Abstract
DNA double strand breaks are generated by genotoxic agents and by cellular endonucleases as intermediates of several important physiologic processes. The cellular response to genotoxic DNA breaks includes the activation of transcriptional programs known primarily to regulate cell cycle checkpoints and cell survival1–5. DNA double strand breaks are generated in all developing lymphocytes during the assembly of antigen receptor genes, a process that is essential for normal lymphocyte development. Here we demonstrate that these physiologic DNA breaks activate a broad transcriptional program. This program transcends the canonical DNA double strand break response and includes many genes that regulate diverse cellular processes important for lymphocyte development. Moreover, the expression of several of these genes is regulated similarly in response to genotoxic DNA damage. Thus, physiologic DNA double strand breaks provide cues that can regulate cell-type-specific processes not directly involved in maintaining the integrity of the genome, and genotoxic DNA breaks could disrupt normal cellular functions by corrupting these processes.
The generation of complete antigen receptor genes in developing lymphocytes requires assembly of the second exon through the process of V(D)J recombination6. This process is initiated by the recombinase activating gene (Rag) endonuclease, composed of Rag1 and Rag2, which introduces DNA double strand breaks (DSBs) at the border of two gene segments and their flanking Rag recognition sequences (recombination signals or RSs)7. Rag cleavage occurs in cells at the G1 phase of the cell cycle, and the resulting pairs of coding and signal DNA ends are processed and joined by the non-homologous end-joining (NHEJ) pathway of DNA DSB repair, generating a coding joint and a signal joint respectively8. All developing lymphocytes must generate and repair several Rag DSBs as they assemble the genes encoding the heterodimeric B and T cell receptors.
The cellular response to genotoxic DSBs is initiated by the ataxia telangiectasia mutated (Atm) serine threonine kinase kinase1–3. A central feature of this response is the activation of transcriptional programs, such as those initiated by p53, which primarily function to promote cell cycle arrest and apoptosis of cells with extensive, or persistent, DNA breaks9. Whether physiologic DSBs generally activate DSB responses that include these transcriptional programs has been unclear. In this regard, stabilization of p53 was observed only in NHEJ-deficient, and not wild type, thymocytes undergoing V(D)J recombination, suggesting that Rag DSBs may only activate transcriptional programs in a small fraction of developing lymphocytes where these breaks are not efficiently repaired and thus pose a hazard to the cell10. Indeed, the unopposed activation of p53 in response to transient Rag DSBs could lead to the unwanted apoptosis of developing lymphocytes that would have otherwise undergone successful antigen receptor gene assembly. Alternatively, all Rag DSBs could generally activate transcriptional pathways, including those that promote survival. In this regard, genotoxic DSBs have been shown to activate NFκB, and, although the genes that are regulated by NFκB in this setting have not been identified, in other settings NFκB regulates the expression of genes with anti-apoptotic activity5, 11, 12. Moreover, NFκB promotes the expression of genes involved in diverse cell-type-specific processes, raising the intriguing possibility that genotoxic, and perhaps physiologic, DSBs activate genetic programs with broader cellular functions13.
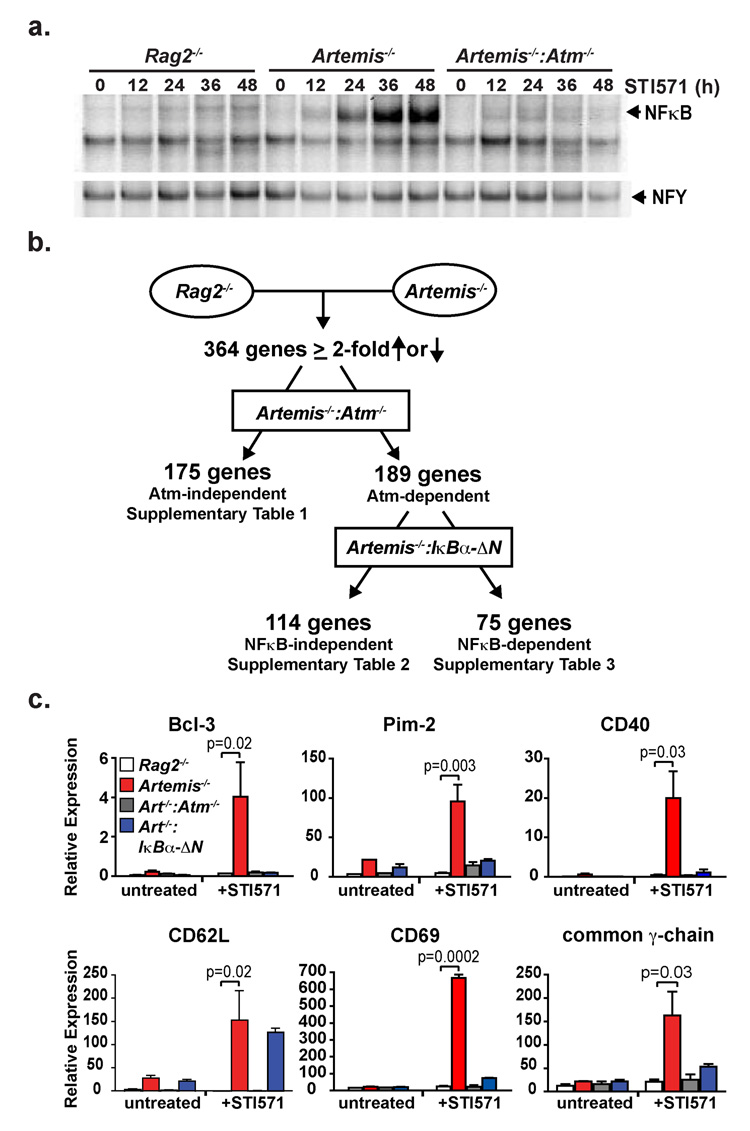
Initially we wished to determine whether Rag DSBs are capable of activating NFκB. To this end, we generated several NHEJ-deficient (Artemis−/−,, Ku70−/− and Scid) v-abl transformed pre-B cell lines, hereafter referred to as abl pre-B cells. G1 cell cycle arrest and Rag expression can be induced in these cells with the v-abl kinase inhibitor, STI57114, 15. As these cells are NHEJ-deficient, Rag induction results in the accumulation of un-repaired DSBs at the immunoglobulin (Ig) light (L) chain κ locus (Supplementary Fig. 1, data not shown). These DSBs lead to activation of NFκB as evidenced by the increased nuclear translocation of NFκB (primarily p50/p65) in STI571 treated NHEJ-deficient (Artemis−/−, Ku70−/− and Scid) abl pre-B cells as compared to STI571 treated Rag2−/− abl pre-B cells (Fig. 1a and Supplementary Fig. 2a and 2b). This NFκB activation was partially dependent on Atm as it was significantly diminished in Artemis−/−:Atm−/− abl pre-B cells and in Artemis−/− abl pre-B cells treated with an Atm inhibitor (Fig. 1a and Supplementary Fig. 2c). As was observed for genotoxic DSBs, NEMO is required for the optimal activation of NFκB in response to Rag DSBs (Supplementary Fig. 2d) 12, 16, 17.
