Abstract
Clinically relevant antibiotics that target the ribosomal peptidyl transferase center (PTC), a highly conserved ribosomal region, exert their inhibitory action by exploiting the flexibility of PTC nucleotides, which trigger modulations of the shape of the antibiotic binding pocket. Resistance to these antibiotics was observed clinically and in vitro. Based on the crystal structures of the large ribosomal subunit from eubacterium suitable to represent pathogens in complex with these antibiotics, it was found that all nucleotides mediating resistance to PTC antibiotics cluster on one side of the PTC. Over half of the nucleotides affecting resistance reside in regions of lower sequence conservation, and are too distal to make Van der Waals interactions with the bound drugs. Alterations of the identity of these nucleotides may not lethally affect ribosome function, but can hamper antibiotic binding through changes in the conformation and flexibility of specific PTC nucleotides. Comparative analysis revealed properties likely to lead to cross-resistance and enabled their parameterization. As the same nucleotides are frequently involved in resistance to more than a single family of antibiotics, the common pattern explains medically observed cross-resistance to PTC antibiotics and suggests the potential for a wider clinical threat.
Keywords: peptidyl transferase center, chloramphenicol, linezolid pleuromutilins, streptograminsA
Ribosomes, the universal nanomachines translating the genetic code into proteins, are riboprotein assemblies that play a key role in cell vitality. Hence, they are targeted by various antibiotics that exert their antimicrobial effects by interacting predominantly with ribosomal RNA (rRNA), the main constituent of the ribosomal functional regions. Clinical usefulness requires discrimination between pathogenic eubacteria and mammalian cells. The high conservation of the ribosomal functional sites (1) implies severe limitations on antibiotics selectivity. Typically, selectivity in binding of ribosomal antibiotics is governed by the identity of a single or a few nucleotides. An example is the MLSB group (macrolides, lincosamides, and streptograminsB) (2) that act by blocking the nascent protein exit tunnel (3–8). Nucleotide 2058 (Escherichia coli nomenclature throughout), the main determinant of the binding of antibiotics belonging to this family, is an adenine in eubacteria and a guanine in eukaryotes and archaea. Consequently, MLSB resistance mechanisms are typically based on minimizing drug binding by A→G mutation or methylation by erm methyltransferase genes (2). However, although A2058G mutation confers resistance, G2058A mutation, does not always lead to inhibitory action, because additional structural elements that influence the shape of the antibiotics binding pocket determine the antibiotic effectiveness (9–11).
Although the peptidyl transferase center (PTC) is one of the most conserved regions of the ribosome, several antibiotics bind to the eubacterial PTC with high affinity and great specificity. Parameters allowing selectivity and effectiveness of PTC antibiotics were revealed by analyzing crystal structures of complexes of large ribosomal subunits from the eubacteria Deinococcus radiodurans, D50S, and the archaeon Haloarcula marismortui, H50S, with the clinically useful antibiotics from the phenicols, lincosamides, pleuromutilins, streptograminsA, oxazolidinones families (9, 10, 12–18), as well as with the yet to be used clinically methymycin (19) and lankacidins (unpublished work). These indicated that chloramphenicol, linezolid, and methymycin hamper A site tRNA binding; pleuromutilins, streptograminsA, and lankacidin hamper A and P site tRNAs accommodation; and clindamycin interferes with peptide bond formation. The structures of D50S/pleuromutilins revealed an elaborate binding mode, a unique inhibitory mechanism and a strategy for acquiring selectivity despite the extremely high sequence conservation (12, 16). Thus, pleuromutilins selectivity is determined by nucleotides residing remote from the PTC and, hence, are less conserved; pleuromutilins binding triggers an induced-fit mechanism that exploits the flexibility of nucleotides residing in and around the PTC, particularly U2585 and U2506, for tightening the binding pocket (16).
Cross-resistance is the tolerance to a specific antibiotic by strains that are resistant to antibiotics from other families. In many cases these share approximately the same binding site but may use different interactions with the binding pocket. It is rather common in the MLSB group. Thus, erythromycin resistance by A2058 methylation that was first observed after short exposure to submicromolar drug concentrations in strains of Staphylococcus aureus (20) was soon detected in strains that became tolerant to the other MLSB group members (1). Data on cross-resistance between PTC antibiotics (21–25), which stimulated recent attempts to overcome it (e.g., refs. 26–28), indicated the existence of common resistant patterns. Consistently, overlapping binding sites were detected in crystal structures of PTC antibiotics with D50S (3, 16, 18, 19) and H50S (17) (Fig. 1). Specifically, crystal structures of complexes of 4 pleuromutilins, obtained at clinically relevant concentrations, show that their common tricyclic cores bind in almost identical fashion to a tight pocket, and that nucleotide U2504 appears to meditate conformational rearrangements at its binding surface (12, 16). As mutations conferring resistance to the pleuromutilin tiamulin cluster in the vicinity of this nucleotide (29), similar mutations are likely to affect the entire family (16).
Fig. 1.
