Abstract
Identifying the factors responsible for the structuring of genetic diversity is of fundamental importance for biodiversity conservation. However, arriving at such understanding is difficult owing to the many factors involved and the potential interactions between them. Here, we present an example of how such interactions can preclude us from arriving at a complete characterization of the demographic history and genetic structure of a species. Ctenomys rionegrensis is a species with restricted dispersal abilities and, as such, should exhibit an isolation by distance (IBD) pattern, which previous studies were unable to uncover. It was therefore concluded that this species underwent a recent population expansion. Using a novel hierarchical Bayesian method, we show that the inability to detect the IBD pattern is due to the interaction between elevation and geographical distance. We posit that populations in low areas suffer periodic floods that may reduce local population sizes, increasing genetic drift, a process that masks the effect of distance on genetic differentiation. Our results do not refute the possibility that the populations of C. rionegrensis underwent a recent population expansion but they indicate that an alternative scenario described by a metapopulation model at or near migration-drift equilibrium cannot be excluded either.
Keywords: genetic structure, isolation by distance, population expansion, Bayesian methods
1. Introduction
For many decades, population geneticists have been interested in quantifying the relative contribution of different evolutionary forces in shaping the genetic variation observed among living organisms. Genetic variation is structured in space, being influenced by evolutionary factors such as genetic drift, selection, migration and mutation, which in turn must operate within the historical and biological context of each species. Understanding the processes responsible for the structuring of genetic variation is of fundamental importance in the fields of biodiversity conservation, improvement of agricultural crops and breeds, and identification of disease genes in humans and economically important species. Arriving at such understanding, however, is difficult owing to the many interactions that may exist between demographic and evolutionary processes and between the different environmental factors influencing genetic structuring. In particular, it is frequently difficult to distinguish scenarios of recent population expansion from those of near equilibrium between migration and genetic drift. It is generally assumed that these two scenarios have contrasting effects on the pattern of spatial apportionment of genetic variation (Matocq et al. 2000). For example, in the case of species with restricted dispersal abilities that are at migration-drift equilibrium, we expect to observe a correlation between the genetic distance between pairs of populations and the geographical distance that separates them, this is the so-called isolation by distance (IBD) pattern. Its absence can be interpreted as evidence for a non-equilibrium situation where the species has recently colonized its current distribution (Slatkin 1993). This conclusion is, however, valid only for distant locations. Indeed, when the range expansion is rapid with a single ancestral population giving rise to all extant demes at the same time t in the past, it is still possible to detect IBD among nearby populations (cf. fig. 2, Slatkin 1993). Nevertheless, when range expansion is more gradual, the IBD pattern among nearby demes may become too weak to be detected. This would be the case if, for example, there is a stepwise colonization process where at time t in the past a single population gives rise to one neighbouring deme and all subsequent colonizations originate from the most recently colonized deme only (Good's model in Slatkin 1993). In this latter case, differentiation between the ancestral deme and the descendant demes will increase as the age of the descendant demes decreases (cf. fig. 8, Slatkin 1993). Thus, it may be possible to identify a gradual range expansion by looking at the effect that the distance from the ancestral population (a surrogate for the age of the descendant population) has on genetic differentiation. If it is unclear which of all the extant demes is the ancestral population, then it is possible to use latitude and longitude under the assumption that colonization progressed steadily from one extreme of the range to the other (e.g. from the southeast to the northwest), as Foll & Gaggiotti (2006) have done in the case of humans.
Another potential explanation for an apparent absence of IBD pattern that has not yet been evaluated is that an environmental factor that has not been taken into consideration precludes the detection of the effect of geographical distance. For example, environmental perturbations that lead to periodic decreases in population size (e.g. floods, droughts, epidemics) can greatly increase the effect of genetic drift in a subset of the subpopulations. Thus, two nearby populations may become genetically differentiated despite being geographically close. Standard population genetics models do not allow us to uncover such effects, but recent developments in the field of statistical genetics have made this possible. Here, we present an example of a situation where the effect of an important environmental factor (elevation) hides the effect of distance on genetic differentiation and, therefore, precludes us from accurately characterizing the demographic history of a population when using standard methods.