Figure 1. Rag DSBs activate a broad genetic program.
(a) NFκB EMSA of nuclear lysates from Rag2−/−, Artemis−/− and Artemis−/−:Atm−/− abl pre-B cells treated with STI571 for the indicated number of hours. NFY EMSA is shown as a control. Results are representative of three experiments. (b) Schematic of gene expression changes in response to Rag DSBs by microarray analyses. The data are reported in Supplementary Tables 1–3. (c) RT-PCR analysis of mRNA isolated from Rag2−/− (white bars), Artemis−/− (red bars), Artemis−/−:IkBα-ΔN (blue bars) and Artemis−/−:Atm−/− (grey bars) abl pre-B cells treated with STI571 for 0 or 48 hours. Mean and standard deviation from two experiments. P values calculated using a one-tailed t-test.
To determine whether the activation of NFκB, and possibly other transcription factors, by Rag DSBs leads to alterations in gene expression, we carried out gene expression profiling on three independently derived Rag2−/− and Artemis−/− abl pre-B cells treated with STI571 for 48 hours (Supplementary Fig. 3). These analyses revealed that the expression of 364 genes was changed ≥2-fold in all three Artemis−/− abl pre-B cells relative to the mean expression level in the three Rag2−/− cells (Fig. 1b, Supplementary Fig. 3a and Supplementary Tables 1, 2 and 3). A number of the gene expression changes were confirmed through quantitative RT-PCR and protein expression analyses (Fig. 1c and Supplementary Fig. 4 and 5). These changes are not due to Artemis deficiency per se, as they were also observed in response to Rag DSBs generated in Ku70−/− and Scid (DNAPKcs-deficient) abl pre-B cells, but not in STI571-treated Artemis−/−:Rag2−/− abl pre-B cells (Supplementary Fig. 6). We conclude that Rag DSBs regulate the expression of a large cohort of genes. A significant portion of this cohort is dependent on the activation of Atm by Rag DSBs as evidenced by analyses of Artemis−/− :Atm−/− abl pre-B cells (Fig. 1, Supplementary Figs. 3–5 and 7 and Supplementary Tables 2 and 3). Remarkably, however, approximately half of the genes are Atm-independent, demonstrating that proteins other than Atm can initiate responses to Rag DSBs in G1-phase cells. Furthermore, analyses of Artemis−/− abl pre-B cells expressing a dominant negative form of IκBα (IκBα-ΔN), which inhibits NFκB activation, revealed that 75 of the Atm-dependent gene expression changes were also NFκB-dependent (Fig. 1b and 1c, Supplementary Fig. 3 and 4, and Supplementary Table 3). As only some of the Rag DSB-dependent genes are dependent on NFκB, additional transcriptional pathways must be integrated into the Rag DSB response. Thus, Rag DSBs promote a broad genetic program, in part, through the activation of Atm and NFκB.