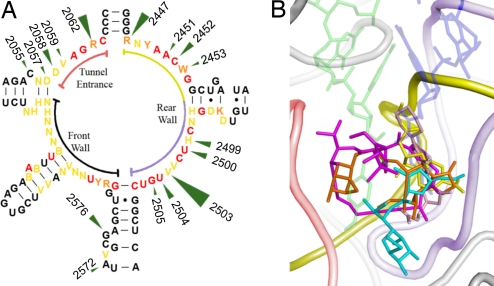
The antibiotics binding pockets within the PTC. (A) 2D diagram of the 23S RNA at the vicinity of the PTC. Green arrows indicate nucleotides mediating resistance to PTC antibiotics. Arrow size is proportional to the number of different classes of antibiotics that are being affected. Relations between E. coli cell vitality and nucleotide alterations (30) are color coded. Absolutely essential nucleotide, which cannot be mutated are shown in red. Nucleotides that can be replaced by a single nucleotide are shown in orange. Nucleotides that can be replaced by 2 or 3 other nucleotides are shown in yellow. Nomenclature used as established (31). Specific sections of the 2D diagram have colors identical to the colors of their corresponding regions in the 3D structure shown in B. (B) The 3-dimensional positions of PTC antibiotics showing their overlapping positions. The antibiotics chloramphenicol, clindamycin, retapamulin, dalfopristin, and linezolid are shown in yellow, cyan, orange, magenta, and pink, respectively. A site tRNA 3′ end and the derived P site tRNA (32, 33) are shown in transparent blue and green, respectively. The 23S rRNA is shown in red, yellow, and blue, as their corresponding sections in the 2D representation (A). 23S rRNA segments not shown in A are colored gray. The black arc in A is not shown in B because the latter is shown from the front wall direction.
Here, we report results of comprehensive structural analyses of the spatial distributions of nucleotides that affect resistance to PTC antibiotics. Because, so far, there are no crystal structures of ribosomes from genuine pathogens, we based the analyses mainly, but not exclusively, on structures of ribosomes from the eubacteriuim D. radiodurans, which proved to be a suitable model for ribosomes of several pathogens, in complexes of antibiotics belonging to 5 PTC families. The identification of structural elements allowing alteration of the binding sites by remote mutations revealed common characteristics governing feasible mechanisms by which cross-resistance between compounds of diverse chemical nature could be acquired.
Results
The spatial distribution of nucleotides associated with resistance to PTC antibiotics identified by various noncrystallographic methods [supporting information (SI) Table S1] mapped onto D50S crystal structure (Fig. 2 and Fig. S1), indicates that almost all of the nucleotides mediating resistance are clustered in a distinct region, although the PTC surroundings offer various nucleotides for antibiotics binding, some of which can be mutated without losing cell vitality (30). This region is located farthest from the intersubunit interface and stretches into the entrance to the nascent protein exit tunnel (Fig. 2 and Fig. S1). The only outlier is the highly flexible nucleotide A2062, which is located closer to the subunit interface and was detected in different orientations in complexes of various antibiotics, namely chloramphenicol (3), streptograminA (14), lankacidin (unpublished work), and methymycin (19).
Fig. 2.
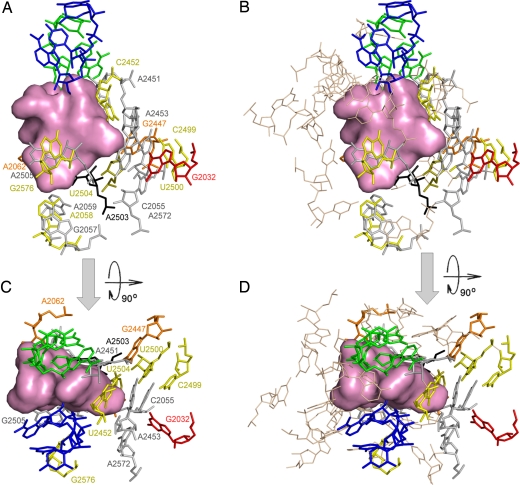
Nucleotides shown to undergo mutations or methylations that confirm PTC antibiotic resistance or reduce susceptibility. Nucleotides are colored according to the number of affected classes (1, 2, 3, 4, and 5 classes are represented by gray, yellow, orange, red, and black, respectively). The pink surface shows the total volume occupied by the PTC antibiotics: clindamycin (lincosamides), dalfopristin (streptograminsA), retapamulin (pleuromutilins), chloramphenicol (phenicols), and linezolid (oxazolidinones). For orientation, the A site tRNA 3′ end and the derived P site tRNA 3′ end (32, 33) are shown in blue and green, respectively. Images were taken from the direction of the L7/12 stalk (A and B) and from the top of the cavity leading to the PTC (C and D). Nucleotides located in the vicinity of the PTC within a distance <8 Å from the corresponding antibiotic, and are not involved in known resistance determinants, are either shown as wheat-colored lines (B and D) or excluded (A and C).
For describing the locations of the nucleotides mediating antibiotics resistance, the PTC was divided by an artificial plane into 2 regions: one contains components participating in resistance mutations, and the other consists of nucleotides that are not involved in resistance. This artificial plane is defined by 2 perpendicular imaginary axes, namely the imaginary 2-fold symmetry axis (32, 33) and the line connecting the bases of nucleotides G2553 and G2251 (Fig. S1); the 2 nucleotides that are located at the boundaries of the space consumed by the rotatory motion of the translocating A-tRNA 3′ end from the A to the P site, and are engaged in Watson–Crick pairing with both tRNAs (Fig. S1). Thus, the translocation of A site tRNA required for nascent chain elongation is performed by 2 correlated motions: sideways progression of most of the tRNA molecule together with the mRNA by 1 codon at the time, and a rotatory motion of the aminoacylated 3′ end of the A site tRNA around the bond connecting it to the rest of the tRNA molecule. This bond coincides with the rotation axis of a pseudo-symmetrical region, comprising 180 and located in and around the PTC (32, 33) within the otherwise asymmetric ribosome. When viewing from the subunit interface, the PTC wall associated with PTC antibiotics resistance is located behind the site of peptide bond formation (called the PTC “rear wall”). It is the main constituent of PTC region that navigates and guides the translocation of the A-tRNA 3′ end from the A to the P site, by creating extensive transient interactions of its backbone.
