Abstract
Insertional mutagenesis by retroviral vectors has emerged as a serious impediment to the widespread application of hematopoietic stem cell gene transfer for the treatment of hematologic diseases. Here we report the development of a 77-bp element, FII/BEAD-A (FB), which contains the minimal enhancer blocking components of the chicken β-globin 5′HS4 insulator and a homologous region from the human T-cell receptor α/δ BEAD-1 insulator. With a new flow cytometry-based assay, we show that the FB element is as effective in enhancer blocking activity as the prototypical 1.2-kb 5′HS4 insulator fragment. When incorporated into the residual U3 region of the 3′ long terminal repeat (LTR) of a self-inactivating (SIN) gammaretroviral vector, the FB element was stably transferred to the 5′ LTR during reverse transcription, flanking the integrated transgene expression cassette. Notably, using a recently established in vitro insertional mutagenesis assay involving primary murine hematopoietic cells, we found that SIN gammaretroviral vectors as well as SIN lentiviral vectors containing the FB element exhibited greatly reduced transforming potential—to background levels under the experimental conditions used—compared to their unshielded counterparts. These results suggest that the FB element-mediated enhancer blocking modification is a promising approach to dramatically improve the safety of retroviral vectors for therapeutic gene transfer.
Keywords: Self-inactivating retroviral vectors, Insertional mutagenesis, Genotoxicity, Enhancer blocking elements, CCCTC-binding factor (CTCF)
INTRODUCTION
Murine leukemia virus-based gammaretroviral vectors have been widely used in hematopoietic stem cell gene therapy clinical trials. A drawback of gammaretroviral vectors is the risk of insertional activation of neighboring proto-oncogenes which could consequently lead to clonal dominance or malignant transformation of the hematopoietic target cells [1, 2]. The risk of insertional mutagenesis with replication-defective gammaretroviral vectors first became evident in a gene therapy trial for the X-linked form of severe combined immunodeficiency when several of the successfully treated patients developed leukemia as a result of insertional activation of the LMO2 proto-oncogene [3]. Similarly, hematopoietic clonal dominance was documented in a gene therapy trial for chronic granulomatous disease, where the expansion of gene-modified cells in two patients was associated with insertions into, or near one or more of, the MDS1-EVI1, PRDM16 or SETBP1 genes [4]. Recent preclinical studies with hematopoietic cells have provided additional evidence of the potentially dangerous consequences of gammaretroviral vector integration events [5–12], further underscoring the necessity of improved vector design for safe therapeutic gene transfer.
Since most safety concerns associated with insertional mutagenesis have to do with retroviral-induced transcriptional activation of genes, obtaining a low copy number of vector integrants per cell is clearly desirable [6]. Besides reducing the frequency of multiple integration events per cell, several safety modifications of the vector backbone can be envisioned [1, 2, 13, 14]. Murine leukemia virus transcriptional activation of cellular gene expression results from the strong enhancer-promoter sequences in the long terminal repeats (LTRs), with cellular gene activation being observed even over very large distances (e.g., as far as 548 kb from the integration site) [5]. Deletion of the two copies of the LTR enhancer-promoter sequences in the self-inactivating (SIN) gammaretroviral vector format [15, 16] – which utilizes an internal enhancer-promoter to drive transgene expression – might be expected to reduce insertional side effects. Although there is experimental data to support this prediction [8, 17], a recent study nonetheless reported induction of hematopoietic clonal imbalance and leukemia by a SIN gammaretroviral vector [12]. Therefore, additional strategies, such as inclusion of enhancer blocking elements to shield the genome from the internal enhancer of a SIN vector [18–20], are needed to further minimize genotoxic impact. Moreover, transcription termination is especially leaky in the SIN design [21]. In addition to promoting mRNA nuclear export, the eukaryotic splicing process results in enhanced 3′ end formation and polyadenylation [22]. Therefore, engineering introns within the vector may reduce RNA 3′ readthrough [23–25]. An alternative or complementary approach is the inclusion of cis-acting RNA transport elements such as the woodchuck hepatitis virus posttranscriptional regulatory element, which also augments RNA 3′ end processing and polyadenylation [23, 26]. In this regard, a safety-modified version of this posttranscriptional regulatory element devoid of potentially oncogenic promoter sequences and open reading frames that were present in the original version has recently been described [27].
Taking all of the factors discussed above into account, we evaluated the impact of retroviral vector design on combinatorial insertional mutagenesis after high-copy gene transfer. Using a recently established cell culture assay that detects insertional transformation by serial replating of primary hematopoietic cells [8, 17], we show that SIN gammaretroviral vectors as well as SIN lentiviral vectors harboring strong internal enhancer-promoters exhibit transforming activity and that a significant reduction in genotoxic potential can be obtained by flanking the vector backbones with a novel enhancer blocking element.
MATERIALS AND METHODS
Plasmid Construction
The reporter plasmid pRL-TKGFP-CMVE which was used to determine the enhancer blocking activity of various test fragments was constructed by modifying the pRL-TK plasmid (Promega, Madison, WI) as the following. The Rluc reporter gene of pRL-TK plasmid was removed by NheI-XbaI digestion and replaced with a green fluorescent protein (GFP) gene by blunt-end ligation, allowing GFP expression from the herpes simplex virus thymidine kinase (TK) promoter. As an internal control, a similar expression cassette utilizing the TK promoter driving the DsRed.T4 gene was generated and blunt-end ligated into the BamHI site of the pRL-TKGFP plasmid in the opposite orientation. Next, two copies of the full-length 1.2-kb 5′HS4 insulator were removed from plasmid pJC13-1, which contains four copies of the element [28, 29](provided by G. Felsenfeld, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA) as a 2.4-kb EcoRI-BamHI fragment and subcloned upstream of the TK-GFP cassette at the BglII site by blunt-end ligation. The human cytomegalovirus (CMV) immediate early region enhancer was removed from the CA promoter of the SIN gammaretroviral vector MSinSB [25] as a 400-bp EcoNI-NcoI fragment and inserted at a unique ClaI site between the TK-GFP and TK-DsRed.T4 expression cassettes by blunt-end ligation. This manipulation recreated a unique ClaI site between the TK-GFP expression cassette and the CMV enhancer for insertion of enhancer blocking elements. To evaluate the enhancer blocking properties of the composite FB element, FII and BEAD-A sequences were generated by hybridization and ligation of synthetic oligonucleotides (FII upper strand: 5′-GGCCGCGAATTCTGAAAGACCCCACCTGTAGGTTTGGCAAGCCCAGGGATGTACGTCCCTAACCCGCTAGGGGGCAGCA-A-3′, FII lower strand: 5′-CTAGTTGCTGCCCCCTAGCGGGTTAGGGACGTACATCCCTGGGCTTCCCAAACCTACAGGTGGGGTCTTTCAGAATTCGC-3′), and (BEAD-A upper strand: 5′-CTAGTCCCAGGCCTGCACTGCCGCCTGCCGGCAGGGGTCCAGTCGCTAGCGCATGCCTGCA-3′, BEAD-A lower strand: 5′-GGCATGCGCTAGCGACTGGACCCCTGCCGGCAGGCGGCAGTGCAGGCCTGGGA-3′). The PCR-amplified FB fragment, a 1.2-kb SacI-SspI fragment containing the full-length 1.2-kb chicken β-globin 5′HS4 insulator or a 250-bp PCR-amplified 5′HS4 insulator core fragment were then individually inserted into the ClaI site.
