic resistances or to metabolize toxic compounds (4). Genomic islands (GEIs) are the most recent addition to the suite of known mobile DNA elements in prokaryotes. Large-scale prokaryotic genome sequencing efforts have revealed a wide diversity and abundance of GEIs, suggesting ancient origins and multiple families of different GEI-types (5–13). Like other mobile DNA, GEIs contribute to bacterial survival, differentiation, and adaptation in particular niches by providing, e.g., virulence factors (7, 14, 15), host-cell adhesion (16), iron uptake (17, 18), antibiotic resistance (19, 20), toxin production (10), aromatic compound metabolism (21, 22), or plant symbiosis (23). Because of their wide distribution, GEI form an important model to test various hypotheses on HGT in general. One of the key questions that have escaped much attention concerns the regulatory decisions controlling HGT at the level of the individual bacterial cell. This seems surprising given the typical low frequencies (1% or less) for HGT in bacterial populations (24), and suggests that cells, despite their clonality, are undergoing some sort of phenotypic variation into transfer-proficient and transfer-silent subpopulations. The phenomenon of phenotypic bifurcation in clonal bacterial populations, or “bistability,” is well-known from competence development and sporulation in Bacillus subtilis (25, 26), but has not before been implicated in HGT.
GEI have an intricate life-style of their own, which can be concluded from detailed studies on a number of elements, such as SXT of Vibrio cholerae (27), ICEHin1056 of Haemophilus influenzae (20, 28), SaPI from Staphylococcus aureus (15), and PAPI-1 from Pseudomonas aeruginosa (29). Similar to prophages but in contrast to plasmids, GEIs are normally integrated in the host's chromosome. At low frequencies they can excise (27, 29–31), and transfer to a new cell either by cotransduction (15) or by conjugation (28). The GEI can subsequently reintegrate via site-specific recombination mediated by the integrase, usually at one or more specific target sites in the chromosome (30, 32). Not all GEI detected by sequencing have a “complete” life-style but may lack one or more functional aspects, which has been interpreted as evolutionary regression (2). Those GEI that are capable of excision, conjugative transfer, and integration have been grouped with other mobile DNA, such as conjugative transposons as integrative and conjugative elements (or ICEs) (33).
The model for HGT we study here is the clc element (designated as ICEclc) (21). ICEclc is a 103-kb self-transferable GEI first described in Pseudomonas knackmussi strain B13, where it is present in two copies (34). The most prominent phenotype encoded by ICEclc is the capacity conferred on the host to use the aromatic compounds 3-chlorobenzoate and 2-aminophenol as unique carbon and energy substrates. ICEclc is a representative of a large set of syntenic GEIs present in Gamma- and Betaproteobacteria (20, 21, 28) (Fig. S1). The process of ICEclc self-transfer begins with activation of the promoter of the intB13 integrase gene (Pint), which is located downstream of the tRNAGly integration site (31, 35), leading to an excised and covalently closed ICEclc molecule (Fig. S2). The excised ICEclc molecule is supposed to self-transfer via a GEI-type IV secretion system in analogy to ICEHin1056 for which this mechanism was recently discovered (28). In the new host cell, ICEclc site-specifically recombines with the 3′ 18-bp of a tRNAGly gene, leading to integration of the element in the host's chromosome and restoration of the tRNAGly gene. Site-specific recombination is catalyzed by the IntB13 integrase, which may be temporarily overexpressed from the strong constitutive promoter in the excised ICEclc molecule (32, 35). Putative regulatory elements implicated in intB13 expression in the integrated ICEclc have been located at the ICEclc's end opposite to the intB13 position (35). One of those was a gene named inrR (i.e., integrase regulator) (Fig. S2). Here, we demonstrate that InrR is positively controlling intB13 expression, and, therewith, ICEclc excision and transfer. Single-cell reporter gene studies showed that expression of both intB13 and of inrR is limited to a small subpopulation of cells, typical for bistability.
