Abstract
A primary antioxidant enzyme in mitochondria, Manganese superoxide dismutase (MnSOD) plays a critical role in the survival of aerobic life. It is well-documented that compared to normal cell counterparts, MnSOD level is decreased in neoplastic transformed cells but is increased in aggressive cancers. However, the underlying mechanism for the observed dysregulation of MnSOD in cancer is unknown. We have previously identified a unique set of mutations located in the promoter region of the SOD2 gene in several types of cancer cells. We found that a C to T transition at −102 and an insertion of A at −93 downregulate MnSOD transcription by interrupting the formation of a single-stranded loop that is essential for a high level of promoter activity. Here, we demonstrate that the additional downstream mutation, C to G transversion at −38, creates a binding site for the transcription factors specificity protein 1 (Sp1) and activating protein 2 (AP-2). The promoter function is regulated by the relative levels of Sp1 and AP-2. In cytokine-induced expression of the SOD2 gene, Sp1 cooperates with a transcriptional complex containing nuclear factor κB (NF-κB) and nucleophosmin (NPM). The presence of AP-2 attenuates this induction. Our results suggest that the high level of MnSOD observed in aggressive cancer cells may be due, in part, to the absence of AP-2 transcriptional repression.
Keywords: Mutation, MnSOD, Sp1, AP-2, NF-κB
Introduction
MnSOD, a nuclear encoded mitochondrial primary antioxidant enzyme, catalyzes superoxide radicals into molecular oxygen and hydrogen peroxide, which are further reduced into water by peroxide metabolizing enzyme systems (1). Oxidative stress is thought to be one of the important pathogenic factors of cancer development and abnormal levels of MnSOD in cancer have been documented. MnSOD expresses at a lower level in many types of transformed and neoplastic cells, suggesting that loss of MnSOD activity may be a general characteristic of tumorigenesis (reviewed in 2). Consistent with this possibility, overexpression of MnSOD has been shown to suppress tumor growth in nude mice and to inhibit the metastasis of transplanted tumors (3). MnSOD mimetic pretreatment also significantly reduces DMBA/TPA-induced skin tumor incidence (4).
In contrast to the association between reduction of MnSOD and tumorigenesis, evidence that a high level of MnSOD is correlated with NF-κB activation is found in various aggressive tumors. Activation of the NF-κB pathway, leading to the induction of prosurvival proteins including MnSOD, has been implicated in the resistance of cancer cells to chemo- and radio-therapies (reviewed in 5). Consequently, suppressing MnSOD by inhibiting the NF-κB pathway results in enhanced radiosensitivity of prostate cancer cells (6, 7). Although some studies have implicated MnSOD in the differential regulation of cancer cells, the mechanistic link between MnSOD reduction and induction at different stages of cancer remains unknown.
Human MnSOD is an 88.6 kDa tetrameric protein containing a Mn2+ associated with each subunit. It is encoded by the human SOD2 gene located at 6q25 and is highly conserved in mammals (8). The human SOD2 gene is activated by a proximal promoter region that is characterized by the absence of TATA- or CAAT-box and the presence of multiple CpG islands (9). Transcription factors Sp1 and AP-2 can directly bind to the CpG islands to differently regulate promoter activity (10). Sp1 plays a central role in transcriptional activation of the SOD2 gene, which is consistent with observations from studies of many other human genes that contain TATA- or CAAT-less promoters. In contrast, AP-2 plays a negative role in transcriptional regulation of the SOD2 gene by attenuating Sp1 function (11).
MnSOD expression is rapidly up-regulated in response to oxidative stress. Numerous studies have demonstrated that MnSOD is induced in various types of cells and tissues by toxic stimuli and treatments, such as tumor necrosis factor alpha (TNF-α), interleukin-1 beta (IL-1β), 12-O-tetradecanoylphorbol-13-acetate (TPA) (12), dinitrophenol (13), paraquat (14), UV (15) and ionizing radiation (6). A second intronic enhancer element (I2E) containing a NF-κB binding site has been identified in both human and mouse sod2 genes and is responsible for cytokine-mediated induction of MnSOD (16, 17). Interaction between the Sp1-based promoter and NF-κB-based enhancer is thought to be essential for transcriptional induction. A recent insight from our investigations demonstrated that NPM functions from a distance as a coordinator between the proximal promoter and NF-κB enhancer (12, 18).
In an effort to determine the cause for the deregulation of MnSOD expression in cancer, we have identified several mutations located in the promoter region of the SOD2 gene of several cancer cell lines (19). One of these mutations was confirmed in molecular epidemiological studies (20). In the present study, we conducted cell-based transcriptional analysis using both cancer and transformed cell lines to delineate the transcriptional mechanism altered by these mutations. The results suggest that the mutations in the SOD2 promoter facilitate an AP-2-dependent modification of MnSOD expression in cancer cells.
Results
Mutations identified in cancer cells alter SOD2 promoter activity
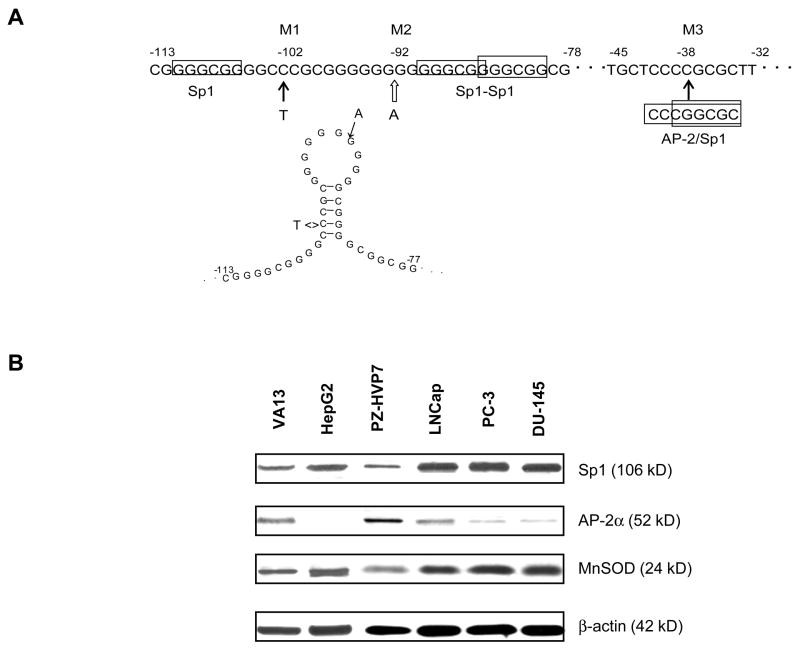
We previously identified three mutations in the proximal promoter region of the SOD2 gene form human cancer cell lines (19). As illustrated in Fig. 1A, these mutations contain C to T transition at −102 (M1), A insertion at −93 (M2), and C to G transversion at −38. Transcription factor database searching indictes that the M3 creates a binding site for both Sp1 and AP-2. In addition, although M1 and M2 are unlikely to change the binding motif for known transcription factors, the sequence change apparently interrupts the formation of a single-stranded loop, which has been shown to be an important structure for the promoter function (12). To verify that Sp1 and AP-2 regulate MnSOD expression, the levels of the three proteins were measured using immunoblots. In cancer cells, high levels of MnSOD correlate to high levels of Sp1 and low levels of AP-2, while in transformed cells, low levels of MnSOD are associated with high levels of AP-2 and low levels of Sp1 (Fig. 1B). To elucidate the effect of an individual mutation on transcriptional regulation, M3 was separated from M1-2 to generate promoter-reporter constructs that contain only M1-2 or M3. Because transcription factors Sp1 and AP-2 have been shown to directly regulate SOD2 promoter activity (11), VA13, low levels of constitutive Sp1 and HepG2, AP-2α deficient cells (21) were transfected with the resulting reporter constructs to examine the regulation of transcription by the mutated promoter sequences. The results show that the presence of M1-2 resulted in transcriptional reduction in both cell lines (Fig. 1C and D). Interestingly, the presence of M3 differentially modulates promoter activity in the two cell lines, i.e., decreases activity in VA13 cells and increases activity in HepG2 cells. The combined effect from the three mutations also resulted in down-regulation of transcription in VA13 cells but up-regulation of transcription in HepG2 cells.
FIGURE 1.
Transcriptional modification by the mutations identified in the SOD2 promoter region from cancer cells. A, positions of the mutations in the promoter region. A transition of C to T at −102 (M1) and an insertion of A at −93 (M2) are located in an 11G unpaired loop that has been shown to play a positive role in transcription of the human SOD2 gene (12). A transversion of C to G at −38 (M3) creates overlapped binding sites for Sp1 and AP-2. The sequence numbers shown are related to the transcription-initiation site (+1), and binding sites for Sp1 and AP-2 are indicated by boxes. B, relative levels of Sp1, AP-2 and MnSOD in several transformed and cancer cell lines were measured by immunoblots. C and D, the promoter-reporter constructs (0.5 nM) were co-transfected with a β-gal internal control (0.1 nM) into VA13 (C) and HepG2 (D) cells. The promoter activity was determined by β-gal-normalized luciferase reporter responses. * indicates significant differences in the promoter activity compared to the wild-type promoter at P<0.01.
