Abstract
CHIP (carboxy terminus of Hsc70-interacting protein) an E3 ubiquitin ligase that binds to Hsp70 and Hsp90, promotes degradation of several Hsp90-regulated signaling proteins and disease-causing proteins containing expanded glutamine tracts. In polyglutamine disease models, CHIP has been considered a primary protection factor by promoting degradation of these misfolded proteins. Here, we show that two CHIP substrates, the glucocorticoid receptor (GR), a classic Hsp90-regulated signaling protein, and the expanded glutamine androgen receptor (AR112Q), are degraded at the same rate in CHIP−/− and CHIP+/+ mouse embryonic fibroblasts after treatment with the Hsp90 inhibitor geldanamycin. CHIP−/− cytosol has the same ability as CHIP+/+ cytosol to ubiquitinate purified neuronal nitric oxide synthase (nNOS), another established CHIP substrate. To determine whether other E3 ubiquitin ligases that bind to Hsp70 (Parkin) or Hsp90 (Mdm2) act on CHIP substrates, each E3 ligase was co-expressed with the GR, nNOS, AR112Q or Q78 ataxin-3. CHIP lowered the levels of all four proteins, Parkin acted on nNOS and Q78 ataxin-3 but not on the steroid receptors, and Mdm2 did not affect any of the co-expressed proteins. Moreover, both CHIP and Parkin co-localized to aggregates of the expanded glutamine AR formed in cell culture and in a knock-in mouse model of spinal and bulbar muscular atrophy. These observations establish that CHIP does not play an exclusive role in regulating the turnover of Hsp90 client signaling proteins or expanded glutamine tract proteins, and show that the Hsp70-dependent E3 ligase Parkin acts redundantly to CHIP on some substrates.
INTRODUCTION
The Hsp90/Hsp70-based chaperone machinery that forms signaling protein•Hsp90 heterocomplexes (reviewed in Ref. 1) is also part of the cellular defense against unfolded proteins (2). Studies with these Hsp90 ‘client’ proteins have shown that assembly of heterocomplexes with Hsp90 stabilizes the proteins, and, when heterocomplex assembly is blocked by specific Hsp90 inhibitors like geldanamycin and radicicol, the signaling proteins undergo rapid degradation through the ubiquitin/proteasome pathway (3). The ubiquitination is mediated by chaperone-dependent E3 ubiquitin ligases, and one of these ligases called CHIP (carboxy terminus of Hsc70-interacting protein) has been most studied with regard to the targeting of Hsp90 client proteins for ubiquitination (4).
CHIP is a 35-kDa member of the RING-domain family of E3 ubiquitin ligases that binds through an amino-terminal tetratricopeptide repeat (TPR) domain to both Hsc/Hsp70 and Hsp90 (5,6) and through a carboxy-terminal U-box to the UBCH5 family of E2 ubiquitin-conjugating enzymes (7). Because the TPR domain of CHIP binds to both Hsp90 and Hsp70, both chaperones have been thought to target substrates for degradation (4,6). However, ubiquitination following geldanamycin treatment must be targeted by Hsc/Hsp70. This conclusion is based on the fact that the chaperone machinery assembles signaling protein•Hsp90 heterocomplexes in a stepwise fashion through an initial ATP-dependent priming interaction with Hsp70 followed by a second ATP-dependent interaction with Hsp90 (1). The interaction with Hsp90 is blocked by geldanamycin, leaving the Hsp70-bound client protein to be ubiquitinated in a CHIP-dependent manner. Hsp70 and Hsp40, a cochaperone that increases the ATPase activity of Hsp70, are required for the ubiquitin-dependent degradation of many proteins (8,9), and the constitutive Hsc70 and inducible Hsp70 appear to act equivalently in promoting proteasome-dependent degradation (10).
Overexpression of CHIP has been shown to increase the degradation of many well-studied Hsp90-regulated signaling proteins, such as the glucocorticoid receptor (GR) (6), p53 (11) and ErbB2 (12); thus, CHIP is often regarded as the most important E3 ligase involved in chaperone-dependent ubiquitination and degradation. However, there are many E3 ligases (13), and it is not known how many of them function in a chaperone-dependent manner, although some clearly do (14). Parkin, for example, is an E3 ubiquitin ligase (15) that is targeted to substrate by Hsp70 (16), and mutations in Parkin are responsible for autosomal recessive Parkinson disease (PD) (17). In another example, the E3 ligase Mdm2 binds to Hsp90 (18), and inhibition of Mdm2 by Hsp90 contributes to mutant p53 stabilization (19). There is some evidence that E3 ligases may function in a redundant manner to ubiquitinate Hsp90 client proteins. For example, interest has focused on the controlled regulation of wild-type p53 by Mdm2. Yet, CHIP can also induce p53 ubiquitination, and in cells depleted of CHIP, the amount of p53 increases, suggesting that CHIP-dependent degradation plays a role in maintaining low concentrations of p53 under physiological conditions (11).
The possible redundancy of E3 ligase activity is of particular importance in considering therapeutic approaches to certain neurodegenerative conditions involving the accumulation of aberrant proteins, including PD and the polyglutamine expansion disorders, such as Huntington disease (HD), spinal and bulbar muscular atrophy (SBMA), and several autosomal-dominant spinocerebellar ataxias (e.g. SCA1, SCA3). Some of the proteins that aggregate in these diseases form heterocomplexes with Hsp90 and Hsp70, including α-synuclein (PD) (20), HD (21) and the expanded glutamine androgen receptor (AR) of SBMA (22). Inhibition of Hsp90 by geldanamycin prevents the aggregation of these proteins in animal models of PD (23), HD (24) and SBMA (25). Overexpression of Hsp70 or Hsp40 decreases α-synuclein or polyglutamine protein levels and improves viability in cellular models of these diseases (26–28). Overexpression of these chaperones also ameliorates the disease phenotype in Drosophila and mouse models (29–32, reviewed in Ref. 33). CHIP is found in aggregates of α-synuclein (Lewy bodies), huntingtin, AR, ataxin-1 and ataxin-3 (34–38), and overexpression of CHIP suppresses aggregation and protein levels in cellular disease models (34,35,37,38). The notion that CHIP is a critical mediator of the neuronal response to misfolded proteins is buttressed by the observations that overexpression of CHIP in a Drosophila model of SCA1 (37) and a mouse model of SBMA (39) suppresses toxicity, and that HD transgenic mice haploinsufficient for CHIP display an accelerated disease phenotype (35). Thus, CHIP presents as a potential therapeutic target for these neurodegenerative diseases. However, this possibility would be markedly reduced if there was a redundancy of actions among several E3 ligases, because therapeutics targeting each active ligase would have to be combined in a multidrug protocol. The possibility of redundancy in E3 ligase action is suggested by reports that overexpression of either CHIP (38) or Parkin (16) increases ubiquitination of polyglutamine-expanded ataxin-3 and reduces its cellular toxicity. In both cases, the effect of the E3 ligase is promoted by Hsp70 (16,38), and it is this chaperone common to both ubiquitination pathways that may ultimately be the best therapeutic target.