The RMSinFB and RMSinOFB gammaretroviral vectors used in this study were derived from MSinSB [25] in several steps as follows. First, the MSinSB 3′ LTR was removed by EcoRI-BclI digestion and replaced with PCR amplified fragments that generated a deleted 3′ LTR that eliminated 31 bp upstream of the U3/R boundary inclusive of the TATA box as well as the entire U5 region which were present in the MSinSB 3′ LTR. The SV40 late poly(A) signal which was present in the R region of the MSinSB 3′ LTR was removed from this location and subcloned into a ClaI site downstream of the 3′ LTR immediately upstream of a bovine growth hormone poly(A) signal. The FB fragment was then inserted into the deleted U3 region of the 3′ LTR between NheI and PstI sites. Next, the U3 enhancer-promoter sequence of the 5′ LTR was substituted with a 230-bp RSV enhancer-promoter, PCR-amplified from the RSV-neo plasmid using 5′RSV (5′-ATCGGCGCGCCAATGTAGTCTTATGCAATACT-3′) and 3′RSV (5′-TTTATTGTATCGAGCTAGGC-3′) primers, which was inserted into EcoRV-AscI-digested plasmid, giving rise to RMSinFB. Finally, a 587-bp EcoRI fragment containing the optimized posttranscriptional regulatory element (OPRE) [27] from the pMP71-GFP-OPRE plasmid (a gift from Axel Schambach, Hannover Medical School, Hannover, Germany) was blunt-end ligated into the XhoI site of the RMSinFB vector downstream of the GFP coding region to yield RMSinOFB.
The SIN simian immunodeficiency virus (SIV)-derived lentiviral vector pCL20cSLFR MSCV-GFP (referred to hereafter as SIV-MSCV) is derived from a nonpathogenic isolate of simian immunodeficiency virus (provided by Arthur Nienhuis, St Jude Children’s Research Hospital, Memphis, TN, USA) [30]. To insert the FB element into the deleted U3 region of the 3′ LTR of SIV-MSCV, a 316-bp NotI-XhoI fragment containing the SIV-MSCV 3′ LTR was first subcloned into the pBSP-IRES-GFP plasmid between the NotI and XhoI sites, giving rise to the pBSP-3′LTR plasmid [19]. The FB fragment was then PCR amplified from RMSinOFB using FB-S (5′-AGATCTGCATTCCCAGGGATGTACGTCCC-3′) and FB-AS (5′-AGATCTCTGCAGGCATGCGCTAGC-3′) primers, digested with BamHI and cloned into the BglII site at the beginning of the deleted U3 region of the 3′ LTR in pBSP-3′LTR (pBSP-3′LTR-FB). SIV-MSCV-FB was generated by substituting the NotI-XhoI 3′ LTR-containing fragment of SIV-MSCV with the corresponding NotI-XhoI fragment from pBSP-3′LTR-FB. Finally, a 587-bp EcoRI fragment containing the OPRE from the pMP71-GFP-OPRE plasmid was blunt-end ligated into the NotI site of the SIV-MSCV-FB vector downstream of the GFP coding region to yield SIV-MSCV-OFB.
Cell Lines
NIH3T3 fibroblasts (ATCC No. CRL-1658; American Type Culture Collection, Manassas, VA, USA), Phoenix-Eco packaging cells (ATCC No. SD 3444) and 293T/17 cells (ATCC No. CRL-11268) were grown in Dulbecco’s modified Eagle medium (Mediatech, Inc., Herndon, VA, USA) supplemented with 4.5 g/l glucose, 4 mM L-glutamine, 50 IU/ml penicillin, 50 μg/ml streptomycin and 10% heat-inactivated fetal bovine serum (Cambrex BioScience Walkersville, Inc., Walkersville, MD, USA). K562 human erythroleukemic cells (ATCC No. CCL-243) were cultured in Iscove’s modified Dulbecco’s medium (Mediatech) plus 10% heat-inactivated fetal bovine serum, L-glutamine (2 mM), penicillin (50 IU/ml) and streptomycin (50 μg/ml). All cells were maintained at 37°C in a humidified incubator containing 5% CO2.
In Vitro Enhancer Blocking Assay
Circular plasmids of the various constructs were transiently transfected into 293T/17 cells, and plasmids were linearized and electroporated into K562 cells for stable expression. 293T/17 cells were transiently transfected with the plasmid DNAs (20 μg) by calcium phosphate coprecipitation as described previously [31]. For stable expression, K562 cells were electroporated with linearized plasmid DNA (20 μg) followed by cell sorting on a FACSAria instrument (BD Biosciences, San Jose, CA, USA) after 10 days of culture [32]. Forty-eight hours following the transient transfection of 293T/17 cells and 2–4 weeks after stable transfection of K562 cells, GFP expression was measured by flow cytometry and normalized with respect to DsRed.T4 fluorescence.
Production of Retroviral Vector Particles
Ecotropic vector particles were generated by transient transfection of Phoenix-Eco cells with the retroviral vector DNA using calcium phosphate coprecipitation as described previously [25, 33]. Vector conditioned medium was collected after 48 hours, centrifuged at 2000g to remove cellular debris, and filtered through a 0.45-μm-pore-size filter (Nalge Nunc International, Rochester, NY, USA) before being aliquoted and frozen at −80°C for future transductions. Amphotropic vector particles were generated by transient transfection of 293T cells with retroviral vector DNA and an amphotropic packaging plasmid using calcium phosphate coprecipitation. Vesicular stomatitis virus (VSV)-G pseudotyped SIV vector particles were generated by transient transfection of 293T/17 cells with vector (SIV-MSCV, SIV-MSCV-FB or SIV-MSCV-OFB) and packaging (pCAG-SIVgprre and pCAG4-RTR-SIV) plasmids (provided by Arthur Nienhuis) plus the VSV-G envelope plasmid pMD. G as described previously [30, 31]. When required, ecotropic vector conditioned medium was concentrated by overnight centrifugation at 9000g. Vector titers were determined on NIH3T3 cells by flow-cytometric analysis of GFP fluorescence using a FACSCalibur analyzer (BD Biosciences) as described previously [19, 23, 25].