We use as case study a species of South American rodents of the genus Ctenomys, commonly known as tuco-tucos, which is the most speciose among all subterranean rodent genera (Reig et al. 1990; Cook & Lessa 1998; Lessa & Cook 1998; Castillo et al. 2005). Populations of Ctenomys are composed of semi-isolated demes, occupying patches of habitat where soil hardness and particle size provide suitable conditions for burrowing activities (Busch et al. 2000). Species-specific habitat requirements, such as those related to body size differences (see Lessa & Thaeler 1989) and other factors intrinsically associated with the species' biology (i.e. pelage crypticity, Krupa & Geluso 2000), strongly influence the type of soil subterranean rodents use to build their burrows, affecting the connectivity among local populations and resulting phylogeographic patterns. Ctenomys shares with other subterranean rodents restricted dispersal capabilities, small local effective population numbers, high territoriality and, in some cases, socially structured mating systems (Reig et al. 1990; Lacey 2000; Massarini & de Freitas 2005).
Several lines of evidence suggest that the Río Negro tuco-tuco, Ctenomys rionegrensis, has undergone a recent population expansion into its current Uruguayan distribution from a more restricted range (D'Elia et al. 1998; Wlasiuk et al. 2003). Wlasiuk et al. (2003) found no correlations between genetic distances based on allozymes, cytochrome b sequences or microsatellites with each other, as well as between any of these and geographical distances. Morphological differentiation appears to show a similar pattern (D'Anatro & Lessa 2006). Additionally, Wlasiuk et al. (2003) carried out several analyses that specifically test for signatures of population expansion using mtDNA and, although non-significant, they were consistent in suggesting such a scenario. Moreover, Fu's test uncovered a significant excess of low-frequency haplotypes, which could be explained by either a departure from the neutral mutation model or a recent expansion. Pleistocene and Holocene marine transgressions may have played an important role in such demographic changes since it is known that these perturbations reached the area of the present distribution of C. rionegrensis in Uruguay (Alonso 1978; Sprechmann 1978). Thus, the prevailing view is that populations of C. rionegrensis have differentiated relatively recently and, therefore, there has been insufficient time for a pattern of IBD to establish. An alternative view that has not been evaluated is that the population may be at or near equilibrium and that the signature of differentiation can be associated to distance and other so far undisclosed factors; their interaction could then preclude us from detecting the pattern of IBD.
Recent advances in statistical genetics have led to the development of Bayesian approaches that allow us to investigate potential interactions between environmental factors involved in the genetic structuring of populations. Here, we use a recently developed method (Foll & Gaggiotti 2006) to try to disentangle the effects of demographic history and those of various environmental factors. Using this approach, we expand previous analyses of C. rionegrensis populations and uncover an IBD pattern that could be suggestive of a near equilibrium situation. Additionally, we show that genetic differentiation in this population is multifactorially determined.
2. Material and methods
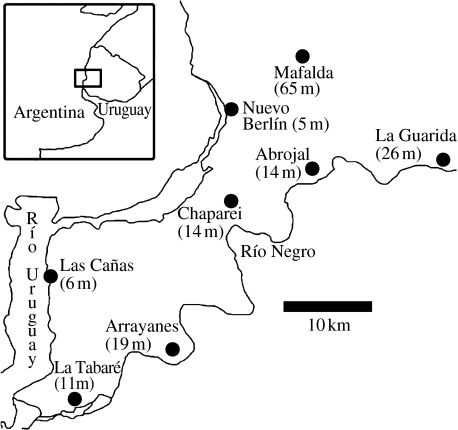
The genetic data used in our analyses consist of the 11 microsatellite loci published by Wlasiuk et al. (2003) for C. rionegrensis (see their appendix). These data were collected from eight sampling locations that cover the geographical range of this species in Uruguay (figure 1). In terms of environmental data, we used the area of each population's patch, estimated from a LANDSAT ETM+ image of the region, distance between populations, and elevation above sea level obtained from a digital elevation model of the region (table 1). This latter factor was considered as a proxy for the effect of floods, which can lead to local population bottlenecks and increased genetic drift. We also considered latitude (noted y) and longitude (noted x) expressed using the Universal Transversed Mercator planar transformation in order to correct for the curvature of the Earth. The effect of distance was modelled using the average distance between each focal population and all other populations, which is a measure akin to connectivity.