In this work, we examine the degradation of two Hsp90-regulated signaling proteins (the GR and neuronal nitric-oxide synthase (nNOS)) and two polyglutamine-expanded proteins (AR112Q and Q78 ataxin-3) in the presence of three E3 ligases (CHIP, Parkin, Mdm2) to assess the redundancy of E3 ligase action. The GR is the most studied Hsp90 client protein, forms stable heterocomplexes with Hsp90 (1) and undergoes profound geldanamycin- (40) and CHIP-induced (6) proteasomal degradation. In contrast, nNOS undergoes very dynamic cycling into heterocomplexes with Hsp90 such that only trace amounts of the chaperone are co-immunoadsorbed with the enzyme (41). nNOS is inherently more stable than classic Hsp90 clients such as the GR. Nonetheless, both Hsp90 inhibitors (41) and CHIP overexpression (42) promote a modest increase in its rate of proteasomal degradation. Similarly, poly Q-expanded AR degrades upon geldanamycin treatment (22,25) and CHIP overexpression induces its ubiquitination and degradation (39). CHIP is also implicated in the degradation of poly Q-expanded ataxin-3 and co-localizes to nuclear aggregates in transfected cells (38). Notably, overexpression of either CHIP (38) or Parkin (16) lowers the levels of poly Q-expanded ataxin-3, resulting in fewer aggregates and diminished toxicity, suggesting that multiple E3 ligases function in redundant manner. Consistent with this notion, overexpression of E4B, a ubiquitin chain assembly factor (E4) that also has E3 ubiquitin ligase activity, promotes the degradation of poly Q-expanded ataxin-3 and suppresses neurodegeneration in a Drosophila model of SCA3 (43).
We report here that the treatment of CHIP−/− mouse embryonic fibroblasts (44) with the Hsp90 inhibitor geldanamycin promotes the degradation of endogenous GR and exogenously expressed AR112Q at the same rate as in CHIP+/+ cells. Furthermore, CHIP−/− cytosol has the same activity in promoting ubiquitination of purified nNOS as CHIP+/+ cytosol. Thus, there must be E3 ligases that function in a redundant manner to CHIP. To determine whether the other E3 ligases that bind Hsp70 (Parkin) or Hsp90 (Mdm2) provide this redundant activity, each of these E3 ligases was co-expressed in human embryonic kidney (HEK) cells with GR, nNOS, AR112Q or Q78 ataxin-3. We found that in each case CHIP lowered the protein levels but Mdm2 did not, suggesting that Mdm2 is either inactive on these substrates or less effective than CHIP. Parkin lowered the levels of nNOS and Q78 ataxin-3 but did not affect GR or AR112Q, suggesting that it acts redundantly to CHIP on some substrates. Similar observations were made for AR112Q and Q78 ataxin-3 in MN-1 mouse motor neuron-neuroblastoma hybrid cells, demonstrating the relevance of the findings to protein degradation processes in neuronal cells. Both CHIP and Parkin co-localized with AR112Q to ligand-dependent aggregates in cell culture and localized to intranuclear aggregates in skeletal muscle in a knock-in mouse model of SBMA, and both CHIP and Parkin were co-immunoadsorbed with AR112Q expressed in neuronal cells. Our data establish that CHIP does not play an exclusive role in regulating the turnover of Hsp90 client proteins or poly Q-expanded proteins, and indicate that the Hsp70-dependent E3 ligase Parkin acts redundantly to CHIP on some substrates.
RESULTS
We first wanted to determine whether CHIP+/+ and CHIP−/− mouse embryonic fibroblasts produce the same levels of the chaperones that bind CHIP through its TPR domain. The immunoblots of Fig. 1A confirm the absence of CHIP protein in CHIP−/− cells and show the same levels of Hsp70 and Hsp90 as in CHIP+/+ cells. The unliganded GR is a client protein whose stability is highly dependent upon the formation of heterocomplexes with Hsp90, and the treatment of cells with Hsp90 inhibitor geldanamycin results in rapid GR degradation (40). Because CHIP has been shown to promote GR ubiquitination and degradation (6), we examined the effect of geldanamycin treatment on endogenous GR in CHIP+/+ and CHIP−/− cells. Remarkably, geldanamycin at low concentrations caused the same rapid loss of GR in the presence and in the absence of CHIP (Fig. 1B and C).
Figure 1.
Geldanamycin-induced degradation of the GR occurs at the same rate in CHIP−/− as in CHIP+/+ cells. (A) Aliquots of CHIP+/+ and CHIP−/− cytosols were immunoblotted for Hsp90, Hsp70 and CHIP. (B) Time course of geldanamycin-induced GR degradation. CHIP+/+ and CHIP−/− cells were treated with 0.3 µm geldanamycin for the indicated times, GR immune pellets were prepared, and pellet proteins were immunoblotted for GR. (C) CHIP+/+ and CHIP−/− cells were treated with 0.1 or 0.3 µm geldanamycin (GA) for 6 h and assayed for the GR as above.
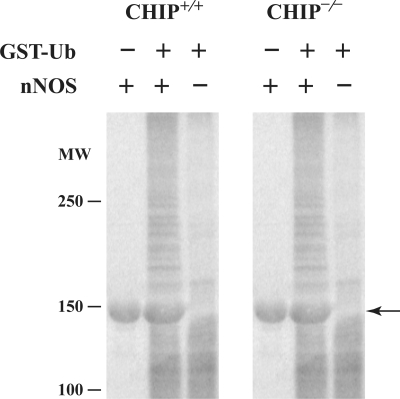
Inasmuch as CHIP−/− cells are able to carry out degradation of an Hsp90 client protein as well as CHIP+/+ cells, other E3 ubiquitin ligases must function in redundant fashion. To directly assay the relative ubiquitinating activity of these two cell lines on a known CHIP substrate, we examined the ubiquitination of nNOS in an in vitro system similar to the one we have described previously (42). In this assay, purified nNOS was incubated with GST-Ubiquitin and cytosol from CHIP+/+ or CHIP−/− cells as a source of E1, E2 and E3 ubiquitinating enzyme activity. In the presence of each cytosol, nNOS- and GST-Ubiquitin-dependent high-molecular-weight conjugates are produced to a similar degree (Fig. 2). Thus, CHIP null cells have the ability to ubiquitinate and degrade Hsp90-regulated signaling proteins, consistent with a redundancy of E3 ligase action.
Figure 2.
Cytosols from CHIP+/+ and CHIP−/− cells have the same nNOS ubiquitinating activity. Aliquots of cytosol containing the same amount of protein from each cell type were incubated for 1.5 h at 37°C with or without GST-Ubiquitin (GST-Ub) and purified nNOS as indicated. The samples were Western blotted by probing with anti-GST IgG. The large amount of unaltered nNOS (indicated by arrow) is visualized in a nonspecific manner by the immunoblotting procedure.