Transduction and In Vitro Transformation of Murine Bone Marrow Cells
The bone marrow (BM) cell replating assay was performed as described [8, 17] with the following modifications. BM cells were harvested from wild-type C57BL/6 mice or coagulation factor VIII-deficient BALB/c mice (BAL-FVIII-KO; breeding pairs provided by David Lillicrap, Queen’s University, Kingston, Ontario, Canada) [34] at 6- to 8-weeks of age 4 days after intraperitoneal injection of 5-fluorouracil (150 mg/kg body weight) and cultured in Iscove’s modified Dulbecco’s medium supplemented with 10% heat-inactivated fetal bovine serum and conditioned medium containing recombinant murine IL-3, recombinant murine IL-6 and recombinant murine stem cell factor [35]. BM processing, transduction and GFP expression analysis were carried out as described in detail previously [16, 25, 33, 35]. After 48 hours of prestimulation, BM cells were transferred to plates coated with full-length human fibronectin (BD Biosciences) and transduced for 3 consecutive days (4 hours each day) by incubation with vector conditioned medium and 8 μg/ml polybrene. For SIN gammaretroviral vectors, MSinSB, RMSinFB and RMSinOFB, one or two extra rounds of transductions were performed in order to obtain similar BM transduction efficiencies as obtained with the parental MSGV1 LTR vector and the SIV lentiviral vectors [8, 17]. BM cells were cultured for 2 weeks, during which time cells were passaged every 3 days. After bulk culture, 100 cells were plated into each well of a 96-well plate and cultured in Iscove’s modified Dulbecco’s medium supplemented with 10% heat-inactivated fetal bovine serum and conditioned medium containing recombinant murine IL-3. Two weeks later, the plates were examined and the replating frequency determined based on the number of wells which contained proliferating cell populations. All animal procedures were carried out in accordance with Institutional Animal Care and Use Committee guidelines.
Nucleic Acid Analysis
Southern and Northern blot analyses were carried out as described previously [19, 23, 25]. A 1.1-kb NheI-ApaI fragment of pYX-Asc-Prdm16 (Cat. No. FL1002; Invitrogen Corp., Carlsbad, CA, USA) and a 0.9-kb EcoRI-HindIII fragment of pBluescript KS(+)-Evi1 [36] were used to detect Prdm16 and Evi1 mRNA expression, respectively.
Statistical Analysis
Data from experiments are expressed as mean values plus/minus standard deviation. The Student’s paired t-test was used to compare differences between the indicated groups. P < 0.05 was considered significant.
RESULTS
Composite FII/BEAD-A (FB) Element Can Effectively Block the Activity of a Strong Enhancer
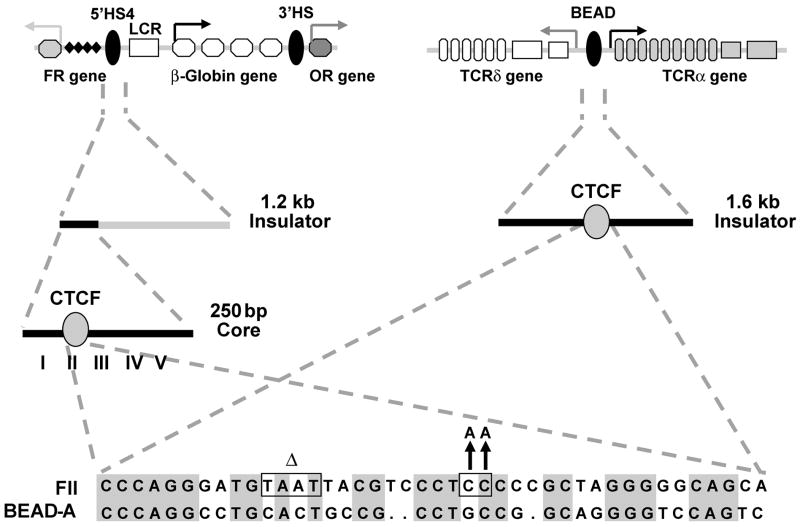
Extensive work by Felsenfeld and colleagues has characterized a 1.2-kb element referred to as the chicken β-globin 5′HS4 (5′ DNase I–hypersensitive site 4) insulator (exhibiting both enhancer blocking and chromatin barrier properties) that separates the regulatory elements of the chicken globin gene cluster from those of an independently regulated folate receptor gene (Fig. 1, left) [18, 28, 29, 37–39]. When a 5′ 250-bp “core” enhancer-blocking component of the 5′HS4 insulator [29] was further dissected into five DNase I-footprinted regions (FI–FV), only the FII region was shown to be necessary and sufficient for enhancer blocking activity [37]. Further analysis of the FII region revealed that the eleven-zinc finger DNA-binding protein, CCCTC-binding factor (CTCF), was responsible for this function [37]. Based on these findings plus the observation that increasing the number of copies of fragments containing the FII region of the chicken β-globin 5′HS4 insulator resulted in a corresponding “stepwise” increase in enhancer blocking activity [37], we generated a composite enhancer blocker (denoted FB for FII/BEAD-A) by joining a 42-bp FII region-containing fragment to a 39-bp sequence (BEAD-A) corresponding to a homologous CTCF binding site with enhancer blocking properties identified in the 1.6-kb BEAD-1 (blocking element alpha/delta 1) insulator that separates the differentially regulated human T cell receptor (TCR) α and δ genes from each other’s enhancers (Fig. 1, right) [37, 40]. Because direct repeats are frequently unstable in retroviral vectors [25, 41, 42], the sequence similarity between the FII and BEAD-A enhancer blocking elements was reduced from 62% to 55% by introducing functionally neutral ‘CC’ to ‘AA’ transversion mutations in the FII element that eliminate a consensus Sp1 binding site and by deleting a nonessential ‘TAAT’ motif, as previously described [37] (Fig. 1, bottom).
Figure 1.
Schematic diagram of the chicken β-globin gene locus (left) and the human T cell receptor (TCR) α/δ locus (right) with the positions of the 5′HS4 and BEAD-1 (BEAD) insulators indicated, respectively. The sequences of the FII and BEAD-A enhancer blocking components comprising the FB element are shown at the bottom. During efforts to identify the protein responsible for enhancer blocking activity (CTCF), Felsenfeld and colleagues previously demonstrated that mutation of a consensus Sp1 site (functionally neutral ‘CC’ to ‘AA’ transversion mutations mentioned in the text) had no effect on CTCF binding or enhancer blocking activity; likewise, these investigators found that deletion of the ‘TAAT’ motif within a sequence having homology to a binding site for the yeast α2 homeodomain repressor protein and which overlaps a partial match to the binding site of the Drosophila suppressor of Hairy-wing protein, had no effect on CTCF binding or enhancer blocking activity. Abbreviations: FR, folate receptor gene; LCR, locus control region; OR, odorant receptor gene. Adapted from refs. [18, 37, 38]. See text for details.