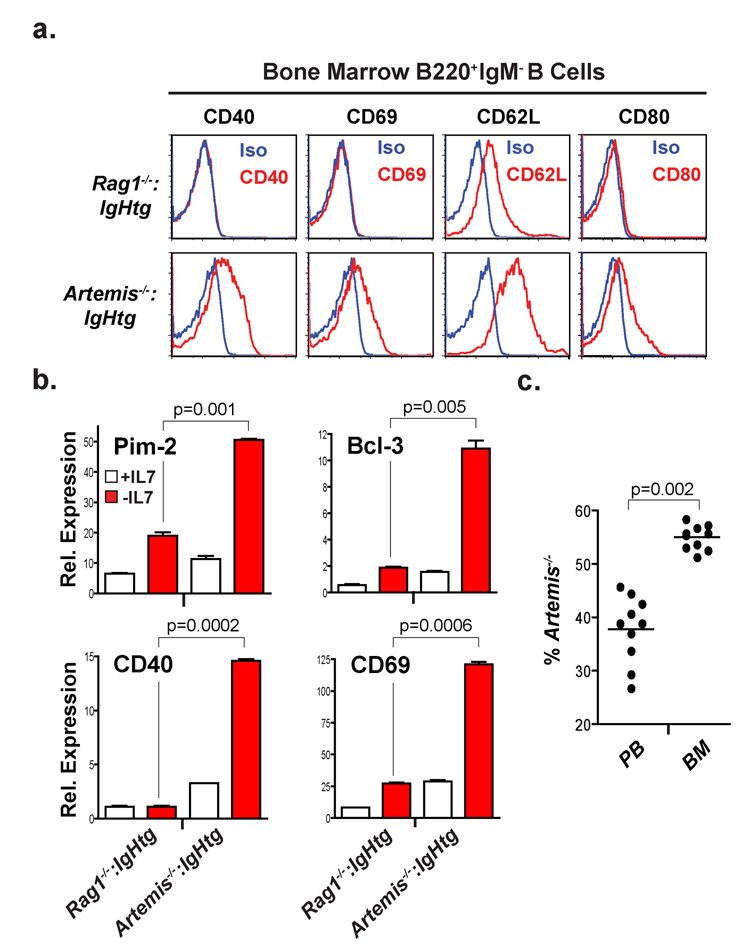
A similar genetic program is activated by Rag DSBs in developing B cells in vivo. This is evidenced by gene expression profiling of purified B220+:IgM− bone marrow B cells, which are primarily pre-B cells, from Artemis−/− and Rag1−/− mice expressing an IgH transgene (Artemis−/−:IgHtg and Rag1−/−:IgHtg, respectively) (Supplementary Fig. 8 and Supplementary Table 4). Of the 364 Rag-DSB-responsive genes identified through the analysis of Artemis−/− abl pre-B cells, 109 exhibited significant changes in expression when comparing Artemis−/−:IgHtg and Rag1−/−:IgHtg pre-B cells (Fig. 1b, Supplementary Fig. 8 and Supplementary Tables 1–4). The asynchronous nature of Rag DSB induction in developing pre-B cells is likely responsible, in part, for the reduced magnitude and number of the gene expression changes in primary Artemis−/−:IgHtg pre-B cells, as compared to the Artemis−/− abl pre-B cells. Importantly, these gene expression changes do lead to significant increases in the expression of the proteins they encode (Fig. 2a and Supplementary Fig. 9). That these genes are Rag DSB-responsive was further demonstrated through the analysis of Artemis−/−:IgHtg and Rag1−/−:IgHtg bone marrow IL-7 pre-B cell cultures. Withdrawal of IL-7 from Artemis−/−:IgHtg, but not Rag1−/−:IgHtg, bone marrow IL-7 cultures leads to an increased accumulation of un-repaired IgLκ coding ends and a robust up-regulation in the expression of many of the genes that are regulated by Rag DSBs in abl pre-B cells (Fig. 2b, Supplementary Fig. 10 and data not shown)18. Together, these findings demonstrate that Rag DSBs activate a broad genetic program in developing pre-B cells. Importantly, these DNA breaks also promote gene expression changes in pro-B and pro-T cells, suggesting that transcriptional programs are activated by Rag DSBs in all developing lymphocytes (Supplementary Fig. 11).
Figure 2. Rag DSB-dependent gene expression changes in developing B cells in vivo.
(a) Flow cytometric analyses of CD40, CD69 CD80 and CD62L expression by B220+:IgM− bone marrow B cells (primarily pre-B cells) from Rag1−/−:IgHtg and Artemis−/− :IgHtg mice (gated in Supplementary Fig. 8). Histograms for specific antibodies (red) and isotype controls (blue) are shown. (b) RT-PCR of gene expression in bone marrow cultures derived from Rag1−/−:IgHtg and Artemis−/−:IgHtg mice before (+IL7, white bar) and 48 hours after (−IL7, red bar) the removal of IL7. Mean and standard deviation from two experiments. P values calculated using a one-tailed t-test. (c) Percentage of Artemis−/− abl pre-B cells in the peripheral blood (PB) and bone marrow (BM) of Rag-deficient mice after co-injection of STI571-treated Artemis−/− and Rag-2−/− abl pre-B cells at a 1:1 ratio. The mean (line) of each set of mice analyzed is indicated and P values were calculated using a two-tailed Wilcoxon matched pairs test.
The genetic program induced by Rag DSBs includes genes that are active in multiple cellular processes. Several have functions that would be part of canonical DNA damage responses; for example, the anti-apoptotic proteins Pim2 and Bcl3 could antagonize pro-apoptotic pathways, such as those initiated by p53, promoting the survival of developing lymphocytes with Rag DSBs19–21. Strikingly, this program also includes genes that have no known role in canonical DNA damage responses; instead, these genes participate in diverse processes, many of which are important for lymphocyte development and function. These include common γ chain, SWAP-70, Notch 1, CD69, CD40, CD80 (B7.1), and CD62L (L-selectin). Notably, CD62L, SWAP-70 and CD69 have known functions in lymphocyte homing and migration, suggesting that these gene expression changes could regulate the localization of developing lymphocytes with Rag DSBs22–26. Indeed, we observed a biased accumulation of Artemis−/− abl pre-B cells with Rag DSBs, as compared to Rag2−/− abl pre-B cells, in the bone marrow after co-injection of these cells into Rag-deficient mice (Fig. 2c and Supplementary Fig. 12). A similar bias was not observed in the peripheral blood of these mice (Fig. 2c). Moreover, inhibition of Atm signaling in Artemis−/− abl pre-B cells led to the diminished localization of these cells in the bone marrow (Supplementary Fig. 12c). Similar results were obtained from analyses of independently derived Artemis−/− and Rag2−/− abl pre-B cells (data not shown). Lymphocytes traverse different microenvironments as they develop in the thymus and bone marrow; our findings suggest that one of the functions of the Rag DSB-dependent genetic program may be to modulate this migration27–30.