Results
P. knackmussi Strains with Deletions in inrR Are Impaired in ICEclc Activation.
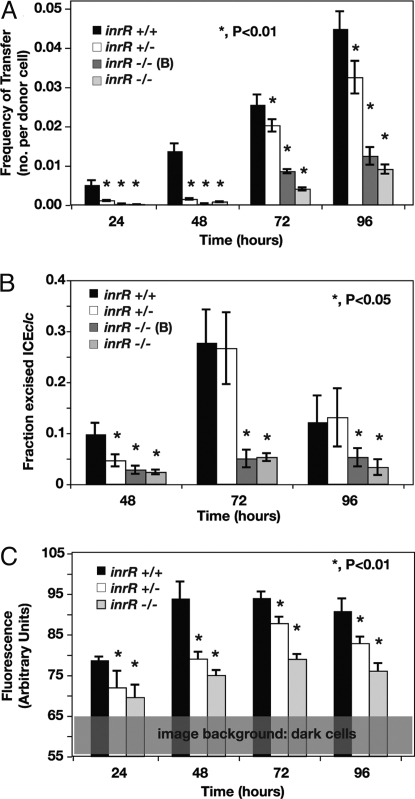
A 156-bp in-frame deletion of one or both inrR genes in either of the ICEclc copies on the chromosome of Pseudomonas knackmussii strain B13 was generated by double homologous recombination and marker exchange to avoid any polar effects on downstream located genes (Fig. S3 and SI Text). Various effects were observed in B13 strains with the inrR in frame deletion that were consistent with a decrease or loss of ICEclc mobility. First, the frequency of ICEclc transfer from strain B13 as the donor to Pseudomonas putida UWC1 as the recipient decreased from 1.4 ± 0.2 × 10−2 transconjugants per recipient after 48 h for wild-type B13 to up to 2 orders of magnitude when inrR was deleted (Fig. 1A). Double inrR deletions produced fewer transconjugants (e.g., 3.1 ± 0.2 × 10−4 after 48 h) than the single deletion (1.4 ± 0.2 × 10−3), suggesting partial complementation of transfer by one of the copies. Second, the amount of excised ICEclc DNA was reduced in populations of single and double inrR deletion mutants, with the double mutant being more severely affected than the single deletion (Fig. 1B). As the excised form is a prerequisite for subsequent transfer, reduced transfer rates, and decreased amounts of the ICEclc excised form are in agreement. Third, expression from the intB13 integrase gene in individual cells was significantly reduced in the inrR mutants. This was inferred from measurements of enhanced green fluorescent protein (egfp) expression in individual cells of strain B13 and the inrR mutants, equipped with a single copy chromosomal transcriptional fusion of the egfp gene to the Pint promoter (Fig. 1C, 2A). These results, therefore, were all in agreement and led us to conclude that InrR is implicated in activation (or derepression) of intB13 expression. Contrary to our expectations, purified N-terminal His6-tagged InrR did not detectably bind DNA fragments comprising the Pint promoter region between the integration site (attR) and the start of the intB13 gene in electrophoretic mobility assays (data not shown). Although we cannot exclude that N-terminal His6-tagged InrR is folded differently as wild-type InrR, the absence of detectable DNA binding to the Pint promoter suggests that the mode of action of InrR is not that of a classical transcription activator.
Fig. 1.
Effects of inrR deletions on ICEclc behavior. (A) Transfer frequencies of ICEclc from P. knackmussii strain B13 or one of the inrR deletion mutants to P. putida UWC1 as recipient. (B) Fraction of the ICEclc excised form per integrated copy in cultures grown with 3-chlorobenzoate minimal medium at different incubation time points. (C) 95% percentile of the cumulative distribution of fluorescence values of individual cells in cultures grown with 3-chlorobenzoate medium. Time 24 h corresponds to the beginning of stationary phase.
Bistable Expression of intB13 and inrR.