Figure 1.
Sampling localities for C. rionegrensis in Uruguay. The elevation of each population is given in parenthesis next to the name of each sampling site.
Table 1.
Environmental variables and posterior mode of population specific FSTs for the eight populations of C. rionegrensis in western Uruguay.
| population | area (km2) | distance (km) | elevation (m) | FST (HPDI) |
|---|---|---|---|---|
| 1. Chaparei | 0.13 | 24.06 | 14 | 0.258 (0.185; 0.369) |
| 2. Abrojal | 0.40 | 27.26 | 14 | 0.412 (0.301; 0.538) |
| 3. La Guarida | 0.08 | 38.47 | 26 | 0.687 (0.538; 0.813) |
| 4. La Tabaré | 0.07 | 44.62 | 11 | 0.479 (0.374; 0.595) |
| 5. Las Cañas | 0.09 | 39.09 | 6 | 0.601 (0.474; 0.743) |
| 6. Arrayanes | 1.58 | 28.76 | 19 | 0.185 (0.127; 0.272) |
| 7. Mafalda | 0.03 | 34.31 | 65 | 0.281 (0.199; 0.389) |
| 8. Nuevo Berlín | 0.10 | 26.99 | 5 | 0.984 (0.892; 1.000) |
We used the hierarchical Bayesian method of Foll & Gaggiotti (2006), as implemented in the programme Geste, to test for the effect that different environmental factors may have on the genetic structure of C. rionegrensis. This method estimates the genetic differentiation between each local population and the overall metapopulation (as measured by a local population FST value) using the approach first put forward by Balding & Nichols (1995) and relates it to environmental factors using a generalized linear model. For the moment, the software that implements this method allows for a maximum of two factors, thus we carried out multiple analyses, each considering two factors at a time. The consideration of two factors and their interaction leads to nine alternative models, and the method provides posterior probabilities for each one of them using a reversible jump Markov chain Monte Carlo (MCMC) approach. The model with the highest posterior probability is the one that best explains the data. In all cases, we used 10 pilot runs of 2000 iterations to obtain the parameters of the proposal distributions used by the MCMC. We also used an additional burn in of 5×104 iterations and a thinning interval of 50. All estimates were obtained using a sample size of 20 000.
3. Results
We first investigated the hypothesis of a gradual population expansion. If this hypothesis is correct, we can assume a fission model in which successive founder events would lead to a gradual increase in genetic differentiation between local and ancestral populations as the distance between them (a proxy for deme age; see §1) increases. Such an effect can be uncovered by a Geste analysis using latitude and longitude as factors (Foll & Gaggiotti 2006). Results show that posterior probabilities are fairly similar for models 1 (constant factor only) and 2 (longitude only) and somewhat higher for model 4 (latitude only). The overall posterior probability for models that include an effect of longitude only (2 and 3) is 0.27. The corresponding value for models that include latitude only (4 and 5) is 0.34. Finally, the posterior probability for models that include both factors (6–9) is 0.15, while that of the null model is 0.24. The effect of longitude is very weak (modal value for αx=−0.008, highest posterior density interval (HPDI)=(−1.17; 1.14)) while that of latitude is somewhat stronger (modal value for αy=0.24, HPDI=(−0.79; 1.52)).