To evaluate the enhancer blocking activity of the composite FB element, we developed the following flow cytometry-based assay. A plasmid was constructed, based on an earlier design [28], in which enhancer blocking activity is measured by down-modulation of expression of a GFP reporter gene when a test fragment is inserted between a herpes simplex virus TK promoter-driven GFP expression cassette and a downstream human CMV immediate early region enhancer. To control for variations in transfection efficiency, GFP expression levels are compared to those of an analogous CMV enhancer-TK promoter-driven DsRed.T4 red fluorescent protein reporter gene expression cassette contained in the same construct. To allow use of circular plasmids in transient transfection experiments and eliminate influences of flanking chromatin structure on GFP expression in stable transfection experiments with linearized plasmids, two copies of the full-length 1.2-kb 5′HS4 insulator were introduced upstream of the GFP expression cassette [28, 38]. For comparison purposes, constructs were made that contained the FB element, the full-length 1.2-kb 5′HS4 insulator or the 250-bp 5′HS4 core element inserted between the CMV enhancer and the GFP expression cassette (Fig. 2A).
Figure 2.
Composite FB element can effectively block the activity of the CMV enhancer in an in vitro enhancer blocking assay. (A) Schematic diagram showing the reporter plasmids used in the flow-cytometry based enhancer blocking assay. Abbreviations: DSR4, DsRed.T4; CMV-E, human CMV immediate early region enhancer; FB, the FII enhancer blocking component of the chicken β-globin 5′ HS4 insulator and the BEAD-A homologous region from the human T-cell receptor α/δ BEAD-I insulator; TK, herpes simplex virus thymidine kinase promoter; PA, SV40 late poly(A) signal. Other abbreviations as described in the text. (B) The constructs shown on the left were used in either transient transfections of 293T cells (upper) or stable transfections of K562 cells (lower). Relative GFP expression (normalized to DsRed.T4 expression) and the percentage of enhancer blocking for each construct are shown for triplicate determinations.
The various constructs were transiently transfected into human 293T embryonic kidney cells or stably transfected into human K562 erythroid cells. Forty-eight hours following transient transfection of 293T cells and 2–4 weeks after stable transfection of K562 cells, GFP fluorescence was measured by flow cytometry and normalized with respect to DsRed.T4 fluorescence. Insertion of the full-length 1.2-kb 5′HS4 insulator mediated 80 ± 6% and 70 ± 10% block of the CMV enhancer on GFP expression in 293T and K562 cells, respectively (P < 0.0001) (Fig. 2B), compared to the control construct lacking any enhancer blocking element. Interestingly, insertion of the FB element resulted in levels of enhancer blocking activity that were similar to and even exceeded those of the full-length 1.2-kb 5′HS4 insulator in both transiently transfected 293T cells (85 ± 4%) and stably electroporated K562 cells (76 ± 5%), respectively (P < 0.0001) (Fig. 2B). In accord with previous data [28, 29, 37], the 250-bp coreelement of the 5′HS4 insulator was significantly less effective in enhancer blocking activity, resulting in only 33 ± 16% and 29 ± 15% blockage of CMV enhancer action on GFP expression in 293T and K562 cells, respectively (P < 0.01).
FB Element with Imperfect Direct Repeats is Stably Transmitted in a Retroviral Vector Backbone and does not Negatively Impact Transduction Efficiency or Transgene Expression
We previously described a murine stem cell virus (MSCV) [43]-based LTR gammaretroviral vector, MSGV1 [44] (Fig. 3A), which is currently in use in a gene therapy clinical trial investigating the ability of genetically engineered peripheral blood lymphocytes to mediate tumor regression in patients with metastatic melanoma [45]. A derivative of this vector, MSGV1-C2, containing two tandem copies of the 250-bp core element of the 5′HS4 insulator in the U3 region of the LTR was found to delete one copy of the element at high frequency during viral replication [25]. We were interested, therefore, in determining whether the imperfect direct repeats of the composite FB enhancer blocking element might be functional and faithfully transmitted in the context of a retroviral vector backbone. To this end, we first constructed the SIN gammaretroviral vector RMSinFB (Fig. 3A). RMSinFB contains a 334-bp deletion within the U3 region of the 3′ LTR that begins at an NheI site 35 bp downstream of the 5′ end of the U3 region and extends to the U3/R boundary, which completely removes all enhancer-promoter sequences. The FB enhancer blocking element was inserted into the NheI site of the deleted 3′ LTR. To prevent restoration of wild-type U3 sequences by homologous recombination during transfection of packaging cells, the U3 region of the 5′ LTR was replaced with a 230-bp fragment of the Rous sarcoma virus enhancer-promoter [46]. The internal promoter (designated the CA promoter) that drives transgene expression in RMSinFB was obtained from the SIN gammaretroviral vector MSinSB [25] (Fig. 3A). The CA promoter is a modified version of the composite CAG promoter [47] that consists of the CMV immediate early region enhancer (that was used to evaluate the FB element in the flow cytometry-based enhancer blocking assay)linked to the chicken β-actin promoter. To increase vector RNA 3′ end formation, the safety-modified woodchuck hepatitis virus posttranscriptional regulatory element (termed OPRE) [27]was inserted into the 3′ untranslated region of the RMSinFB vector [23], giving rise to RMSinOFB. To also evaluate the activity of FB enhancer blocker in the context of a lentiviral vector backbone, we constructed a series of SIN SIV-derived lentiviral vectors in parallel. The parental vector SIV-MSCV utilizes the MSCV LTR present in MSGV1 as an internal enhancer-promoter to drive GFP expression [30]. SIV-MSCV-FB, which contains the FB element in the deleted U3 region of the 3′ LTR, was derived from SIV-MSCV in several steps (see Materials and Methods); the OPRE was subsequently inserted into the 3′ untranslated region of SIV-MSCV-FB, yielding SIV-MSCV-OFB (Fig. 3A). All vectors include a GFP transgene.
Figure 3.