Mutation at −38 creates binding motifs for Sp1 and AP-2
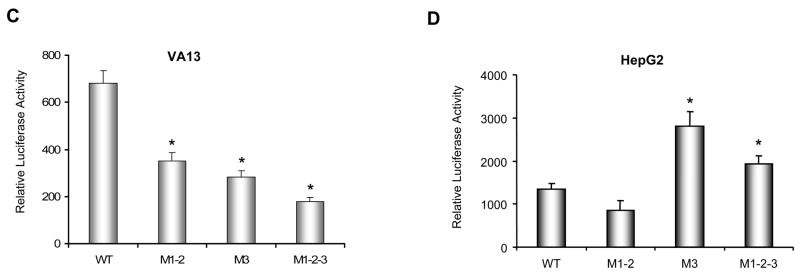
To determine how mutations affect transcriptional regulation by Sp1 and AP-2, Sp1 and AP-2α expression constructs were co-transfected with promoter-reporter constructs into HepG cells. Increasing Sp1 level in HepG2 cells markedly elevates promoter activity. In contrast, expression of AP-2α in HepG2 cells significantly suppresses both constitutive and Sp1-activated promoter activities (Fig. 2A). The presence of M1-2 reduces promoter activity. In contrast, the presence of M3 leads to an enhanced effect by Sp1 and a repressed effect by AP-2α. The repressive effect of AP-2 on the wild-type or the mutant promoter was further analyzed by transfecting various concentrations of the AP-2α expression construct into the HepG2 cells (Fig. 2B). The promoter activity is reduced in a dose-dependent manner when the cells express AP-2α. Importantly, AP-2α has a greater negative effect on the mutant promoter than on the wild-type promoter.
FIGURE 2.
Mutations mediate transcriptional modification via change of AP-2 and Sp1 binding sites. A, effects of Sp1 and AP-2 on transcriptional regulation. Expression constructs for Sp1 and AP-2α were co-transfected with the promoter-reporter constructs into HepG2 cells, and effects of Sp1, AP-2α or Sp1 plus AP-2α on the promoter activity were determined. Fold-increases by Sp1 or fold-decreases by AP-2α in the promoter activities compared to vector only control are indicated above the histograms. B, AP-2-mediated transcriptional repression. Different concentrations of AP-2α expression construct were co-transfected with the promoter-reporter constructs into HepG2 cells. The increased AP-2α was verified by immunoblots. The effect of AP-2α on the reporter activity driven by different promoters was determined. The vector DNA was added to make an equal mol concentration of total DNA in each transfection. * indicates significant differences in the promoter activity compared to vector only in (A) or to the wild-type promoter in (B) at P<0.01. C, alteration of Sp1 and AP-2 binding activities by the mutation at −38. Nuclear extracts from AP-2α-transfected HepG2 cells were incubated with probe containing the wild-type (WT) or cancer-type (M3) at −38. 100-fold self- and nonself-polynucleotides were added to identify the specific bindings for Sp1 and AP-2 and are indicated by arrows. Supershift bands (SS) by Sp1 and AP-2α antibodies are indicated by open arrows. Sequences of probes are shown on the bottom and the M3 position is underlined.
To verify the effect of M3, electrophoretic mobility shift assay (EMSA) was performed to determine whether Sp1 and AP-2α bind to the promoter region containing M3. A promoter fragment was radiolabeled to generate both wild-type and M3 probes. AP-2α protein was ectopically expressed in HepG2 cells. Nuclear extracts from the transfected cells were probed and specific bindings of Sp1 and AP-2 were detected by the respective antibodies (Fig. 2C). As shown in Fig. 2C, in comparison with the wild-type probe without detectable binding activity, the M3 probe shows both Sp1 and AP-2 binding activities with HepG2 nuclear extracts. Importantly, the AP-2 binding activity is dose-dependently increased as the concentration of AP-2α is increased. Thus, the results of EMSA are consistent with the reporter assay, suggesting that the effect of M3 on transcription is dependent on the relative level of AP-2 and Sp1.
Promoter mutations affect transcriptional induction
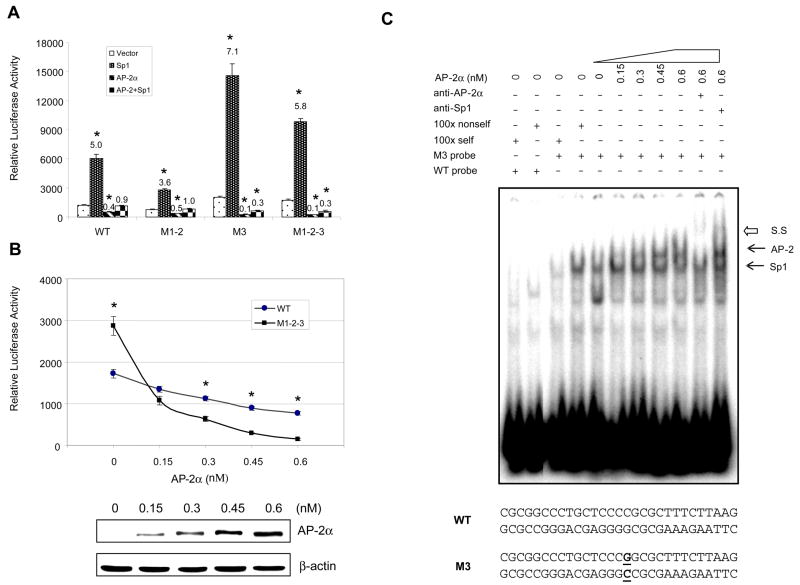
To detect whether the mutant promoter also affects transcriptional induction of the SOD2 gene, enhancer-promoter-reporter plasmids containing mutations in the promoter region were constructed. After transfection of the resulting reporter constructs, cells were simultaneously treated with TNF-α, IL-1β, and TPA (TIT). Transcriptional induction was estimated by normalizing the treatment groups with the corresponding untreated groups. The induction fold decreases when reporter constructs containing the mutant promoter are used in VA13 cells (Fig. 3A). However, the induction response in HepG2 cells is differentially regulated by an individual mutation in the promoter region. While the response by the M1-2-containing promoter is decreased, the response by the M3-containing promoter is increased (Fig. 3B). These results are consistent with the results from the promoter analysis shown in Fig. 1, suggesting that mutations also modulate transcriptional induction through promoter-enhancer coordination.
FIGURE 3.
Induction of the human SOD2 gene by TPA and cytokines. The I2E enhancer region was linked to the promoter region containing mutations in the luciferase reporter construct. A and B, transcriptional induction in VA13 and HepG2 cells. The generated constructs (0.5 nM) were co-transfected with β-gal (0.1 nM) into VA13 (A) and HepG2 (B) cells and followed by TIT treatment. The fold induction compared to untreated control in each enhancer-promoter-reporter construct was estimated by normalized luciferase reporter responses as shown above the histograms. * indicates significant differences in the luciferase activity compared to the wild-type promoter (I2E-P7) in both untreated and TIT-treated groups at P<0.01.