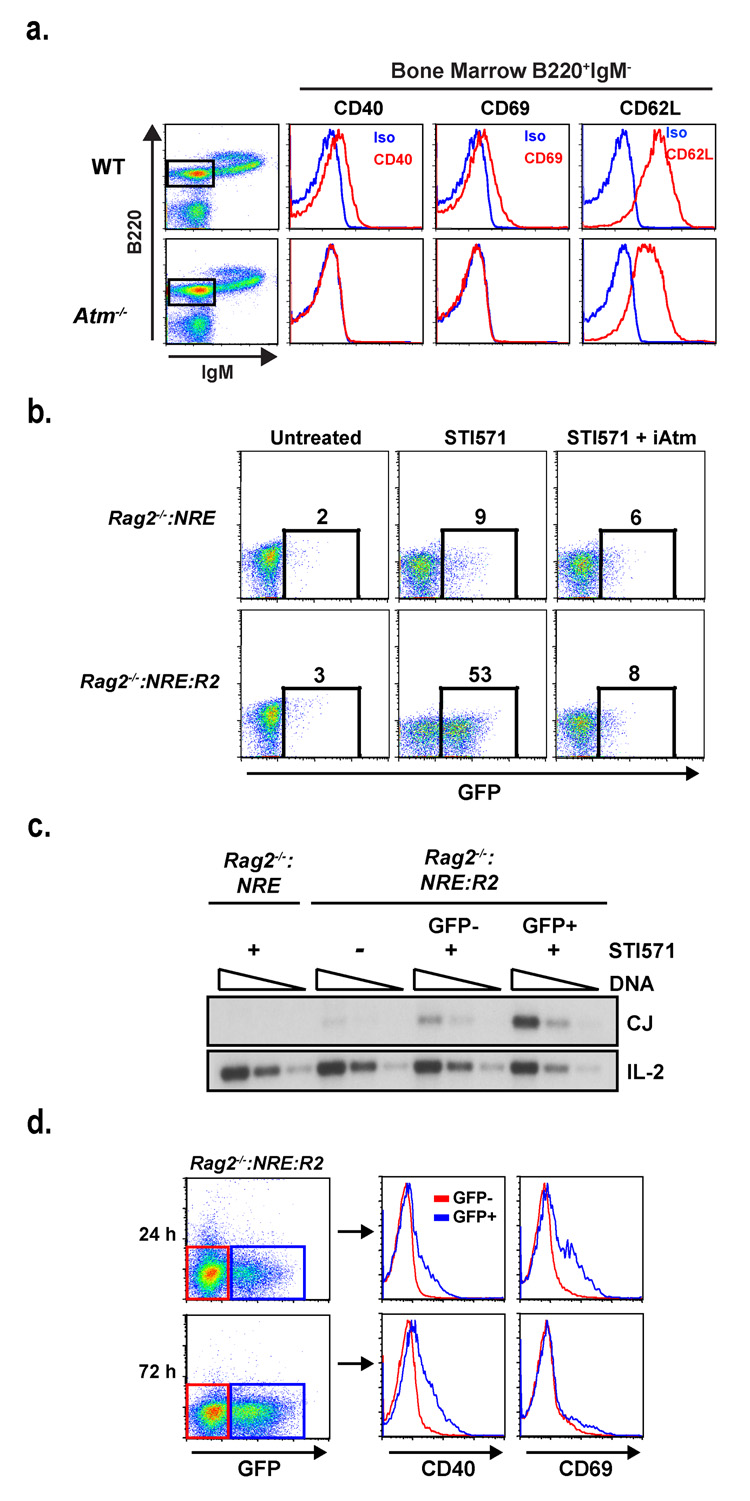
We have established that persistent Rag DSBs in NHEJ-deficient lymphocytes activate a diverse genetic program in vitro and in vivo. In wild-type lymphocytes, most Rag DSBs are repaired rapidly; thus, for this program to regulate processes that are generally important for development, it must also be initiated by these transient DSBs. In this regard, EMSA analyses did not reveal a marked increase in NFκB activation in wild type abl pre-B cells undergoing V(D)J recombination; however, we reasoned that this analysis might not be sensitive enough to detect the activation of NFκB in response to transient Rag DSBs (Supplementary Fig. 13). In agreement with this notion, we find that NFκB-dependent, Rag DSB-responsive genes are up-regulated in an Atm-dependent manner in wild type developing B cells undergoing V(D)J recombination (Fig. 3a, Supplementary Figs. 9 and 14).
Figure 3. NFκB activation in response to transient Rag DSBs.
(a) Flow cytometric analyses of CD40, CD69 and CD62L expression by B220+:IgM− bone marrow B cells (gated cells in dot plot) from WT and Atm−/− mice. Histograms for the specific antibodies (red) and isotype control (blue) are shown. (b) Flow cytometric analysis of GFP expression in Rag2−/−:NRE and Rag2−/−:NRE:R2 cells treated with STI571 for 48 hours in the presence or absence of the Atm inhibitor KU-55933 (iAtm). The percentage of GFP+ cells is indicated. Results are representative of three experiments. (c) Rag2−/− :NRE and Rag2−/−:NRE:R2 abl pre-B cells were un-treated (−) or treated with STI571 for 48 hours (+) and GFP+ and GFP− Rag2−/−:NRE:R2 abl pre-B cells isolated by flow cytometric cell sorting. Serial 4-fold dilutions of genomic DNA from all cells was assayed for pMX-DELCJ rearrangement by PCR (see Supplementary Fig. 15b). Results are representative of two experiments. (f) Flow cytometric analysis of CD40 and CD69 protein expression on GFP+ (blue histograms) and GFP− (red histograms) Rag-2−/− :NRE:R2 cells treated with STI571 for 24 or 72 hours. Results are representative of two experiments.
