SUMMARY
dsRNA binding proteins (dsRBPs) facilitate Dicer functions in RNAi. C. elegans RDE-4 facilitates cleavage of long dsRNA to siRNA, while human TRBP functions downstream to pass siRNA to RISC. We show that these distinct in vivo roles are reflected in in vitro binding properties. RDE-4 preferentially binds long dsRNA, while TRBP binds siRNA with an affinity that is independent of dsRNA length. These properties are mechanistically based in the fact that RDE-4 binds cooperatively, via contributions from multiple domains, while TRBP binds non-cooperatively. Our studies offer a paradigm for how dsRBPs, which are not sequence-specific, discern dsRNA length. Additionally, analyses of the ability of RDE-4 deletion constructs and RDE-4/TRBP chimeras to reconstitute Dicer activity suggest RDE-4 promotes activity using its dsRBM2 to bind dsRNA, its linker region to interact with Dicer, and its C-terminus for Dicer activation.
Keywords: Cooperativity, RNA interference, Dicer, C. elegans, siRNA
INTRODUCTION
The RNase III enzyme Dicer is a key enzyme in the RNA interference (RNAi) pathway, and in all organisms studied thus far, functions in association with double-stranded RNA-binding proteins (dsRBPs).1 For example, human Dicer associates with TRBP and PACT,2-4 Drosophila Dicer-2 associates with R2D2,5,6 and Caenorhabditis elegans DCR-1 associates with RDE-4.7 All of these accessory dsRBPs have very similar domain structures: two N-terminal dsRNA-binding motifs (dsRBMs) and a C-terminus that contains a third degenerate dsRBM.
Despite similarities in domain structure, these dsRBPs have different roles in RNAi. Drosophila Dicer-2 does not require R2D2 to cleave dsRNA in vitro or in vivo, but downstream of this step, a complex of Dicer-2 and R2D2 is essential for loading siRNA into the RNA-induced silencing complex (RISC).5,6,8 Likewise, human Dicer processes dsRNA without TRBP and PACT in vitro,9,10 and while there are some conflicting data,11 the primary roles for TRBP and PACT appear to be after the production of siRNAs, in facilitating their incorporation into RISC.2-4,12 In contrast, C. elegans RDE-4 is required for DCR-1-mediated cleavage of dsRNA to siRNA, but is not required in subsequent steps.7,13,14 This is emphasized by the observation that rde-4 mutant worms are incapable of RNAi when injected with long dsRNA, but this defect can be bypassed by the injection of siRNAs.14
dsRBPs bind dsRNA indiscriminant of sequence.15 However, the different functions of dsRBPs in RNAi require that some bind long dsRNA, while others bind short siRNA, raising the possibility that dsRBPs can discriminate dsRNA based on length. Consistent with this idea, RDE-4 forms stable complexes with long dsRNA in vivo, but does not stably interact with siRNA.7 Similarly, with purified components, RDE-4 preferentially binds long dsRNA.13 The latter study indicated that RDE-4 binds dsRNA cooperatively. This suggests a simple model to explain how dsRBPs discriminate dsRNA based on length, invoking classic studies of sequence-independent proteins that bind to nucleic acid lattices.16,17 According to this paradigm, cooperativity favors binding to long dsRNA, a nucleic acid lattice that has multiple binding sites and thus maximizes cooperative interactions. Here we report further studies that support this model, involving studies of RDE-4 as well as a second dsRBP, human TRBP. We show that human TRBP, whose in vivo function requires binding to siRNA rather than long dsRNA, binds siRNA with high affinity, but is not cooperative. Using a comprehensive set of RDE-4 truncations, we dissect the functions of RDE-4’s domains using in vitro binding studies, and assays for reconstitution of Dicer activity in extracts of rde-4 mutant C. elegans. Our studies indicate dsRBM2 of RDE-4 is most important for binding dsRNA, but multiple domains contribute to cooperativity. While RDE-4’s ability to bind dsRNA is important for facilitating cleavage of dsRNA by Dicer, the linker region also plays an important role, possibly mediating direct interactions with Dicer.
RESULTS
To investigate the contribution of the different domains of RDE-4 to cooperativity, we first overexpressed and purified RDE-4 variant proteins that lacked, or contained mutations in, one or more domains (Fig. 1a). These variants were named according to their domain content. For example, the RDE-4 variant that lacks dsRBM1 (R1) and the linker region (L), but has dsRBM2 (R2) and the C-terminus (C), is called R2C; when a variant contained only the N- or C-terminal region of a domain, the included region was indicated by a subscript (e.g., LcR2). In addition, for comparison, we sought a non-cooperative dsRBP. The cooperativity of RDE-4 is consistent with its in vivo function, which requires preferential binding to long dsRNA over short siRNA. We reasoned that dsRBPs that act later in RNAi, after a dsRNA has been cleaved into siRNAs, would not need to bind long dsRNA and thus would lack cooperativity. The human dsRBP TRBP was a good candidate for a non-cooperative dsRBP, since it acts downstream of siRNA production, facilitating incorporation of siRNA into RISC.2,3,12
FIGURE 1.
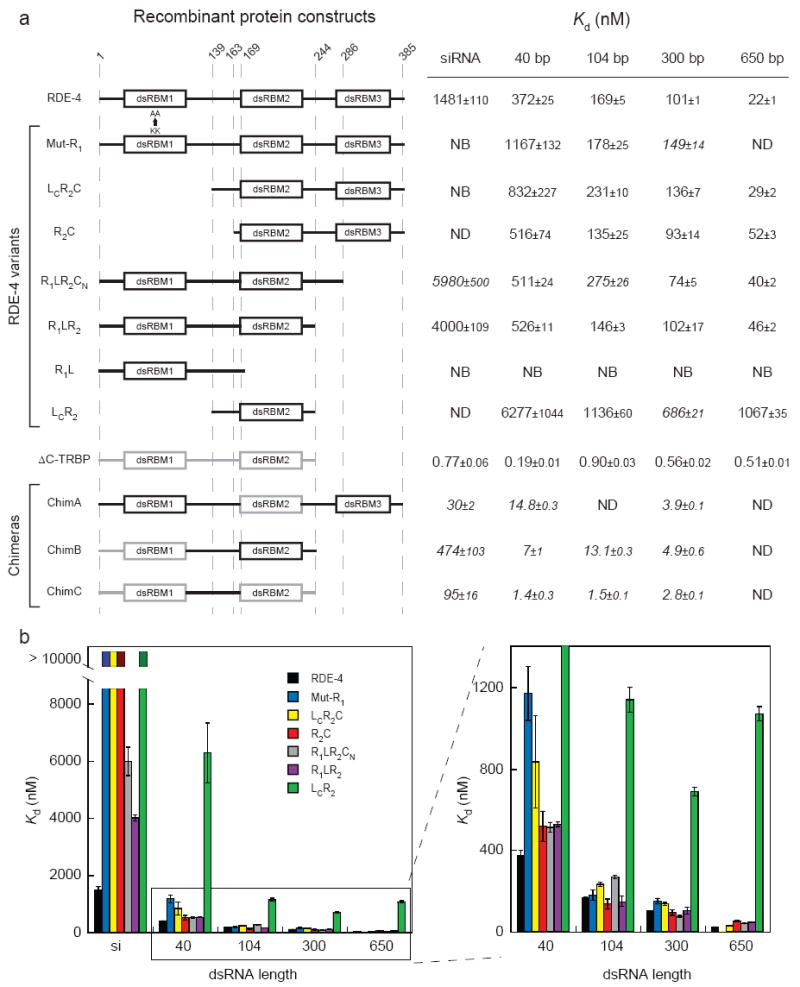
Affinity of dsRBP variants for dsRNA of varying length (a) Domain structures are shown on the left. Numbers at the top of dotted lines indicate amino acid positions with reference to RDE-4; for amino acids included in chimeric constructs, see Materials and Methods. Black lines, RDE-4 domains; gray lines, TRBP domains. As indicated, in Mut-R1, lysines 89 and 90 of dsRBM1 are mutated to alanines. Values for the dissociation constant (Kd) were calculated from gel-shift experiments using 32P-labeled dsRNAs (see Fig. 2 legend and Materials and Methods); data represent average values of multiple experiments (3 ≤n ≤ 5) ± standard error of curve fitting, except those in italics, where n = 2. NB, no binding up to 5μM (10 μM for R1L); ND, not determined. (b) Kd values observed for different lengths of dsRNA as tabulated in a are compared for the different RDE-4 variants, colored as indicated; data for dsRNA ≥ 40 bp are expanded on the right.