Transcription factors regulate induction
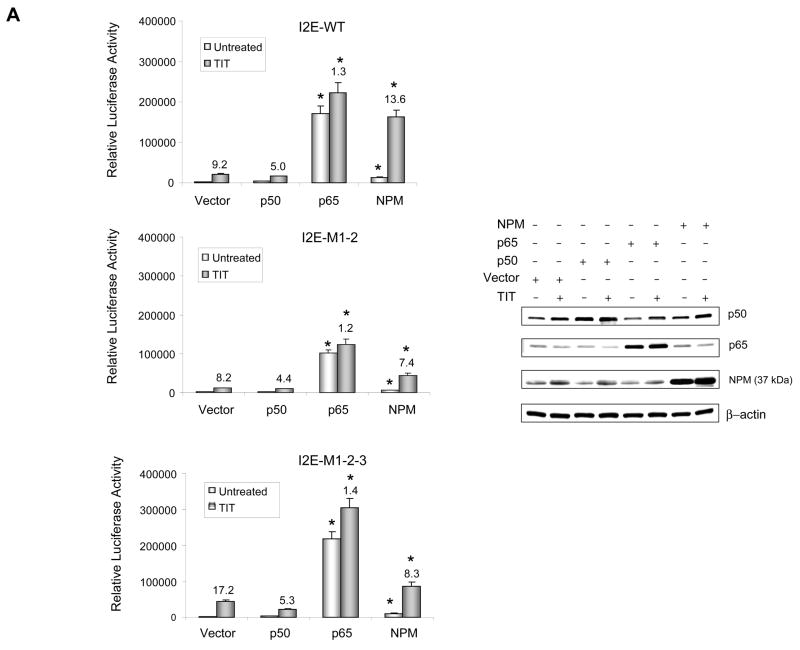
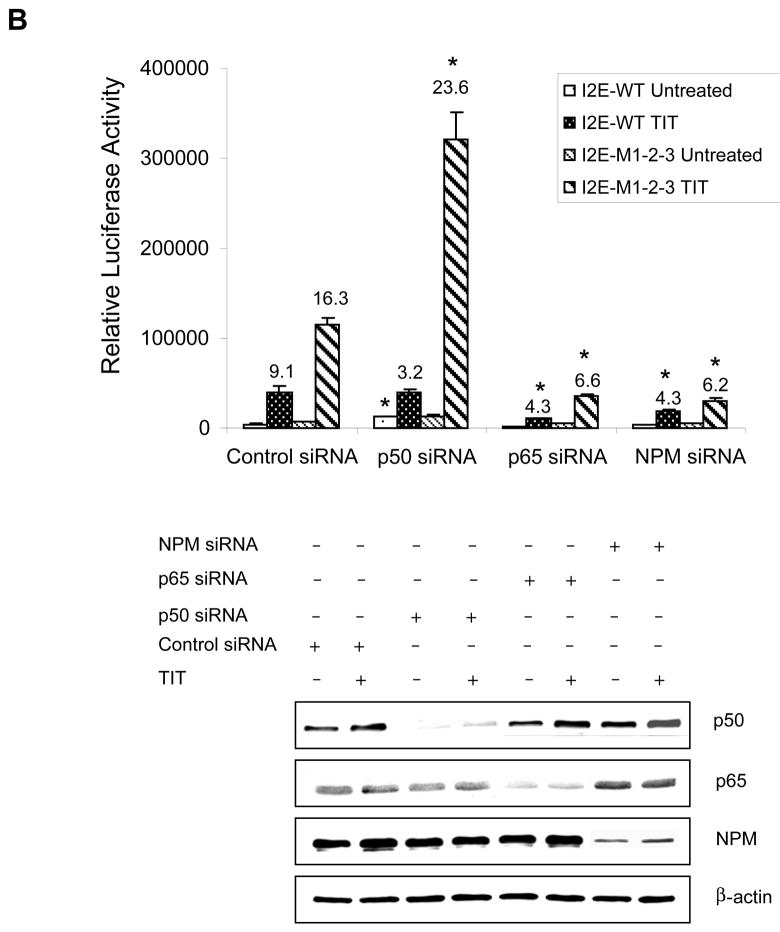
To determine whether NF-κB affects mutation-mediated alteration in transcriptional induction, the effects of p50, p65 and NPM were investigated. Expression constructs of those proteins were co-transfected with enhancer-promoter-reporter constructs into HepG2 cells and followed by TIT treatment. The expressed proteins were verified by immunoblots (Fig. 4A). Reporter analysis indicates that overexpression of p50 or p65 results in increased constitutive transcriptional level, but it does not significantly alter TIT-mediated induction. High levels of p50 and p65 do not change transcriptional patterns affected by mutations. Notably, a high level of NPM enhances both constitutive and inducible transcription. The presence of mutations appears to reduce NPM-mediated transcriptional activation, which is consistent with our previous finding that M1-2 interrupts the formation of a single-stranded loop structure in the promoter region (12). To confirm the effect of each protein, short interference RNA (siRNA) molecules were co-transfected with reporter constructs into HepG2 cells. The expression of siRNA-based knock-down was confirmed by immunoblots and the effect of siRNA on transcriptional induction was determined by reporter responses. Because the three mutations occur in cancer cells as a common mutation set in the promoter region (19), the M1-2-3 was used to serve as the cancer-type promoter in subsequent experiments. Reduction of p50 results in increased constitutive transcription but not in inductive transcription. Reduction of p65 and NPM represses both constitutive and inducible transcriptions. Compared to the wild-type promoter, reduction of p50 enhances transcription by the presence of mutations, whereas reduction of p65 and NPM reduces transcription (Fig. 4B). These results indicate that the transcriptional enhancers NF-κB and NPM have no direct effect on regulating promoter activity through mutations, but they do affect transcriptional induction by interacting with promoter-binding proteins.
FIGURE 4.
The effects of p50, p65, and NPM on transcriptional stimulation. A, overexpression of p50, p65 and NPM in HepG2 cells. The enhancer-promoter-reporter constructs (0.3 nM) were co-transfected with expression constructs of p50, p65, NPM, or vector only (0.5 nM) plus β-gal control (0.1 nM) into HepG2 cells, followed by TIT treatment. The increased levels of p50, p65, and NPM were determined by immunoblots (left panel) and the effects of these proteins on transcriptional stimulation were determined (right panel) using the wild-type (I2E-WT, top), mutations in the loop structure (I2E-M1-2, middle) and the cancer-type (I2E-M1-2-3, bottom). B, down-regulation of p50, p65, and NPM in HepG2 cells. The reporter constructs (0.3 nM) were co-transfected with siRNAs for p50, p65, NPM and a control siRNA (5 nM) plus β-gal control (0.1 nM). Reduction in levels of the siRNA targeted proteins was measured by immunoblots (bottom) and induction folds were determined by reporter assay. Induction folds compared to untreated controls are indicated above the histogram bars. * indicates significant differences in the luciferase activity compared to vector only control (A) and control siRNA (B) in both untreated and TIT-treated groups at P<0.01.
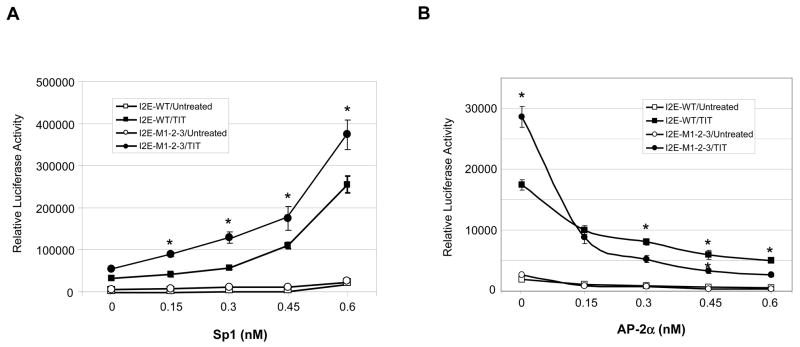
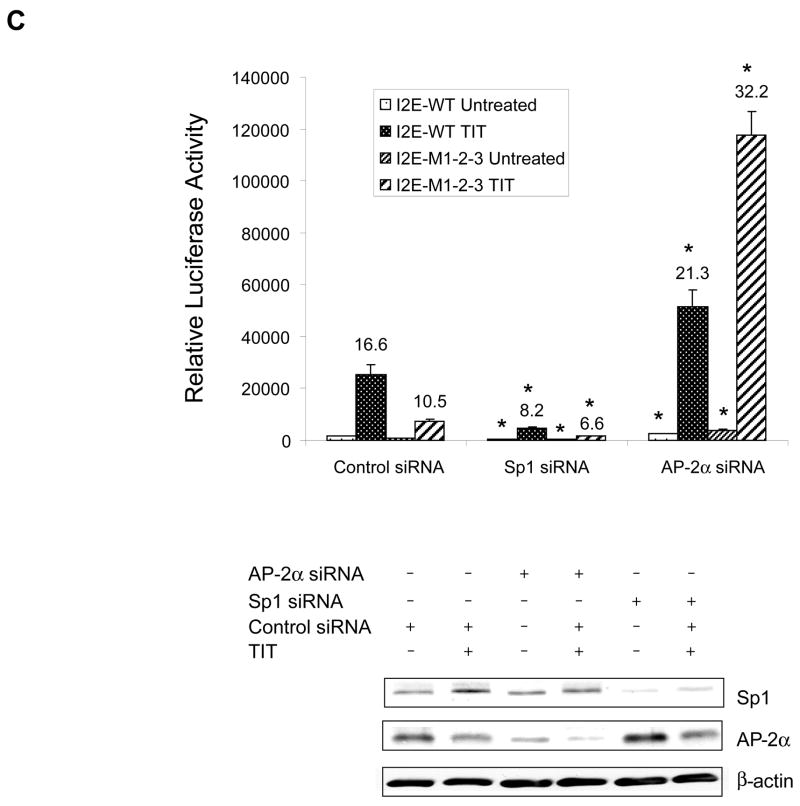
Since Sp1 and AP-2 can directly regulate promoter function, the roles of Sp1 and AP-2 in mutation-mediated alterations in transcriptional induction were further investigated. Expression of Sp1or AP-2α levels in HepG2 cells shows a Sp1-dependent increase or an AP-2α-dependent decrease in transcriptional induction (Fig. 5A and B). The effects of siRNA-based knock-down of Sp1 and AP-2α in VA13 cells, as confirmed by immunoblots, consistently show a decrease in transcriptional induction when Sp1 is reduced but an increase in transcriptional induction when AP-2α is reduced (Fig. 5C). Importantly, the effects of Sp1 and AP-2α are enhanced by the presence of mutations. Together, these results indicate that the presence of mutations affects both constitutive and inductive transcription via the changed Sp1 and AP-2 binding motifs.