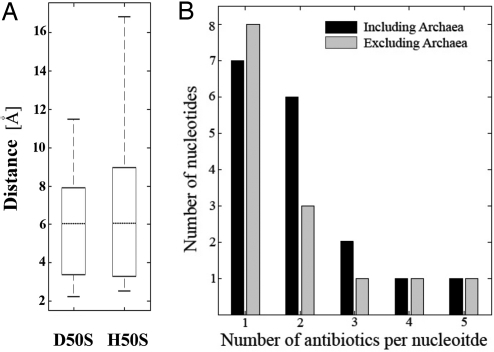
Fig. 3A shows that, for clinically relevant complexes, half of the nucleotides mediating resistance to PTC antibiotics are located at distances of 6–12 Å from the affected antibiotic. For instance, pleuromutilins resistance is acquired by mutations of remotely located nucleotides of the 23S RNA as well as of residues of r-protein L3 (12, 16, 29). Fig. 3B shows that approximately half of the nucleotides involved in resistance to PTC antibiotics affect 2 or more different families. In some cases resistance to drugs from up to 5 different antibiotics families are associated with same nucleotide (Table S1 and Fig. 3B). Similar results were obtained when resistance mutations observed for the archaeon Halobacterium halobium (34–36) were included (Fig. 3B).
Fig. 3.
Overlaps of resistance determinants and distances between antibiotics binding sites and nucleotides mediating resistance. (A) Boxplot representation of nucleotide-antibiotic distances determined for D50S and H50S complexes. Dotted lines show medians; the upper and lower horizontal lines of the boxes stand for upper and lower quartiles (namely cutoffs for 25% and 75% of the data). Top and bottom external horizontal lines show the maximal and minimal values. (B) Resistant mutations or methylations observed in bacterial strains including (black bars) or excluding (gray bars) archaea (34–36). x axis indicates the number of different classes of antibiotics that are being affected. y axis stands for the number of nucleotides characterized for this observation.
Confining the resistance space to a single region in the PTC (Fig. 2 and Fig. S1), regardless of the bacterial species and the used methodology indicates a functional distinction between the 2 sides of the PTC. Limiting the mutations to the PTC rear wall is consistent with the finding that the rear wall backbone, rather than its bases, plays a crucial functional role in guiding the translocation of A site tRNA 3′ end from A to P site during peptide bond formation (32, 33).
Discussion
Resistance to PTC Antibiotics Is Frequently Acquired by Mutating Remote Nucleotides.
Because the PTC is highly conserved, the mechanisms for acquiring resistance are based on altering the conformation and the flexibility of nucleotides residing remotely. This trend is represented by Fig. 3A, which shows that approximately half of the nucleotides mediating antibiotic resistance reside at distances >6 Å from the affected bound drug. Because such distances are too long for direct interactions with the drugs, resistance mechanisms to PTC antibiotics appear to be assisted by additional nucleotides capable of forming remote interactions that, in turn, alter the conformation and the flexibility of the binding pocket surface.
Because altering the identity of PTC nucleotides in the immediate vicinity of the antibiotics is unfavorable, a common mechanism for acquiring resistance to PTC antibiotics is mediated by altering remote nucleotides (Fig. 3A). Indeed, proximity limitations would have severely limited the nucleotides pool, which in principle can be increased significantly (e.g., by approximately the power of 3) with respect to the distance between the altered nucleotide and the antibiotic binding site. However, because of the requirement to trigger progressive conformational rearrangements, there is an upper limit for the nucleotides that can be included in the pool. Therefore, nucleotides residing at relatively large distances (> ≈10Å) from the binding site are less likely to significantly alter its conformation. Hence, the advantage of increasing the pool of potential nucleotides by including nucleotides residing far from the binding site is compromised by their minute contribution to resistance, consistent with the existence of relatively fewer such cases. This effect can be somewhat compensated by altering more than a single nucleotide, as observed for pleuromutilins (29).
Resistance acquired by remote mutations has also been observed in other antibiotics families. For example, erythromycin resistance can result from mutations in r-proteins L4 and L22 (37, 38) which do not interact directly with the bound drug (3). However, because resistance to macrolides can be acquired by alterations of nucleotides that interact with the drug, such as A2058, it seems that remotely acquired resistance to macrolides is an alternative mechanism, contrary to resistance to PTC antibiotics that is typically mediated by remote interactions.
U2504 at the Crossroad of Remote Mutations Networks That Hamper Binding of PTC Antibiotics.