We cloned, overexpressed and purified TRBP as a maltose-binding protein (MBP) fusion for comparative binding studies. Despite retaining the ability to bind dsRNA, a large fraction of the full-length recombinant TRBP formed soluble aggregates. As an alternative, a stable, well-behaved C-terminal deletion variant of TRBP (ΔC-TRBP) was purified and used in our binding studies (Fig. 1a). Similar binding affinities were observed for the full-length aggregates and ΔC-TRBP (data not shown). This result is consistent with previous studies that demonstrated the C-terminus of TRBP does not contribute to dsRNA-binding properties.18,19
Comparison of dsRNA binding properties of RDE-4 and TRBP
We used gel-shift assays to compare binding of RDE-4, its variants, and ΔC-TRBP, to various lengths of dsRNA. These data are tabulated in Figure 1a, with representative data shown in Figure 2. Consistent with previous studies, the affinity of RDE-4 for dsRNA increased markedly with successive increases in dsRNA length (Fig. 1a, b). In contrast, ΔC-TRBP bound dsRNA with an affinity that was not dependent on length. In fact, differences in the affinity of ΔC-TRBP for the various dsRNA substrates were not significant (p ≥0.16, t-test), except for the slight increase in affinity for the 40 bp dsRNA; possibly there is a feature of this substrate that increases binding affinity. Notably, ΔC-TRBP exhibited a significantly higher affinity for dsRNA, with a Kd that was more than an order of magnitude lower than that of RDE-4 binding to a 650 bp dsRNA (Fig. 1a). Non-cooperative proteins bind nucleic acids based solely on their intrinsic binding affinity without contributions from cooperativity (see Fig. 5) and therefore should exhibit similar affinities regardless of RNA length.17 Thus, as predicted, ΔC-TRBP bound dsRNAs non-cooperatively.
FIGURE 2.
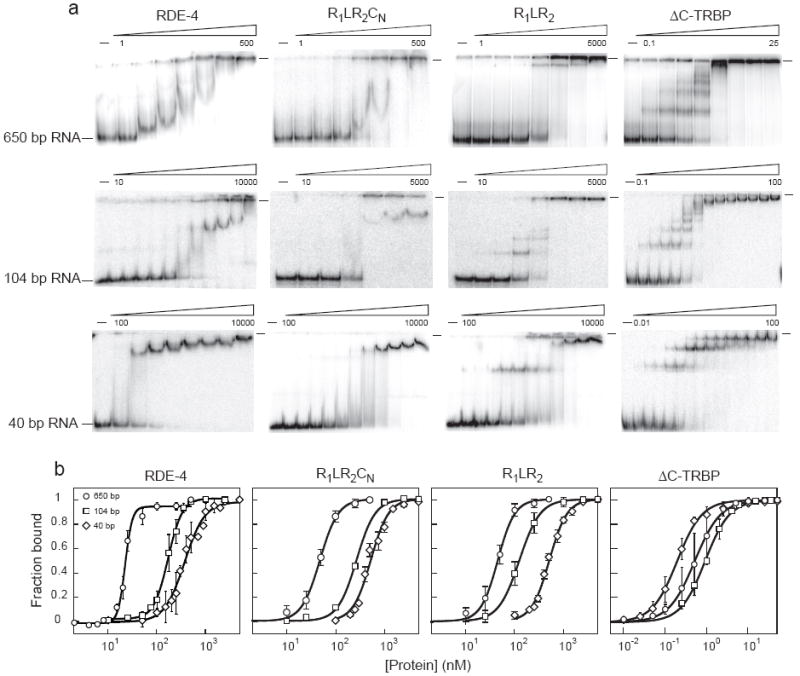
Comparison of RDE-4 binding properties to C-terminal deletion constructs and ΔC-TRBP. (a) Increasing amounts of recombinant proteins were added to 0.5-1 fmole 32P-labeled 650 bp dsRNA (top panel), 104 bp dsRNA (middle panel), and 40 bp dsRNA (bottom panel) and complex formation was analyzed by native gel electrophoresis. Beginning and ending protein concentrations are noted above gels with adjacent lanes representing a 2 or 4-fold difference in protein concentration. As more of the C-terminus is deleted (left to right), the banding pattern changes to look more like the non-cooperative binding pattern of ΔC-TRBP. (b) Multiple gel-shift assays as in a were performed to derive binding isotherms. ○ = 650 bp dsRNA, □ = 104 bp dsRNA, ◇ = 40 bp dsRNA. Radioactivity in gels was quantified to determine fraction bound = (1-[dsRNA]free)/[dsRNA]total. Data points with error bars (standard deviation) represent average values (3 ≤n ≤5, except for R1LR2CN/104bp where n = 2) and were fit using the Hill formalism (see Materials and Methods). Note that all three RDE-4 constructs show a marked increase in affinity for longer dsRNA, whereas ΔC-TRBP exhibits a relatively constant affinity for all dsRNAs tested (see Fig. 1). The wells of each gel are marked by a dash at the top right. In some assays, but not all, radioactive material remained in the well; Kd values derived from either situation were identical.
FIGURE 5.
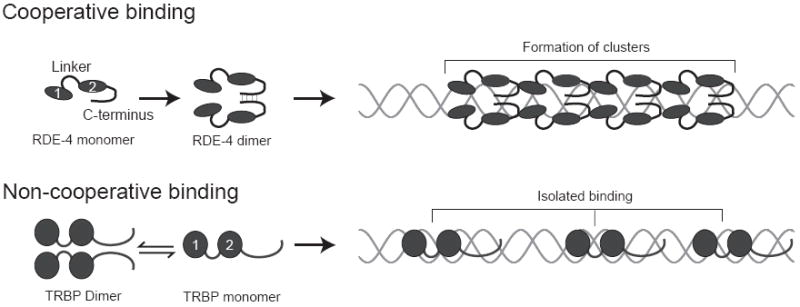
Schematics illustrating cooperative binding versus isolated binding. RDE-4 is shown with dsRBM1 and dsRBM2 (ovals) followed by a C-terminus that mediates dimerization. Our data indicate RDE-4 binds cooperatively, thus forming protein clusters along the dsRNA (top panel). TRBP is represented similarly, with spheres representing dsRBM1 and dsRBM2. Consistent with our data, TRBP is shown binding non-cooperatively at isolated sites (bottom panel). Note that while in vitro studies show that TRBP is a dimer in solution,41,42 it is a monomer in the active Dicer complex.12 We have illustrated RDE-4 interacting with dsRNA as a dimer, but it is possible that it is a monomer while facilitating Dicer function.