FIGURE 5.
The effects of Sp1 and AP-2 on transcriptional stimulation. A and B, overexpression of Sp1 and AP-2α in HepG2 cells. The enhancer-promoter-reporter constructs (0.3 nM) were co-transfected with expression constructs of Sp1 and AP-2α at indicated concentrations plus β-gal control (0.1 nM), followed by TIT treatment. Vector DNA was added to maintain an equal mol concentration of total DNA in each transfection. Effects of these proteins on transcriptional stimulation were determined. * indicates significant differences in the luciferase activity compared to the wild-type (I2E-P7) in TIT-treated group at P<0.01. C, down-regulation of Sp1 and AP-2α in VA13 cells. The reporter constructs (0.3 nM) were co-transfected with siRNAs of Sp1, AP-2α and a siRNA control (5 nM) plus β-gal control (0.1 nM). Reduction in levels of the siRNA targeted proteins was measured by immunoblots (bottom) and the inducted folds were determined by reporter assay. Induction folds compared to untreated controls are indicated above the histograms. * indicates significant differences in the luciferase activity compared to control siRNA in both untreated and TIT-treated groups at P<0.01.
Mutation at −38 alters transcriptional modification
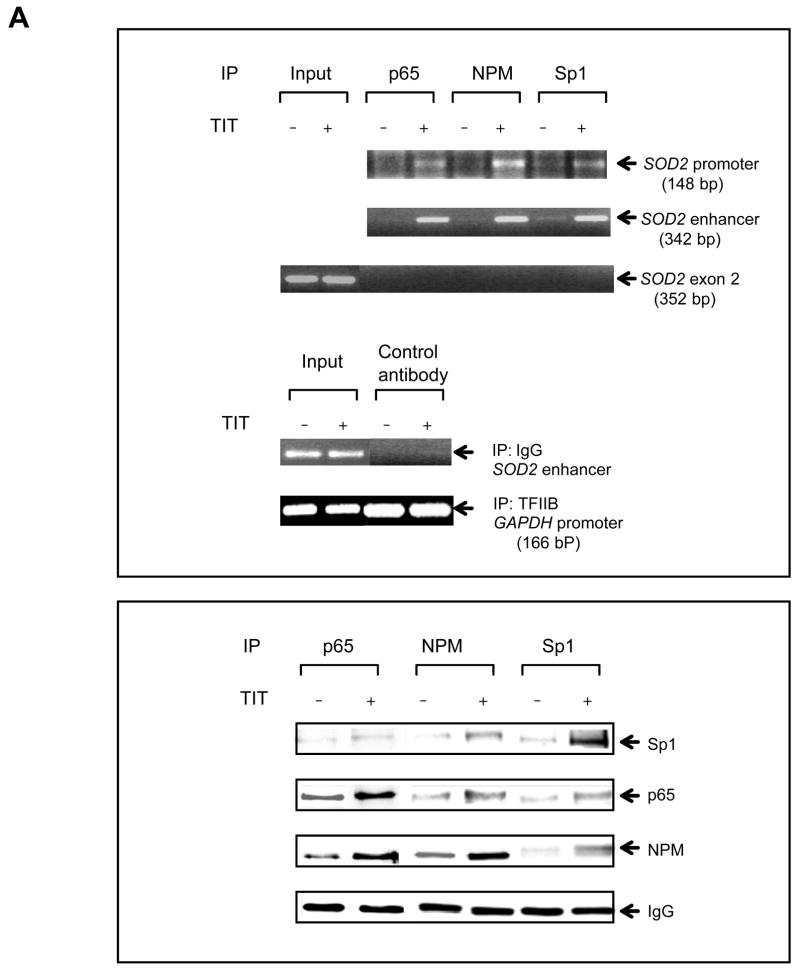
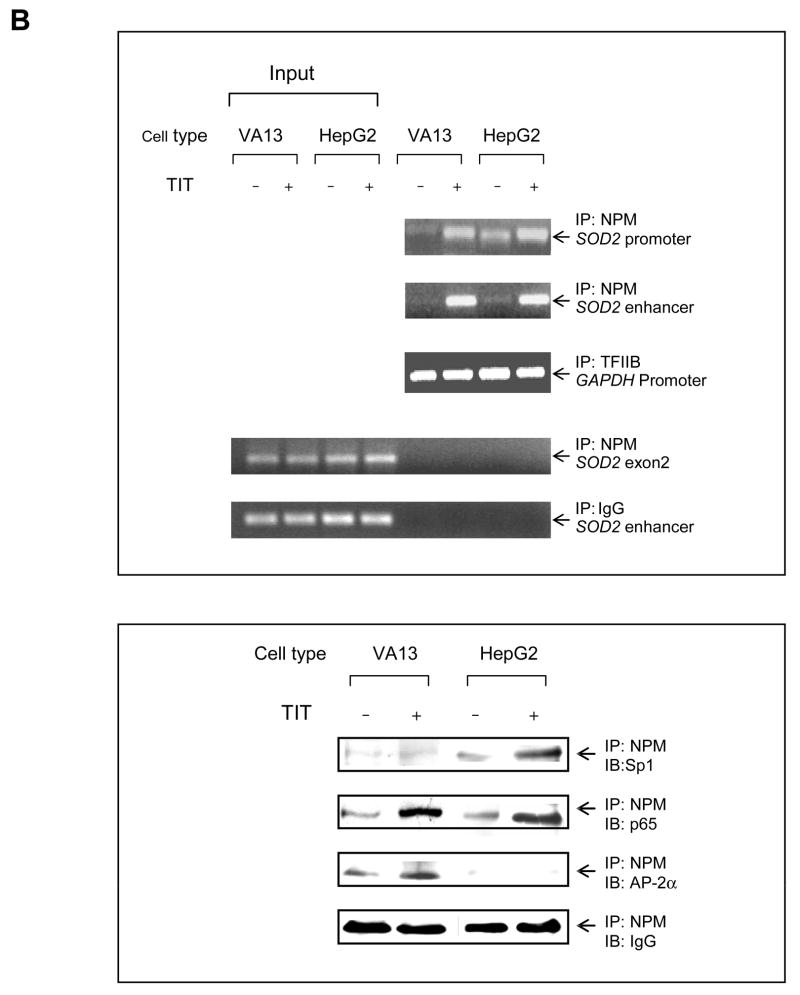
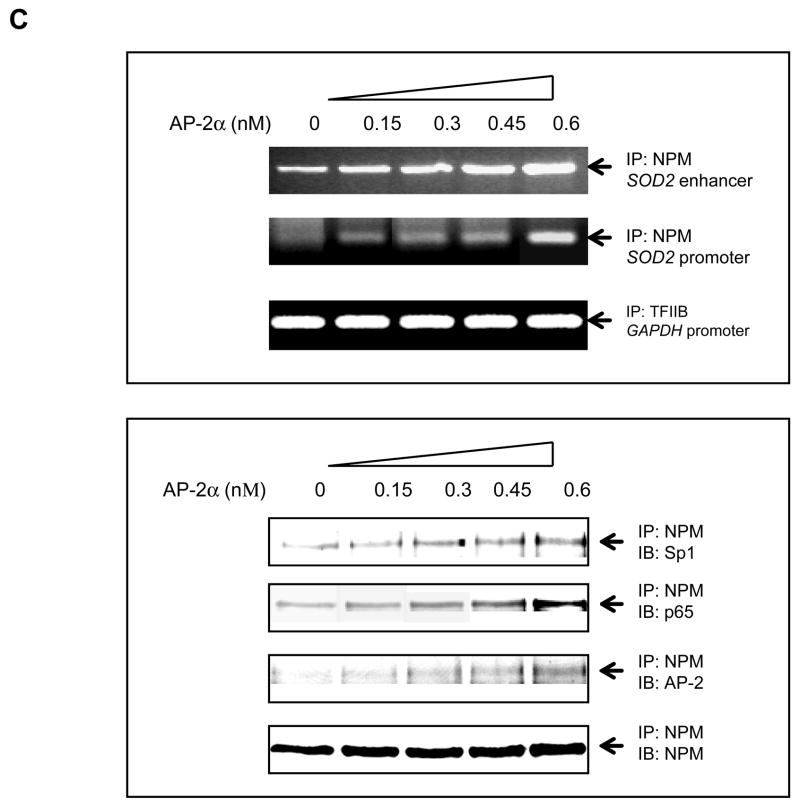
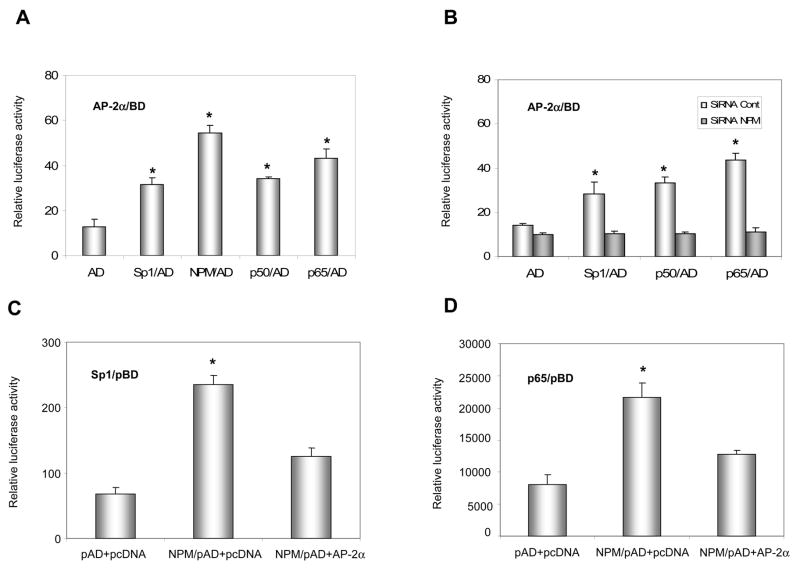
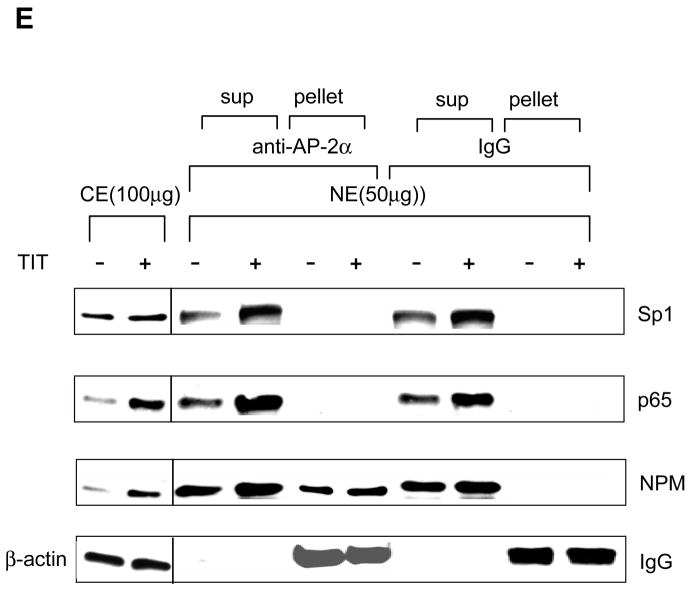
DNA-protein and protein-protein interactions were investigated using chromatin immunoprecipitation (ChIP) and two-hybrid systems to explore the molecular mechanism by which AP-2 modifies MnSOD expression in cancer cells. First, chromatin isolated from HepG cells was immunoprecipitated using antibodies to p65, Sp1 and NPM with IgG and TFIIB antibodies as controls. The contents of DNA and proteins in the precipitated chromatins were quantified by PCR and immunoblots, respectively. As shown in the top panel of Fig. 6A, both SOD2 enhancer and promoter regions are precipitated whereas no precipitation of the enhancer region is observed in the IgG control. No amplification is observed in the SOD2 exon2 region that was used as an untargeted control. The levels of the precipitated enhancer and promoter fragments are increased in response to TIT treatment, but are not increased in the GAPDH promoter fragment precipitated by the TFIIB antibody which served as a loading control. The levels of p65, Sp1, and NPM proteins in the precipitated chromatins are also increased in response to TIT treatment but are not increased in the IgG control. Second, chromatins from both VA13 and HepG2 cells were immunoprecipitated by the NPM antibody. The SOD2 enhancer-promoter regions as well as Sp1 and p65 proteins were quantified. The enhancer and promoter regions pulled-down from the two cell lines are increased in response to treatment. As predicted, AP-2α in VA13 cells is pulled-down and its level is also responsive to treatment, but HepG2 cells are not responsive (Fig. 6B). Because AP-2 is absent in HepG2 cells, this result suggests that AP-2 may be involved in the transcriptional complex that contains NF-κB, NPM and Sp1. Third, to determine whether AP-2 interacts with these proteins in the chromatin, AP-2α was expressed in HepG2 cells and chromatin was precipitated by the NPM antibody. Both SOD2 enhancer and promoter regions in the pulled-down chromatins are increased in an AP-2α dose-dependent manner. The levels of AP-2α, p65, and Sp1 in the chromatins consistently increase in the same manner (Fig. 6C). Fourth, a two-hybrid system was used to verify the interaction between AP-2α and other proteins. An AP-2α-Gal4 fusion protein containing a Gal4 Binding Domain (BD) was activated by p65-, NPM-, or Sp1-linked VP16 Activation Domain (AD) (Fig. 7A). Activation through interaction between AP-2α and Sp1, or between AP-2α and p65, is abolished when NPM is reduced by its specific siRNA, indicating that the AP-2α directly