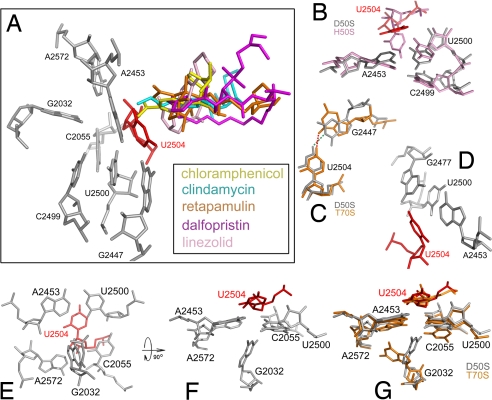
U2504, which plays pivotal roles in resistance to PTC antibiotics, belongs to the binding pockets of phenicols (3), lincosamides (3, 13), pleuromutilins (12, 16) (Fig. 4 and S2a-r) and oxazolidinones (17, 18). Mutations of U2504 were shown to promote resistance to pleuromutilin in the veterinary pathogens Brachyspira pilosicoli and Brachyspira hyodysenteriae (29) and to linezolid in the archaeon H. halobium (36). However, because it resides close to the PTC center, in the first layer of the PTC nucleotides that define the binding pocket surface, its alteration is expected to cause serious problems or be impossible. Consequently, altering neighboring nucleotides that can remotely affect 2504 may circumvent its essentiality. A mechanism by which U2504 is being perturbed by mutations of proximal nucleotides was suggested for tiamulin resistance (29). This mechanism was later extended to be the general mechanism for resistance and selectivity of the pleuromutilin family, based on comparative crystallographic studies (16). Likewise, linezolid resistance was shown to be acquired by a mutation in nucleotide G2576 (G2576U) (23, 37, 39) that is located >6 Å away from the bound drug (17), but can affect the conformation of nucleotide U2504.
Fig. 4.
Remote mutations that affect the conformation and/or the flexibility of U2504 by a network of interactions. (A) PTC antibiotics chloramphenicol (yellow), clindamycin (cyan), retapamulin (orange), dalfopristin (magenta) and linezolid (pink) bind in close proximity to U2504 (red). (B–G) Shown are selected interactions within the networks around U2504. rRNA is gray, pink, or orange for D50S, H50S, or T70S, respectively. Wherever drawn, U2504 of D50S is red. Images of D50S, T70S, and H50S were generated from their coordinates (PDB ID codes 1NKW, 2J01, and 1S72, respectively).
Notably, in many cases, albeit not in all eubacteria, U2504 is a pseudouridine (40, 41). Nevertheless, regardless of being uridine or pseudouridine, in all known structures of eubacterial ribosomes, U2504 makes similar interactions with its neighboring nucleotides (e.g., 2447) (42–46). Interestingly, although this posttranslational modification is not necessary for smooth function of eubacterial ribosomes, it was linked to resistance to PTC antibiotics, because E. coli strains deficient of it are more susceptible to tiamulin, clindamycin, and linezolid (47). The elevated susceptibility by the loss of posttranscriptional modification can be attributed, in part, to the centrality of this nucleotide in the binding pocket of PTC antibiotics. The same rationale, namely, the centrality of nucleotide U2504, explains why the key role played by this nucleotide in PTC resistance mechanism is independent of the presence or absence of the additional potential interactions that may exist in pseudouridine compared with unmodified nucleotide.
Second Layer Nucleotides.
The conformation and flexibility of nucleotide 2504 is dictated by 5 nt residing within the second layer from the PTC wall, namely U2500, A2453, C2055, A2572 and G2447. The base pair U2500:A2453 forms a planar barrier that prevents U2504 from tilting away from the PTC (42, 44) (Fig. 4B and Fig. S2b). Thus, disruption of this pairing should release this planar barrier and enable U2504 to flip away from the PTC, in accord with the appearance of resistance to linezolid in clinical S. aureus isolates carrying a mutation U2500A (48) and in H. halobium strains carrying either of the induced mutations U2500C, A2453G, or A2453C (36), all of which are predicted to destabilize the U2500:A2453 base pair.
G2447 is an additional nucleotide residing in the second layer that forms close contacts (≈3Å) with U2504 (Fig. 4 C and D and Fig. S2 c–h). In D50S its interactions with U2504 involve a hydrogen bond between O4 carbonyl of the uracil base of U2504 and N3 of G2447 (Fig. 4C and Fig. S2e). In the structures of Thermus thermophilus ribosomes (T70S) (44, 46) (Fig. 4C and Fig. S1e) and of E. coli (E70S) (45) O4 of U2504 faces the carbonyl O6 of G2447 base. In all structures G2447 stacks to U2500 (Fig. 4D and Fig. S2f) and thus restrains its conformation and indirectly facilitates further stabilization of U2504 by U2500. Hence, it is not surprising that although G2447 does not interact with any PTC antibiotics, its mutation G2447U (Fig. S2h) confers resistance to linezolid in E. coli (49) and Mycobacterium smegmatis (50), as well as to tiamulin in E. coli (23) and B. hyodysenteriae (29). Furthermore, E. coli and Bacillus stearothermophilus ribosomes carrying the mutation G2447A (Fig. S2g) are not impaired by chloramphenicol in peptide bond formation in vitro (51), as positions 1 and 6 in this purine undergo a polar inversion in which both H bond donors become acceptors and vice versa. The conversion from purine to pyrimidine, followed the mutation G2447U, should result in exclusion of all interactions between these 2 nt (Fig. S2h), as well as lose the stacking interaction with U2500.