Composite FB element does not adversely affect performance of SIN gammaretroviral and lentiviral vectors. (A) Schematic representation of gammaretroviral (MSGV1, MSinSB, RMSinFB, RMSinOFB) and lentiviral (SIV-MSCV, SIV-MSCV-FB, SIV-MSCV-OFB) vectors. Plasmid forms of the vectors are illustrated. All vectors include a GFP transgene under the transcriptional control of the LTR (MSGV1), an internal CA promoter (SIN gammaretroviral vectors) or an internal MSCV LTR promoter (SIN lentiviral vectors). Arrows depict the initiation sites of LTR-, the CA-, or the MSCV-promoter directed RNA synthesis. The deletion in the U3 region of the 3′ LTR in the SIN configuration is indicated by ΔU3. Other abbreviations: B (black type), bovine growth hormone poly(A) signal; B (white type), rabbit β-globin poly(A) signal; CMV, cytomegalovirus immediate early enhancer-promoter; cPPT, central polypurine tract; FB, composite FII/BEAD-A element; OPRE, safety-modified woodchuck hepatitis virus posttranscriptional regulatory element; RRE, Rev-responsive element; S, SV40 late poly(A) signal. Vector titers were measured on NIH3T3 cells and are shown for triplicate determinations (right). *Significantly higher than the RMSinFB titer (P < 0.001). **Significantly higher than the SIV-MSCV-FB titer (P < 0.01). (B) Comparison of GFP fluorescence intensity levels (MFI values) of human 293T cells transduced with the indicated gammaretroviral and lentiviral vectors. Cells were transduced once at low MOI (~1) in triplicate and evaluated by flow cytometry 10 days later. Only the transductions with less than 10% positive cells were analyzed to minimize the effects of multiple integrations. *Significantly higher expression level than the MSGV1 vector (P = 0.003), the MSinSB vector (P < 0.02) and the RMSinFB vector (P < 0.01). **Significantly higher expression level than the SIV-MSCV vector (P < 0.001) and the SIV-MSCV-FB vector (P < 0.001).
We produced replication-defective gammaretroviral and lentiviral vector particles and measured the titers on murine NIH3T3 cells (Fig. 3A, right). Importantly, inclusion of the FB element did not negatively affect titers in either vector platform, as was previously the case for SIN lentiviral vectors harboring the full-length 1.2-kb 5′HS4 insulator [19]. On the other hand, addition of the OPRE to the FB element-containing vectors resulted in significant increases in titers for both gene delivery systems: the SIN gammaretroviral vector RMSinOFB exhibited titers (2.2 ± 0.6 x 106 TU/ml) that were >4-fold higher than its OPRE-minus counterpart RMSinFB (0.5 ± 0.2 x 106 TU/ml; P < 0.001) comparable to the parental LTR vector MSGV1 (2.1 ± 0.3 x 106TU/ml), whereas the titers of the SIN lentiviral vector SIV-MSCV-OFB (25.0 ± 1.0 x 106 TU/ml) were ~30% higher than its OPRE-minus counterpart SIV-MSCV-FB (19.1 ± 2.0 x 106 TU/ml; P < 0.01).
When vector performance was assessed on human 293T cells, the levels of transgene expression achieved by the FB element-containing SIN gammaretroviral and lentiviral vectors were comparable to or greater than those achieved with the respective FB element-negative versions (Fig. 3B). This is a noteworthy observation because insertion of the full-length 1.2-kb 5′HS4 insulator into the MSCV LTR was previously found to dramatically decrease the level of expression from an internal (phosphoglycerate kinase) promoter [48].
To document the stability of the FB element, genomic DNA was isolated from RMSinOFB vector-transduced 293T cells. Using a pair of primers designed to specifically amplify the 5′ LTR sequences, a 455-bp DNA fragment of expected size was PCR-amplified from RMSinOFB-transduced cells (Fig. 4). By comparison, a 670-bp DNA fragment of expected size was PCR-amplified from 293T cells transduced with the MSGV1 vector bearing intact LTRs. Sequencing of the 455-bp DNA fragment confirmed the presence and proper transmission of the FB element from the RMSinOFB 3′ LTR to the 5′ LTR. Furthermore, the absence of any smaller PCR products indicated that, unlike the frequent deletion of one copy of the duplicated 250-bp core element of the 5′HS4 insulator observed previously with the MSGV1-C2 vector [25], the short imperfect direct repeats of the composite FB enhancer blocker were retained in the vast majority of RMSinOFB vector integrants.
Figure 4.
Composite FB element is stable in the context of a SIN gammaretroviral vector. Shown are PCR analyses demonstrating proper transmission of the FB element in the RMSinOFB vector. Genomic DNAs from untransduced (293T), MSGV1-transduced (MSGV1) and RMSinOFB-transduced (RMSinOFB) 293T cells were analyzed by PCR using a forward primer complementary to sequences at the beginning of the U3 region and a reverse primer recognizing sequences downstream of the 5′ LTR. The positions of the primers are indicated by arrows underneath the schematic representations of the 5′ ends of the integrated forms of the MSGV1 and RMSinOFB vectors. The sizes of the PCR DNA products from RMSinOFB (455 bp) and MSGV1 (670 bp), which were verified by DNA sequencing, are indicated.
Incorporation of the FB Element in SIN Gammaretroviral and Lentiviral Vectors Significantly Reduces their Transforming Potential
To evaluate whether the FB enhancer blocking element might reduce insertional mutagenesis in the context of SIN gammaretroviral and lentiviral vectors, we assessed in vitro transforming potential of primary murine BM progenitors [7, 8, 17]. BM cells were isolated from 4-day 5-fluorouracil-treated mice and transduced with a series of vectors at equivalent MOIs to achieve similar copy numbers per cell. Transduced BM cells were cultured in vitro for 2 weeks before being replated in 96-well plates. Two weeks later, the 96-well plates were scored for the number of wells with proliferating cell populations, some of which were expanded for further analysis [8, 17].