interacts with NPM (Fig. 7B). Since NPM is able to interact with Sp1 and p65 (12), AP-2α may indirectly interact with Sp1 or p65 in the presence of NPM. To probe this possibility, the two-hybrid system was performed by linking Sp1- or p65-Gal4BD to NPM-VA16AD in the AP-2α transfected HepG2 cells. The results show that AP-2α interrupts both Sp1-NPM and p65-NPM interactions (Fig. 7C and D). Finally, immunoprecipitation (IP) was used to verify direct interaction between AP-2 and NPM. Nuclear extracts from the TIT-treated or untreated VA13 cells were precipitated by AP-2α antibody or IgG control. Supernatants and pellets were collected and immunoblotted with antibody to Sp1, NPM or p65. As shown in Fig. 7E, compared to the control IgG pulled-down samples, the AP-2α antibody can only pull- down NPM from the extracts, as no Sp1 or p65 was found in the immunopellets. These results suggest that AP-2 interaction with NPM may interfere with NPM function as a transcription enhancer.
FIGURE 6.
Interaction between transcription factors in chromatin. A, interaction between p65, Sp1 and NPM. Chromatins from the treated and untreated HepG2 cells were precipitated using p65, Sp1, NPM, TFIIB or IgG antibody. The SOD2 enhancer and promoter regions were analyzed by PCR (top panel). A fragment of the exon2 of the SOD2 gene and the GAPDH promoter region were amplified as an untargeted control and a loading control. IgG-precipitated product served as a negative antibody control. Additionally, p65, Sp1, and NPM in the precipitated chromatin were quantified by immunoblots normalized with IgG (bottom panel). B, NPM interacting with p65, Sp1 or AP-2. Chromatins from the treated and untreated VA13 and HepG2 cells were precipitated using NPM, TFIIB or IgG antibody. The enhancer and promoter regions (top panel) and Sp1, AP-2α and NPM (bottom panel) were quantified with several controls as described above. C, effect of AP-2 on interaction between p65, NPM and Sp1. Increasing concentrations of AP-2 were transfected into HepG2 cells and chromatin from the transfected cells was precipitated using NPM, TFIIB or IgG antibody. The enhancer and promoter regions were quantified (top panel) and effect of AP-2α on precipitation of p65 and Sp1 was quantified by immunoblots (bottom panel) with the loading controls.
FIGURE 7.
The effect of protein-protein interaction on transcription. A, detection of AP-2α-Sp1, AP-2α-NPM, and AP-2α-p65 interactions. AP-2α is linked to the GAL4 binding domain (AP-2/BD) and Sp1, NPM, and p65 were attached to the VA16 activation domain (Sp1/AD, NPM/AD, and p65/AD). The generated fusion proteins with BD and AD tags (0.5 nM for each construct) were transfected into HepG2 cells. Reactivity via switching the AD to BD was estimated by relative luciferase response. B, effect of NPM on AP-2α-Sp1 or AP-2α-p65 interaction. NPM was knocked-down from HepG2 cells by co-transfecting 5 nM NPM siRNA with the two-hybrid constructs and the effect of the reduced NPM on the interactions was determined. C and D, the effect of AP-2α on Sp1-NPM and p65-NPM interactions. AP-2α was overexpressed in HepG2 cells by co-transfecting 0.3 nM AP-2 expression construct with the two-hybrid system of Sp1/BD to NPM/AD (C) or p65/BD to NPM/AD (D). The effect of expressed AP-2α on the two-hybrid reactions was analyzed. Significant differences (P<0.01) in the two-hybrid reaction compared to the AD vector only control are indicated by arrows. E, the role of interaction between AP-2 and NPM in transcription regulation. IP was performed to pull-down nuclear extracts (NE) from the TIT-treated or untreated VA13 cells using AP-2α antibody and IgG control. The amounts of Sp1, NPM and p65 in the supernatants and pellets were quantified by immunoblots using the relevant antibodies. Cell extracts (CE) were used to indicate the identical proteins that respond to TIT treatment. β-actin or IgG served as loading control for the section of CE or EC, respectively.