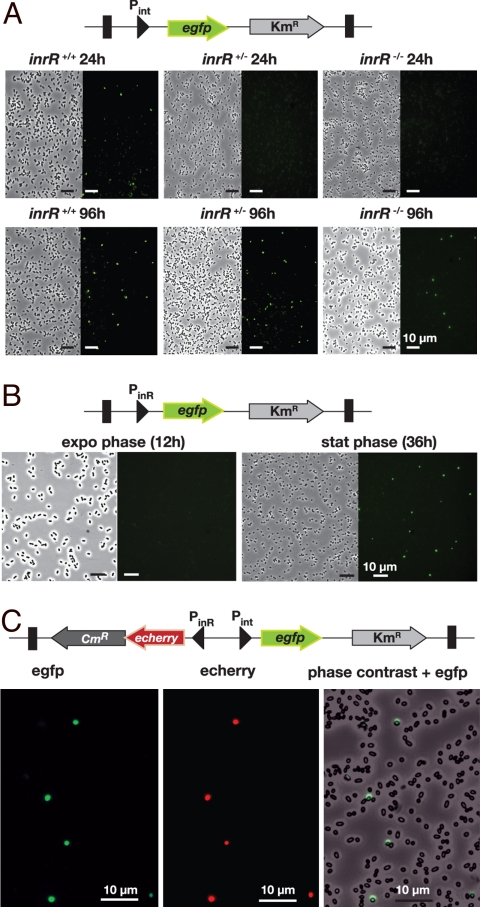
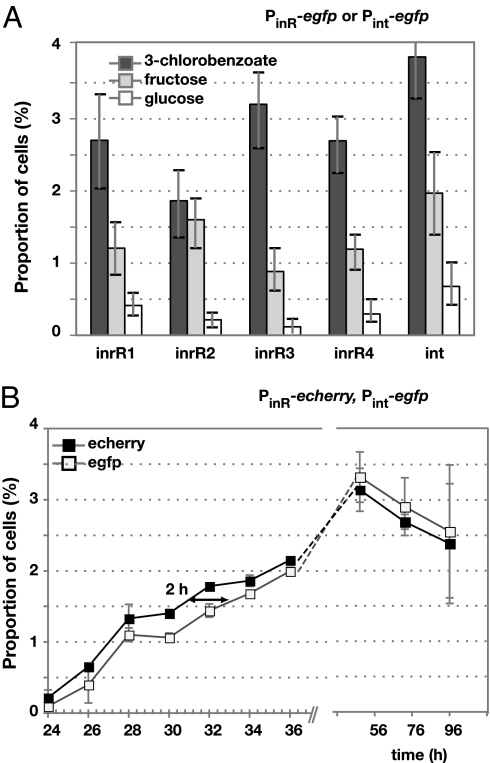
P. knackmussii B13 cells with a single-copy Pint-egfp fusion did not produce egfp expression detectable with epifluorescence microscopy during exponential phase. Interestingly, in stationary phase ≈3% of cells within a population displayed egfp-specific fluorescence (Figs. 2A and 3A). This proportion was significantly higher when cells were grown with 3-chlorobenzoate than with fructose or glucose (Fig. 3A). These results indicated that intB13 was expressed in a bistable manner and that, most likely, ICEclc became excised only in those cells.
Fig. 2.
Stochastic expression of the Pint and PinR promoters in P. knackmussii strain B13 cultures from single copy chromosomal transcriptional fusions to the egfp and/or echerry gene. (A) Egfp expression from Pint in strain B13 or the single and double inrR deletion derivatives. (B) P. knackmussii strain B13 cells equipped with single copy PinR-egfp. (C) Colocalization of echerry and egfp fluorescence in stationary phase cells (28 h) of P. knackmussii strain B13 with a single copy Pint-egfp, PinR-echerry fusion. Shown are phase-contrast micrographs at 1,000× magnification and corresponding epifluorescence images for egfp or echerry.
Fig. 3.
Bistable stochastic expression of egfp and/or echerry from PinR or Pint in P. knackmussii strain B13. (A) Subpopulation sizes expressing egfp from PinR (four randomly picked clones with different chromosomal insertion site of the reporter gene fusions) or Pint in stationary phase (76 h) after growth on different carbon substrates. Error bars denote calculated 95% confidence intervals on the subpopulation size. (B) Bistable echerry expression from PinR preceeds that of egfp from Pint in P. knackmussii cells by ≈ 2 h (see Table 1).
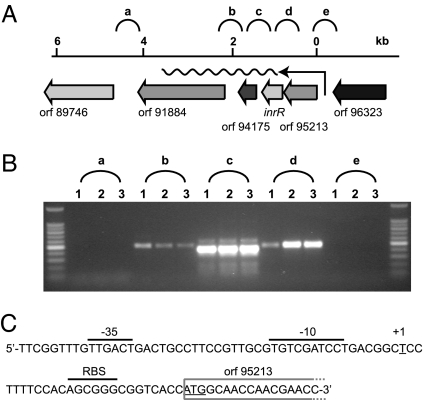
To further understand the bistable nature of intB13 gene expression, we focused on inrR expression itself. Reverse transcriptase-PCR analysis of total RNA from wild-type strain B13 grown with 3-chlorobenzoate revealed that inrR is the second gene in a 4-gene operon (Fig. 4A). The transcriptional start site was located upstream of the gene ORF95213 (Fig. 4B). The sequence in this area showed clear features for a stationary phase expressed promoter (Fig. 4C), which we named PinR. Consequently, it seemed likely that inrR expression would be controlled by an RpoS-like sigma factor and take place primarily in stationary phase. Transcriptional gene fusions between the PinR promoter and the egfp gene were constructed and inserted in single copy in P. knackmussii B13. Single-cell epifluorescence analysis demonstrated that the PinR promoter indeed became active during early stationary phase and not during exponential growth (Fig. 2B). Egfp expression from PinR was not significantly different in the single or the double inrR deletion mutant of strain B13, indicating that InrR does not autoregulate its own expression (Table S1). Most strikingly, however, and similar to B13 cells carrying a single-copy Pint-egfp fusion, only a small proportion of cells in the population expressed egfp under the control of PinR (Fig. 2B). This proportion was again significantly higher when cells were grown with 3-chlorobenzoate than with fructose or glucose (Fig. 3A).