The differences between the cooperative RDE-4, and the non-cooperative TRBP, were quite evident in their gel-shift patterns (Fig. 2). Discrete mobility-shift intermediates were observed for ΔC-TRBP binding to dsRNA of all lengths, indicating stable isolated binding events (Figs. 2a, 5). In contrast, the gel-shift patterns for RDE-4 did not show intermediates, consistent with cooperative protein-protein interactions that preclude isolated binding events. Binding of RDE-4 showed a sharp transition, with complete binding occurring over a narrow range of protein concentration (Fig. 2b). This positive cooperativity was evidenced by Hill coefficients ≥ 2 (Table 1). Cooperativity was not observed with ΔC-TRBP and binding occurred over a broad concentration range with Hill coefficients ≃ 1 (Table 1).
Table 1.
Summary of Hill coefficients
| Protein variant | dsRNA length
|
||||
|---|---|---|---|---|---|
| siRNA | 40 bp | 104 bp | 300 bp | 650 bp | |
| RDE-4 | 1.2±0.1 | 2.1±0.3 | 3.0±0.2 | 3.0±0.1 | 6.0±1.0 |
| Mut-R1 | NB | 2.2±0.4 | 2.8±0.9 | 2.3±0.4 | ND |
| LCR2C | NB | 1.3±0.3 | 2.9±0.3 | 2.4±0.2 | 1.8±0.2 |
| R2C | ND | 2.2±0.5 | 1.7±0.3 | 1.6±0.2 | 2.3±0.2 |
| R1LR2CN | 1.6±0.4 | 2.4±0.2 | 2.1±0.3 | 3.2±0.7 | 2.3±0.2 |
| R1LR2 | 1.1±0.1 | 2.4±0.1 | 2.0±0.1 | 1.6±0.4 | 2.6±0.2 |
| R1L | NB | NB | NB | NB | NB |
| LCR2 | ND | 1.1±0.2 | 3.1±0.4 | 5.0±0.4 | 3.8±0.4 |
| ΔC-TRBP | 0.8±0.4 | 1.5±0.1 | 1.5±0.1 | 1.1±0.1 | 1.4±0.1 |
| ChimA | 1.6±0.1 | 1.9±0.1 | ND | 1.7±0.1 | ND |
| ChimB | 0.7±0.1 | 0.7±0.1 | 1.5±0.1 | 1.4±0.2 | ND |
| ChimC | 0.9±0.1 | 1.2±0.3 | 1.1±0.1 | 2.5±0.1 | ND |
NB = No binding at protein concentrations up to 5 μM (10 μM for R1L)
ND = Not determined
RDE-4’s cooperativity is mediated by multiple domains
Surprisingly, except in two cases, all RDE-4 variants exhibited cooperativity as evidenced by an increase in affinity with each progression to a longer dsRNA (Fig. 1a, b). While there were differences in magnitude, we observed that cooperative interactions were possible with RDE-4 variants that lacked, or contained mutations in, dsRBM1 (Mut-R1, LCR2C, R2C), the linker (LCR2C, R2C), and the C-terminus (R1LR2CN, R1LR2). The two exceptions were R1L, which could not bind dsRNA at all, and LCR2. LCR2 showed a very low affinity for dsRNA of all lengths, and only showed an increase in affinity when dsRNA length was increased from 40bp to 104bp (Fig. 1a, b). While LCR2 showed minimal cooperativity, its ability to bind dsRNA emphasizes the importance of dsRBM2 in dsRNA binding. Other variants containing only a single dsRBM, such as R1L (Fig. 1), and a variant containing only dsRBM3 that was assayed in our previous study,13 were unable to bind dsRNA.
LCR2C and R2C, which differ only in the C-terminal portion of the linker (LC), show almost identical properties (Fig. 1) suggesting the linker contributes little to cooperativity. Overall our analysis of the RDE-4 variants indicates that if dsRBM2 is present to allow dsRNA binding, either the N-terminal domain containing dsRBM1, or the C-terminus containing the degenerate dsRBM3, can confer cooperativity.
While either the N-terminal or C-terminal domain of RDE-4 was sufficient to confer cooperativity, variants lacking either domain exhibited cooperativity that was less than that of the full-length RDE-4 (Fig. 1, Table 1). For example, consistent with their increased affinity for binding to longer dsRNA (Fig. 1), the RDE-4 variants R1LR2CN and R1LR2 showed positive cooperativity, although less than the wildtype protein (Fig. 2b, Table 1). This suggested the C-terminus of RDE-4 was not solely responsible for cooperativity, but that it makes contributions to this property. This was emphasized by subtle differences in the gel-shift patterns of RDE-4 and the C-terminal deletion variant R1LR2 (Fig. 2a). Construct R1LR2CN, which is missing two-thirds of the C-terminus, resembled full-length RDE-4 in both affinity (Figs. 2b, 1a) and gel-shift patterns (Fig. 2a). However, when 42 more amino acids were deleted from the C-terminus, as in construct R1LR2, the binding affinities appeared near wildtype (Figs. 2b, 1a), but gel-shift patterns showed distinct intermediates as observed with the non-cooperative TRBP (Fig. 2a). Cooperative proteins bind in clusters along the dsRNA lattice.17 Perhaps deleting the C-terminus has altered this mode of contiguous binding, allowing the resolution of discrete intermediates.
dsRBM2 mediates binding to long dsRNA, while both motifs are required for binding siRNA
The observation of gel-shift intermediates with R1LR2 emphasized that the truncations and mutations in our RDE-4 variants affected cooperativity, but it seemed likely that some variants also had differences in the RNA-protein interactions important for intrinsic affinity. The latter should be most apparent in binding to short dsRNA when cooperative interactions are minimal. Indeed, for most variants studied, affinity was near wildtype with long dsRNA, but decreased dramatically with very short dsRNA (Fig. 1b). For example, binding of LcR2C and Mut-R1 exhibited near wildtype affinities for a 104bp, 300bp and 650bp dsRNA (Figs. 1a, 3b). However, with both variants, affinity decreased somewhat with a shorter, 40bp dsRNA, and binding to a 20bp siRNA was undetectable even at concentrations as high as 4 μM (Figs. 1, 3a, c). Another variant, R2C, which lacked dsRBM1 as well as the entire linker, gave similar results (Fig. 1). These data suggested that dsRBM1 is particularly important for binding shorter dsRNA such as siRNA, where cooperative interactions are minimal. However, as mentioned, we found that the RDE-4 variant, R1L, which consists of dsRBM1 and the complete linker region, was unable to bind any dsRNA at concentrations as high as 10 μm (Fig. 1a). Together, our data indicate that interactions with long dsRNA are mediated primarily by dsRBM2, but both dsRBMs are required for binding short dsRNA.
FIGURE 3.