To directly determine whether transient Rag DSBs activate transcriptional pathways, we developed a sensitive single-cell assay for NFκB activation. To this end, a retroviral reporter, pMSCV-NRE-GFP, that contains five tandem NFκB responsive elements (NREs) driving expression of the green fluorescent protein (GFP) cDNA on the anti-sense strand was generated (Supplementary Fig. 15a). pMSCV-NRE-GFP was introduced into Rag2−/− abl pre-B cells that contained the pMX-DELCJ retroviral V(D)J recombination substrate, hereafter referred to as Rag2−/−:NRE cells (Fig. 3b and Supplementary Fig. 15)14. These cells do not express significant levels of GFP in response to STI571, but they do up-regulate GFP in an ATM- and NFκB-dependent manner in response to DSBs induced by ionizing radiation (Fig. 3b and Supplementary Fig. 16).
To determine whether transient Rag DSBs activate NFκB, Rag2−/−:NRE cells were transduced with a retrovirus encoding Rag2 (hereafter referred to as Rag2−/− :NRE:R2 cells), which permits efficient V(D)J recombination after induction of Rag1 expression (Fig. 3c and data not shown). As STI571-treated cells are not dividing, GFP produced in response to transient Rag DSBs will persist, marking any cell that has activated NFκB (Fig. 3b and Supplementary Fig. 15c). Strikingly, after 48 hours of STI571 treatment, 53% of Rag2−/−:NRE:R2 cells were GFP+ (Fig. 3b). This NFκB activation was Atm-dependent, as an Atm inhibitor abrogated GFP expression (Fig. 3b). If NFκB activation was stimulated by transient Rag DSBs, then there should be a higher level of completed rearrangements in the GFP+ cells. Indeed, PCR analyses revealed a 5-fold higher level of pMX-DELCJ rearrangement (coding joint, CJ, formation) in the GFP+ cells as compared to GFP− cells (Fig. 3c). Moreover, the GFP+ cells exhibited increased expression of genes regulated by Rag DSBs, including Pim-2, Bcl-3, CD40 and CD69 (Figs. 3d and Supplementary Fig. 17). CD69 expression in the GFP+ population was transient, similar to what was observed in response to persistent Rag DSBs (Fig. 3d and Supplementary Figs. 4a and 5b). Together, these data demonstrate that transient Rag DSBs generated during normal V(D)J recombination activate DNA DSB responses, including Atm and NFκB, which lead to the initiation of a diverse genetic program in all developing lymphocytes.
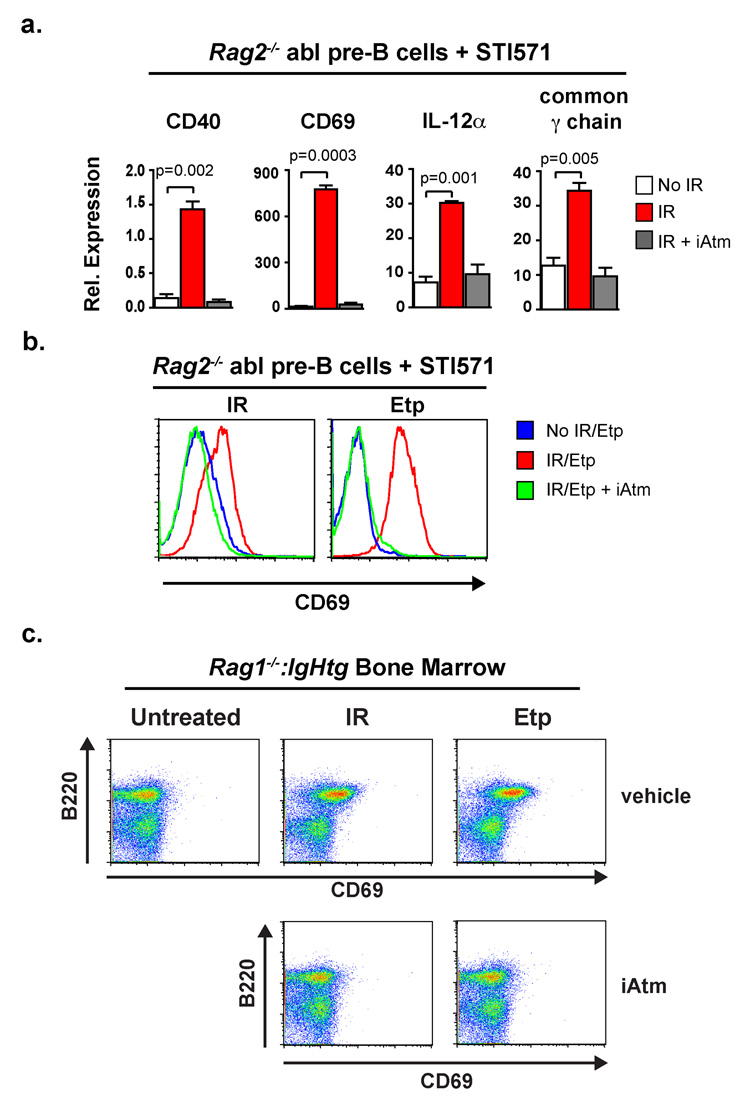
As physiologic Rag DSBs activate a broad genetic program, we reasoned that genotoxic DSBs may also activate components of this program in developing lymphocytes. Indeed, treatment of STI571-treated Rag2−/− abl pre-B cells with either ionizing radiation (IR) or etoposide, which both generate DNA DSBs, led to the Atm-dependent induction of many genes that are also induced by Rag DSBs (Fig. 4a and b, Supplementary Fig. 18). Furthermore, treatment of bone marrow from Rag1−/−:IgHtg mice with either ionizing radiation or etoposide leads to a B-cell-specific (B220+), Atm-dependent induction of CD69 expression (Fig. 4c). These findings demonstrate that, in developing B cells, genotoxic DSBs can induce lymphocyte-specific gene expression changes that are normally induced by Rag DSBs.