Fig. 4.
Operon structure and transcription start site determination for the inrR transcript. (A) Gene localization and direction of the 4-gene operon containing inrR. (B) RT-PCR products with primers targeting regions a-e indicated in a from total RNA isolated from P. knackmussii strain B13 grown with 3-chlorobenzoate in stationary phase (Table S2). (C) Sequence of the region upstream of ORF95213 and indication of the 5′-end of the transcript (as “+1”). Corresponding −10 and −35 regions and the ribosome binding site (RBS) are overligned. Note the extended −10 TG motif typical for σs promoters (39).
Colocalization of Bistable inrR and intB13 Expression.
We then asked the question whether the reason for bistability at the Pint promoter was in fact a consequence of bistable expression at the PinR promoter. A further derivative of strain B13 was created in which egfp was produced from Pint and echerry from PinR, both on the same minitransposon and inserted in single copy (Fig. 2C). These two autofluorescent proteins have emission spectra that are sufficiently well separated to not interfere with one another even at low expression levels (36). Indeed, in stationary phase cultures grown with 3-chlorobenzoate, echerry colocalized with egfp in the same cell. This indicates that cells that express inrR also express intB13 (Fig. 2C). These cells, most likely, are the ones, which will activate ICEclc excision and continue to transfer should a suitable recipient be present. In fact, the proportion of cells expressing egfp from Pint in stationary phase (3–5%) equals the maximum transfer frequency observed for ICEclc (≈ 4 × 10−2 per donor, Fig. 1A), suggesting that all such cells engage in successful transfer when presented with a suitable recipient cell.
Echerry and egfp measurements of individual cells cultured with 3-chlorobenzoate in minimal medium further showed that echerry formation preceded egfp by ≈2 h (Fig. 3B and Table 1). To avoid that slight differences in maturation rate of egfp and echerry (36) would be responsible for this time effect, we repeated the experiment with a strain in which the promoter-reporter gene fusions were reversed. In this case, the egfp fluorescence (now controlled by PinR) appeared first (Fig. S4 and Table 1), followed in the same cell by echerry (expressed from Pint). This strongly suggests, therefore, that the PinR promoter (and thus inrR) is expressed first, after which InrR could promote expression from Pint (and thus intB13). Because at this point they are in stationary phase, the cells do not further divide and any egfp or echerry measured is the result of expression in those particular cells and not of inheritance by cell division. Because close to 100% of all individual cells that express echerry (from PinR) also express egfp (from Pint) in stationary phase, this would mean that a robust bistable signal propagation is generated (Table 1).
Table 1.