Whereas U2500, A2453, and G2447 limit potential motions of U2504 base, both C2055 and A2572, located ≈3 Å away from it, block its ribose sugar from shifting away from the PTC (Fig. S2 i and j). The involvement of C2055 and A2572 in restraining U2504 conformation and flexibility is further supported by the tiamulin-resistant isolate of B. hyodysenteriae that carries both the mutations A2572U and C2055A (29) (Fig. S2j), although they are located 6–8 Å away from the bound drug. The purine to pyrimidine conversion after A2572U mutation releases the sterical hindrance of U2504 ribose sugar. In contrast, pyrimidine to purine conversion caused by the mutation C2055A can provide stacking interface that stabilizes U2504 in an altered conformation, with its base tilted away from the PTC, as observed for H50S (43), where nucleotide 2055 is an adenine. Thus, the different environments of U2504 in the eubacteria D50S and the archaea H50S, which is closer to eukaryotes in this aspect, explain the selectivity of pleuromutilins (16).
Third Layer Nucleotides.
Alteration in the PTC surface can be performed also by third layer nucleotides. Among them nucleotide 2032, a highly conserved guanine in bacteria (>94%), is involved in resistance to PTC antibiotics of 4 different families although it reside 6–8Å from each of the bound drugs. Thus, the mutation G2032A (Fig. 4 E–G and Fig. S2 n–r) confers resistance to the antibiotics chloramphenicol, clindamycin (52), and linezolid (49) in E. coli but not in T. thermophilus (53). The same mutation in B. hyodysenteriae, together with L3 r-protein mutation Asn148Ser, confers resistance to tiamulin (29). In addition, elevated minimum inhibitory concentrations (MICs) of linezolid were observed for E. coli strains carrying the mutations G2032U and G2032C (49).
The base of G2032 in T70S is tilted by 90° with respect to the corresponding nucleotide in D50S (Fig. 4G and Fig. S2m), indicating its flexibility. In T70S carbonyl O6 of G2032 forms a hydrogen bond with O2′ hydroxyl of nucleotide A2453 and its secondary amine N1 forms a hydrogen bond with O4′ of A2572 (Fig. S2n). G2032A mutation should lead to hydrogen bond acceptor imine at position N1 of adenine instead of the donor amine at position N1 of guanine. Additionally, the acceptor O6 carbonyl will be replaced by a primary amine that can act as a donor (Fig. S2o). These 2 polar inversions are likely to result in local repulsions that will force nucleotides A2453 and A2572 to adopt a slightly altered U2504 conformation. In D50S the secondary amine N1 of G2032 is located within a short distance (≈2.5Å) from the pyrimidine ring carbonyl oxygen of the proximal C2055 (Fig. S2p). Hence, in the G2032A mutant the secondary amine hydrogen in position 1 of the guanine base is replaced by the imine lone pair of electrons located at the same position in adenine (Fig. S2q). This substitution can result in repulsion between the imine nitrogen of the mutated A2032 and the carbonyl oxygen of C2055 pyrimidine ring. These interactions are likely to indirectly perturb the conformation and flexibility of the proximal U2504.
C2499, a 100% conserved nucleotide in eubacteria, is also located in the third layer. Assuming that the conformation of 2032 in B. hyodysenteriae resembles its conformation in D50S, the C2499A mutation should stabilize the mutated nucleotide A2032 (originally guanine) by a favorable polar attraction between the primary amine at position 6 of A2032 and the imine nitrogen of the nearby mutated A2499 (originally cytosine) (Fig. S2r). The need for mutation in 2499 for the stabilization of 2032 in a conformation capable of pushing 2055 toward the PTC is further supported by the difference in the conformations of 2032 in T70S and D50S (Fig. 4G and Fig. S2m). It suggests that the unfavorable interactions with 2055, caused by the mutation G2032A without the compensating mutation C2499A, may stabilize 2032 in the T70S conformation. Therefore, the role of 2499 in resistance involves stabilizing the mutated 2032 in a conformation enabling repulsion of 2055, which ought to push 2504 toward the PTC and restrain its flexibility. Mutations in 2032 were observed in eubacteria, together with mutations of 2504 or 2499, as well as mutation in L3 Asn148Ser (29). Whereas G2032A+U2504G double mutation should lead to resistance, mainly because of U2504G mutation, G2032A+C2499A should stimulate the mechanism proposed above (29). The contribution of G2032A and C2499A double mutation for drug resistance in the later case is further supported by the reduced MIC of an identical bacterial strain that carries the same mutation in L3 protein but not the G2032A/C2499A double mutation (29). The single example for mutation in 2032 causing linezolid resistance without the involvement of 2499 also fits with the general trait of resistance mediated by remote nucleotides.
The strategic position of U2504 at the A site and the cleft formed between it and the PTC rear wall serve as a hot spot for antibiotic binding (Fig. 4A and Fig. S2a), but is less suitable to mediate resistance owing to its high conservation. However, U2504 possesses a significant level of flexibility, which seems to be used for acquiring pleuromutilin selectivity via remote interactions with less conserved components (16). The similarities in the binding modes of all PTC antibiotics suggest that this mechanism can be extended to contribute to the selectivity of all 5 classes of clinically useful PTC antibiotics, in accord with the findings showing that the conformation of nucleotide 2504 affects the binding and resistance of PTC antibiotics even when the drugs are not in direct contact with it. Further support for this suggestion is the finding that 2504 acquires bacterial-like conformation in H50S when binding linezolid, despite the huge differences (3 orders of magnitude) in drug concentrations required for obtaining crystallographically suitable complexes of H50S (17) compared with D50S (18).