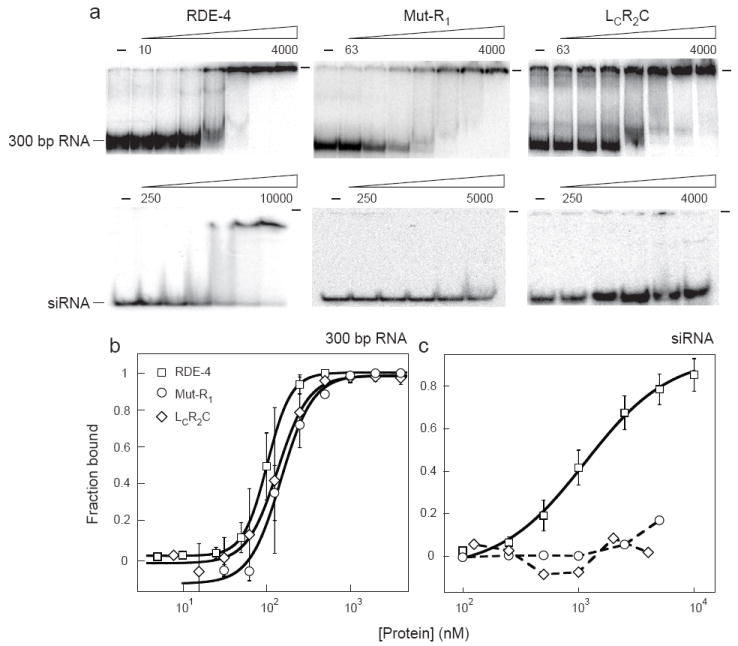
Comparison of RDE-4 binding properties to variants lacking, or with mutations in, dsRBM1. (a) Increasing concentrations of recombinant proteins were added to 0.5-1 fmole 32P-labeled 300 bp dsRNA (top panel) or 20 bp siRNA (bottom panel) and complex formation was analyzed by native gel electrophoresis. Beginning and ending protein concentrations (nM) are noted above gels with adjacent lanes representing a 2 or 4-fold difference in protein concentration. The wells of each gel are marked by a dash at the top right. (b,c) Multiple gel-shift assays as in a were performed to derive binding isotherms for RDE-4 and variants (symbols as indicated) for binding to 300bp dsRNA (b) and siRNA (c). Data points with error bars (standard deviation) represent average values (2 ≤ n ≤ 4) and were fit as in Figure 2b.
We also observed that, similar to constructs lacking dsRBM1, RDE-4 variants missing sequences in the C-terminus (R1LR2CN and R1LR2) exhibited near wildtype affinities for longer dsRNA, but a marked decrease in affinity for siRNA (Fig. 1). However, for these variant proteins the reduction in affinity for siRNA was less severe. While binding to siRNA was undetectable for N-terminal variants (Mut-R1 and LcR2C), C-terminal variants (R1LR2CN and R1LR2) showed an approximate 3-fold reduction as compared to RDE-4 (Fig. 1). We also found that removal of both domains, as in LcR2, compromised affinity for dsRNA of all lengths (Fig. 1), suggesting that one or both of these domains, in addition to conferring the ability to bind short dsRNA, also make contributions to the affinity for long dsRNA. The R1LR2CN, R1LR2 and LCR2 C-terminal deletion constructs differ from RDE-4 in that they exist as stable monomers in solution (data not shown), whereas RDE-4 is a dimer.13 Possibly dimerization, in conjunction with dsRBM1, is required for binding short dsRNAs.
Transposing dsRNA binding properties using protein chimeras
Our analysis of RDE-4 and TRBP showed that these proteins were quite distinct in their properties, possibly reflecting their unique in vivo roles. RDE-4 has a relatively low affinity for dsRNA, but its intrinsic cooperativity allows it to make tight interactions with long dsRNA. By comparison, TRBP has a high affinity for binding dsRNA, thus allowing it to bind tightly to dsRNA of any length, without cooperativity (Fig. 1a, Table 1). We found that almost any part of one protein, when inserted into the other, created a chimeric protein with attributes of both RDE-4 and TRBP. For example, replacing RDE-4’s dsRBM2 with that of TRBP, gave a protein, ChimA, that had the high affinity binding characteristic of TRBP, with an increase in affinity for longer dsRNAs as observed for RDE-4 (Fig. 1a). Similarly, ChimB and ChimC exhibited constant high affinity for dsRNAs ≥ 40bp, but lower affinity for siRNA as observed with RDE-4 (Fig. 1a). Interestingly, ChimB exhibited high affinity binding despite a previous report suggesting dsRBM1 of TRBP binds dsRNA weakly.20 Because we found dsRBM1 of RDE-4 did not bind dsRNA at all (Fig. 1a), it is likely that its replacement, even with a low affinity dsRBM such as dsRBM1 of TRBP, affects overall affinity. Finally, our analysis of the chimeric proteins emphasized that while RDE-4’s N- and C-terminal domains could confer cooperativity to TRBP sequences (ChimA), its linker and dsRBM2 were unable to confer this property to TRBP (ChimB, ChimC).
The linker region of RDE-4 is essential for reconstitution of Dicer activity
C. elegans Dicer has not been amenable to overexpression and purification, but extracts prepared from C. elegans embryos have Dicer activity and can process dsRNA into siRNA.21 Using this system, we showed that extracts prepared from rde-4 mutant C. elegans lack detectable Dicer activity, but activity can be restored by the addition of recombinant RDE-4.13 This “add-back” system is useful for delineating how the various domains of RDE-4 facilitate Dicer’s function in RNAi, and we previously showed that reconstitution is dependent on the C-terminal 100 amino acids of RDE-4, a domain that also mediates RDE-4 homodimerization.13
To further define which domains of RDE-4 are required for siRNA production, we tested the ability of our new protein variants to reconstitute Dicer activity in rde-4(ne299) extracts. A 650 bp dsRNA, internally labeled with 32P, was incubated with extract +/- recombinant protein, and siRNA production was monitored by resolving reaction products on denaturing gels. The various protein constructs exhibited slightly different affinities for dsRNA (Fig. 1a), so we tested their ability to reconstitute Dicer activity over a range of concentrations (Fig. 4a, 100 nM; 4b, 500 nM; data not shown). Data from multiple analyses were quantified by determining the fraction of total RNA that appeared as siRNA, and reported relative to the reconstitution observed with full-length RDE-4 at 100 nM (Fig. 4e, f).
FIGURE 4.
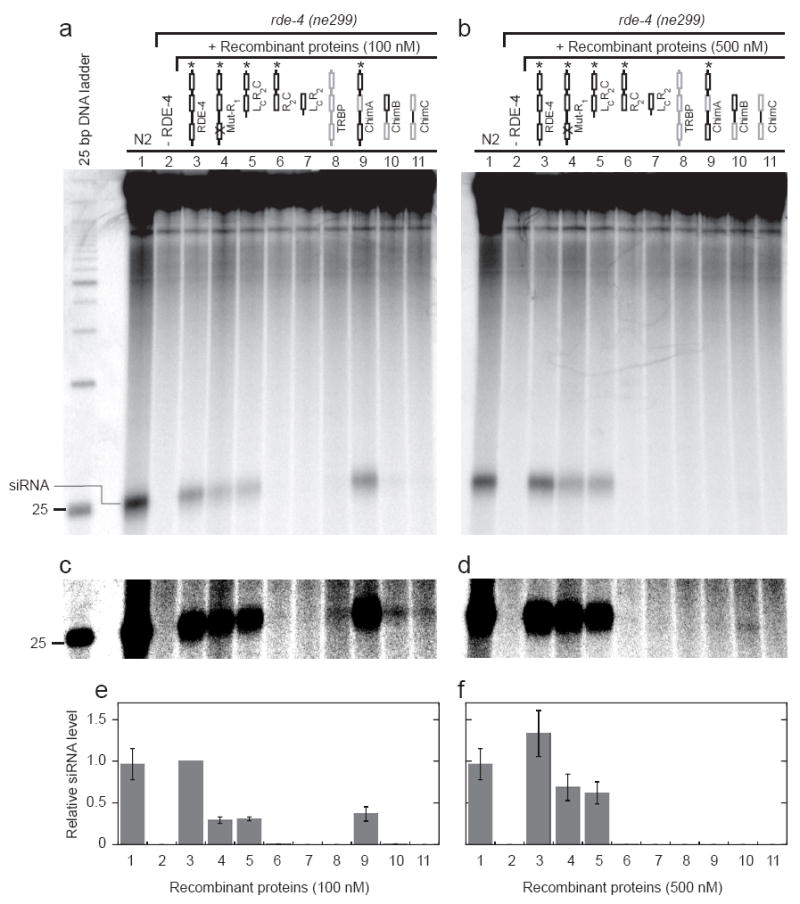
In vitro reconstitution of siRNA production using C. elegans extracts and recombinant RDE-4 variants. (a) Extracts prepared from wildtype (N2) or rde-4 (ne299) embryos were incubated for 1 h ± 100 nM recombinant proteins as indicated. The autoradiogram shows reaction products separated by electrophoresis on a 15% denaturing polyacrylamide gel. As indicated, 23 nt siRNAs migrate slightly slower than the 25 nt DNA marker. (b) As in a, but with the addition of 500 nM recombinant proteins. (c,d) Same as a and b, but with increased contrast to illustrate small amount of siRNA production for ΔC-TRBP, ChimB and ChimC. (e,f) Multiple reconstitution assays as in a and b were quantified; bar height represents siRNA levels upon reconstitution with the various recombinant proteins (1-11, as in a-d) at 100 (e) and 500 (f) nM. From a western blot analysis using an antibody to RDE-4, we estimate N2 reactions contained 25-50 nM RDE-4, consistent with the complete reconstitution obtained with addition of 100 nM recombinant RDE-4. Data represent average values from 3-6 independent experiments. siRNA levels are reported relative to full-length RDE-4 at 100 nM, which was set to 1.