Discussion
ROS is an important mediator in the initiation and promotion of neoplastic growth (22). Aberrant expression and/or dysfunction of antioxidant enzymes are thought to contribute to a high level of ROS in cancer. As a primary antioxidant enzyme in the mitochondria, MnSOD is important for maintenance of cellular redox balance. With rare exceptions, MnSOD activity is reduced in over 80 different types of human and rodent neoplastic cells, including both spontaneous tumors and tumors induced by chemicals or viruses (2, 23, 24). The finding that overexpression of MnSOD leads to suppression of cancer phenotypes in vitro and in vivo further supports the hypothesis for the loss-of-function of MnSOD in the early stages of cancer development (25, 26). The reduction of MnSOD activity in neoplastic and transformed cells is regulated at the transcriptional level (27). Methylation of the CG-rich promoter region is involved in the reduction of MnSOD (28). Increased methylation of CpG islands in the SOD2 promoter region is correlated with decreased histone acetylation, leading to reduction of MnSOD in breast cancer cells but not in their normal cell counterparts (29). In addition to transcriptional regulation, a T to C mutation was identified in the exon2 of the SOD2 gene, which is associated with risk of prostate and breast cancers. The mutation leads to a change of Val to Ala at the −9 position of the mitochondrial targeting sequence, which affects the transport of MnSOD into mitochondria (30).
While a reduced level of MnSOD has been found in neoplastic and transformed cells, numerous reports demonstrate that the level of MnSOD is high in many types of aggressive tumor tissues (6, 31–35). In some cases, increased MnSOD expression is correlated with dysfunction of p53 (31, 36, 37). The high constitutive level of MnSOD in these tumors renders them chemo- and radio-resistant (7, 36, 38). To address the question of how MnSOD is differentially regulated in cancer, the present study shows that mutations identified in cancer cells may contribute to the regulation of MnSOD levels, i.e., down-regulation of MnSOD in transformed cells but up-regulation of MnSOD in cancer cells. The down- and up-regulation of MnSOD in different types of cells are critically dependent on AP-2 level. Our results suggest that AP-2-dependent transcriptional repression/derepression may serve as a novel mechanism for regulation of MnSOD in cancer development, which explains the seemingly conflicting information reported by previous studies with different systems.
AP-2, a transcription factor binding to the CpG islands in the enhancer or the promoter regions of many genes in mammalian cells, contains isoforms of AP-2α, AP-2β, AP-2γ, AP-2δ, and AP-2ε. AP-2 mediates either activation or repression of its target genes (39). For down-regulation of its target genes, AP-2 has been shown to diminish Sp1-dependent transcription via competing with Sp1 for binding to the GC-rich promoters (11, 40, 41). The present study shows that the effect of mutations on transcription regulation of the SOD2 gene is dependent on the relative levels of Sp1 and AP-2. Thus, high levels of MnSOD in some cancer cells may be due to low levels of AP-2. In addition to AP-2 directly interacting with its cis-element, it has been reported that AP-2 regulates its target genes through interacting with other transcription factors, such as Rb, Myc, or p53 (42). Notably, AP-2α directly interacts with p53 via p53 binding sites, resulting in up-regulation of the cyclin-dependent kinase inhibitor p21WAF/CIP1 (43). Recently, Liu et al. demonstrated that recruitment of NPM to an AP-2 binding site leads to an alleviation of gene repression by retinoic acid and AP-2α (44). The present study shows that AP-2α may repress TIT-mediated transcriptional induction of the SOD2 gene by interrupting NPM function. Thus, AP-2α can suppress MnSOD expression either by directly competing with Sp1 for the binding site or by interacting with NPM.
AP-2 is developmentally regulated, retinoic acid-inducible, and necessary for normal development. It participates in the regulation of various cell processes, including apoptosis, cell growth and cell differentiation (45). Down-regulation of AP-2 in cancer has been reported in melanoma, breast, prostate, lung, and colon cancers, indicating that the loss of AP-2 is associated with a malignant phenotype (46). Overexpression of AP-2 has been shown to suppress tumorigenicity (47), suggesting that AP-2 may function as a tumor suppressor gene. It has been demonstrated that AP-2 inhibits tumor cell growth by inducing expression of cell-cycle inhibitor p21WAF1/CIPI through the p53-dependent (48) or p53-independent pathway (49). AP-2 also suppresses cell proliferation by inhibiting transaction of Myc or induces apoptosis by mediating RB-mediated activation of the bcl-2 gene (50).
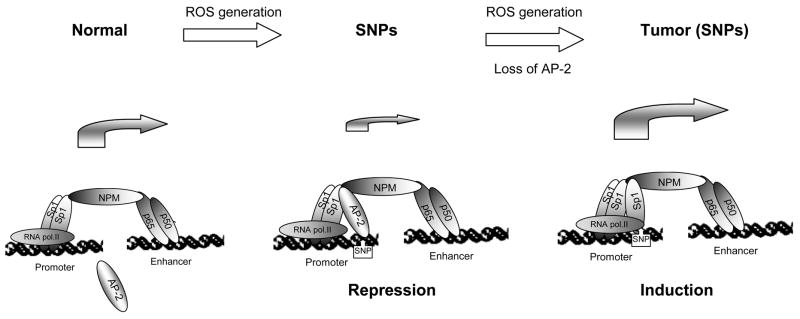
Our study, which shows that Sp1/AP-2 ratio plays an important role in the expression of the SOD2 gene, reveals a novel mechanism by which MnSOD is altered in cancer cells. Loss of AP-2 in tumor cells alters Sp1/AP-2 ratio, resulting in up-regulation of MnSOD. A high level of MnSOD in advanced cancer cells may cause resistance to chemo- and radio-therapeutics. The results obtained from the present study extend to demonstrate a potential role of mutations in tumor cells and predict up-regulation of MnSOD by derepression of AP-2, as depicted in Fig. 8. Genetic instability mediated by ROS may contribute to the formation of mutations in the SOD2 promoter region, which enhances AP-2-based transcriptional repression of the SOD2 gene. During malignancy progression, expression of AP-2 is attenuated, resulting in transcriptional activation of the SOD2 gene by Sp1. Thus, the present study identifies the first molecular event that may explain the cause for the reduction or the induction of MnSOD in various types of cancer. This finding may be beneficial for the development of novel strategies to prevent cancer progression and to enhance cancer therapy.
FIGURE 8.
A hypothetical model for the effect of mutations on transcriptional regulation of the human SOD2 gene in cancer.
Materials and Methods
Plasmid construction
A luciferase reporter gene driven by the human SOD2 promoter (P7) or by the human SOD2 intron 2 element with the P7 promoter (I2E-P7) was constructed prior to the study (17). In this study, mutations identified in cancer cells were separated into two reporter constructs, M1-2 and M3, as shown in Fig. 1A. A 102 bp Pvu II-Hind III fragment containing wild-type sequences was subcloned into the reporter constructs containing the promoter with the M3 to generate the M1-2 reporter constructs. Additionally, a fragment containing the M3 replaced the wild-type sequence to generate the M3 reporter constructs. The generated constructs were confirmed by DNA sequencing. Moreover, plasmids used in this study for expressing transcriptional regulators were constructed prior to the study (12).
Cell transfection and treatments
Cell lines used in this study were obtained from American Type Culture Collection and grown in the recommended media: SV40-transformed human lung fibroblast WI38 (VA13), human hepatocellular carcinoma (HepG2), papillomavirus-transformed human prostate epithelium (PZ-HPV-7), and human prostate carcinoma/adenocarcinoma (LNCap, PC-3 and DU145). For transfection, cells were plated in 12-well culture plates at a density of 5 × 105 cells in 1ml culture medium per well. The cells were cultured overnight and then transfected with the reporter constructs or co-transfected with expression constructs of p50, p65, Sp1, AP-2α, and NPM. A β-galactosidase (β-gal) expression construct was co-transfected to control cell transfection efficiency. Lipofectamine 2000 reagent (Invitrogen) was used to perform cell transfection according to the manufacturer’s protocol. For induction of MnSOD, the cells were treated with three inducers for 12 hours: 100 u/ml recombinant human TNF-α (R&D Systems, Inc.), 2 ng/ml recombinant human IL-1β (Endogen) and 100 mM TPA (Sigma). For reporter assay, the cells were washed with 1 × PBS and lysed in Passive Lysis Buffer (Promega). Activity of the luciferase reporter was measured using a luciferase assay kit (Promega) with a TD-20/20 luminometer. β-gal activity was measured using chlorophenol red-a-D-glactopyranoside monosodium (Roche Molecular Biochemicals) as a colorimetric substrate with SpectraMax (Molecular Devices Corp.) at 470 mM. Transcription activity was estimated by β-gal-normalized luciferase responses.