An additional nucleotide that can affect U2504 is its covalently attached neighbor A2503 (m2A in E. coli) (54), which is prone to methylation in eubacterial strains caring the resistance Cfr gene (21, 22, 25). Supporting the suggestion that A2503 induces resistance indirectly is the finding that although chloramphenicol does not interact with A2503, alterations of A2503 lead to resistance probably through altered conformations of U2504 and G2061, in line with the finding that nucleotides that shape the antibiotics binding pockets determine the usefulness of bound antibiotics (9–11).
Although most of the resistance mechanisms mediated by indirect contacts seem to hinge on U2504, in a few cases PTC antibiotics are hindered remotely by other nucleotides. Among these are the mutation G2576U that was suggested to hamper linezolid binding indirectly by nucleotides U2505 and U2506 (17, 23) in addition to U2504, as well as the resistance to the streptograminA virginiamycin M1 in the archaeon H. halobium by mutation in nucleotide A2059 (34). An open issue is the case of chloramphenicol, for which resistance by nucleotides 2057, 2058, and 2062, located at the exit tunnel, was reported (35, 52, 55) similar to chloramphenicol location in the ribosome of archaeon H. marismortui (13) that possesses sequence-resembling ribosomes from eukaryote, contrary to chloramphenicol binding site in the PTC of the pathogen model D. radiodurans (3).
Resistance to Various PTC Antibiotics Mediated by the Same Nucleotides.
The involvement of the same nucleotides in resistance to several antibiotic families of different chemical natures (Table S1, Figs. 1, 2, and 3B, and Fig. S1) occurs presumably because of the overlapping binding sites of these drugs. Because only a limited pool of nucleotides belonging to the PTC rear wall and the tunnel entrance is used for acquiring resistance, the probability of inducing resistance to more than a single antibiotic family by altering a given nucleotide is fairly high. This effect is further enhanced by the potential flexibility and the central location of U2504, which amplifies its possible involvement in resistance to various PTC antibiotics by indirect perturbation of its conformation and flexibility.
Resistance and/or reduced susceptibility to different antibiotics include (i) E. coli carrying the mutation C2032A that confers resistance to chloramphenicol and clindamycin (52); (ii) G2057A in other E. coli strains that causes resistance to chloramphenicol and erythromycin (55); (iii) A2058G and A2058U in E. coli acquiring reduced susceptibility to chloramphenicol and resistance to clindamycin and erythromycin (52); (iv) A2062C that appears together with the mutation Ser20Asn in L4 r-protein confirmed resistance to both streptogramins and macrolides in an isolate of S. pneumoniae (35).
Cross-resistance in the PTC was observed for linezolid and tiamulin in S. aureus following the mutation G2576U in all copies of 23S genes (23). An example for potential clinical threat of cross-resistance in the PTC is the multidrug resistance phenotype mediated by the cfr rRNA methyltransferase. This gene encodes a methyltransferase that modifies the PTC nucleotide A2503, and is responsible for resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptograminA antibiotics, namely the PhLOPSA group (21, 22). Remarkably, a clinical isolate of S. aureus was shown to carry this gene together with the ermB gene on the same chromosome (25) and in the same operon (24). The expression of this operon, designated mlr for modification of large ribosomal subunit, resulted in a strain resistant to all clinically relevant antibiotics that target the large ribosomal subunit.
To conclude, cross-resistance is of great importance in light of the increasing use of diverse antibiotics families hitting the same, or a similar, target. Structural studies on resistance mechanisms to various clinically useful families of PTC antibiotics revealed that all share overlapping positions in the vicinity of the PTC rear wall and nucleotide U2504. Common traits of resistance to several antibiotic families showed that almost all of the 23S nucleotides mediating resistance cluster within a defined small region of the PTC confined by its rear wall. Furthermore, approximately half of the nucleotides implicated in PTC antibiotic resistance are not directly interacting with the bound drugs, but reshape the binding pocket indirectly via networks of remote interactions, most of which through the flexible nucleotide U2504.
The significant number of nucleotides associated with multiple resistance phenotypes for PTC antibiotics indicates a linkage between structure, function, and resistance to several antibiotics, some with immense clinical value. Hence, the common traits of resistance revealed by this study may be useful in the design of preferred chemical moieties. Thus, it is likely that rather than being confined to the PTC rear wall, designing new PTC antibiotics with capabilities to dock to additional PTC components provides a feasible strategy for decreasing the probability for cross-resistance between PTC antibiotics.
Materials and Methods
To perform general structural and statistical analysis, we used a database holding information on the nucleotides involved in resistance or reduced susceptibility to PTC antibiotics (Table S1). Because not every nucleotide associated with resistance to >1 family of antibiotics indicates cross-resistance, the analysis was based on a large database containing information from various resistance strains obtained in several different laboratories by diverse methods. This database includes information for each nucleotide, the antibiotics families affected by it, and the distance between it and the most proximal atoms of the bound antibiotic (excluding hydrogens). Structural analysis was based on crystal structures of complexes of D50S with antibiotics, including the phenicol chloramphenicol, the lincosamide clindamycin, the streptograminA dalfopristin, the pleuromutilin tiamulin, and the oxazolidinone linezolid. Crystal structures of D50S with other pleuromutilins, methymycin, and lankacidin were not included as currently no resistance data are available for them. Biochemical data on antibiotics resistance in the archaeon H. halobium, and crystallographic data obtained for complexes of PTC antibiotics with the large ribosomal subunit from the archaeon H. marismortui that may resemble eukaryotes, were also included. Because all known pathogens are eubacteria, distances and data analyses were performed separately for eubacteria and for archaea. To enable excluding or including resistance determinants revealed solely for archaea information about the source was included. Calculations and database analysis were performed by MATLAB (MathWorks). In silico mutagenesis and structural analysis were performed by coot (56) and PyMol (57). PyMol was also used for image rendering. PDB ID codes are shown when relevant.