As shown in Figure 4a and b, siRNAs were readily detected after incubation of 650 bp dsRNA in extracts prepared from wildtype worms (N2, lane 1), but were absent after incubation in extracts prepared from rde-4(ne299) worms (lane 2). Addition of 100 nM recombinant, full-length RDE-4 to rde-4(ne299) extracts rescued cleavage of 650 bp dsRNA to siRNA (Fig. 4a, e, lane 3), and a further increase in siRNA was observed at 500 nM (Fig. 4b, f, lane 3), suggesting 100 nM was not saturating. Similarly, the dsRBM1 mutant, Mut-R1, and the N-terminal deletion variant, LCR2C, both of which were unable to stably bind siRNA (Fig. 3a, c), retained the ability to reconstitute Dicer activity (Fig. 4a, b lanes 4 and 5). However, both proteins showed a marked decrease in siRNA levels when compared to full-length RDE-4 (Fig. 4a, b, e, f, lanes 3, 4 and 5). These data indicate that dsRBM1 of RDE-4 is not essential for Dicer cleavage of dsRNA in extracts. However, the lower levels of reconstitution suggest that dsRBM1 may either facilitate this process or aid in siRNA stabilization.
In contrast to the reconstitution observed with LCR2C, variant R2C was unable to support siRNA production (Fig. 4a-f, lane 6), suggesting that the linker region contained in LCR2C is critical for Dicer activity. Importantly, both N-terminal variants LCR2C and R2C bound 650 bp dsRNA with near wildtype affinities (Fig. 1) and eluted as stable dimers from a Superdex 200 gel filtration column (data not shown). These data suggest that the linker region deleted in R2C is required for siRNA production in a manner independent of dsRNA binding and dimerization, perhaps by mediating protein-protein interactions with Dicer. However, assays with the deletion variant, LcR2 (Fig. 4a-f, lane 7), combined with previous studies of the C-terminal deletion variants, R1LR2CN and R1LR2,13 indicate that the linker region is not sufficient for reconstitution in the absence of the C-terminus. Taken together, our data suggest RDE-4 has three regions that are essential for facilitating cleavage of dsRNA by Dicer: its dsRBM2 that allows binding to dsRNA, its linker region that may promote protein-protein interactions with Dicer, and the C-terminus that is required for homodimerization of RDE-4.
We next tested for C. elegans Dicer reconstitution using human TRBP and our RDE-4/TRBP chimera constructs. It was previously shown that the C-terminus of TRBP mediates interactions with human Dicer.3 Therefore, in our reconstitution assays we utilized full-length TRBP, which in our hands formed soluble aggregates but retained dsRNA binding ability (data not shown). While TRBP was unable to restore wildtype levels of Dicer activity (Fig. 4a-f, lane 8), somewhat surprisingly, at 100nM, the human dsRBP was able to reconstitute minimal activity in C. elegans rde-4(ne299) extracts (Fig. 4c, lane 8). In contrast, the ΔC-TRBP construct, similar to the RDE-4 constructs lacking the C-terminus, showed no reconstitution (data not shown). Surprisingly, ChimA, which contains the important C-terminal and linker domains of RDE-4, but replaces dsRBM2 with the second dsRBM of TRBP, gave moderate restoration of Dicer activity at 100 nM (Fig. 4a, e, lane 9). Lastly, ChimB and ChimC showed low but detectable reconstitution activity at 100 nM (Fig. 4c, lanes 10-11), suggesting the high affinity dsRBMs of TRBP, in combination with the RDE-4 linker, marginally overcome the deficiency seen in the absence of the C-terminus.
At 500 nM, however, the activities observed for TRBP and the RDE-4/TRBP chimeras were either undetectable (TRBP, ChimA, ChimC, Fig. 4b, d, f, lanes 8, 9, 11) or greatly diminished (ChimB, Fig. 4b, d, f, lane 10). This result is reminiscent of observations made in our previous study,13 where the addition of high concentrations of full-length RDE-4 was observed to inhibit reconstitution of rde-4(ne337) extracts, as well as siRNA production in a wildtype (N2) extract. Similar to our interpretation in those studies, high concentrations of TRBP and the RDE-4/TRBP chimeras may titrate a limiting factor, and this may occur at lower concentrations of these proteins because of their higher binding affinities compared to full-length RDE-4.
DISCUSSION
A general model for how dsRBPs discriminate dsRNA length
Early theoretical work put forth that cooperative nucleic acid-binding proteins exhibit observed affinities (Kobs) that are products of the intrinsic affinity of an isolated protein-nucleic acid interaction (Kint) and the cooperativity parameter, ω (Kobs = Kintω).17 The cooperativity parameter, ω, is a unitless factor that specifies the relative affinity of an additional ligand for a contiguous versus an isolated binding site.17 Therefore, high ω values result in protein cluster formation along a nucleic acid lattice, presumably due to stabilizing interactions between adjacently bound proteins.17 RDE-4 exhibits an approximate 67-fold increase in affinity in going from 20 bp (siRNA) to 650 bp dsRNA (Fig. 1). If siRNA is the minimal substrate for RDE-4, the affinity observed for siRNA would represent the Kint for RDE-4, and using the relationship of Kobs = Kintω, 67 would approximate the cooperativity parameter, ω. This value suggests RDE-4 is moderately cooperative, for example, by comparison to the highly cooperative ssDNA-binding protein, T4 gene 32, which has an ω value of ~ 1000.22 However, 67 is likely an underrepresentation of the true value of ω for RDE-4, since “end effects” from the finite-length lattices used in our studies lead to a loss of cooperative interactions.16,23 More detailed analyses are required to dissect the exact values of Kint and ω for RDE-4.