Figure 4. Genotoxic DSBs promote changes in expression of lymphocyte-specific genes.
(a) RT-PCR analysis of gene expression in Rag2−/− abl pre-B cells 2 hours after receiving 0 Gy (white bars) or 4 Gy of IR in the presence (grey bars) or absence (red bars) of the Atm inhibitor KU-55933 (iAtm). All cells were treated with STI571 for 24 hours prior to IR. Results are the mean and standard deviation from two experiments. P values were calculated using a one-tailed t-test. (b) Flow cytometric analysis of CD69 expression on STI571-treated Rag2−/− abl pre-B cells treated with IR as described in (a) or 2 hours after being treated with 5µM etoposide (Etp). Results are representative of three experiments. (c) Flow cytometric analysis of B220 and CD69 expression on bone marrow cells from Rag1−/−:IgHtg mice 2 hours after irradiation (0.5 Gy) or etoposide treatment (5µM) in the presence or absence of the Atm inhibitor KU-55933. Results are representative of two experiments.
We have shown that physiologic DNA DSBs generated during antigen receptor gene assembly activate a broad genetic program in developing lymphocytes. Importantly, this program is initiated by transient Rag DSBs that are rapidly repaired during normal V(D)J recombination in all developing lymphocytes. Thus, we propose that Rag DSBs provide cellular cues that regulate processes, such as migration and homing, that are integral components of normal lymphocyte development. As Atm is required to activate a portion of this Rag DSB-dependent genetic program, we expect that defects in the expression of these genes contribute to the lymphopenia observed in ataxia-telangiectasia.
Our findings establish for the first time that DNA DSBs generated during physiologic processes can regulate the cell-type-specific expression of genes that participate in functions not directly involved in maintaining genomic stability or suppressing malignant transformation. We speculate that DNA DSBs generated in other physiologic settings, such as immunoglobulin class switch recombination, meiosis, and DNA replication, may have similar effects. Furthermore, genotoxic DSBs generated by agents such as ionizing radiation and chemotherapeutic drugs could corrupt normal cellular processes through the aberrant activation of these cell-type-specific genetic programs. Thus, the impact of DNA DSB responses on cell physiology and function is likely more pervasive than previously appreciated.
Methods Summary
Standard methodologies employed for cell culture, retroviral vector generation, western blotting, Southern blotting, EMSA, gene expression profiling, PCR and flow cytometry are described in detail in the Methods Section. All v-abl-transformed pre-B cell lines were generated from mice harboring the EµBcl-2 transgene as previously described14. Flow cytometric analyses were performed on a FACSCaliber (BD Biosciences) with the normalized geometric mean fluorescence intensity (MFI) calculated as the difference between the geometric mean fluorescence for the specific antibody and the isotype control. Flow cytometric cell sorting was performed using a FACSVantage (BD Biosciences). To generate pMSCV-NRE-GFP, the NFκB responsive elements and TATA box from pNF-κB-Luc (Stratagene) were amplified using the following oligonucleotides: pNRE-f: CCAAACTCATCAATGTATCTTATCATG; pNRE-r: TACCAACAGTACCGGAATGC. This PCR product was cloned upstream of a cDNA encoding enhanced GFP (Clontech) on the bottom strand of pMSCV containing an IRES upstream of the Thy1.2 cDNA. Gene expression profiling was performed using the Affymetrix 430 v2.0 mouse genome microarray. To assay abl pre-B cell localization, cells were treated for 48 hrs with STI571, labeled with 0.1 mM CFSE (Invitrogen) or 5 mM SNARF (Invitrogen) at 37°C for 15 minutes in PBS at 2–5 × 106/mL, washed 3 times, and re-suspended in DMEM at a concentration of 80 × 106/mL. Labeled cells were then mixed in a 1:1 ratio, and a total of 40 × 106 were injected into the tail vein of each Rag-deficient mouse. Bone marrow and peripheral blood was harvested 30–60 minutes post-injection and analyzed by FACS for presence of CFSE- and SNARF-labeled abl pre-B cells.
Methods
Mice
The Atm−/−, Scid, Ku70−/−, Artemis−/−, VH147 IgH transgenic and EµBcl-2 transgenic mice have been described previously14, 31–33. Mice were bred and maintained under specific pathogen-free conditions at the Washington University School of Medicine and were handled in accordance to the guidelines set forth by the Division of Comparative Medicine of Washington University.