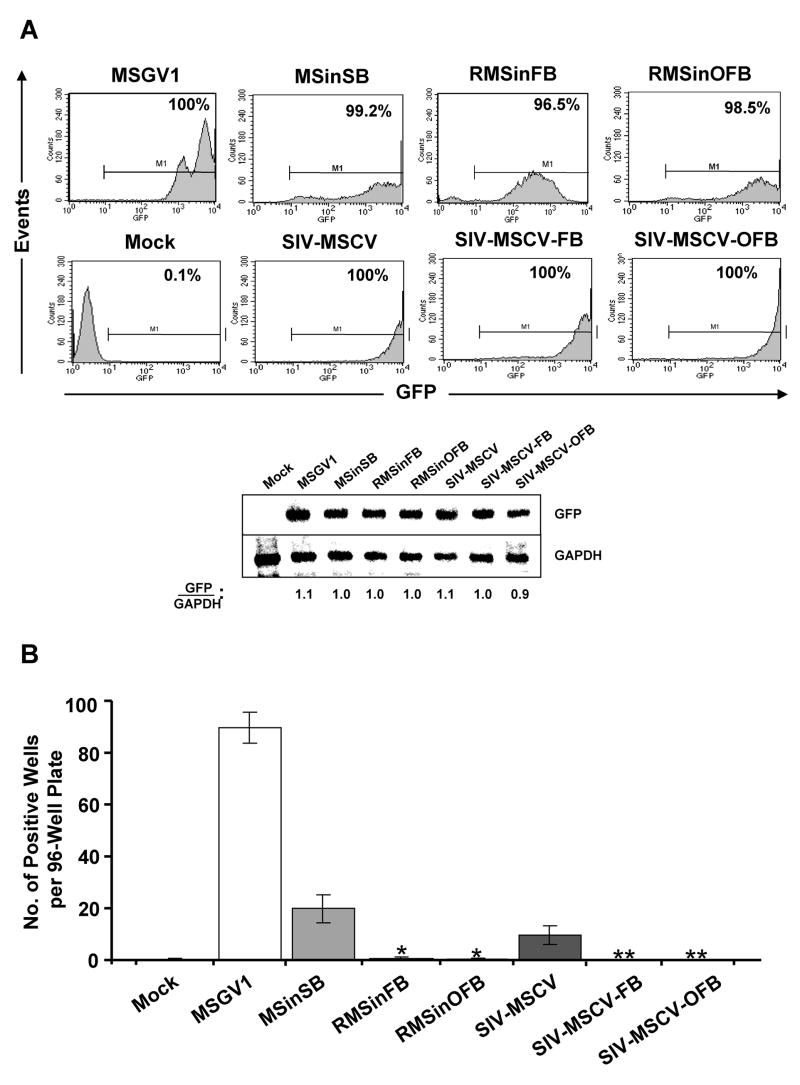
In an initial series of experiments (Supplementary Fig. S1), ecotropic RMSinOFB vector particles were used to transduce 106 BM cells from wild-type C57BL/6 mice at an MOI of 5 for three consecutive days. Controls included ecotropic pseudotypes of the LTR vector MSGV1 and the SIN vector MSinSB, and VSV-G-pseudotyped SIN lentiviral vector SIV-MSCV. One week after the last transduction, transduction efficiencies and relative transgene copy numbers were determined by flow cytometry and Southern blot analyses, respectively. Flow cytometric analysis of BM cells for GFP expression demonstrated that similar levels of transduction were obtained with all of the vectors: MSGV1, 89.9%; MSinSB, 80.6%; RMSinOFB, 82.8%; and SIV-MSCV, 78.0% in the representative experiment shown in Supplementary Fig. S1A. For Southern blotting, genomic DNA from pools of transduced BM cells was digested with NcoI and EcoRI (NcoI and NotI for SIV-MSCV) – restriction enzymes that cleave the vectors at either end of the GFP gene – and hybridized with a GFP-specific probe. The blot was then stripped and rehybridized with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific probe. When normalized to GAPDH, the analysis confirmed that the vectors were all present at relatively similar copy numbers (Supplementary Fig. S1A).
After 2 weeks of expansion in culture, 100 cells were plated per well in 96-well plates; 2 weeks later, proliferating cell populations were scored as an estimate of the transforming (immortalizing) potential by insertional mutagenesis with a particular vector [7, 8, 17]. As shown in Supplementary Fig. S1B, nearly half of the wells in the 96-well plates (42.5 ± 11.9; n = 4) that received MSGV1-transduced BM cells had proliferating cell populations. Wells with proliferating cells were randomly selected and the cultures were further expanded. Northern blot analysis of an expanded culture that was established as an immortalized cell line revealed overexpression of the proto-oncogene Prdm16 (see Supplementary Fig. S1C), in accordance with previous work [7, 12]. Significantly fewer wells (6.5 ± 3; n = 4) contained proliferating cells following transduction with the MSinSB vector. Interestingly, a significant number of positive wells (5.8 ± 1.5; n = 4) was also observed on plates that received BM cells transduced with the SIN lentiviral vector SIV-MSCV. Notably, only an occasional single positive well per 96-well plate (0.5 ± 0.6; n = 4), which is similar to background levels obtained with untransduced BM cells and which could not be expanded, was observed on plates that received RMSinOFB-transduced BM cells (P = 0.008).
In a further series of experiments, we extended the in vitro assay to a disease-specific genetic background and determined whether the FB element could provide protection against vector-induced genotoxicity at higher MOIs. Using BM cells from coagulation factor VIII-deficient BAL-FVIII-KO mice (hemophilia A disease model) [34], 3 rounds of transduction were performed using an MOI of 10. Furthermore, since positive wells were obtained upon replating in the previous experiment after gene transfer with the SIN SIV-MSCV lentiviral vector, we evaluated the blocking activity of the FB element in the context of the a SIN lentiviral vector backbone. The use of higher MOI led to ~100% transduction of the BM cells with all vectors as determined by flow cytometric analysis of GFP expression 1 week after the last transduction (Fig. 5A); Southern blotting demonstrated approximately similar vector copy numbers in all cell populations (Fig. 5A). As in the earlier experiments, after 2 weeks of expansion in bulk culture, 100 cells were plated into each well of a 96-well plate and the plates were scored for proliferating cell populations 2 weeks later. As shown in Fig. 5B, nearly all of the wells in the 96-well plates (89.5 ± 6.1; n = 6) that received MSGV1-transduced BM cells had proliferating cell populations. Again, significantly fewer wells (19.8 ± 5; n = 6) contained proliferating cells following transduction with the MSinSB vector. Notably, only occasional positive wells per 96-well plate, which could not be expanded, were observed on plates that received RMSinFB- or RMSinOFB-transduced BM cells: 0.5 ± 0.8 and 0.3 ± 0.5, respectively (n = 6; P < 0.001). Asignificant number of positive wells (9.7 ± 4; n = 6) were again observed on plates that received BM cells transduced with the unshielded SIN lentiviral vector SIV-MSCV; in contrast, we did not obtain any positive wells on plates that received SIV-MSCV-FB- or SIV-MSCV-OFB- transduced BM cells.
Figure 5.
Incorporation of FB element in SIN gammaretroviral and lentiviral vectors blocks in vitro transformation of murine BM cells. (A) Flow cytometric histograms of GFP fluorescence 1 week after the last transduction with the indicated vectors at an MOI of 10 (x 3). Shown in each case is the percentage of GFP-positive transduced BM cells. Below the histograms is a Southern blot analysis of NcoI/EcoRI (for gammaretroviral vectors) or NcoI/NotI (for lentiviral vectors)-digested genomic DNAs hybridized with a GFP probe (top panel) and a GAPDH probe (bottom panel) showing the relative vector copy number per cell population. The results presented are representative of two independent experiments. (B) Replating ability of vector-transduced BM cells. After retroviral transduction, BM cells were expanded as mass cultures for 2 weeks in IL-3-conditioned medium. During this period, cell density was adjusted to 1 x 106 cells/ml every 3 days. Expanded BM cells were subsequently transferred into 96-well plates (n = 6 per vector) at a density of 100 cells/well. Two weeks later, the number of wells that contained proliferating cell populations were scored. Under these conditions, the majority of untransduced BM cells failed to survive replating or had ceased proliferating by this time. *Significantly fewer positive wells than MSinSB-transduced samples (P = 0.008). **Significantly fewer positive wells than SIV-MSCV-transduced samples (P < 0.05).