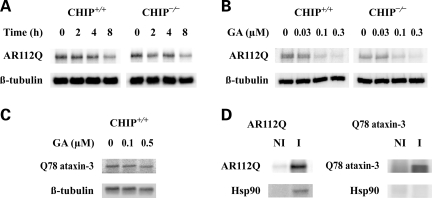
We next sought to determine whether there is functional redundancy of E3 ligases that mediate degradation of misfolded proteins, as we observed for signaling proteins. We initially examined the effect of CHIP deletion on the degradation of the poly Q-expanded AR, the disease-causing protein of SBMA. CHIP+/+ and CHIP−/− cells expressing AR112Q were treated with geldanamycin to assess time- and dose-dependent AR degradation. Like the endogenous GR, the level of exogenously expressed AR112Q decreases similarly in both cell types upon geldanamycin treatment (Fig. 3A and B). We conclude that other E3 ligases function redundantly with CHIP to promote the degradation of the poly Q AR. To examine whether similar redundancy mediates the degradation of other misfolded proteins, we expressed an expanded glutamine (Q78) form of ataxin-3. In contrast to the effects on the AR, no geldanamycin-induced degradation of Q78 ataxin-3 was observed, even in CHIP+/+ cells following 24 h drug treatment (Fig. 3C). Also, immunoadsorption of AR112Q from cytosol is accompanied by co-adsorption of Hsp90, whereas immunoadsorption of Q78 ataxin-3 is not (Fig. 3D). Thus, it appears that Q78 ataxin-3 is not an Hsp90 client protein, indicating that the presence of an expanded glutamine tract is not sufficient to determine the assembly of heterocomplexes with Hsp90.
Figure 3.
Geldanamycin-induced degradation of expressed AR112Q in CHIP+/+ and CHIP−/− cells. (A) Cells expressing AR112Q were treated for the indicated times with 0.3 µm geldanamycin, and cell lysates were immunoblotted for AR112Q and β-tubulin. (B) Cells expressing AR112Q were incubated for 6 h with the indicated concentrations of geldanamycin, and cell lysates were immunoblotted for AR112Q and β-tubulin. (C) CHIP+/+ cells expressing Q78 ataxin-3 were incubated for 24 h with the indicated concentrations of geldanamycin, and cell lysates were immunoblotted for ataxin-3 (anti-Myc) and β-tubulin. (D) Cytosols were prepared from MN-1 cells expressing AR112Q or Q78 ataxin-3. Non-immune (NI) pellets and immune (I) pellets for the expressed proteins were prepared, and pellet proteins were immunoblotted for Hsp90 and the respective expanded glutamine tract protein.
Because there are numerous E3 ligases [more than 400 potential E3s in the human genome (45)] that could potentially function in the absence of CHIP, we focus here on the two, Mdm2 and Parkin, that have been reported to interact with the major chaperones in the machinery that forms signaling protein•Hsp90 heterocomplexes. To confirm their association with Hsp70 and/or Hsp90, each E3 ligase was immunoprecipitated from cytosol of HEK cells in which they were overexpressed. As shown in Fig. 4A, both Hsp90 and Hsp70 co-immunoprecipitated with CHIP, consistent with the reported interaction of the CHIP TPR domain with each of these chaperones (5,6). Also, as previously reported (16), Hsp70 co-immunoprecipitated with Parkin, but Hsp90 did not (Fig. 4A). Under the same conditions, neither Hsp90 nor Hsp70 co-immunoprecipitated with Mdm2 (Fig. 4A). Our failure to detect an Mdm2•Hsp90 complex in cytosol under conditions where other E3 ligase–chaperone interactions are seen suggests that their interaction under native conditions is likely to be comparatively weak. To demonstrate Mdm2 binding to purified Hsp90 in vitro, Mdm2 must first be converted into zinc-dependent homo-oligomers (18), and it is unlikely that this reflects the native complex with Hsp90 in cells.
Figure 4.
Co-expression of E3 ligases with GR and nNOS. (A) Aliquots of cytosol from HEK cells transiently expressing CHIP, Mdm2 or Parkin were immunoadsorbed with nonimmune antibody (NI) or antibody (immune, I) directed against Mdm2 or myc tag (CHIP, Parkin). Co-immunoprecipitated proteins were visualized by western blot using antibodies against E3 ligase, hsp90 and hsp70. (B) Effects of E3 ligases on GR. HEK cells were transfected with rat GR cDNA (0.5 µg) and different amounts of E3 cDNA (0, 1, 2 and 3 µg), harvested 48 h later, and lysates were immunoblotted for GR, the indicated E3 and β-tubulin for a loading control. (C) Effects of E3 ligases on nNOS. HEK cells were co-transfected with cDNAs for rat nNOS and E3 ligases as above. Bar graphs in (B) and (C) show the relative amount of signaling protein in the presence of 3 µg E3 ligase expression vector as a percent of the vector plasmid control (mean ± SEM for three to seven experiments, *P < 0.01 by unpaired Student’s t-test).
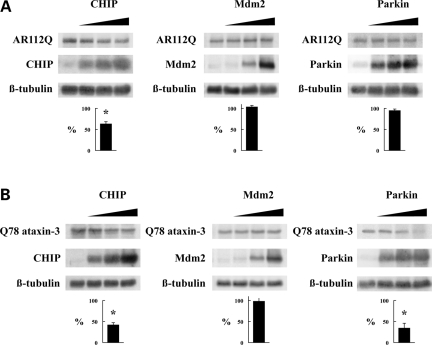
To compare the effects of CHIP, Mdm2 and Parkin on signaling proteins, each E3 ligase was co-expressed in HEK cells with the GR (Fig. 4B) or nNOS (Fig. 4C). The levels of both of these signaling proteins were decreased by co-expression of CHIP. Co-expression of Mdm2, however, did not alter their levels, suggesting that neither protein is a substrate for Mdm2 under these conditions. In contrast to the effects of Mdm2, Parkin co-expression decreased the levels of nNOS (Fig. 4C) but did not alter the levels of the GR (Fig. 4B). These data indicate that a subset of signaling proteins can be targeted for degradation by both CHIP and Parkin.
As was found with signaling proteins, co-expression of CHIP with the poly Q-expanded proteins AR112Q (Fig. 5A) or Q78 ataxin-3 (Fig. 5B) decreased the protein levels, whereas co-expression of Mdm2 did not. Co-expression of Parkin also decreased the Q78 ataxin-3 levels (Fig. 5B) but did not alter the AR112Q levels (Fig. 5A). These findings are consistent with a report showing that Parkin reduces the aggregation and toxicity of a poly Q-expanded ataxin-3 fragment by complexing with this fragment and Hsp70 to enhance ubiquitination (16). Taken together, our data suggest that CHIP and Parkin function redundantly to promote the degradation of a subset of misfolded proteins, similar to their actions on Hsp90 client proteins.
Figure 5.
Co-expression of E3 ligases with AR112Q and Q78 ataxin-3. HEK cells were co-transfected with AR112Q (A) or Q78 ataxin-3 (B) and the indicated E3 ligase, lysates were prepared 48 h later and immunoblotted for the indicated proteins. Bar graphs show the relative amount of polyglutamine protein in the presence of 3 µg E3 ligase expression vector as a percent of the vector plasmid control (mean ± SEM for three to four experiments, *P < 0.01 by unpaired Student’s t-test).