Immunoblotting analysis
To quantify protein levels, 50–100 μg of cell extracts were fractionated on SDS-PAGE 8% (w/v) polyacrylamide gel, transferred to nitrocellulose membranes and blotted with primary antibodies to the MnSOD, p50, p65, Sp1, AP-2α, NPM, and β-actin. The primary antibodies to MnSOD, NPM and β-actin were obtained from Upstate Biotech., NeoMarkers and Sigma, respectively. The primary antibodies to p50, p65, Sp1, AP-2α and all secondary antibodies were purchased from Santa Cruz Biotech. A goat anti-mouse IgG-HRP conjugated secondary antibody was used to quantify Sp1, AP-2, NPM, and β-actin, and a goat anti-rabbit IgG-HRP conjugated secondary antibody was used for p50, p65 and MnSOD. Human β-actin was used as an internal control to normalize other proteins. Immunoblots were visualized by an enhanced chemiluminescence detection system (ECL, Amersham Pharmacia Biotech.).
IP
Fifty μg nuclear extracts were incubated overnight with 1 μg AP-2α and control IgG antibodies at 4 °C. Subsequently, 20 μl of protein A/G agarose (Santa Cruz Biotech.) were added to the mixture and the extracts were incubated for 4 h at 4 °C. Immunocomplexes were precipitated by centrifugation for 5 min at 2500 rmp at 4 °C. The supernatants were collected and the pellets were washed four times with RIPA buffer (9.1 mM/L Na2HPO4, 1.7 mM/L NaH2PO4, 150 mM/L NaCl, 0.5% sodium deoxycholate, 1% v/v NP-40, and 1% SDS, pH 7.2) and resuspended in 20 μl of 2x sample loading buffer. The supernatants and pellets were fractionated by the SDS-PAGE gel and blotted with primary antibodies to Sp1, p65, NPM, and IgG.
Nuclear extraction and EMSA
AP-2α expression construct was transfected into HepG2 cells and nuclear proteins were extracted from the transfected cells as described previously (11). A fragment (−53 to −25) containing the M3 or wild-type was labeled with [γ-32P] ATP by T4 DNA kinase (New England Biolabs). The probes were purified on 20% (w/v) polyacrylamide gel and quantified by scintillation counting (Beckman). Ten μg of the nuclear extracts were incubated with 0.5 pM probes on ice for 30 min and then separated on 4% (w/v) polyacrylamide gel in 0.5x TBE electrophoresis buffer. A 100-fold concentration of cool probe (self) and 100-fold concentration of 30-bp oligonucleotides containing sequences of the multiple cloning sites in the pGL3 vector (nonself) were included for probe competition. To detect supershift by the specific antibody, 2 μg Sp1 and AP-2α antibodies were added to the nuclear extracts 1 hr prior to incubation with the probes. After electrophoresis, gels were dried and scanned by Typhoon 8600.
ChIP
A ChIP-IT system (Active Motif) was used to investigate interaction between transcription factors and the SOD2 enhancer and promoter regions, according to the manufacturer’s protocol. Briefly, chromatins extracted from TIT-treated or AP-2α-transfected cells were pulled- down using antibodies to NPM, p65, Sp1 and AP-2α. After multiple washings, DNA and proteins were purified from the immunoprecipitated chromatins. A quantitative PCR procedure was used to quantify the SOD2 promoter and enhancer regions. In the control group, a fragment from the SOD2 exon2 was amplified as an untargeted control; the GAPDH promoter region pulled-down by the TFIIB antibody was amplified as a loading control; and chromatins pulled-down by IgG served as the negative antibody control. PCR primer pairs and conditions for each target have been described (12). In addition, immunoblotting analysis was used to quantify the associated proteins in the ChIP preparations.
RNA interference
Short interference RNA (siRNA) was used to selectively knock-down Sp1, AP-2α, p50, p65 and NPM. The cultured cells were transfected or co-transfected with 0.1μM siRNA to target those proteins in a serum-reduced Opti-MEM medium (Invitrogen). The siRNA molecules were purchased from Santa Cruz Biotech. After siRNA transfection, the targeted proteins were measured by immunoblotting with the protein-specific antibodies and effects of the siRNA targeting on regulation of transcription were estimated by reporter responses.
Two-hybrid analysis
A mammalian hybridization system (Promega) was used to detect protein-protein interactions. Tested proteins were previously subcloned in either pBIND or pACT to tag them with either the Gal4-binding fusion domain or the VP16-activation fusion domain; the procedure for the two hybridizations has been described (11, 12).
Statistical analysis
Multiple independent cell transfections and reporter assays were performed. Statistical significances between the different constructs or treatments were analyzed using one-way Anova and Tukey’s Multiple Comparison Test followed by data analysis with Graphpad Prism version 4.0. Differences in the comparison tests lower than P<0.01 were considered to be insignificant.
Acknowledgments
We would like to thank Dr. Robert Tjian, University of California Berkeley; Dr. Brett T. Spear and Dr. Vivek M. Rangnekar, University of Kentucky, for providing the human Sp1, AP-2α, NF-κB p65 and p50 expression constructs used in this study.
Grant support: National Institutes of Health grants CA 49797 and CA 73599 to Daret K. St. Clair.
References
- 1.Fridovich I. The biology of oxygen radicals. Science. 1978;201:875–80. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- 2.Oberley LW, Buettner GB. Role of superoxide dismutase in cancer: a review. Cancer Res. 1979;39:1141–9. [PubMed] [Google Scholar]
- 3.Oberley LW. Mechanism of the tumor suppressive effect of MnSOD overexpression. Biomed Pharmacother. 2005;59:143–8. doi: 10.1016/j.biopha.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, Chaiswing L, Oberley TD, et al. A mechanism-based antioxidant approach for the reduction of skin carcinogenesis. Cancer Res. 2005;65:1401–5. doi: 10.1158/0008-5472.CAN-04-3334. [DOI] [PubMed] [Google Scholar]
- 5.Pani G, Colavitti R, Bedogni B, et al. Mitochondrial superoxide dismutase: a promising target for new anticancer therapies. Curr Mes Chem. 2004;11:1299–308. doi: 10.2174/0929867043365297. [DOI] [PubMed] [Google Scholar]
- 6.Josson S, Xu Y, Fang F, et al. RelB regulates manganese superoxide dismutase gene and resistance to ionizing radiation of prostate cancer cells. Oncogene. 2006;25:1554–9. doi: 10.1038/sj.onc.1209186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Fang F, St Clair DK, et al. Suppression of RelB-mediated manganese superoxide dismutase expression reveals a primary mechanism for radiosensitization effect of 1alpha,25-dihydroxyvitamin D(3) in prostate cancer cells. Mol Cancer Ther. 2007;6:2048–56. doi: 10.1158/1535-7163.MCT-06-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuda Y, Higashiyama S, Kijima Y, et al. Human liver manganese superoxide dismutase. Purification and crystallization, subunit association and sulfhydryl reactivity. Eur J Biochem. 1990;194:713–20. doi: 10.1111/j.1432-1033.1990.tb19461.x. [DOI] [PubMed] [Google Scholar]
- 9.Wan XS, Devalaraja MN, St Clair DK. Molecular structure and organization of the human manganese superoxide dismutase gene. DNA Cell Biol. 1994;13:1127–36. doi: 10.1089/dna.1994.13.1127. [DOI] [PubMed] [Google Scholar]
- 10.Yeh CC, Wan XS, St Clair DK. Transcriptional regulation of the 5′ proximal promoter of the human manganese superoxide dismutase gene. DNA Cell Biol. 1998;17:921–30. doi: 10.1089/dna.1998.17.921. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y, Porntadavity S, St Clair DK. Transcriptional regulation of the human manganese superoxide dismutase gene: the role of specificity protein 1 (Sp1) and activating protein-2 (AP-2) Biochem J. 2002;362:401–12. doi: 10.1042/0264-6021:3620401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Fang F, Dhar SK, et al. The role of a single-stranded nucleotide loop in transcriptional regulation of the human sod2 gene. J Biol Chem. 2007;282:15981–94. doi: 10.1074/jbc.M608979200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dryer SE, Dryer RL, Autor AP. Enhancement of mitochondrial, cyanide-resistant superoxide dismutase in the livers of rats treated with 2,4-dinitrophenol. J Biol Chem. 1980;255:1054–7. [PubMed] [Google Scholar]
- 14.Krall J, Bagley AC, Mullenbach GT, et al. Superoxide mediates the toxicity of paraquat for cultured mammalian cells. J Biol Chem. 1988;263:1910–4. [PubMed] [Google Scholar]