DISCUSSION
The genotoxic risk of integrating gammaretroviral-based gene transfer vectors has been shown to depend on vector architecture such that it could be reduced by using a SIN design [8, 17]. However, insertional mutagenesis still remains a concern for SIN vectors [8, 12, 17], especially in gene therapy settings where high levels of transgene expression—achieved with strong internal enhancer-promoter sequences or by multiple copy gene transfer [6]—are required to obtain clinical benefit. In this study, we demonstrated that the hematopoietic transforming activity of SIN gammaretroviral vectors as well as SIN lentiviral vectors harboring strong internal enhancer-promoters could be virtually eliminated—to background levels under the experimental conditions used—by incorporation of the novel 77-bp FB enhancer blocking element in the deleted U3 region of the 3′ LTR.
Flanking the SIN retroviral vector backbone with enhancer blocking elements derived from chromatin insulators was previously proposed as a strategy to prevent insertional mutagenesis [1, 2, 13, 14]. We and others introduced the full-length 1.2-kb chicken β-globin 5′HS4 insulator [28] into the deleted U3 regions of SIN lentiviral vectors [19, 20]. In our hands though this modification resulted in a substantial drop in vector titer due either to topological or size constraints, the latter of which in particular makes this modification especially difficult to widely implement in gammaretroviral vectors [48]. For this reason, we subsequently tested whether two copies of the 250-bp 5′HS4 core inserted in tandem into the U3 region of a gammaretroviral vector 3′ LTR would be faithfully propagated to target cells. Unfortunately, molecular analysis of the genomic DNA extracted from stably transduced cells indicated that one copy of the core was deleted at high frequency during viral replication [25]. Consequently, we posited that the sequences of the short FII variant and BEAD-A elements might be sufficiently different to allow their stable transmission when introduced in tandem into the U3 region of a SIN retroviral vector 3′ LTR [41, 42]. Sequence analysis of PCR-amplified genomic DNA from cells transduced with the RMSinOFB vector having the composite FB element in this position confirmed that it was successfully transferred to the 5′ LTR. By extension, it is possible that additional protection from retroviral insertional genotoxicity might be achieved by combining other sufficiently diverged CTCF-binding sites [49] with the FB element (e.g., such as that from the human amyloid β protein promoter [37]).
A somewhat unexpected observation to come from this study is that the SIV-MSCV SIN lentiviral vector exhibited substantial in vitro transforming capacity, approaching that of the MSinSB SIN gammaretroviral vector. Because of differences in genomic integration site preferences [50, 51], SIN lentiviral vectors have been believed to have low oncogenic potential, and there is some experimental evidence in support of this idea [52]. On the other hand, a recent study described the emergence of dominant clones in short-term cultures of human T cells transduced with a human immunodeficiency virus-based SIN lentiviral vector having the same strong internal enhancer-promoter as SIV-MSCV [20], while another recent investigation reported that this vector as well as a β-globin SIN lentiviral vector were capable of up-regulating endogenous gene expression in primary murine hematopoietic cells as far as 321 kb away from the integration site [53]. Of note, in other work, a high incidence of integration-associated tumorigenesis was observed following in utero and neonatal gene transfer in mice with SIN lentiviral vectors based on equine infectious anaemia virus containing the CMV immediate early region enhancer-promoter [54]. In this regard, it has recently been suggested that the lack of oncogenic potential observed in one study with a SIN lentiviral vector [52] is largely the result of the weak enhancer of the internal promoter that was used (phosphoglycerate kinase) [17]. In accord with this notion and in support of our findings, Baum and colleagues recently described in vitro transforming capacity of a SIN lentiviral vector containing the strong spleen focus-forming virus LTR as an internal enhancer-promoter [55]. Thus, when considered in the context of hematopoietic stem cell gene therapy applications, based on the results presented herein, it seems reasonable to propose that SIN lentiviral vectors harboring strong internal enhancer-promoter sequences (indeed, we would argue all integrating vectors) will also benefit from an analogous FB element-mediated enhancer blocking modification.
Supplementary Material
Acknowledgments
We thank Sara Karandish for technical assistance. This work was supported in part by National Institutes of Health Grants R01HL65519, R01HL66305 and R24RR16209, and by an Elaine H. Snyder Cancer Research Award and a King Fahd Endowed Professorship (to R.G.H.) from The George Washington University Medical Center.
References
- 1.Baum C, Kustikova O, Modlich U, et al. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum Gene Ther. 2006;17:253–263. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- 2.Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 4.Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 5.Kustikova O, Fehse B, Modlich U, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- 6.Modlich U, Kustikova OS, Schmidt M, et al. Leukemias following retroviral transfer of multidrug resistance 1 (MDR1) are driven by combinatorial insertional mutagenesis. Blood. 2005;105:4235–4246. doi: 10.1182/blood-2004-11-4535. [DOI] [PubMed] [Google Scholar]
- 7.Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Modlich U, Bohne J, Schmidt M, et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calmels B, Ferguson C, Laukkanen MO, et al. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seggewiss R, Pittaluga S, Adler RL, et al. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood. 2006;107:3865–3867. doi: 10.1182/blood-2005-10-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XB, Beard BC, Trobridge GD, et al. High incidence of leukemia in large animals after stem cell gene therapy with a HOXB4-expressing retroviral vector. J Clin Invest. 2008;118:1502–1510. doi: 10.1172/JCI34371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modlich U, Schambach A, Brugman MH, et al. Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16. Leukemia. 2008;22:1519–1528. doi: 10.1038/leu.2008.118. [DOI] [PubMed] [Google Scholar]
- 13.Hawley RG. Progress toward vector design for hematopoietic stem cell gene therapy. Curr Gene Ther. 2001;1:1–17. doi: 10.2174/1566523013348904. [DOI] [PubMed] [Google Scholar]