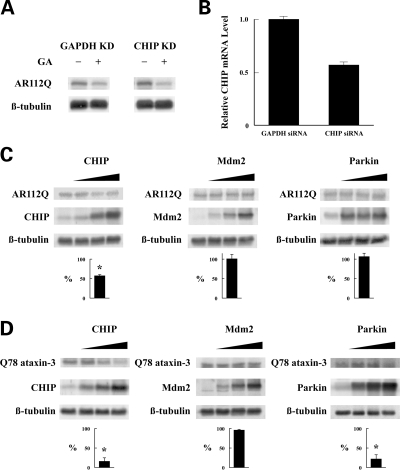
Because the data of Figs 1–5 are derived in non-neuronal cell lines, experiments with AR112Q and Q78 ataxin-3 were performed in MN-1 motor neuron-neuroblastoma hybrid cells to assess the relevance of our findings to protein degradation processes occurring in neurons. As shown in Fig. 6A, treatment with geldanamycin causes the same decrease in the level of expressed AR112Q in CHIP siRNA knockdown cells as in control (GAPDH knockdown) cells. Because our antibodies do not visualize mouse CHIP on immunoblotting, CHIP knockdown was confirmed by TaqMan assay of CHIP mRNA (Fig. 6B). As was found in non-neuronal cells, co-expression of CHIP with AR112Q in MN-1 neuronal cells decreased the AR112Q levels, whereas co-expression of Mdm2 or Parkin did not (Fig. 6C). Also, consistent with effects in non-neuronal cells, both CHIP and Parkin decreased the Q78 ataxin-3 levels in MN-1 neuronal cells but Mdm2 did not (Fig. 6D).
Figure 6.
Effects of CHIP siRNA knockdown and co-expression of E3 ligases in MN-1 mouse neuronal cells. (A) MN-1 cells transfected with AR112Q cDNA and either mouse CHIP siRNA or mouse GAPDH siRNA (as a control) were grown for 48 h and then treated for a further 6 h with geldanamycin (GA). (B) CHIP mRNA expression was determined by TaqMan assay on MN-1 cells treated with CHIP or GAPDH siRNAs as in (A). MN-1 cells co-transfected with AR112Q (C) or Q78 ataxin-3 (D) and the indicated E3 ligase were lysed after 48 h and immunoblotted for the indicated proteins. Bar graphs in (C) and (D) show the relative amount of polyglutamine protein in the presence of 3 µg E3 ligase expression vector as a percent of the vector plasmid control (mean ± SEM for three experiments, *P < 0.01 by unpaired Student’s t-test).
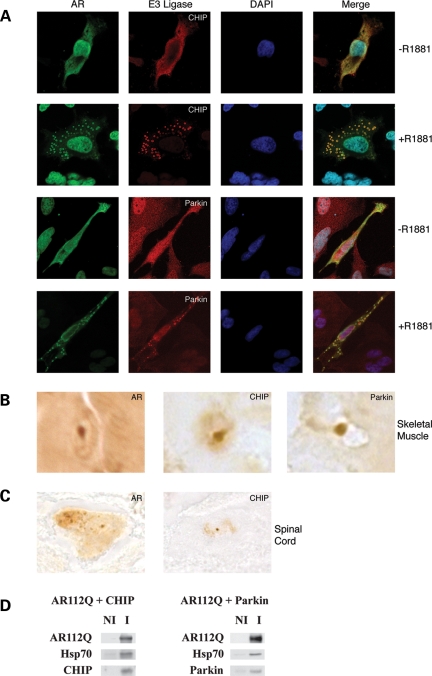
CHIP is present in aggregates of the poly Q-expanded AR (39) and ataxin-3 (38), and this co-localization has been offered as additional evidence for the importance of CHIP as the E3 ligase involved in the ubiquitination of these proteins. To determine whether multiple E3 ligases co-localize with the poly Q-expanded AR, cells exogenously expressing AR112Q were treated with the synthetic androgen R1881. In this system, AR aggregates form in the cell cytoplasm in an androgen-dependent manner. As shown in Fig. 7A, both endogenous CHIP and Parkin co-localize with AR112Q aggregates, implying that these E3 ligases moved with AR112Q and its associated chaperones in response to ligand treatment. Similarly, both CHIP and Parkin were identified in nuclear aggregates in skeletal muscle of a knock-in mouse model of SBMA (46) (Fig. 7B). Neuronal intranuclear inclusions in spinal cord of aged mice also stained for AR and CHIP (Fig. 7C). Parkin immunoreactivity, however, was not detected in spinal cord aggregates, reflecting the fact that Parkin antibodies failed to stain formalin-fixed, paraffin-embedded tissue. Nonetheless, our data establish that multiple E3 ligases co-localize with poly Q-expanded AR in models of SBMA. Also, both E3 ligases were co-immunoadsorbed with Hsp70-bound AR112Q from cytosols of MN-1 cells exogenously expressing AR112Q and CHIP or Parkin, respectively (Fig. 7D).
Figure 7.
CHIP and Parkin localize to aggregates in SBMA cell and mouse models. (A) HeLa cells transiently expressing AR112Q were treated for 24 h with the AR ligand R1881 (10 nm) or vehicle control. Localization of AR (green) and the indicated E3 ligase (red) was determined by confocal microscopy (original magnification ×630). (B) Punctate intranuclear staining of AR, CHIP and Parkin was identified in frozen sections of skeletal muscle from AR113Q knock-in male mice by immunohistochemistry (original magnification ×1000). (C) AR and CHIP were located in aggregates in spinal cord neuronal nuclei from AR113Q knock-in male mice (original magnification ×1000). Parkin antibodies failed to stain formalin-fixed, paraffin-embedded tissue. (D) CHIP and Parkin co-immunoprecipitate with AR112Q. Cytosol prepared from MN-1 cells exogenously expressing AR112Q and CHIP or Parkin were immunoadsorbed with anti-AR, and pellet proteins were immunoblotted for AR, Hsp70 and the E3 ligase.
DISCUSSION
Here, we have shown that CHIP−/− cells have the same ability to degrade the GR and AR112Q as CHIP+/+ cells following geldanamycin treatment (Figs 1 and 3). Furthermore, CHIP−/− cytosol has the same ability as CHIP+/+ cytosol to ubiquitinate purified nNOS (Fig. 2). Taken together, these observations establish that other E3 ligases act redundantly to CHIP to promote the degradation of signaling and poly Q-expanded proteins. We also show that the level of Q78 ataxin-3 was not affected by geldanamycin and that Hsp90 did not co-immunoprecipitate with Q78 ataxin-3 (Fig. 3), indicating that this protein is not an Hsp90 client. Consistent with this interpretation, an analysis of chaperones in aggregates produced in cells expressing truncated ataxin-3 protein showed the presence of Hsp70 but not Hsp90 (47). In contrast, Hsp90 is present in aggregates of both poly Q-expanded huntingtin and AR (21,22). Interestingly, the poly Q-expanded AR has been shown to be more sensitive to geldanamycin-induced degradation than wild-type AR (25), suggesting that the presence of an expanded Q tract increases the protein stabilization by Hsp90. The fact that Q78 ataxin-3 is not in a complex with Hsp90 and does not respond to geldanamycin shows that the presence of an expanded Q tract is not sufficient for heterocomplex assembly with, or stabilization by, Hsp90.