Proportion of cells expressing both PinR-echerry and Pint-egfp or PinR-egfp and Pint-echerry in cultures of P. knackmussii strain B13
| Incubation time,* h | PinR-echerry, Pint-egfp |
PinR-egfp, Pint-echerry |
||||||
|---|---|---|---|---|---|---|---|---|
| Proportion echerry† | Proportion egfp† | Percentage of common per echerry‡ | Percentage of common per egfp‡ | Proportion echerry† | Proportion egfp† | Percentage of common per echerry‡ | Percentage of common per egfp‡ | |
| 24 | 0.22 (±0.11) | 0.09 (±0.06) | 33 | 75 | 0.09 (±0.12) | 0.48 (±0.28) | 18 | 12 |
| 26 | 0.65 (±0.05) | 0.40 (±0.26) | 58 | 96 | 0.42 (±0.16) | 1.1 (±0.08) | 68 | 26 |
| 28 | 1.3 (±0.19) | 1.1 (±0.09) | 83 | 100 | 0.7 (±0.3) | 1.8 (±0.2) | 70 | 28 |
| 30 | 1.4 (±0.06) | 1.1 (±0.07) | 70 | 96 | 2.2 (±0.4) | 2.9 (±0.6) | 58 | 48 |
| 32 | 1.8 (±0.01) | 1.4 (±0.09) | 80 | 98 | — | — | — | — |
| 34 | 1.9 (±0.08) | 1.7 (±0.01) | 88 | 98 | — | — | — | — |
| 36 | 2.2 (±0.02) | 2.0 (±0.06) | 89 | 97 | 2.5 (±1.2) | 3.8 (±0.8) | 76 | 41 |
| 48 | 3.1 (±0.30) | 3.3 (±0.36) | 97 | 92 | 3.9 (±0.5) | 3.9 (±0.3) | 81 | 80 |
| 72 | 2.7 (±0.10) | 2.9 (±0.41) | 96 | 90 | — | — | — | — |
| 96 | 2.4 (±0.85) | 2.5 (±0.94) | 100 | 93 | — | — | — | — |
*P. knackmussii B13 (PinR-echerry, Pint-egfp) or (PinR-egfp, Pint-echerry) cells cultured in batch on minimal medium with 5 mM 3-chlorobenzoate as sole carbon and energy substrate. Incubation time denotes time of sampling since inoculation start; 24 h corresponds to early stationary phase. Cell samples immediately imaged under epifluorescence microscopy at two separate wavelengths for egfp and echerry.
†Proportion of cells with fluorescence in echerry or egfp filter above background (Fig. S4 B-D) compared to total number of cells in sample (n > 1'500 cells). Values are average from triplicate samples of two independent P. knackmussii B13 (PinR-echerry, Pint-egfp) or three (PinR-egfp, Pint-echerry) clones (i.e., having different mini-Tn insertion positions).
‡Percentage of cells having both echerry and egfp intensities above background per number of those having echerry or egfp.
Discussion
We are unaware of another study describing bistable control of GEI-transfer or HGT in general. Our results suggest a series of stochastic events underlying bistability formation. The first detected event occurs at the level of inrR expression, bifurcating the population in cells that produce InrR or not. Bistability dictates that (small) cell-to-cell stochastic expression differences (here in inrR) would be amplified and temporarily locked; something, which can be achieved by mechanisms such as a positive or a double-negative transcriptional feedback loop (37). The nature of this amplification mechanism at the level of PinR is currently not clear, but may be less in amplitude than, e.g., the ComK-comKp autoregulatory loop, because only 3% of all cells engage in InrR-IntB13 bistability [compared with ≈20% for competence (25)]. Because inrR mutants still showed bistable egfp expression from PinR we conclude it is unlikely that InrR itself is provoking feedback transcription amplification, although it might somehow influence its own activity. The “locked” state of the bistability is apparent from the results that cells in which inrR is expressed propagate bistable expression at intB13. However, because previous results indicated the implication of a repressor on intB13 expression (35), we postulate that the control of Pint actually comprises a second mechanism for bistable control, involving a balance between two counteracting factors (Fig. S5). Cells that have a high amount of InrR would be able to tip the balance in favour of activation at the Pint promoter, whereas cells expressing InrR at very low amounts would repress Pint. Because InrR does not seem to act as a classical DNA binding protein, it may impose its function by direct protein–protein interaction on the presumed transcriptional repressor for intB13 expression (Fig. S5). Not only are the population proportions and individual cells expressing inrR (seen with PinR-egfp or -echerry fusions) the same as those expressing intB13, but both increase after growth with 3-chlorobenzoate compared with fructose or glucose (Fig. 3A). This indicates that the bistable switch is stochastic but can be modulated by external conditions.