These results provide evidence for a gradual range expansion. However, the genetic signal is somewhat weak, which suggest that enough time may have elapsed since colonization to approach migration-drift equilibrium. If this is the case, C. rionegrensis may be described by a metapopulation model that is at or near migration-drift equilibrium. The absence of IBD pattern could be the result of an interaction between distance and some other undisclosed factor. Thus, we investigated the effect of habitat patch area and elevation and their interaction with the average distance among populations. We carried out analyses that considered all possible pairwise combinations of these three factors. Only the analysis that considered elevation and distance provided a strong and unambiguous result showing an effect on the genetic structuring (table 2). More specifically, the overall posterior probability for the models that considered both factors and their interaction (8 and 9) was 0.67; all other models have a much lower posterior probability. Models that include the effect of distance only have a posterior probability of 0.05, while those that include elevation only add up to 0.20. Thus, neither factor by itself explains genetic structuring well; instead, they act concurrently and in particular through their interaction. The very strong effect of the interaction is illustrated by the dramatic decrease in the variance that remains unexplained by the model, being 1.22 for the model that includes elevation only, 1.51 for the one that considers distance only, 1.20 for the model with both factors but no interaction and 0.74 for the model with the interaction. The correlation between elevation and distance is extremely low (r=0.08), thus the observed result is not an artefact resulting from the mimicking of a quadratic model in one of the two explicative factors.
Table 2.
Posterior probabilities associated with the nine alternative models arising in the analysis that considers elevation above sea level and distance. (σ2 is the posterior mode of unexplained variance associated to each model. Values in parenthesis correspond to the 95 per cent highest posterior probability interval. The model with the highest probability and the lowest variance is highlighted in italics).
| model | probability | σ2 (HPDI) |
|---|---|---|
| 1. constant | 0.048 | 1.62 (0.40; 6.36) |
| 2. elevation | 0.161 | 1.22 (0.34; 5.51) |
| 3. constant+elevation | 0.036 | 1.50 (0.53; 6.89) |
| 4. distance | 0.040 | 1.51 (0.51; 6.17) |
| 5. constant+distance | 0.008 | 1.61 (0.66; 7.31) |
| 6. elevation+distance | 0.030 | 1.20 (0.43; 6.57) |
| 7. constant+elevation+distance | 0.006 | 1.51 (0.50; 7.15) |
| 8. elevation+distance+elevation×distance | 0.586 | 0.74 (0.20; 3.60) |
| 9. constant+elevation+distance+elevation × distance | 0.086 | 0.87(0.17; 4.39) |
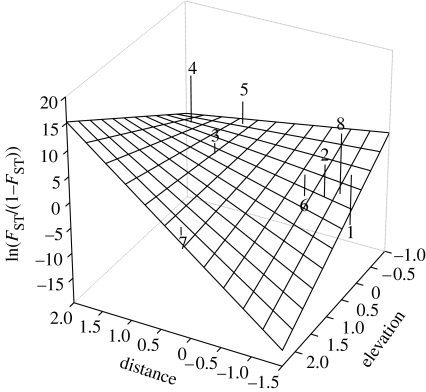
The magnitude of the elevation effect is measured by αE=−1.26; HPDI=(−2.345; −0.2056), which is negative, indicating that genetic differentiation increases with decreasing elevation. The effect of distance is positive (αD=1.38; HPDI=(0.00406; 2.69)), indicating that, as expected, genetic differentiation increases with geographical distance (figure 2). Finally, the effect of the interaction is much stronger than the effect of either factors alone (αE×D=3.26; HPDI=(0.6621; 5.994)). This interaction indicates that distance manifests its effect only among populations at higher elevation because strong genetic drift at low elevation hides the IBD pattern (figure 2). In order to verify this assertion, we carried out an additional analysis excluding the two lowest populations (Las Cañas and Nuevo Berlin, table 1). The results differ dramatically from those of the analysis that included all demes. More specifically, the posterior probability of models that include the interaction (8 and 9) becomes negligible (the overall posterior probability for them decreased to 0.045) while that of models that include only distance increased to 0.41. Additionally, the posterior probability of models that include elevation decreased to 0.16 and that of models that include both factors increased to 0.07. Finally, the probability of the null model increased to 0.32, but it is still lower than that of models that include distance only. These results indicate that the IBD pattern is present but is hidden by the strong genetic drift affecting low-altitude populations.