The co-expression experiments shown in Figs 4–6 demonstrate that CHIP promotes the degradation of two signaling proteins, GR and nNOS, and two poly Q-expanded proteins, AR112Q and Q78 ataxin-3. Consistent with our observations, CHIP has been implicated in the turnover of the unliganded GR (6), and there is good evidence that CHIP is the major E3 ligase involved in GR down-regulation after chronic exposure of HT22 cells to glucocorticoid (48). In contrast to the effects of CHIP, we found that co-expression of Mdm2 with each of these four substrates had no effect on protein levels. However, Mdm2 has been implicated in GR turnover, but only when the GR is complexed with p53 (49,50). Similarly, several RING finger E3 ligases have been shown to bind to the AR and modulate its activity (51,52). There are a number of reports of Mdm2-mediated ubiquitination of the AR leading to its proteasomal degradation (53–55), but this occurs only when the receptor is phosphorylated by Akt (53).
Although co-expression of Parkin did not alter the steroid receptor levels, it did cause a marked decrease in both nNOS and Q78 ataxin-3 levels. Therefore, this Hsp70-dependent E3 ubiquitin ligase acts redundantly to CHIP on a subset of substrates. While co-expression of Parkin did not affect the AR112Q levels (Fig. 5), like CHIP, it co-immunoadsorbs with AR112Q and it clearly co-localizes to androgen-dependent AR112Q aggregates in cell culture and nuclear aggregates in SBMA knock-in mice (Fig. 7). This suggests that Parkin, like CHIP, moves with the AR and its associated Hsp70 into aggregates. The failure of Parkin to act on AR112Q in co-expression experiments may reflect our inability to attain the critical ratio of Parkin:AR112Q that is necessary to promote the degradation. In contrast to AR, nNOS levels were quite sensitive to Parkin co-expression. This effect is particularly interesting in that a study of Parkin mutants that cause autosomal recessive juvenile PD also suggests a role for Parkin in determining the nNOS levels. Expression of Parkin mutants that lack ligase activity resulted in elevated levels of nitrated proteins, reactive nitrogen species and nNOS protein (56). The authors suggest that the presence of mutant Parkin in the substantia nigra may increase nitric oxide production and oxidative stress, sensitizing pigmented neurons to death following other insults (56).
Our data establish that CHIP does not play an exclusive role in regulating the turnover of Hsp90 client proteins or poly Q-expanded proteins. Furthermore, our findings suggest that Parkin functions in a redundant manner to CHIP on a subset of proteins to regulate degradation. It may be that other E3 ligases also function redundantly to CHIP. Both CHIP and Parkin bind to Hsp70, and it is likely that Hsp70 detects early stages of unfolding, thereby targeting these proteins for ubiquitination and degradation. The CHIP TPR domain also binds to Hsp90, but there is no evidence that Hsp90-bound CHIP is involved in the ubiquitination of Hsp90 client proteins. Mdm2 is the only other E3 ligase that has been reported to interact with Hsp90 (18), and it does not promote Hsp90 client protein degradation under the same conditions as the Hsp70-binding E3 ligases CHIP and Parkin. Indeed, in all the cases we know of forming a complex with Hsp90 inhibits client protein degradation by the ubiquitin/proteasome pathway (1). Thus, compounds that inhibit Hsp90 are lead drugs of great promise in the treatment of polyglutamine diseases. Considering that the Hsp70-dependent E3 ligases may act in a redundant manner, and that overexpression of Hsp70 or Hsp40 decreases polyglutamine protein levels and improves outcome in both cellular and animal models of polyglutamine diseases (30–32), Hsp70 is also an appropriate target for drug development. Thus, compounds that act like Hsp40 to stimulate the ATPase activity of Hsp70 (57,58) may constitute an important approach to the treatment of polyglutamine diseases.
MATERIALS AND METHODS
Materials
CHIP+/+ and CHIP−/− mouse embryonic fibroblasts (44) and the cDNA for CHIP (7) were provided by Dr. Cam Patterson (University of North Carolina). Rabbit anti-CHIP antibody was from Affinity Bioreagents (Golden, CO, USA). The N27F3-4 anti-72/73-kDa Hsp monoclonal IgG (anti-Hsp70) and the AC88 monoclonal IgG against Hsp90 were from StressGen Biotechnologies (Victoria, BC, Canada). Affinity-purified IgG against nNOS was from BD Transduction Laboratories (Lexington, KY, USA). The mouse monoclonal IgGs against Parkin and Mdm2 were from Cell Signaling Technology (Danvers, MA, USA) and Calbiochem, respectively. The FiGR monoclonal IgG used to immunoadsorb the mouse GR was provided by Dr. Jack Bodwell (Dartmouth Medical School, Lebanon, NH, USA) and the BuGR2 monoclonal used to immunoblot the mouse GR was from Affinity Bioreagents (Golden). The mouse monoclonal IgG against the AR (N-20) and the monoclonal anti-Myc IgG were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The mouse monoclonal anti-GST and anti-β-tubulin in ascites fluid were from Sigma (St. Louis, MO, USA). The p6R HA GR plasmid encoding the rat GR was provided by Dr. Jorge Iniguez (University of Michigan Medical School, Ann Arbor, MI, USA), and the cDNA for rat nNOS was provided by Dr. Solomon Snyder (The Johns Hopkins Medical School, Baltimore, MD, USA). The plasmid encoding AR112Q was from Dr. Kenneth Fischbeck (N.I.H.), and the cDNA for Q78 Myc-tagged human ataxin-3 was from Dr. Randall Pittman (University of Pennsylvania Medical School, Philadelphia, PA, USA). The cDNA for Myc-tagged human Parkin was provided by Dr. Ted Dawson (Johns Hopkins Medical School), and the cDNA for human Mdm2 was from Dr. Arnold Levine (Rockfeller University). Nucleofector and nucleofection reagents were from Amaxa GmbH (Köln, Germany).