The InrR control mode might actually be a much more common mechanism. For example, currently >60 orthologous members of InrR exist in GenBank (Fig. S6), most of which have been detected in putative GEI-regions of bacterial genomes. Furthermore, the large number of genome regions highly related to the functional core of ICEclc (Fig. S1), as recognized in ref. 20, is a strong indication that the bistable regulation mode for ICEclc transfer might be a rather common way for GEIs of this family to decide on excision and transfer. In addition, even though this was not looked at specifically with the help of single-cell reporter gene fusions, the behavior of other GEI-types suggests bistable control mechanisms as well. For example, excision and transfer of SXT of V. cholerae is controlled by a repressor-activator loop similar as for phage Lambda, which has been predicted to produce bistability (37). This loop implicates SetR, a repressor that undergoes RecA-dependent cleavage during SOS-response, upon which the transcription factors SetC and SetD can activate expression of the integrase and transfer genes (27). Interestingly, SOS activation leads to an increase of SXT transfer from Escherichia coli from ≈10−4 to 10−2 per donor (27), which is in the same order of frequency as observed for ICEclc. Also ICEMlSymR7A from M. loti excises at a maximum frequency of ≈1% in stationary phase, whereas observed transfer frequencies are in the order of 10−4 per donor (30). Finally, the element PAPI-1 in P. aeruginosa excises in a proportion of 0.16% in a population of cells in stationary phase and transfers at frequencies between 10−4 and 10−6 per donor (29). All of these results are thus in agreement with a more general hypothesis of bistable control of GEI transfer, albeit probably generated by different mechanisms in the case of SXT, ICEMlSymR7A, PAPI-1, and ICEclc.
Our results explain why not all cells in a population engage in GEI transfer. Despite being seemingly “random” in a population of cells, decisions on ICEclc transfer appear to be governed by highly complex signaling chains within individual cells. For the time being, the most likely explanation is that the choice for ICEclc excision in an individual cell is stochastic. Because excision is a requirement for ICEclc transfer, the stochastic decision for excision forms a key event in transfer control, albeit not the sole. For instance, expression of the transfer apparatus and DNA mobilization enzymes must be simultaneously ensured. The source for stochasticity may originate in transcriptional noise at the inr promoter, which is amplified to bistability and then subsequently faithfully transmitted to the intB13 promoter. At present the nature and magnitude of the noise at the inr promoter (e.g., intrinsic or extrinsic) could not be reliably investigated because of the low frequency of the bistable population expressing InrR and IntB13 (Fig. S4 B–D). We are convinced that the comprehension of such stochastic mechanisms is important for understanding the conditions favoring GEI horizontal transfer, which in the long term may provide better control over desired and undesired bacterial adaptation processes.
Materials and Methods
Bacterial Strains and Culture Conditions.
Bacterial strains used in this study are described in SI Text. P. knackmussii B13 strains were cultured in liquid medium under batch conditions at 30 °C and shaking. Cultures were typically growing exponentially between 6 and 18 h after inoculation, whereas stationary phase (i.e., cessation of growth) was reached after 24 h. Increase in culture turbidity at 600 nm was followed during growth to estimate the onset of the stationary phase and exact sampling times for epifluorescence microscopy.
ICEclc Self-Transfer.
Self-transfer was tested by mixing donors (P. knackmussii B13 or one of the inrR deletion derivatives) and recipient (P. putida UWC1) on membrane filters for 24, 48, 72, or 96 h as described in ref. 38. Transconjugants (P. putida UWC1 with ICEclc) were selected on minimal medium plates with 5 mM 3-chlorobenzoate as sole carbon and energy source (to select for ICEclc) and 50 μg per ml rifampicin (resistance marker of the recipient). Transfer frequencies were expressed as number of transconjugant colonies per number of donors.
Mutant Construction.
To produce deletions in inrR on either of the two ICEclc copies in P. knackmussii strain B13 we used homologous recombination with a cloned gene containing an internal deletion of 156 base-pairs produced with the PCR. The inrR-derivative was cloned on a nonreplicative plasmid for Pseudomonas (Fig. S3). Single recombinants (i.e., those in which the whole plasmid was integrated) in strain B13 were selected by resistance to tetracycline and purified. Double recombinants were enriched and plated, after which individual colonies were screened by the PCR for the appropriate integration and absence of plasmid replicon. Strangely, strain B13 with a double inrR mutation was not able to grow with 3-chlorobenzoate and cultures developed a strong black color as a result of photopolymerization of chlorocatechol, a metabolic intermediate from 3-chlorobenzoate. However, mutants growing with 3-chlorobenzoate developed spontaneously in the culture flasks. Purified double inrR mutants displayed growth rates similar as P. knackmussii B13 wild-type and no longer produced black color with 3-chlorobenzoate as growth substrate. InrR mutants are marked inrR+/− (one interrupted copy), inrR−/− (two interrupted copies) or inrR−/−“B,” the double inrR mutant strain that accumulated dark color when incubated with 3-chlorobenzoate.
Single-Copy Promoter Gene Fusions.