Figure 2.
Three-dimensional plot of elevation above sea level, distance and genetic differentiation ln[FST/(1−FST)] for eight populations of C. rionegrensis in western Uruguay. Numbers identify local populations as per table 1.
We can compare the results of the analysis that considers latitude and longitude with those of the one that includes elevation and distance by focusing on the estimates of the variance that remain unexplained by the models with the highest posterior probability. In the case of the latitude/longitude analysis, the variance that remains unexplained by the best model is 1.52 (HPDI=(0.52; 6.32)). On the other hand, the respective estimate for the elevation/distance analysis that includes all populations is 0.74 (HPDI=(0.2; 3.60)). The much lower variance of the latter suggests that the near equilibrium scenario is a better description of the demographic history of C. rionegrensis.
4. Discussion
The results presented here illustrate very clearly the importance of considering the effect of interactions between environmental factors in studies of population genetic structure. In the specific case of the example we consider, the interaction between geographical distance and elevation above sea level hides the pattern of IBD that is expected in species with limited dispersal capabilities. Populations of C. rionegrensis in western Uruguay are located near rivers and, therefore, populations in low areas are potentially subjected to periodic floods that may reduce local population sizes and increase the effect of genetic drift; a process that masks the effect of distance on genetic differentiation. Figure 2 shows that the effect of distance does increase with elevation, a pattern that is captured by the interaction term of the generalized linear model implemented in the hierarchical Bayesian approach used for our analyses.
It is worth emphasizing that it is the consideration of the interaction between environmental factors that allow us to identify the effect of geographical distance on genetic differentiation. In this regard, we note that we carried out a multivariate Mantel test to estimate the correlation between pairwise genetic and geographical distances with difference in elevation partialled out and the results were non-significant. This highlights the usefulness of new Bayesian methods that allow us to consider complex models and to investigate the effect of factors that are population specific (such as elevation) and cannot be measured in terms of pairwise differences between populations.
The structuring of neutral genetic variation in natural populations is influenced by its demographic history. Thus, the analysis of genetic data can in principle allow us to distinguish between alternative hypotheses concerning the processes that have led to current spatial patterns of genetic structuring. However, achieving this goal is not straightforward because the differences between the genetic signatures left by alternative demographic processes are sometimes subtle. As explained in §1, a scenario of migration-drift equilibrium in species with limited dispersal will lead to a clear IBD pattern (Slatkin 1993). However, an alternative non-equilibrium scenario can also lead to IBD but only among nearby populations. This small-scale IBD can be discernible in the case of species that underwent a rapid range expansion, but it would be very difficult to uncover if the range expansion was gradual. Nevertheless, in this latter case, we expect to observe a progressive genetic differentiation between the ancestral deme and the descendant demes as the distance between them (a surrogate for the age of the descendant population) increases.
Our analyses suggest that the observed genetic structure of C. rionegrensis could be described by a metapopulation model that is approaching migration-drift equilibrium after having undergone a gradual range expansion. However, the alternative scenario of a rapid recent expansion cannot be completely excluded because the geographical scale considered in this study (64 km between the two most distant populations) may be too small, in which case a rapid population expansion could also have led to the observed IBD pattern. In this regard, we note that at least one of the many population expansion tests carried out by Wlasiuk et al. (2003) is significant, and although the others had a poor fit, they all were consistent in suggesting population expansion. Nevertheless, whether the population expansion of C. rionegrensis in western Uruguay is recent or not, our analysis clearly demonstrates that this species exhibits an IBD pattern.