Cooperative binding represents an important biophysical property of RDE-4 that is directly related to its biological function – binding to long dsRNA substrates but dissociating from siRNA products. In contrast, the primary function of the human dsRBP, TRBP, is downstream of siRNA production,2,3 eliminating the need for preferential binding to long dsRNA. Indeed, we found that TRBP bound all dsRNA lengths with similar high affinity (Fig. 1a), as predicted for a non-cooperative protein where ω = 1. Thus, the different binding properties of RDE-4 and TRBP likely reflect their in vivo functions. Here it is important to point out that the exact role of TRBP during RNAi remains a bit unclear. A complex of TRBP, Dicer and Ago2, known as the RISC-loading complex (RLC), loads siRNA into the mRNA cleaving enzyme, Ago2.12,24 TRBP, which recruits Dicer to Ago2, confers siRNA-binding ability to the RLC.2 Together with data demonstrating TRBP-independent dsRNA cleavage by Dicer,2,4,9,10,25 these data support a role for TRBP downstream of siRNA production. However, two recent studies report modest stimulation of in vitro Dicer cleavage activity upon the addition of TRBP.11,25 Importantly, the data suggest this stimulation occurs by a different mechanism compared to that by which RDE-4 facilitates Dicer function. The stimulatory effect does not appear related to binding of the dsRNA substrate, but rather, due to stabilization of an active conformation of Dicer.25
Cooperative dsRNA binding by RDE-4: an act of multiple domains
At least two factors contribute to binding of dsRNA by RDE-4, dsRNA-protein interactions and cooperative protein-protein interactions. When the latter are present, an increase in affinity is observed as dsRNA length increases, since multiple RDE-4 molecules form long clusters along the dsRNA (Fig. 5). When cooperative interactions are absent, as with TRBP (Fig. 5), or unavailable, as with RDE-4 binding a short siRNA substrate, the observed affinity is based solely on dsRNA-protein interactions.
In hopes of defining the domain responsible for cooperative protein-protein interactions in RDE-4, we assayed deletion variants of RDE-4 for dsRNA binding ability (Fig. 1a). Similar approaches successfully defined the cooperativity domain of T4 Gene 32 protein, where deletion of the first 21 amino acids abrogated preferential binding to long single-stranded DNA.26 Surprisingly, most of our RDE-4 variants bound longer dsRNA with higher affinity (Fig. 1), suggesting that cooperativity is not a characteristic conferred by a single domain of RDE-4, but the combined effect of multiple domains. However, in general, RDE-4 variants showed a dramatic decrease in affinity for short dsRNAs, with dsRBM1 mutants showing no detectable siRNA binding (Figs. 1, 3). Interestingly, similar findings were reported for dsRBM1 mutants of the Drosophilia dsRBP, R2D2,6 suggesting that the requirement for multiple dsRBMs in binding short dsRNAs is an evolutionarily conserved phenomenon.
Deleting either the N- or C-terminus of RDE-4 not only decreased siRNA affinity, but also affected Hill coefficients (Table 1), and in the case of R1LR2, gel shift patterns (Fig. 2a). Possibly, disrupting either of these domains interrupts potential cooperative protein-protein interactions, thus lowering the probability of saturating the dsRNA lattice. Alternatively, our data may indicate that cooperativity arises not from protein-protein interactions, but instead from the production of highly favorable binding sites adjacent to bound proteins, possibly due to slight perturbations of the dsRNA structure. If such perturbations made it more favorable for contiguously bound proteins to propagate, compared to the initiation of a new, isolated binding event, binding would also be cooperative.27 In this scenario RDE-4’s cooperativity would arise solely from dsRNA binding, thus explaining our inability to completely disrupt cooperativity with the N–terminal and C-terminal truncations. However, the inability of variant LCR2 to bind cooperatively suggests that at least two dsRBMs (i.e., dsRBM2 + dsRBM1 or dsRBM2 + dsRBM3) are required for the putative dsRNA structure perturbations. Future studies will be required to differentiate between these two possibilities.
Domain requirements in RDE-4 for facilitating Dicer activity
C. elegans has only one Dicer that must process both pre-miRNAs and dsRNAs in the miRNA and RNAi pathways, respectively.28,29 To date, it is not known whether or how C. elegans Dicer discriminates between these two types of substrates. Together with previously published results, our in vitro cleavage data indicate that RDE-4 is essential for processing long dsRNAs (Fig. 4),7,13,14 but not miRNAs.30,31 We predict that the interaction between RDE-4 and Dicer, combined with RDE-4’s cooperativity, provide substrate specificity to Dicer by directing it to long dsRNAs. These two attributes of RDE-4 are critical for RNAi in C. elegans.
Our data indicate that the N-terminus, including dsRBM1, of RDE-4 is dispensable for reconstituting siRNA production (Fig. 4, lanes 4 and 5). Consistent with our results, RDE-4 variants lacking dsRBM1 retain interactions with Dicer as monitored by differential cytolocalization assays (DCLA),32 and are able to rescue RNAi in rde-4 mutant worms to near wildtype levels (D. Blanchard and A. Fire, personal communication). Why RDE-4 contains two dsRBMs when dsRBM1 appears to be dispensable for promoting Dicer cleavage activity is unclear. However, recent reports demonstrate that the structure of the double-stranded siRNA or miRNA products of Dicer influences the selection of the appropriate downstream Argonaute protein, and suggest that RDE-4 is a potential mediator of this selection process.33-36 We find dsRBM1 of RDE-4 is required for siRNA binding (Fig. 3), and it would be interesting to see if this motif functions in downstream steps of RNAi.
Our reconstitution assays also demonstrated that the linker region of RDE-4 is critical for siRNA production (Fig. 4, lane 6). Since our experiments indicate the linker does not affect dsRNA binding or RDE-4 dimerization, we propose that it mediates interactions with Dicer. This idea is supported by results of DCLA experiments, which indicate that both the linker and dsRBM2 are required for stable interactions with Dicer (D. Blanchard and A. Fire, personal communication). We found the protein construct ChimA, which replaces dsRBM2 of RDE-4 with the second dsRBM of TRBP, supports moderate reconstitution of siRNA production (Fig. 4a, e lane 9). Since it seems unlikely that dsRBM2 of human TRBP is able to interact directly with C. elegans Dicer, we propose the requirement of dsRBM2 is indirect, and mediated by its ability to bind dsRNA. However, we cannot rule out that the conserved amino acid residues between the dsRBMs of TRBP and RDE-4 allow for direct ChimA/Dicer interactions.
Most surprisingly, we found that in the absence of RDE-4, TRBP facilitated a low level of cleavage activity by C. elegans Dicer (Fig. 4c, lane 8). How the human protein is able to facilitate C. elegans Dicer activity in our studies is unknown. Perhaps TRBP’s ability to promote an active Dicer conformation is recapitulated in our extracts.25 Similar low-level reconstitution was seen with ChimB and ChimC (Fig. 4c, lanes 10-11). Based on the published data,37,38 we predict that these proteins are dimers in solution. Therefore, the low level of Dicer activity supported by ChimB and ChimC suggests that the presence of the linker region of RDE-4, high affinity for dsRNA, and possibly homodimerization, enables minimal Dicer activity in the absence of wildtype RDE-4.