Cell culture and DNA transfection
For experiments with the endogenous GR, CHIP+/+ and CHIP−/− murine fibroblasts were grown in Dulbecco’s minimum Eagle’s medium (DMEM) supplemented with 10% calf serum in 75 cm2 flasks and harvested by trypsinization. Because these cells are highly resistant to transfection by conventional means, AR112Q and Q78 ataxin-3 transfections were carried out using the Amaxa Nucleofector system. Briefly, CHIP+/+ and CHIP−/− mouse embryonic fibroblasts were grown to 70% confluence and detached by trypsinization. The pellets of 4 × 106 cells were resuspended in 100 µl of Amaxa Nucleofector solution V together with 2.5 µg of plasmid DNA. Following electroporation with Nucleofector program M-31, the cells were replated into 6-well plates. At 16 h post-transfection, the medium was replaced with fresh medium containing geldanamycin. At indicated times, the cells were washed once with phosphate-buffered saline and harvested by adding 1 ml of sodium dodecyl sulfate (SDS) sample buffer and gentle rocking for 5 min. Cell lysates were boiled for 5 min and 100 µl aliquots were submitted to SDS–polyacrylamide gel electrophoresis and immunoblotting. HEK 293T cells were cultured in minimum essential medium supplemented with 10% fetal bovine serum. Transient transfections of 293T cells were carried out with the use of a standard calcium phosphate method in 6-well plates as described previously (59). MN-1 cells were cultured in DMEM supplemented with 10% calf serum, and transfections were carried out with the Amaxa Nucleofector system. Indicated amounts of CHIP, Parkin or Mdm2 cDNA and GR, nNOS, AR112Q or Q78 ataxin-3 cDNA were transfected into 50–70% confluent cells such that the total amount of cDNA was kept constant with vector plasmid. Cells were transfected for 48 h, washed with phosphate-buffered saline, and cytosols were prepared.
Gel electrophoresis, western blotting and immunoprecipitation
To prepare cytosol, cells were washed twice with Hanks BSS, resuspended in 100 µl of HEM buffer (10 mm HEPES, pH 7.35, 1 mm EDTA, 20 mm sodium molybdate) containing 1 tablet/10 ml of Complete Mini protease inhibitor mix and 100 µm phenylmethylsulphonylfluoride, and ruptured by sonication. Sonicates were centrifuged at 300 000g for 10 min, and supernatants were aliquoted, flash frozen and stored at −80°C. Aliquots (20 µg of protein) of cytosols were resolved on 9% SDS–polyacrylamide gels and transferred to nitrocellulose membranes for immunoblotting. Immunoreactive bands were visualized by reaction with a [125I]-labeled counterantibody and scanning with a Typhoon Trio+ imaging system that calculates the relative density of each band. In co-transfection experiments, the amount of signaling protein or polyglutamine protein expressed in several experiments in the presence of 3 µg E3 ubiquitin ligase cDNA is expressed as a percentage of the control co-transfections with vector plasmid (bar graphs in Figs 4–6). To enhance the detection of endogenous GR in CHIP+/+ and CHIP−/− cells, aliquots (1 mg protein) of cytosol were immunoadsorbed to protein A-Sepharose with FiGR antibody in TEGM buffer (10 mm TES, pH 7.6, 50 mm NaCl, 4 mm EDTA, 10% glycerol, 20 mm sodium molybdate) for 2 h at 4°C. Immunopellets were washed four times with TEGM buffer and immunoadsorbed GR was resolved on 8% polyacrylamide gels and immunoblotted with BuGR2 antibody.
In vitro ubiquitination of nNOS
Rat nNOS was expressed in Sf9 insect cells using a recombinant baculovirus and purified by 2′,5′-ADP-sepharose and gel-filtration chromatography as described previously (42). To conjugate ubiquitin to nNOS, purified nNOS (13 µg) was incubated with 0.3 µl of cytosol from CHIP+/+ or CHIP−/− cells (8.3 mg protein/ml), GST-tagged ubiquitin (70 µM), 2 mm dithiothreitol, 0.7 µm ubiquitin aldehyde (to inhibit deubiquitination), 0.2 mm MG132 (to inhibit proteasome activity), and an ATP-regenerating system consisting of 4 mm ATP, 20 mm creatine phosphate, 10 mm MgCl2, and 20 units/ml creatine phosphokinase, expressed as final concentrations, for 1.5 h at 37°C in a total volume of 40 µl of 50 mm Tris–HCl, pH 7.5. After incubation, 60 µl of sample buffer was added, and the sample was loaded for western blotting.
Immunohistochemistry and immunofluorescence
For immunohistochemistry, skeletal muscle from 12- to 20-week-old mice was frozen in isopentane chilled by liquid nitrogen and cut in 7 mm thick sections. Spinal cord was harvested from 24-month-old mice, fixed in formalin, embedded in paraffin and sectioned at 5 µm. Antigen retrieval was achieved by boiling in glycine buffer (pH 3.5) for 10 min. Staining with primary antibodies against AR, CHIP and Parkin was visualized using a Vectastain ABC kit (Vector Laboratories). Images were captured using an Olympus BX41 microscope, X40 lens and Insight digital camera. For immunofluorescence, HeLa cells transfected with an AR112Q expression vector were grown in chambered slides in phenol-red free DMEM containing 10% charcoal stripped calf serum. Twenty-four hours after transfection, cells were treated with 10 nm R1881 or vehicle for 24 h, then fixed in methanol and stained. For visualization of AR, CHIP and Parkin, we used secondary antibodies conjugated to Alexa Fluor 594 and 488 (Molecular Probes). Confocal images were captured using a Zeiss LSM 510 microscope and an X63 water immersion lens.
siRNA knockdown of CHIP
MN-1 mouse embryonic motor neuron-neuroblastoma hybrid cells (60) were suspended in Nucleofector solution V (Amaxa) and mixed with 2.7 µg ON-TARGETplus SMART pool mouse CHIP (NM_019719) siRNA (Dharmacon, L-063143-01) or mouse GAPDH (NM_008085) siRNA (D-001830-20) as a control and AR112Q cDNA. The suspension was electroporated using program G-04 on the Amaxa Nucleofector. Cells were plated in 6-well plates, and 48 h later, cells were treated for 6 h with geldanamycin, after which cell lysates were collected for western blot.
CHIP expression analysis
Total RNA isolated from MN-1 cells with Trizol (Invitrogen) served as a template for cDNA synthesis using the High Capacity cDNA Archive kit from Applied Biosystems. Gene-specific primers and probes labeled with a fluorescent reporter dye and quencher were purchased from Applied Biosystems. TaqMan assays were performed using 5 ng aliquots of cDNA. Replicate tubes were analyzed for the expression of 18S rRNA using VIC-labeled probe. CT values were determined by an ABI Prism 7900HT sequence detection system, and relative expression levels were calculated using the standard curve method of analysis.
FUNDING
This work was supported by a McKnight Foundation Neuroscience of Brain Disorders Award (to A.P.L.) and by National Institutes of Health grants GM77430 (to Y.O.) and NS055746 (to A.P.L.).