Our estimates of population-specific FSTs can be used to characterize the genetic distinctiveness of each local population. The larger the local FST is, the more genetically different this population is from the metapopulation as a whole. Poor connectivity and/or small effective sizes lead to high FST. The two populations located in low elevation areas (Nuevo Berlín and Las Cañas) have the largest and third largest FSTs, most probably due to the fact that, as already mentioned, they may have small population sizes due to recurrent flooding. The distinctiveness of these two populations at the molecular level is also manifested at the morphological level since they are only composed of individuals with melanic pelage. This correspondence between high population-specific FST and melanic colour fixation gives additional support to the hypothesis that drift alone is responsible for the fixation of this coat type.
The lack of detection of IBD pattern in species with low dispersal capabilities is not uncommon (e.g. Dixon et al. 2007; Florin & Hoglund 2007). As mentioned before, a commonly invoked explanation for this observation is that the species is not yet at the migration-drift equilibrium because it has recently undergone a spatial expansion. Other examples of this situation are studies on pocket gophers (Alvarez-Castañeda & Patton 2004), other tuco-tucos (Mora et al. 2006) and mustelids (Congdon et al. 2000). Another invoked explanation is that Euclidean distance does not take into account the effect of landscape structure. Thus, there has been a growing interest in using landscape ecology measures based on the least cost path (LCP) model (Coulon et al. 2004), to better explain observed patterns of genetic differentiation (Spear et al. 2005; Broquet et al. 2006; Vuilleumier & Fontanillas 2007). LCP is supposed to reflect relative movement difficulties through different habitat types and, therefore, requires the assumption of hypotheses on how the effect of different landscape attributes can limit or facilitate dispersal. Another problem of LCP measures is that they cannot incorporate the effect of independent parallel pathways connecting sampling locations (McRae 2006). To overcome this problem, McRae (2006) has proposed a distance metric based on circuit theory. However, as is the case for LCP-based metrics, the resistance metric he proposed incorporates the effect of many environmental factors simultaneously and, therefore, does not allow us to evaluate the relative importance of each one of them separately. Thus, although LCP and resistance metric approaches can be useful for descriptive purposes, they do not allow us to test hypothesis about the factors responsible for the genetic structuring of natural populations.
Examples such as the one we present here illustrate the fact that although it is sometimes useful to present equilibrium and non-equilibrium scenarios as mutually exclusive, in reality most species represent intermediate situations along a continuum that extends between these two extreme scenarios. The use of novel hierarchical Bayesian methods in conjunction with more traditional approaches allows us to fully characterize the complex demographic and evolutionary histories of species such as the one we analyse here.
Acknowledgements
This research was supported by grant A05B01 from the programme ECOS-Sud (France/Secyt-Argentina). Additional funding was provided by grants from CONICET (PIP-5844) and FONCYT (PICT-38361) to M.J.K. and from Fond National de la Science to O.E.G. (ACI-Impbio-2004-42-ADGP). M.J.K. is a researcher from CONICET, Argentina. Enrique Lessa and an anonymous reviewer provided very useful comments that greatly improved the original manuscript.