The C-terminus of RDE-4 is important for forming homodimers in solution and for facilitating siRNA production by Dicer in extracts.13 Deleting the C-terminus of RDE-4 also abolishes RNAi in vivo, but not Dicer interactions as monitored by DCLA experiments (D. Blanchard and A. Fire, personal communication). These data indicate the C-terminus of RDE-4 plays a key role in RNAi, independent of direct interactions with Dicer. Possibly, the C-terminus mediates a dimerization event that is necessary for recruiting Dicer in an active form. Alternatively, the C-terminus of RDE-4 may facilitate Dicer activity independent of dimerization, as has been demonstrated for other dsRBP partners. For example, the C-terminus of the human dsRBP, PACT, is not required for high-affinity associations with protein kinase R (PKR), but is required for PKR activation.39,40 Similarly, a recent study25 suggests auto-inhibition of human Dicer is alleviated by interactions with the C-terminus of TRBP.3,4 Interestingly, both TRBP and PACT form homodimers in solution,41,42 yet recent evidence indicates that TRBP exists as a monomer in the active Dicer complex.12 Taken together, the data for PACT and TRBP set a precedent for stimulatory roles of dsRBP C-terminal domains and raise the possibility that the C-terminus of RDE-4 acts in a similar manner.
MATERIALS AND METHODS
Construction, expression and purification of RDE-4 variants, TRBP and chimeras
Truncated RDE-4 constructs were PCR amplified from RDE-4-YEpTOP2GAL1 which encoded wildtype RDE-4.13 The sequences of primers (5’ to 3’) used to construct these variants are listed below.
RDE-4_BamH1_For: CGT CAA GGA GAA AAA ACC CCG GAT CCG TAA CC
RDE-4_169_Xho1_Rev: GTC ATT ACT CGA GTC AAT TCT CGG TTG GCG AAA TAC CAG GTG G
RDE-4_Xho1_Rev: GTC ATT ACT CGA GTC AAT CCG TGA AAT C
RDE-4_139_LR: GCA TCT GAA GTT GAT CGC TGA AAA TAC AGG TTT TCG G
RDE-4_139_RF: CCG AAA ACC TGT ATT TTC AGC GAT CAA CTT CAG ATG C
RDE-4_163_LR: CCC AAT TCT CGG TTG GCG AAA TAC CCT GAA AAT ACA GGT TTT CGG
RDE-4_163_RF: CCG AAA ACC TGT ATT TTC AGG GTA TTT CGC CAA CCG AGA ATT GGG
R4_Rev_Xho1: GTC ATT ACT CGA GTC ACA TAT CAT ATG ATT CCA GAG ATT CGA TAC CG
R1L was constructed using RDE-4_BamH1_For and RDE-4_169_Xho1_Rev primers. For LCR2C and R2C, overlapping PCR technique (“PCR sewing”) was used. In conjunction with two terminal primers, RDE-4_BamH1_for and RDE-4_Xho1_Rev, the following overlapping primers were used to make the truncated constructs: LCR2C, RDE-4_139_LR and RDE-4_139_RF; R2C, RDE-4_163_LR and RDE-4_163_RF. The construct for LCR2 was made using LCR2C cDNA as the template and RDE-4_BamH1_For and R4_Rev_Xho1 as the forward and reverse primers, respectively. Mut-R1 was made using mutant PCR oligos that replace two lysines at positions 89 and 90 by two alanines. Sequences of all recombinant RDE-4 constructs were confirmed by DNA sequencing. All proteins were expressed in the S. cerevisiae BCY123 strain and purified as described.13 For R2C, the N-terminal polyhistidine tag was not removed as this reduced its stability.
Human cDNA was synthesized by reverse transcription of polyA+ RNA using random primers. TRBP sequence was PCR amplified from the cDNA using forward (5’-GGG CCC TCA TGA GTG AAG AGG AGC AAG GCT CC - 3’) and reverse (5’- GGG GGG AAG CTT TCA CTT GCT GCC TGC CAT GAT CTT - 3’) primers containing 5’ BsphI and 3’ HindIII sites, respectively, allowing ligation into a modified pMAL vector (New England Biolabs) that inserted a TEV protease cleavage site followed by an Nco1 (BsphI compatible) restriction site downstream of the MBP ORF. The sequence of the TRBP construct was confirmed by DNA sequencing. BL21-codon plus (DE3) cells were transformed with TRBP plasmid and plated on LB media containing 50 μg/ml ampicillin and 34 μg/ml chloramphenicol. 5 ml of LB containing antibiotics were inoculated with a single transformant and grown overnight at 37 °C. Overnight cultures were diluted into 1 L of LB and grown approximately 3 h until the OD reached 0.5. Expression of TRBP was induced by adding IPTG to a final concentration of 0.3 mM. Induction was for 4 hours. Cells were pelleted by centrifugation and stored at -80 °C.
For purification, cells were resuspended in Lysis buffer (20 mM Tris (pH 8.0), 200 mM NaCl, 1 mM EDTA, 1 mM 2-Mercaptoethanol and 5 % v/v Glycerol) and lysed using a French Press and sonication, and then centrifuged at 30,000 g for 30 min. Supernatant was added to 10 ml of amylose resin and allowed to bind at 4 °C for 1 h. The resin was washed by 3×10 ml of Lysis buffer and bound protein was eluted using Buffer B (20 mM Tris (pH 8.0), 100 mM NaCl, 1 mM EDTA, 1 mM 2-Mercaptoethanol and 5 % v/v Glycerol) and 20 mM Maltose. Eluted protein was loaded onto a 5 ml Hi-Trap Heparin column (Pharmacia) equilibrated with Buffer B. The column was washed with 50 ml of Buffer B and developed with a 25 ml gradient of 100 mM − 1 M NaCl. Purified protein was dialyzed into storage buffer (30 mM Tris (pH 8.0), 100 mM NaCl, 1 mM 2-Mercaptoethanol and 20 % v/v Glycerol) and stored at -80 °C. Soluble ΔC-TRBP was separated from full-length TRBP aggregates by size exclusion chromatography using a Superdex 200 column.
To make ChimA, B and C recombinant constructs, the appropriate DNA fragments were amplified from plasmids containing full-length RDE-4 or TRBP sequences. DNA fragments were ligated using the overlapping PCR method. The amino acid sequence compositions of the chimeras are as follows:
ChimA: RDE-4, 1-168; TRBP, 159-245; RDE-4, 245-385
ChimB: TRBP, 1-95; RDE-4, 106-244
ChimC: TRBP, 1-95; RDE-4, 106-168; TRBP, 159-245
All cDNAs were cloned into YEpTOP2GAL1 vector using BamH1 and Xho1 restriction endonuclease sites. Expression and purification of the chimeric proteins were performed as described for the RDE-4 variants.
RNA preparation
Internally or end 32P-labeled 650, 104, 40 bp dsRNAs were prepared as described.13 Internally 32P-labeled 300 bp dsRNA was transcribed with T7 polymerase from a template encoding sequence from the 6th exon of the C. elegans gene, unc-22. siRNA was generated by annealing chemically synthesized end 32P-labeled 22 nt ssRNAs, resulting in a 20 bp dsRNA with 2 nt 3’ overhang at both ends.13
Gel mobility shift assay
Gel mobility shift assays were performed as described.13 Either internally or end 32P-labeled dsRNAs of 20, 40, 104, 300 or 650 bp were used. Reactions were performed in 10 or 20 μl final vol and mobility shifts were monitored on 5% (650 bp and 300 bp dsRNAs) and 8% (104 bp, 40 bp and 20 bp dsRNAs) native gels at 4 °C. Dissociation constants were calculated using the Hill formalism. Briefly, radioactivity corresponding to dsRNAfree and dsRNAtotal was detected using a Typhoon PhosphorImager and quantified using ImageQuant software (GE Healthcare). All RNAs of slower mobility than dsRNAfree were considered as bound. To obtain Kd values, fraction of RNA bound vs. concentration of recombinant protein (calculated using monomer molecular weight) was plotted using KaleidaGraph software and fit to the following equation:
where, a = amplitude of the binding curve (~1), b = base line (~0), n = Hill coefficient.