- 14.Ramezani A, Hawley TS, Hawley RG. Reducing the genotoxic potential of retroviral vectors. Methods Mol Biol. 2008;434:183–203. doi: 10.1007/978-1-60327-248-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu SF, von Rüden T, Kantoff PW, et al. Self-inactivating retroviral vectors designed for transfer of whole genes into mammalian cells. Proc Natl Acad Sci USA. 1986;83:3194–3198. doi: 10.1073/pnas.83.10.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawley RG, Covarrubias L, Hawley T, et al. Handicapped retroviral vectors efficiently transduce foreign genes into hematopoietic stem cells. Proc Natl Acad Sci USA. 1987;84:2406–2410. doi: 10.1073/pnas.84.8.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zychlinski D, Schambach A, Modlich U, et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- 18.Bell AC, West AG, Felsenfeld G. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 19.Ramezani A, Hawley TS, Hawley RG. Performance- and safety-enhanced lentiviral vectors containing the human interferon-β scaffold attachment region and the chicken β-globin insulator. Blood. 2003;101:4717–4724. doi: 10.1182/blood-2002-09-2991. [DOI] [PubMed] [Google Scholar]
- 20.Evans-Galea MV, Wielgosz MM, Hanawa H, et al. Suppression of clonal dominance in cultured human lymphoid cells by addition of the cHS4 insulator to a lentiviral vector. Mol Ther. 2007;15:801–809. doi: 10.1038/sj.mt.6300103. [DOI] [PubMed] [Google Scholar]
- 21.Zaiss AK, Son S, Chang LJ. RNA 3′ readthrough of oncoretrovirus and lentivirus: implications for vector safety and efficacy. J Virol. 2002;76:7209–7219. doi: 10.1128/JVI.76.14.7209-7219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Ramezani A, Hawley TS, Hawley RG. Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol Ther. 2000;2:458–469. doi: 10.1006/mthe.2000.0190. [DOI] [PubMed] [Google Scholar]
- 24.Kraunus J, Schaumann DH, Meyer J, et al. Self-inactivating retroviral vectors with improved RNA processing. Gene Ther. 2004;11:1568–1578. doi: 10.1038/sj.gt.3302309. [DOI] [PubMed] [Google Scholar]
- 25.Ramezani A, Hawley TS, Hawley RG. Stable gammaretroviral vector expression during embryonic stem cell-derived in vitro hematopoietic development. Mol Ther. 2006;14:245–254. doi: 10.1016/j.ymthe.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zufferey R, Donello JE, Trono D, et al. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schambach A, Bohne J, Baum C, et al. Woodchuck hepatitis virus post-transcriptional regulatory element deleted from X protein and promoter sequences enhances retroviral vector titer and expression. Gene Ther. 2006;13:641–645. doi: 10.1038/sj.gt.3302698. [DOI] [PubMed] [Google Scholar]
- 28.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 29.Chung JH, Bell AC, Felsenfeld G. Characterization of the chicken β-globin insulator. Proc Natl Acad Sci USA. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanawa H, Hematti P, Keyvanfar K, et al. Efficient gene transfer into rhesus repopulating hematopoietic stem cells using a simian immunodeficiency virus-based lentiviral vector system. Blood. 2004;103:4062–4069. doi: 10.1182/blood-2004-01-0045. [DOI] [PubMed] [Google Scholar]
- 31.Ramezani A, Hawley RG. Generation of HIV-1-based lentiviral vector particles. In: Ausubel F, Brent R, Kingston B, et al., editors. Current Protocols in Molecular Biology. New Jersey: John Wiley & Sons, Inc; 2002. pp. 16.22.1–16.22.15. [DOI] [PubMed] [Google Scholar]
- 32.Hawley TS, Telford WG, Ramezani A, et al. Four-color flow cytometric detection of retrovirally expressed red, yellow, green and cyan fluorescent proteins. BioTechniques. 2001;30:1028–1034. doi: 10.2144/01305rr01. [DOI] [PubMed] [Google Scholar]
- 33.Moayeri M, Hawley TS, Hawley RG. Correction of murine hemophilia A by hematopoietic stem cell gene therapy. Mol Ther. 2005;12:1034–1042. doi: 10.1016/j.ymthe.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown BD, Shi CX, Rawle FE, et al. Factors influencing therapeutic efficacy and the host immune response to helper-dependent adenoviral gene therapy in hemophilia A mice. J Thromb Haemost. 2004;2:111–8. doi: 10.1111/j.1538-7836.2004.00552.x. [DOI] [PubMed] [Google Scholar]
- 35.Hawley RG, Hawley TS, Fong AZC, et al. Thrombopoietic potential and serial repopulating ability of murine hematopoietic stem cells constitutively expressing interleukin-11. Proc Natl Acad Sci USA. 1996;93:10297–10302. doi: 10.1073/pnas.93.19.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morishita K, Parker DS, Mucenski ML, et al. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell. 1988;54:831–840. doi: 10.1016/s0092-8674(88)91175-0. [DOI] [PubMed] [Google Scholar]
- 37.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 38.Recillas-Targa F, Bell AC, Felsenfeld G. Positional enhancer-blocking activity of the chicken β-globin insulator in transiently transfected cells. Proc Natl Acad Sci U S A. 1999;96:14354–14359. doi: 10.1073/pnas.96.25.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, et al. Position-effect protection and enhancer blocking by the chicken β-globin insulator are separable activities. Proc Natl Acad Sci U S A. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong XP, Krangel MS. An enhancer-blocking element between α and δ gene segments within the human T cell receptor α/δ locus. Proc Natl Acad Sci U S A. 1997;94:5219–5224. doi: 10.1073/pnas.94.10.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Beveren C, Rands E, Chattopadhyay SK, et al. Long terminal repeat of murine retroviral DNAs: sequence analysis, host-proviral junctions, and preintegration site. J Virol. 1982;41:542–556. doi: 10.1128/jvi.41.2.542-556.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhode BW, Emerman M, Temin HM. Instability of large direct repeats in retrovirus vectors. J Virol. 1987;61:925–927. doi: 10.1128/jvi.61.3.925-927.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hawley RG, Lieu FHL, Fong AZC, et al. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 44.Hughes MS, Yu YY, Dudley ME, et al. Transfer of a TCR gene derived from a patient with a marked antitumor response conveys highly active T-cell effector functions. Hum Gene Ther. 2005;16:457–472. doi: 10.1089/hum.2005.16.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morgan RA, Dudley ME, Wunderlich JR, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schambach A, Mueller D, Galla M, et al. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther. 2006;13:1524–1533. doi: 10.1038/sj.gt.3302807. [DOI] [PubMed] [Google Scholar]
- 47.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 48.Yannaki E, Tubb J, Aker M, et al. Topological constraints governing the use of the chicken HS4 chromatin insulator in oncoretrovirus vectors. Mol Ther. 2002;5:589–598. doi: 10.1006/mthe.2002.0582. [DOI] [PubMed] [Google Scholar]
- 49.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 50.Wu X, Li Y, Crise B, et al. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 51.Hematti P, Hong BK, Ferguson C, et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montini E, Cesana D, Schmidt M, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 53.Hargrove PW, Kepes S, Hanawa H, et al. Globin lentiviral vector insertions can perturb the expression of endogenous genes in β-thalassemic hematopoietic cells. Mol Ther. 2008;16:525–533. doi: 10.1038/sj.mt.6300394. [DOI] [PubMed] [Google Scholar]
- 54.Themis M, Waddington SN, Schmidt M, et al. Oncogenesis following delivery of a nonprimate lentiviral gene therapy vector to fetal and neonatal mice. Mol Ther. 2005;12:763–771. doi: 10.1016/j.ymthe.2005.07.358. [DOI] [PubMed] [Google Scholar]
- 55.Modlich U, Navarro S, Mishra A, et al. Importance of vector content and vector backbone to reduce the risk of insertional mutagenesis. Mol Ther. 2008;16(suppl 1):S360. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.