ACKNOWLEDGEMENTS
We thank Drs Cam Patterson, Jack Bodwell, Jorge Iniguez, Solomon Snyder, Kenneth Fischbeck, Randall Pittman, Ted Dawson and Arnold Levine for providing cells, antibodies and plasmids used in this work.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Pratt W.B., Toft D.O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 2.Sherman M.Y., Goldberg A.L. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 3.Isaacs J.S., Xu W., Neckers L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell. 2003;3:213–217. doi: 10.1016/s1535-6108(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 4.Cyr D.M., Hohfeld J., Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 5.Ballinger C.A., Connell P., Wu Y., Hu Z., Thompson L.J., Yin L.Y., Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connell P., Ballinger C.A., Jiang J., Wu Y., Thompson L.J., Hohfeld J., Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 7.Jiang J., Ballinger C.A., Wu Y., Dai Q., Cyr D.M., Hohfeld J., Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of hsc70 as a target for ubiquitylation. J. Biol. Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 8.Bercovich B., Stancovski I., Mayer A., Blumenfeld N., Laszlo A., Schwartz A.L., Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J. Biol. Chem. 1997;272:9002–9010. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- 9.Lee D.H., Sherman M.Y., Goldberg A.L. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickey C.A., Patterson C., Dickson D., Petrucelli L. Brain CHIP: removing the culprits in neurodegenerative disease. Trends Mol. Med. 2007;13:32–38. doi: 10.1016/j.molmed.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Esser C., Scheffner M., Hohfeld J. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J. Biol. Chem. 2005;280:27443–27448. doi: 10.1074/jbc.M501574200. [DOI] [PubMed] [Google Scholar]
- 12.Zhou P., Fernandes N., Dodge I.L., Reddi A.L., Rao N., Safran H., DiPetrillo T.A., Wazer D.E., Band V., Band H. ErbB2 degradation mediated by the co-chaperone protein CHIP. J. Biol. Chem. 2003;278:13829–13837. doi: 10.1074/jbc.M209640200. [DOI] [PubMed] [Google Scholar]
- 13.Glickman M.H., Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 14.Hatakeyama S., Matsumoto M., Yada M., Nakayama K.I. Interaction of U-box-type ubiquitin-protein ligases (E3s) with molecular chaperones. Genes Cells. 2004;9:533–548. doi: 10.1111/j.1356-9597.2004.00742.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Gao J., Chung K.K.K., Huang H., Dawson V.L., Dawson T.M. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl Acad. Sci. USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai Y.C., Fishman P.S., Thakor N.V., Oyler G.A. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J. Biol. Chem. 2003;278:22044–22055. doi: 10.1074/jbc.M212235200. [DOI] [PubMed] [Google Scholar]
- 17.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the Parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 18.Burch L., Shimizu H., Smith A., Patterson C., Hupp T.R. Expansion of protein interaction maps by phage peptide display using Mdm2 as a prototypical conformationally flexible target protein. J. Mol. Biol. 2004;337:129–145. doi: 10.1016/j.jmb.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Peng Y., Chen L., Changgong L., Lu W., Chen J. Inhibition of Mdm2 by hsp90 contributes to mutant p53 stabilization. J. Biol. Chem. 2001;276:40583–40590. doi: 10.1074/jbc.M102817200. [DOI] [PubMed] [Google Scholar]
- 20.Uryu K., Richter-Landsberg C., Welch W., Sun E., Goldbaum O., Norris E.H., Pham C.T., Yazawa I., Hilburger K., Micsenyi M., et al. Convergence of heat shock protein 90 with ubiquitin in filamentous α-synuclein inclusions of α-synucleinopathies. Am. J. Pathol. 2006;168:947–961. doi: 10.2353/ajpath.2006.050770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitsui K., Nakayama H., Akagi T., Nekooki M., Ohtawa K., Takio K., Hashikawa T., Nukina N. Purification of polyglutamine aggregates and identification of elongation factor-1α and heat shock protein 84 as aggregate-interacting proteins. J. Neurosci. 2002;22:9267–9277. doi: 10.1523/JNEUROSCI.22-21-09267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas M., Harrell J.M., Morishima Y., Peng H.M., Pratt W.B., Lieberman A.P. Pharmacologic and genetic inhibition of hsp90-dependent trafficking reduces aggregation and promotes degradation of the expanded glutamine androgen receptor without stress protein induction. Hum. Mol. Genet. 2006;15:1876–1883. doi: 10.1093/hmg/ddl110. [DOI] [PubMed] [Google Scholar]
- 23.Auluck P.K., Bonini N.M. Pharmacological prevention of Parkinson disease in Drosophila. Nat. Med. 2002;8:1185–1186. doi: 10.1038/nm1102-1185. [DOI] [PubMed] [Google Scholar]
- 24.Hay D.G., Sathasivam K., Tobaben S., Stahl B., Marber M., Mestril R., Mahal A., Smith D.L., Woodman B., Bates G.P. Progressive decrease in chaperone protein levels in a mouse model of Huntington’s disease and induction of stress proteins as a therapeutic approach. Hum. Mol. Genet. 2004;13:1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 25.Waza M., Adachi H., Katsuno M., Minamiyama M., Sang C., Tanaka F., Inukai A., Doyu M., Sobue G. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat. Med. 2005;11:1088–1095. doi: 10.1038/nm1298. [DOI] [PubMed] [Google Scholar]
- 26.Klucken J., Shin Y., Masliah E., Hyman B.T., McLean P.J. Hsp70 reduces α-synuclein aggregation and toxicity. J. Biol. Chem. 2004;279:25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 27.Jana N.R., Tanaka M., Wang G., Nukina N. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum. Mol. Genet. 2000;9:2009–2018. doi: 10.1093/hmg/9.13.2009. [DOI] [PubMed] [Google Scholar]
- 28.Bailey C.K., Andriola I.F.M., Kampinga H.H., Merry D.E. Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum. Mol. Genet. 2002;11:515–523. doi: 10.1093/hmg/11.5.515. [DOI] [PubMed] [Google Scholar]