Cell culture
Three independently derived Rag2−/− (R.1, R.2 and R.3), three independently derived Artemis−/− (A.1, A.2 and A.3), two independently derived Artemis−/−:Atm−/− (AA.1 and AA.2) and individual Scid and Ku-70−/− v-abl-transformed pre-B cell lines containing the EµBcl-2 transgene were generated. The three Artemis−/−:IκBα-ΔN cells (A.3ΔN1, A.3ΔN2, A.3ΔN3) were generated through standard transfection of the A.3 cells with a cDNA encoding an N-terminally truncated IκBα (IκBα-ΔN)34. Expression of the IκBα-ΔN protein in these cells was confirmed by western blotting. STI571 treatments were carried out with 3µM STI571, as previously described14. The KU-55933 Atm inhibitor (Sigma) was used at 15µM, the BAY-11-7085 inhibitor (Calbiochem) was used at 20µM, and etoposide was used at 5µM. Irradiation was carried out with a Cs137 source, at doses of either 0.5 or 4 Gy.
Southern blotting
Southern blot analysis was performed as described previously14, 35.
Western blotting and EMSA
Western blotting was carried out as described previously, using an antibody to Pim-2 (Santa Cruz, 1D12)36. The secondary reagent was horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Zymed). EMSAs were run as described previously, and analyzed using a Li-Cor Odyssey Infrared scanner37. Supershifts were performed using anti-p50 and anti-p65 antibodies (Santa Cruz, sc-114 X and sc-372 X).
Gene expression profiling
All abl pre-B cells were harvested 48 hours following treatment with STI571. RNA was isolated using Qiagen RNeasy technology following the manufacturer’s instructions. Gene expression analysis was conducted using Affymetrix Mouse Genome 2.0 GeneChip® arrays (Mouse 430 v2, Affymetrix, Santa Clara, CA). One µg of total RNA was amplified as directed in the Affymetrix One-Cycle cDNA Synthesis protocol. Fifteen µg of amplified biotin-cRNAs were fragmented and hybridized to each array for 16 hours at 45°C in a rotating hybridization oven using the Affymetrix Eukaryotic Target Hybridization Controls and protocol. Array slides were stained with streptavidin/phycoerythrin utilizing a double-antibody staining procedure and then washed using the EukGE-WS2v5 protocol of the Affymetrix Fluidics Station FS450 for antibody amplification. Arrays were scanned in an Affymetrix Scanner 3000 and data was obtained using the GeneChip® Operating Software (GCOS; Version 1.2.0.037). Data pre-processing, normalization, and error modeling was performed with the Rosetta Resolver system (Version 6.0)38.
In order to identify differentially expressed genes between the Rag2−/− and Artemis−/− abl pre-B cells, an error-weighted analysis of variance (ANOVA) utilizing the Benjamini Hochberg false discovery rate was performed using Rosetta Resolver (www.rosettabio.com). At p-value ≤ 0.05, this analysis yielded 1578 probe sets. Fold changes were generated in Resolver, based on the ratio of each individual cell line relative to the average of the three Rag2−/− cells with 100-fold being the maximum change that Resolver can report. Of the 1578 probes found using the ANOVA analysis, only those that had a fold-change ≥ 2.0 (not including anti-correlated genes) in all ratios for a given genotype were considered for further analysis. A Venn diagram of the resulting ANOVA and fold-change data sets for each genotype comparison to Rag2−/− was then used to determine which expression changes were dependent on Atm and NFκB.
For gene expression profiling of primary developing B cells, B220+ cells were isolated from Rag1−/−:IgHtg and Art−/−:IgHtg bone marrow using Mouse Pan B (B220) Dynabeads from Invitrogen. RNA was isolated using the Qiagen RNeasy Mini Kit. Microarrays were treated as described above for the abl pre-B cells. For the probe sets that were identified as being differentially regulated in response to Rag DSBs in the abl-pre-B cells, we compared the average expression in the two Rag1−/−:IgHtg samples to the each of the two Art−/−:IgHtg samples. The probe sets that were not anti-correlated and changed ≥1.5-fold in the average of these comparisons are included in Supplementary Table 4.
Flow cytometry
Flow cytometric analyses were performed using fluorescein isothiocyanate (FITC)-conjugated anti-CD45R/B220, allophycocyanin (APC)-conjugated anti-IgM, phycoerythrin (PE)-conjugated anti-CD40, PE-conjugated anti-CD62L, PE-conjugated anti-CD69, and the appropriate isotype controls (BD Biosciences).
Analysis of pMX-DELCJ rearrangement
PCR was carried out as described previously on 4-fold dilutions of genomic DNA14. IL-2 PCR primers were IMR42 and IMR43, and the product was probed with IMR042-2, all of which have been described previously1. Primer sequences for pMX-DELCJ are as follows:
pA: CACAGGATCCCACGAAGTCTTGAGACCT
pB: ATCTGGATCCGTGCCGCCTTTGCAGGTGTATC
pD: AGACGGCAATATGGTGGA
RT-PCR gene expression analysis
Quantitative real-time PCR (RT-PCR) was performed using a Stratagene Mx3000P real-time PCR machine and Platinum® SYBR® Green qPCR SuperMix-UDG (Invitrogen). PCRs were carried out using the following program: 95°C, 30 sec; 55°C, 60 sec; 72°C, 30 sec; 40 cycles. Relative expression is calculated as the difference between beta-actin expression (C1) and expression of the gene of interest (C2), using the following equation: relative expression = 2−(C1–C2). C is the PCR cycle where the product detection curve becomes linear. Primers are listed in Supplementary Table 5.
Supplementary Material
Acknowledgements
The authors thank Dr. Jeff Bednarski for critical review of the manuscript, Dr. Frederick W. Alt for generously providing us with the Artemis−/− mice and Dr. Dean Ballard for providing us with the IκBα-ΔN construct. This research is supported by NIH grant AI47829 and the Washington University Department of Pathology and Immunology (B.P.S.); the Department of Pathology and Center for Childhood Cancer Research of the Children's Hospital of Philadelphia, and the Abramson Family Cancer Research Institute (C.H.B.); the intramural research program of the NIH, National Institute of Environmental Health Sciences (R.S.P.). B.P.S. is a recipient of a Research Scholar Award from the American Cancer Society. C.H.B. is a Pew Scholar in the Biomedical Sciences. A.L.B. is supported by a post-doctoral training grant from the NIH.