Expression from Pint or PinrR was measured at single-cell level by introducing a stable single-copy chromosomal transcriptional fusion via minitransposon Tn5 delivery (35). Schematic fusion structures are depicted in Fig. 2 on top of each panel. Appropriate DNA fragments containing either Pint or PinrR were cloned in front of promoterless egfp or echerry genes, and the fusion was subsequently inserted within the miniTn5 delivery vector. The echerry gene is an E. coli optimized codon variant of mcherry (36). For colocalization studies a single miniTn5 was inserted with both Pint-egfp and PinR-echerry or with Pint-echerry and PinR-egfp (SI Text, Fig. 3C). Four independent clones with different insertion position of the miniTn5s were analyzed.
Fluorescence Microscopy.
Culture samples of 10 μL were immediately placed on regular microscope slides, closed with a 50 mm × 0.15 mm coverslip, and imaged within 1–2 min. Fluorescence intensities of ≈1,000 individual cells were digitally recorded on image fields not previously exposed to UV-light to avoid bleaching. The mean pixel intensity of all objects was quantified by an automatic subroutine in the program Metaview (version 6.1; Universal Imaging; Visitron Systems) as described in ref. 31. Fluorescence intensities per cell were expressed as cellular average gray values (AGVs). Subpopulation expression was determined from the 95% percentile in a cumulative ranking of all objects according to their AGV. Bootstrapping served to calculate 95% confidence intervals on percentile determinations. Relative proportions of subpopulations were determined from the intercept between linear regressions on each of the subpopulations plotted in quantile normalized graphs of all individual cells versus their average gray value (Fig. S4 B–D). Images for display were autolevelled and further cropped to the final size with Adobe Photoshop.
DNA and RNA Techniques.
Quantification of the ICEclc circular form in cultures was done by densitrometric analysis of band intensities on Southern-hybridized total DNAs (SI Text). All time points represent those after culture inoculation, whereby 24 h corresponds to early stationary phase. RNA was isolated from stationary phase cultures of P. knackmussii strain B13 grown on MM with 10 mM 3CBA for 48 h at 30 °C, as described in ref. 21. The 5′ end of the transcript including inrR was mapped with the SMART RACE cDNA Amplification Kit (Clontech) according to the manufacturer's protocol (SI Text, Table S2.
Statistical Testing.
Significance of different treatments (e.g., wild-type versus inrR mutant behavior) was examined by pairwise t test (two-sided, P < 0.01). Error bars denote standard deviations from the mean or calculated 95% confidence intervals in triplicate experiments.
Supplementary Material
Acknowledgments.
We thank Itzel Ramos and Dianne Newman for the gift of the echerry gene; Ted Farmer, David Johnsson and Thomas Nyström for discussion and for critical reading the manuscript. This work was supported by grants from the Swiss National Science Foundation (3100-67229, 3100A0-108199).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806164106/DCSupplemental.
References
- 1.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 2.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 3.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 4.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: The agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 5.Vernikos GS, Parkhill J. Resolving the structural features of genomic islands: A machine learning approach. Genome Res. 2008;18:331–342. doi: 10.1101/gr.7004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greub G, Collyn F, Guy L, Roten CA. A genomic island present along the bacterial chromosome of the Parachlamydiaceae UWE25, an obligate amoebal endosymbiont, encodes a potentially functional F-like conjugative DNA transfer system. BMC Microbiol. 2004;4:48. doi: 10.1186/1471-2180-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He JX, et al. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc Natl Acad Sci USA. 2004;101:2530–2535. doi: 10.1073/pnas.0304622101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsiao WW, et al. Evidence of a large novel gene pool associated with prokaryotic genomic islands. PLoS Genet. 2005;1:e62. doi: 10.1371/journal.pgen.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klockgether J, Würdemann D, Reva O, Wiehlmann L, Tümmler B. Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa. J Bacteriol. 2007;189:2443–2459. doi: 10.1128/JB.01688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulsen IT, et al. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol. 2005;23:873–878. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perna NT, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 12.Simpson AJ, et al. The genome sequence of the plant pathogen Xylella fastidiosa. Nature. 2000;406:151–157. doi: 10.1038/35018003. [DOI] [PubMed] [Google Scholar]
- 13.Mathee K, et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci USA. 2008;105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coburn PS, Baghdayan AS, Dolan GT, Shankar N. Horizontal transfer of virulence genes encoded on the Enterococcus faecalis pathogenicity island. Mol Microbiol. 2007;63:530–544. doi: 10.1111/j.1365-2958.2006.05520.x. [DOI] [PubMed] [Google Scholar]