In vitro Dicer activity and reconstitution assays
In vitro Dicer activity and reconstitution assays were performed essentially as described,13 with the following modifications. Extract lysis buffer contained 30 mM HEPES (pH 7.4), 100 mM KOAc, 2 mM MgOAc, 5 mM DTT and 20% glycerol. Dicer activity assays consisted of 20 μl reactions containing 50 μg of extract, 10 fmol of internally 32P-labeled 650-bp dsRNA and a final concentration of 100 or 500 nM of recombinant proteins as specified. The cleavage reactions were performed at 20 °C for 1 h, RNAs were Phenol/CHCl3 extracted, resolved on 15% denaturing polyacrylamide gels, visualized using a Typhoon PhosphorImager and quantified using ImageQuant software. siRNA production was quantified for each recombinant protein by dividing the radioactivity corresponding to siRNA by the total RNA (total radioactivity in entire lane). Values were normalized to the value obtained during reconstitution with full-length RDE-4 (100 nM), which was set to 1 (e.g., Fig. 4e, f). The presence of approximately equimolar amounts of Dicer in N2 and rde-4(ne299) extracts was confirmed by western-blots using an antibody against C. elegans Dicer (data not shown).
Acknowledgments
We thank members of the Bass lab for helpful discussions and A. Krauchuk for technical assistance. We also thank Dr. Andrew Fire for communicating unpublished results and Dr. Craig Mello for antibodies to RDE-4 and C. elegans Dicer. This work was supported by funds to B.L.B. from the National Institutes of Health (GM067106). B.L.B. is a Howard Hughes Medical Institute Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jaskiewicz L, Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- 2.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhatta R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haase AD, Jaskiewicz L, Zhang H, Lainé S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee Y, Hur I, Park S-Y, Kim Y-K, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Q, Rand TA, Kalidas S, Du F, Kim H-E, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Jiang F, Kalidas S, Smith D, Liu Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA. 2006;12:1514–1520. doi: 10.1261/rna.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-Box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 8.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80S complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 9.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Rådmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kok KH, Ng M-HJ, Ching Y-P, Jin D-Y. Human TRBP and PACT directly interact with each other and associate with Dicer to facilitate the production of small interfering RNA. J Biol Chem. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 12.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci U S A. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker GS, Eckert DM, Bass BL. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA. 2006;12:807–818. doi: 10.1261/rna.2338706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parrish S, Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- 15.Saunders LR, Barber GN. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 16.Kowalczykowski SC, Paul LS, Lonberg N, Newport JW, McSwiggen JA, von Hippel PH. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986;25:1226–1240. doi: 10.1021/bi00354a006. [DOI] [PubMed] [Google Scholar]
- 17.McGhee JD, von Hippel PH. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 18.Gatignol A, Buckler C, Jeang KT. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and Drosophila staufen. Mol Cell Biol. 1993;13:2193–2202. doi: 10.1128/mcb.13.4.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta V, Huang X, Patel RC. The carboxy-terminal, M3 motifs of PACT and TRBP have opposite effects on PKR activity. Virology. 2003;315:283–291. doi: 10.1016/s0042-6822(03)00589-0. [DOI] [PubMed] [Google Scholar]
- 20.Daviet L, Erard M, Dorin D, Duarte M, Vaquero C, Gatignol A. Analysis of a binding difference between the two dsRNA-binding domains in TRBP reveals the modular function of a KR-helix motif. Eur J Biochem. 2000;267:2419–2431. doi: 10.1046/j.1432-1327.2000.01256.x. [DOI] [PubMed] [Google Scholar]
- 21.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalczykowski SC, Lonberg N, Newport JW, Paul LS, von Hippel PH. On the thermodynamics and kinetics of the cooperative binding of bacteriophage T4-coded gene 32 (helix destabilizing) protein to nucleic acid lattices. Biophys J. 1980;32:403–418. doi: 10.1016/S0006-3495(80)84964-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Epstein IR. Cooperative and non-cooperative binding of large ligands to a finite one-dimensional lattice. A model for ligand-oligonucleotide interactions. Biophys Chem. 1978;8:327–339. doi: 10.1016/0301-4622(78)80015-5. [DOI] [PubMed] [Google Scholar]
- 24.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Auto-inhibition of human Dicer by its internal helicase domain. J Mol Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spicer EK, Williams KR, Konigsberg WH. T4 gene 32 protein trypsin-generated fragments. Fluorescence measurement of DNA-binding parameters. J Biol Chem. 1979;254:6433–6436. [PubMed] [Google Scholar]
- 27.Kelly RC, Jensen DE, v Hippel PH. DNA “melting” proteins. IV. Fluorescence measurements of binding parameters for bacteriophage T4 gene 32-protein to mono-, oligo-, and polynucleotides. J Biol Chem. 1976;251:7240–7250. [PubMed] [Google Scholar]
- 28.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 29.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 Gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 31.Welker NC, Habig JW, Bass BL. Genes misregulated in C. elegans deficient in Dicer, RDE-4, or RDE-1 are enriched for innate immunity genes. RNA. 2007;13:1090–1107. doi: 10.1261/rna.542107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanchard D, Hutter H, Fleenor J, Fire A. A differential cytolocalization assay for analysis of macromolecular assemblies in the eukaryotic cytoplasm. Mol Cell Proteomics. 2006;5:2175–2184. doi: 10.1074/mcp.T600025-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steiner FA, Hoogstrate SW, Okihara KL, Thijssen KL, Ketting RF, Plasterk RHA, Sijen T. Structural features of small RNA precursors determine Argonaute loading in Caenorhabditis elegans. Nat Struct Mol Biol. 2007;14:927–933. doi: 10.1038/nsmb1308. [DOI] [PubMed] [Google Scholar]
- 36.Jannot G, Boisvert M-EL, Banville IH, Simard MJ. Two molecular features contribute to the Argonaute specificity for the microRNA and RNAi pathways in C. elegans. RNA. 2008;14:829–835. doi: 10.1261/rna.901908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daher A, Longuet M, Dorin D, Bois F, Segeral E, Bannwarth S, Battisti PL, Purcell DF, Benarous R, Vaquero C, Meurs EF, Gatignol A. Two dimerization domains in the trans-activation response RNA-binding protein (TRBP) individually reverse the protein kinase R inhibition of HIV-1 long terminal repeat expression. J Biol Chem. 2001;276:33899–33905. doi: 10.1074/jbc.M103584200. [DOI] [PubMed] [Google Scholar]
- 38.Laraki G, Clerzius G, Daher A, Melendez-Peña C, Daniels S, Gatignol A. Interactions between the double-stranded RNA-binding proteins TRBP and PACT define the Medipal domain that mediates protein-protein interactions. RNA Biol. 2008;5:92–103. doi: 10.4161/rna.5.2.6069. [DOI] [PubMed] [Google Scholar]
- 39.Patel RC, Sen GC. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters GA, Hartmann R, Qin J, Sen GC. Modular structure of PACT: distinct domains for binding and activating PKR. Mol Cell Biol. 2001;21:1908–1920. doi: 10.1128/MCB.21.6.1908-1920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cosentino GP, Venkatesan S, Serluca FC, Green SR, Mathews MB, Sonenberg N. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc Natl Acad Sci U S A. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hitti EG, Sallacz NB, Schoft VK, Jantsch MF. Oligomerization activity of a double-stranded RNA-binding domain. FEBS lett. 2004;574:25–30. doi: 10.1016/j.febslet.2004.07.080. [DOI] [PubMed] [Google Scholar]