References
- 1.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3(3):155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 2.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297(5581):547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 3.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408(6811):433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 4.Innes CL, et al. ATM requirement in gene expression responses to ionizing radiation in human lymphoblasts and fibroblasts. Mol Cancer Res. 2006;4(3):197–207. doi: 10.1158/1541-7786.MCR-05-0154. [DOI] [PubMed] [Google Scholar]
- 5.Rashi-Elkeles S, et al. Parallel induction of ATM-dependent pro- and antiapoptotic signals in response to ionizing radiation in murine lymphoid tissue. Oncogene. 2006;25(10):1584–1592. doi: 10.1038/sj.onc.1209189. [DOI] [PubMed] [Google Scholar]
- 6.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 7.Fugmann SD, et al. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 8.Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 9.Meek DW. The p53 response to DNA damage. DNA Repair (Amst) 2004;3(8–9):1049–1056. doi: 10.1016/j.dnarep.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Guidos CJ, et al. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes Dev. 1996;10(16):2038–2054. doi: 10.1101/gad.10.16.2038. [DOI] [PubMed] [Google Scholar]
- 11.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8(1):49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 12.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115(5):565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 13.Gerondakis S, et al. Unravelling the complexities of the NF-kappaB signaling pathway using mouse knockout and transgenic models. Oncogene. 2006;25(51):6781–6799. doi: 10.1038/sj.onc.1209944. [DOI] [PubMed] [Google Scholar]
- 14.Bredemeyer AL, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442(7101):466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 15.Muljo SA, Schlissel MS. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat Immunol. 2003;4(1):31–37. doi: 10.1038/ni870. [DOI] [PubMed] [Google Scholar]
- 16.May MJ, et al. Selective inhibition of NF-kappaB activation by a peptide that blocks the interaction of NEMO with the IkappaB kinase complex. Science. 2000;289(5484):1550–1554. doi: 10.1126/science.289.5484.1550. [DOI] [PubMed] [Google Scholar]
- 17.Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311(5764):1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 18.Grawunder U, et al. Expression of DNA-dependent protein kinase holoenzyme upon induction of lymphocyte differentiation and V(D)J recombination. Eur J Biochem. 1996;241(3):931–940. doi: 10.1111/j.1432-1033.1996.00931.x. [DOI] [PubMed] [Google Scholar]
- 19.Kashatus D, Cogswell P, Baldwin AS. Expression of the Bcl-3 proto-oncogene suppresses p53 activation. Genes Dev. 2006;20(2):225–235. doi: 10.1101/gad.1352206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amaravadi R, Thompson CB. The survival kinases Akt and Pim as potential pharmacological targets. J Clin Invest. 2005;115(10):2618–2624. doi: 10.1172/JCI26273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18(18):2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 22.Rosen SD. Ligands for L-selectin: homing, inflammation, and beyond. Annu Rev Immunol. 2004;22:129–156. doi: 10.1146/annurev.immunol.21.090501.080131. [DOI] [PubMed] [Google Scholar]
- 23.Feng C, et al. A potential role for CD69 in thymocyte emigration. Int Immunol. 2002;14(6):535–544. doi: 10.1093/intimm/dxf020. [DOI] [PubMed] [Google Scholar]
- 24.Matloubian M, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427(6972):355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 25.Shiow LR, et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440(7083):540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 26.Pearce G, et al. Signaling protein SWAP-70 is required for efficient B cell homing to lymphoid organs. Nat Immunol. 2006;7(8):827–834. doi: 10.1038/ni1365. [DOI] [PubMed] [Google Scholar]
- 27.Ladi E, Yin X, Chtanova T, Robey EA. Thymic microenvironments for T cell differentiation and selection. Nat Immunol. 2006;7(4):338–343. doi: 10.1038/ni1323. [DOI] [PubMed] [Google Scholar]
- 28.Johnson K, et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28(3):335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 29.Tokoyoda K, et al. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 2004;20(6):707–718. doi: 10.1016/j.immuni.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Nagasawa T. Microenvironmental niches in the bone marrow required for B-cell development. Nat Rev Immunol. 2006;6(2):107–116. doi: 10.1038/nri1780. [DOI] [PubMed] [Google Scholar]
- 31.Rooney S, et al. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 32.Gu Y, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 33.Mandik-Nayak L, Racz J, Sleckman BP, Allen PM. Autoreactive marginal zone B cells are spontaneously activated but lymph node B cells require T cell help. J Exp Med. 2006;203:1985–1998. doi: 10.1084/jem.20060701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brockman JA, et al. Coupling of a signal response domain in I kappa B alpha to multiple pathways for NF-kappa B activation. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramsden DA, Gellert M. Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev. 1995;9:2409–2420. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- 36.Khor B, et al. Proteasome activator PA200 is required for normal spermatogenesis. Mol Cell Biol. 2006;26:2999–3007. doi: 10.1128/MCB.26.8.2999-3007.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weaver BK, Bohn E, Judd BA, Gil MP, Schreiber RD. ABIN-3: a molecular basis for species divergence in interleukin-10-induced anti-inflammatory actions. Mol Cell Biol. 2007;27:4603–4616. doi: 10.1128/MCB.00223-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weng L, et al. Rosetta error model for gene expression analysis. Bioinformatics. 2006;22:1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.