- 29.Auluck P.K., Chan H.Y.E., Trojanowski J.Q., Lee V.M.Y., Bonini N.M. Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 30.Warrick J.M., Chan H.Y.E., Gray-Board G.L., Chai Y., Paulson H.L., Bonini N.M. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 31.Chan H.Y.E., Warrick J.M., Gray-Board G.L., Paulson H.L., Bonini N.M. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum. Mol. Genet. 2000;9:2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- 32.Adachi H., Katsuno M., Minamiyama M., Sang C., Pagoulatos G., Angelidis C., Kusakabe M., Yoshiki A., Kobayashi Y., Doyu M., Sobue G. Heat shock protein 70 chaperone overexpression ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model by reducing nuclear-localized mutant androgen receptor protein. J. Neurosci. 2003;23:2203–2211. doi: 10.1523/JNEUROSCI.23-06-02203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muchowski P.J., Wacker J.L. Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 34.Shin Y., Klucken J., Patterson C., Hyman B.T., McLean P.J. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates α-synuclein degradation decisions between proteasomal and lysosomal pathways. J. Biol. Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 35.Miller V.M., Nelson R.F., Gouvion C.M., Williams A., Rodriguez-Lebron E., Harper S.Q., Davidson B.L., Rebagliati M.R., Paulson H.L. CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J. Neurosci. 2005;25:9152–9161. doi: 10.1523/JNEUROSCI.3001-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas M., Dadgar N., Aphale A., Harrell J.M., Kunkel R., Pratt W.B., Lieberman A.P. Androgen receptor acetylation site mutations cause trafficking defects, misfolding, and aggregation similar to expanded glutamine tracts. J. Biol. Chem. 2004;279:8389–8395. doi: 10.1074/jbc.M311761200. [DOI] [PubMed] [Google Scholar]
- 37.Al-Ramahi I., Lam Y.C., Chen H.K., de Gouyon B., Zhang M., Perez A.M., Branco J., de Haro M., Patterson C., Zoghbi H.Y., Botas J. CHIP protects from the neurotoxicity of expanded and wild-type ataxin-1 and promotes their ubiquitination and degradation. J. Biol. Chem. 2006;281:26714–26724. doi: 10.1074/jbc.M601603200. [DOI] [PubMed] [Google Scholar]
- 38.Jana N.R., Dikshit P., Goswami A., Kotliarova S., Murata S., Tanaka K., Nukina N. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J. Biol. Chem. 2005;280:11635–11640. doi: 10.1074/jbc.M412042200. [DOI] [PubMed] [Google Scholar]
- 39.Adachi H., Waza M., Tokui K., Katsuno M., Minamiyama M., Tanaka F., Doyu M., Sobue G. CHIP overexpression reduces mutant androgen receptor protein and ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model. J. Neurosci. 2007;27:5115–5126. doi: 10.1523/JNEUROSCI.1242-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitesell L., Cook P. Stable and specific binding of heat shock protein 90 by geldanamycin disrupts glucocorticoid receptor function in intact cells. Mol. Endocrinol. 1996;10:705–712. doi: 10.1210/mend.10.6.8776730. [DOI] [PubMed] [Google Scholar]
- 41.Bender A.T., Silverstein A.M., Demady D.R., Kanelakis K.C., Noguchi S., Pratt W.B., Osawa Y. Neuronal nitric-oxide synthase is regulated by the hsp90-based chaperone system in vivo. J. Biol. Chem. 1999;274:1472–1478. doi: 10.1074/jbc.274.3.1472. [DOI] [PubMed] [Google Scholar]
- 42.Peng H.M., Morishima Y., Jenkins G.J., Dunbar A.Y., Lau M., Patterson C., Pratt W.B., Osawa Y. Ubiquitylation of neuronal nitric-oxide synthase by CHIP, a chaperone-dependent E3 ligase. J. Biol. Chem. 2004;279:52970–52977. doi: 10.1074/jbc.M406926200. [DOI] [PubMed] [Google Scholar]
- 43.Matsumoto M., Yada M., Hatakeyama S., Ishimoto H., Tanimura T., Tsuji S., Kakizuka A., Kitagawa M., Nakayama K.I. Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J. 2004;23:659–669. doi: 10.1038/sj.emboj.7600081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai Q., Zhang C., Wu Y., McDonough H., Whaley R.A., Godfrey V., Li H.H., Madamanchi N., Xu W., Neckers L., Cyr D., Patterson C. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W., Chanda S.K., Micik I., Joazeiro C.A.P. Methods for the functional genomic analysis of ubiquitin ligases. Methods Enzymol. 2005;398:280–291. doi: 10.1016/S0076-6879(05)98023-3. [DOI] [PubMed] [Google Scholar]
- 46.Yu Z., Dadgar N., Albertelli M., Gruis K., Jordan C., Robins D.M., Lieberman A.P. Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. J. Clin. Invest. 2006;116:2663–2673. doi: 10.1172/JCI28773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chai Y., Koppenhafer S.L., Bonini N.M., Paulson H.L. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J. Neurosci. 1999;19:10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X., DeFranco D.B. Alternative effects of the ubiquitin-proteasome pathway on glucocorticoid receptor down-regulation and transactivation are mediated by CHIP, an E3 ligase. Mol. Endocriol. 2005;19:1474–1482. doi: 10.1210/me.2004-0383. [DOI] [PubMed] [Google Scholar]
- 49.Kinyamu H.K., Archer T. Estrogen receptor-dependent proteasomal degradation of the glucocorticoid receptor is coupled to an increase in Mdm2 protein expression. Mol. Cell. Biol. 2003;23:5867–5881. doi: 10.1128/MCB.23.16.5867-5881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sengupta S., Wasylyk B. Ligand-dependent interaction of the glucocorticoid receptor with p53 enhances their degradation by Hdm2. Genes Dev. 2001;15:2367–2380. doi: 10.1101/gad.202201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang H.Y., Yeh S., Fujimoto N., Chang C. Cloning and characterization of human prostate coactivator ARA54, a novel protein that associates with the androgen receptor. J. Biol. Chem. 1999;274:8570–8576. doi: 10.1074/jbc.274.13.8570. [DOI] [PubMed] [Google Scholar]
- 52.Poukka H., Karvonen U., Yoshikawa N., Tanaka H., Palvimo J.J., Janne O.A. The RING finger protein SNURF modulates nuclear trafficking of the androgen receptor. J. Cell Sci. 2000;113:2991–3001. doi: 10.1242/jcs.113.17.2991. [DOI] [PubMed] [Google Scholar]
- 53.Lin H.K., Wang L., Hu Y.C., Altuwaijri S., Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 2002;21:4037–4038. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaughan L., Logan I.R., Neal D.E., Robson C.N. Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 2005;33:13–26. doi: 10.1093/nar/gki141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cha T.L., Qiu L., Chen C.T., Wen Y., Hung M.C. Emodin down-regulates androgen receptor and inhibits prostate cancer cell growth. Cancer Res. 2005;65:2287–2295. doi: 10.1158/0008-5472.CAN-04-3250. [DOI] [PubMed] [Google Scholar]
- 56.Hyun D.H., Lee M.H., Hattori N., Kubo S.I., Mizuno Y., Halliwell B., Jenner P. Effect of wild-type or mutant parkin on oxidative damage, nitric oxide, antioxidant defenses, and the proteasome. J. Biol. Chem. 2002;277:28572–28577. doi: 10.1074/jbc.M200666200. [DOI] [PubMed] [Google Scholar]
- 57.Fewell S.W., Smith C.M., Lyon M.A., Dumitrescu T.P., Wipf P., Day B.W., Brodsky J.L. Small molecule modulators of endogenous and co-chaperone-stimulated Hsp70 ATPase activity. J. Biol. Chem. 2004;279:51131–51140. doi: 10.1074/jbc.M404857200. [DOI] [PubMed] [Google Scholar]
- 58.Evans C.G., Wisen S., Gestwicki J.E. Heat shock proteins 70 and 90 inhibit early stages of amyloid β-(1–42) aggregation in vitro. J. Biol. Chem. 2006;281:33182–33191. doi: 10.1074/jbc.M606192200. [DOI] [PubMed] [Google Scholar]
- 59.Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. Introduction of DNA into Mammalian Cells. New York: John Wiley & Sons; 1993. [Google Scholar]
- 60.Salazar-Grueso E.F., Kim S., Kim H. Embryonic mouse spinal cord motor neuron hybrid cells. Neuroreport. 1991;2:505–508. doi: 10.1097/00001756-199109000-00002. [DOI] [PubMed] [Google Scholar]