- 15.Ubeda C, Barry P, Penades JR, Novick RP. A pathogenicity island replicon in Staphylococcus aureus replicates as an unstable plasmid. Proc Natl Acad Sci USA. 2007;104:14182–14188. doi: 10.1073/pnas.0705994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobrindt U, et al. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J Bacteriol. 2003;185:1831–1840. doi: 10.1128/JB.185.6.1831-1840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larbig K, et al. Gene islands integrated into tRNAGly genes confer genome diversity on a Pseudomonas aeruginosa clone. J Bacteriol. 2002;184:6665–6680. doi: 10.1128/JB.184.23.6665-6680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchrieser C, Prentice M, Carniel E. The 102-kilobase unstable region of Yersinia pestis comprises a high- pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J Bacteriol. 1998;180:2321–2329. doi: 10.1128/jb.180.9.2321-2329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beaber JW, Hochhut B, Waldor MK. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol. 2002;184:4259–4269. doi: 10.1128/JB.184.15.4259-4269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohd-Zain Z, et al. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J Bacteriol. 2004;186:8114–8122. doi: 10.1128/JB.186.23.8114-8122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaillard M, et al. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J Bacteriol. 2006;188:1999–2013. doi: 10.1128/JB.188.5.1999-2013.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toussaint A, et al. The biphenyl- and 4-chlorobiphenyl-catabolic transposon Tn4371, a member of a new family of genomic islands related to IncP and Ti plasmids. Appl Environ Microbiol. 2003;69:4837–4845. doi: 10.1128/AEM.69.8.4837-4845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan JT, et al. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J Bacteriol. 2002;184:3086–3095. doi: 10.1128/JB.184.11.3086-3095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sørensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. Studying plasmid horizontal transfer in situ: A critical review. Nat Rev Microbiol. 2005;3:700–710. doi: 10.1038/nrmicro1232. [DOI] [PubMed] [Google Scholar]
- 25.Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 26.Smits WK, Kuipers OP, Veening JW. Phenotypic variation in bacteria: The role of feedback regulation. Nat Rev Microbiol. 2006;4:259–271. doi: 10.1038/nrmicro1381. [DOI] [PubMed] [Google Scholar]
- 27.Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 28.Juhas M, et al. Novel type IV secretion system involved in propagation of genomic islands. J Bacteriol. 2007;189:761–771. doi: 10.1128/JB.01327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu X, Gurkar AU, Lory S. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2006;103:19830–19835. doi: 10.1073/pnas.0606810104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramsay JP, Sullivan JT, Stuart GS, Lamont IL, Ronson CW. Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol Microbiol. 2006;62:723–734. doi: 10.1111/j.1365-2958.2006.05396.x. [DOI] [PubMed] [Google Scholar]
- 31.Sentchilo VS, Ravatn R, Werlen C, Zehnder AJB, van der Meer JR. Unusual integrase gene expression on the clc genomic island of Pseudomonas sp. strain B13. J Bacteriol. 2003;185:4530–4538. doi: 10.1128/JB.185.15.4530-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravatn R, Studer S, Zehnder AJB, van der Meer JR. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J Bacteriol. 1998;180:5505–5514. doi: 10.1128/jb.180.21.5505-5514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155:376–386. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Ravatn R, Zehnder AJB, van der Meer JR. Low-frequency horizontal transfer of an element containing the chlorocatechol degradation genes from Pseudomonas sp. strain B13 to Pseudomonas putida F1 and to indigenous bacteria in laboratory-scale activated-sludge microcosms. Appl Environ Microbiol. 1998;64:2126–2132. doi: 10.1128/aem.64.6.2126-2132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sentchilo VS, Zehnder AJB, van der Meer JR. Characterization of two alternative promoters and a transcription regulator for integrase expression in the clc catabolic genomic island of Pseudomonas sp. strain B13. Mol Microbiol. 2003;49:93–104. doi: 10.1046/j.1365-2958.2003.03548.x. [DOI] [PubMed] [Google Scholar]
- 36.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 37.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaillard M, Pernet N, Vogne C, Hagenbüchle O, van der Meer JR. Host and invader impact of transfer of the clc genomic island into Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci USA. 2008;105:7058–7063. doi: 10.1073/pnas.0801269105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becker G, Hengge-Aronis R. What makes an Escherichia coli promoter σs dependent? Role of the -13/-14 nucleotide promoter positions and region 2.5 of σs. Mol Microbiol. 2001;39:1153–1165. doi: 10.1111/j.1365-2958.2001.02313.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.