- 15.Poswig A, Wenk J, Brenneisen P, et al. Adaptive antioxidant response of manganese- superoxide dismutase following repetitive UVA irradiation. J Invest Dermatol. 1999;112:13–8. doi: 10.1046/j.1523-1747.1999.00465.x. [DOI] [PubMed] [Google Scholar]
- 16.Jones PL, Ping D, Boss JM. Tumor necrosis factor alpha and interleukin-1beta regulate the murine manganese superoxide dismutase gene through a complex intronic enhancer involving C/EBP-beta and NF-kappaB. Mol Cell Biol. 1997;17:6970–81. doi: 10.1128/mcb.17.12.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Kiningham KK, Devalaraja MN, et al. An intronic NF-kappaB element is essential for induction of the human manganese superoxide dismutase gene by tumor necrosis factor-alpha and interleukin-1beta. DNA Cell Biol. 1999;18:709–22. doi: 10.1089/104454999314999. [DOI] [PubMed] [Google Scholar]
- 18.Dhar SK, Lynn BC, Daosukho C, St Clair DK. Identification of nucleophosmin as an NF-kappaB co-activator for the induction of the human SOD2 gene. J Biol Chem. 2004;279:28209–19. doi: 10.1074/jbc.M403553200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, Krishnan A, Wan XS, et al. Mutations in the promoter reveal a cause for the reduced expression of the human manganese superoxide dismutase gene in cancer cells. Oncogene. 1999;18:93–102. doi: 10.1038/sj.onc.1202265. [DOI] [PubMed] [Google Scholar]
- 20.Martin RC, Hughes K, Doll MA, et al. Method for determination of (−102C>T) single nucleotide polymorphism in the human manganese superoxide dismutase promoter. BMC Genet. 2004;5:33. doi: 10.1186/1471-2156-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan C, Clemmons DR. Transcription factor AP-2 regulates human insulin-like growth factor binding protein-5 gene expression. J Biol Chem. 1995;270:24844–51. doi: 10.1074/jbc.270.42.24844. [DOI] [PubMed] [Google Scholar]
- 22.Benhar M, Engelberg D, Levitzki A. ROS, stress-activated kinases and stress signaling in cancer. EMBO Rep. 2002;3:420–5. doi: 10.1093/embo-reports/kvf094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oberley LW, Oberley TD. In: Free Radicals, Aging and Degenerative Diseases. Johnson JE Jr, Harman D, Miquel J, editors. Alan R. Liss; New York: 1986. pp. 325–72. [Google Scholar]
- 24.Bravard A, Sabatier L, Hoffschir F, et al. SOD2: a new type of tumor-suppressor gene? Int J cancer. 1992;51:476–80. doi: 10.1002/ijc.2910510323. [DOI] [PubMed] [Google Scholar]
- 25.Oberley LW. Mechanism of the tumor suppressive effect of MnSOD overexpression. Biomed Pharmacother. 2005;59:143–8. doi: 10.1016/j.biopha.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 26.St Clair DK, Zhao Y, Chaiswing L, Oberley TD. Modulation of skin tumorigenesis by SOD. Biomed Pharmacother. 2005;59:209–14. doi: 10.1016/j.biopha.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 27.St Clair DK, Holland JC. Complementary DNA encoding human colon cancer manganese superoxide dismutase and the expression of its gene in human cells. Cancer Res. 1991;51:939–43. [PubMed] [Google Scholar]
- 28.Huang Y, He T, Domann FE. Decreased expression of manganese superoxide dismutase in transformed cells is associated with increased cytosine methylation of the SOD2 gene. DNA Cell Biol. 1999;18:643–52. doi: 10.1089/104454999315051. [DOI] [PubMed] [Google Scholar]
- 29.Hitchler MJ, Wikainapakul K, Yu L, et al. Epigenetic regulation of manganese superoxide dismutase expression in human breast cancer cells. Epigenetics. 2006;1:163–71. doi: 10.4161/epi.1.4.3401. [DOI] [PubMed] [Google Scholar]
- 30.Ambrosone CB, Freudenheim JL, Thompson PA, et al. Manganese superoxide dismutase (MnSOD) genetic polymorphisms, dietary antioxidants, and risk of breast cancer. Cancer Res. 1999;59:602–6. [PubMed] [Google Scholar]
- 31.Nakano T, Oka K, Taniguchi N. Manganese superoxide dismutase expression correlates with p53 status and local recurrence of cervical carcinoma treated with radiation therapy. Cancer Res. 1996;56:2771–5. [PubMed] [Google Scholar]
- 32.Landriscina M, Remiddi F, Ria F, et al. The level of MnSOD is directly correlated with grade of brain tumours of neuroepithelial origin. Br J Cancer. 1996;74:1877–85. doi: 10.1038/bjc.1996.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janssen AM, Bosman CB, Sier CF, et al. Superoxide dismutases in relation to the overall survival of colorectal cancer patients. Br J Cancer. 1998;78:1051–7. doi: 10.1038/bjc.1998.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung-man Ho J, Zheng S, Comhair SA, et al. Differential expression of manganese superoxide dismutase and catalase in lung cancer. Cancer Res. 2001;61:8578–85. [PubMed] [Google Scholar]
- 35.Ria F, Landriscina M, Remiddi F, et al. The level of manganese superoxide dismutase content is an independent prognostic factor for glioblastoma. Biological mechanisms and clinical implications. Br J Cancer. 2001;84:529–34. doi: 10.1054/bjoc.2000.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pani G, Colavitti R, Bedogni B, et al. Mitochondrial superoxide dismutase: a promising target for new anticancer therapies. Curr Mes Chem. 2004;11:1299–308. doi: 10.2174/0929867043365297. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Chaiswing L, Velez JM, et al. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer Res. 2005;65:3745–50. doi: 10.1158/0008-5472.CAN-04-3835. [DOI] [PubMed] [Google Scholar]
- 38.Greenberger JS, Epperly MW. Review. Antioxidant gene therapeutic approaches to normal tissue radioprotection and tumor radiosensitization. In Vivo. 2007;21:141–6. [PubMed] [Google Scholar]
- 39.Eckert D, Buhl S, Weber S, et al. The AP-2 family of transcription factors. Genome Biol. 2005;6:246. doi: 10.1186/gb-2005-6-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Getman DK, Mutero A, Inoue K, Taylor P. Transcription factor repression and activation of the human acetylcholinesterase gene. J Biol Chem. 1995;270:23511–9. doi: 10.1074/jbc.270.40.23511. [DOI] [PubMed] [Google Scholar]
- 41.Chen TT, Wu RL, Castro-Munozledo F, Sun TT. Regulation of K3 keratin gene transcription by Sp1 and AP-2 in differentiating rabbit corneal epithelial cells. Mol Cell Biol. 1997;17:3056–64. doi: 10.1128/mcb.17.6.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Batsché E, Muchardt C, Behrens J, et al. RB and c-Myc activate expression of the E-cadherin gene in epithelial cells through interaction with transcription factor AP-2. Mol Cell Biol. 1998;18:3647–58. doi: 10.1128/mcb.18.7.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wajapeyee N, Somasundaram K. Cell cycle arrest and apoptosis induction by activator protein 2alpha (AP-2alpha) and the role of p53 and p21WAF1/CIP1 in AP-2 alpha-mediated growth inhibition. J Biol Chem. 2003;278:52093–101. doi: 10.1074/jbc.M305624200. [DOI] [PubMed] [Google Scholar]
- 44.Liu H, Tan BC, Tseng KH, et al. Nucleophosmin acts as a novel AP2alpha-binding transcriptional corepressor during cell differentiation. EMBO Rep. 2007;8:394–400. doi: 10.1038/sj.embor.7400909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Hagopian-Donaldson S, Serbedzija G, et al. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–41. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- 46.Pellikainen JM, Kosma VM. Activator protein-2 in carcinogenesis with a special reference to breast cancer--a mini review. Int J Cancer. 2007;120:2061–7. doi: 10.1002/ijc.22648. [DOI] [PubMed] [Google Scholar]
- 47.Ruiz M, Pettaway C, Song R, et al. Activator protein 2alpha inhibits tumorigenicity and represses vascular endothelial growth factor transcription in prostate cancer cells. Cancer Res. 2004;64:631–8. doi: 10.1158/0008-5472.can-03-2751. [DOI] [PubMed] [Google Scholar]
- 48.McPherson LA, Loktev AV, Weigel RJ. Tumor suppressor activity of AP2alpha mediated through a direct interaction with p53. J Biol Chem. 2002;277:45028–33. doi: 10.1074/jbc.M208924200. [DOI] [PubMed] [Google Scholar]
- 49.Zeng YX, Somasundaram K, el-Deiry WS. AP2 inhibits cancer cell growth and activates p21WAF1/CIP1 expression. Nat Genet. 1997;15:78–82. doi: 10.1038/ng0197-78. [DOI] [PubMed] [Google Scholar]
- 50.Decary S, Decesse JT, Ogryzko V, et al. The retinoblastoma protein binds the promoter of the survival gene bcl-2 and regulates its transcription in epithelial cells through transcription factor AP-2. Mol Cell Biol. 2002;22:7877–88. doi: 10.1128/MCB.22.22.7877-7888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]