Supplementary Material
Acknowledgments.
We thank all members of the ribosome group at the Weizmann Institute. Crystallographic data were collected at the Advanced Photon Source (Argonne, IL) and the European Synchrotron Radiation Facility (Grenoble, France). This work was supported by National Institutes of Health Grant GM34360 and by the Kimmelman Center for Macromolecular Assemblies. C.D. was supported by the Adams Fellowship Program of the Israel Academy of Sciences and Humanities and A.Y. holds the Martin and Helen Kimmel Professorial Chair.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810826105/DCSupplemental.
References
- 1.Bashan A, Yonath A. Correlating ribosome function with high-resolution structures. Trends Microbiol. 2008;9:326–335. doi: 10.1016/j.tim.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schleunzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 4.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Berisio R, et al. Structural insight into the antibiotic action of telithromycin against resistant mutants. J Bacteriol. 2003;185:4276–4279. doi: 10.1128/JB.185.14.4276-4279.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berisio R, et al. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat Struct Biol. 2003;10:366–370. doi: 10.1038/nsb915. [DOI] [PubMed] [Google Scholar]
- 7.Schluenzen F, et al. Structural basis for the antibiotic activity of ketolides and azalides. Structure (Camb) 2003;11:329–338. doi: 10.1016/s0969-2126(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 8.Pyetan E, Baram D, Auerbach-Nevo T, Yonath A. Chemical parameters influencing fine-tuning in the binding of macrolide antibiotics to the ribosomal tunnel. Pure Appl Chem. 2007;79:955–968. [Google Scholar]
- 9.Yonath A, Bashan A. Ribosomal crystallography: Initiation, peptide bond formation, and amino acid polymerization are hampered by antibiotics. Annu Rev Microbiol. 2004;58:233–251. doi: 10.1146/annurev.micro.58.030603.123822. [DOI] [PubMed] [Google Scholar]
- 10.Yonath A. Antibiotics targeting ribosomes: Resistance, selectivity, synergism, and cellular regulation. Annu Rev Biochem. 2005;74:649–679. doi: 10.1146/annurev.biochem.74.082803.133130. [DOI] [PubMed] [Google Scholar]
- 11.Bommakanti AS, Lindahl L, Zengel JM. Mutation from guanine to adenine in 25S rRNA at the position equivalent to E. coli A2058 does not confer erythromycin sensitivity in Sacchromyces cerevisae. RNA. 2008;14:460–464. doi: 10.1261/rna.786408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schleunzen F, Pyetan E, Fucini P, Yonath A, Harms JM. Inhibition of peptide bond formation by pleuromutilins: The structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol Microbiol. 2004;54:1287–1294. doi: 10.1111/j.1365-2958.2004.04346.x. [DOI] [PubMed] [Google Scholar]
- 13.Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- 14.Harms JM, Schleunzen F, Fucini P, Bartels H, Yonath A. Alterations at the peptidyl transferase centre of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin. BMC Biol. 2004;2:4. doi: 10.1186/1741-7007-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auerbach T, Bashan A, Yonath A. Ribosomal antibiotics: Structural basis for resistance, synergism and selectivity. Trends Biotechnol. 2004;22:570–576. doi: 10.1016/j.tibtech.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Davidovich C, et al. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc Natl Acad Sci USA. 2007;104:4291–4296. doi: 10.1073/pnas.0700041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ippolito JA, et al. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J Med Chem. 2008;51:3353–3356. doi: 10.1021/jm800379d. [DOI] [PubMed] [Google Scholar]
- 18.Wilson DN, et al. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci USA. 2008;105:13339–13344. doi: 10.1073/pnas.0804276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auerbach T, et al. Structural basis for the antibacterial activity of the 12-membered-ring mono-sugar macrolide methymycin. Biotechnologia. 2008 in press. [Google Scholar]
- 20.Lai CJ, Weisblum B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc Natl Acad Sci USA. 1971;68:856–860. doi: 10.1073/pnas.68.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: Methylation of 23S ribosomal RNA at A2503. Mol Microbiol. 2005;57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 22.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother. 2006;50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller K, Dunsmore CJ, Fishwick CW, Chopra I. Linezolid and tiamulin cross-resistance in Staphylococcus aureus mediated by point mutations in the peptidyl transferase center. Antimicrob Agents Chemother. 2008;52:1737–1742. doi: 10.1128/AAC.01015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith LK, Mankin AS . Transcriptional and translational control of the mlr operon which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob Agents Chemother. 2008;52:1703–1712. doi: 10.1128/AAC.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toh SM, et al. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol Microbiol. 2007;64:1506–1514. doi: 10.1111/j.1365-2958.2007.05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skripkin E, et al. R{chi}-01: A new family of oxazolidinones that overcomes ribosomal-based linezolid resistance. Antimicrob Agents Chemother. 2008;28:28. doi: 10.1128/AAC.01193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lolk L, et al. A click chemistry approach to pleuromutilin conjugates with nucleosides or acyclic nucleoside derivatives and their binding to the bacterial ribosome. J Med Chem. 2008;51:4957–4967. doi: 10.1021/jm800261u. [DOI] [PubMed] [Google Scholar]
- 28.Springer DM, et al. Cyclopentanone ring-cleaved pleuromutilin derivatives. Eur J Med Chem. 2006;42:109–113. doi: 10.1016/j.ejmech.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Pringle M, Poehlsgaard J, Vester B, Long KS. Mutations in ribosomal protein L3 and 23S ribosomal RNA at the peptidyl transferase centre are associated with reduced susceptibility to tiamulin in Brachyspira spp. isolates. Mol Microbiol. 2004;54:1295–1306. doi: 10.1111/j.1365-2958.2004.04373.x. [DOI] [PubMed] [Google Scholar]
- 30.Sato NS, Hirabayashi N, Agmon I, Yonath A, Suzuki T. Comprehensive genetic selection revealed essential bases in the peptidyl-transferase center. Proc Natl Acad Sci USA. 2006;103:15386–15391. doi: 10.1073/pnas.0605970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: Recommendations. Nucleic Acids Res. 1985;13:3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bashan A, et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell. 2003;11:91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 33.Agmon I, Bashan A, Zarivach R, Yonath A. Symmetry at the active site of the ribosome: Structural and functional implications. Biol Chem. 2005;386:833–844. doi: 10.1515/BC.2005.098. [DOI] [PubMed] [Google Scholar]
- 34.Porse BT, Garrett RA. Sites of interaction of streptogramin A and B antibiotics in the peptidyl transferase loop of 23 S rRNA and the synergism of their inhibitory mechanisms. J Mol Biol. 1999;286:375–387. doi: 10.1006/jmbi.1998.2509. [DOI] [PubMed] [Google Scholar]
- 35.Mankin AS, Garrett RA. Chloramphenicol resistance mutations in the single 23S rRNA gene of the archaeon H. halobium. J Bacteriol. 1991;173:3559–3563. doi: 10.1128/jb.173.11.3559-3563.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kloss P, Xiong L, Shinabarger DL, Mankin AS. Resistance mutations in 23 S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J Mol Biol. 1999;294:93–101. doi: 10.1006/jmbi.1999.3247. [DOI] [PubMed] [Google Scholar]
- 37.Gregory ST, Dahlberg AE. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J Mol Biol. 1999;289:827–834. doi: 10.1006/jmbi.1999.2839. [DOI] [PubMed] [Google Scholar]
- 38.Zaman S, Fitzpatrick M, Lindahl L, Zengel J. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in Escherichia coli. Mol Microbiol. 2007;66:1039–1050. doi: 10.1111/j.1365-2958.2007.05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prystowsky J, et al. Resistance to linezolid: Characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob Agents Chemother. 2001;45(7):2154–2156. doi: 10.1128/AAC.45.7.2154-2156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Del Campo M, et al. Number, position, and significance of the pseudouridines in the large subunit ribosomal RNA of Haloarcula marismortui and Deinococcus radiodurans. RNA. 2005;11:210–219. doi: 10.1261/rna.7209905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: Analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- 42.Harms J, et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 43.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 44.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 45.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 46.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 47.Toh SM, Mankin AS. An indigenous posttranscriptional modification in the ribosomal peptidyl transferase center confers resistance to an array of protein synthesis inhibitors. J Mol Biol. 2008;380:593–597. doi: 10.1016/j.jmb.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meka VG, et al . Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23SrRNA gene and loss of a single copy of rRNA. J Infect Dis. 2004;190:311–317. doi: 10.1086/421471. [DOI] [PubMed] [Google Scholar]
- 49.Xiong L, Kloss P, Douthwaite S, Andersen NM, Swaney S, Shinabarger DL, Mankin AS. Oxazolidinone resistance mutations in 23S rRNA of Escherichia coli reveal the central region of domain V as the primary site of drug action. J Bacteriol. 2000;182:5325–5331. doi: 10.1128/jb.182.19.5325-5331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sander P, et al. Ribosomal and non-ribosomal resistance to oxazolidinones: Species-specific idiosyncrasy of ribosomal alterations. Mol Microbiol. 2002;46:1295–1304. doi: 10.1046/j.1365-2958.2002.03242.x. [DOI] [PubMed] [Google Scholar]
- 51.Thompson J, et al. Analysis of mutations at residues A2451 and G2447 or 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc Natl Acad Sci USA. 2001;98:9002–9007. doi: 10.1073/pnas.151257098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Douthwaite S. Functional interactions within 23S rRNA involving the peptidyltransferase center. J Bacteriol. 1992;174:1333–1338. doi: 10.1128/jb.174.4.1333-1338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gregory ST, et al. Mutational analysis of 16S and 23S rRNA genes of Thermus thermophilus. J Bacteriol. 2005;187:4804–4812. doi: 10.1128/JB.187.14.4804-4812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kowalak JA, Bruenger E, McCloskey JA. Posttranscriptional modification of the central loop of domain V in Escherichia coli 23 S ribosomal RNA. J Biol Chem. 1995;270:17758–17764. doi: 10.1074/jbc.270.30.17758. [DOI] [PubMed] [Google Scholar]
- 55.Ettayebi M, Prasad SM, Morgan EA. Chloramphenicol-erythromycin resistance mutations in a 23S rRNA gene of Escherichia coli. J. Bacteriol. 1985;162:551–557. doi: 10.1128/jb.162.2.551-557.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.DeLano WL. The PyMOL Molecular Graphics System. CA: San Carlos; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.