References
- Alonso C. La fauna de moluscos del yacimiento de Playa Pascual con referencia a otros yacimientos estuáricos y marinos del cuaternario de Uruguay. Com. Soc. Malaco. Uruguay. 1978;4:365–383. [Google Scholar]
- Alvarez-Castañeda S, Patton J. Geographic genetic architecture of pocket gopher (Thomomys bottae) populations in Baja California, Mexico. Mol. Ecol. 2004;13:2287–2301. doi: 10.1111/j.1365-294X.2004.02243.x. doi:10.1111/j.1365-294X.2004.02243.x [DOI] [PubMed] [Google Scholar]
- Balding D.J, Nichols R.A. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica. 1995;96:3–12. doi: 10.1007/BF01441146. doi:10.1007/BF01441146 [DOI] [PubMed] [Google Scholar]
- Broquet T, Ray N, Petit E, Fryxell J, Burel F. Genetic isolation by distance and landscape connectivity in the American marten (Martes americana) Landsc. Ecol. 2006;21:877–889. doi:10.1007/s10980-005-5956-y [Google Scholar]
- Busch C, Antinuchi C, del Valle J, Kittlein M, Malizia A, Vassallo A, Zenuto R. Population ecology of subterranean rodents. In: Lacey E, Patton J, Cameron G, editors. Life underground: the biology of subterranean rodents. University of Chicago Press; Chicago, IL: 2000. pp. 183–226. [Google Scholar]
- Castillo A, Cortinas M, Lessa E. Rapid diversification of South American tuco-tucos (Ctenomys; Rodentia, Ctenomyidae): contrasting mitochondrial and nuclear intron sequences. J. Mammal. 2005;86:170–179. doi:10.1644/1545-1542(2005)086<0170:RDOSAT>2.0.CO;2 [Google Scholar]
- Congdon B, Piatt J, Martin K, Friesen V. Mechanisms of population differentiation in marbled murrelets: historical versus contemporary processes. Evolution. 2000;54:974–986. doi: 10.1111/j.0014-3820.2000.tb00097.x. [DOI] [PubMed] [Google Scholar]
- Cook J, Lessa E. Are rates of diversification in subterranean South American tuco-tucos (genus Ctenomys, Rodentia: Octodontidae) unusually high? Evolution. 1998;52:1521–1527. doi: 10.1111/j.1558-5646.1998.tb02035.x. doi:10.2307/2411323 [DOI] [PubMed] [Google Scholar]
- Coulon A, Cosson J, Angibault J, Cargnelutti B, Galan M, Morellet N, Petit E, Aulagnier S, Hewison A. Landscape connectivity influences gene flow in a roe deer population inhabiting a fragmented landscape: an individual-based approach. Mol. Ecol. 2004;13:2841–2850. doi: 10.1111/j.1365-294X.2004.02253.x. doi:10.1111/j.1365-294X.2004.02253.x [DOI] [PubMed] [Google Scholar]
- D'Anatro A, Lessa E. Geometric morphometric analysis of geographic variation in the Rio Negro tuco-tuco, Ctenomys rionegrensis (Rodentia: Ctenomyidae) Mamm. Biol. 2006;71:288–298. doi:10.1016/j.mambio.2006.02.001 [Google Scholar]
- D'Elia G, Lessa E, Cook J. Geographic structure, gene flow, and maintenance of melanism in Ctenomys rionegrensis (Rodentia: Octodontidae) Z. Saugetierkd. 1998;63:285–296. [Google Scholar]
- Dixon J.D, Oli M.K, Wooten M.C, Eason T.H, McCown J.W, Cunningham M.W. Genetic consequences of habitat fragmentation and loss: the case of the Florida black bear (Ursus americanus floridanus) Conserv. Genet. 2007;8:455–464. doi:10.1007/s10592-006-9184-z [Google Scholar]
- Florin A.B, Hoglund J. Absence of population structure of turbot (Psetta maxima) in the Baltic Sea. Mol. Ecol. 2007;16:115–126. doi: 10.1111/j.1365-294X.2006.03120.x. doi:10.1111/j.1365-294X.2006.03120.x [DOI] [PubMed] [Google Scholar]
- Foll M, Gaggiotti O.E. Identifying the environmental factors that determine the genetic structure of populations. Genetics. 2006;174:875–891. doi: 10.1534/genetics.106.059451. doi:10.1534/genetics.106.059451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupa J.J, Geluso K.N. Matching the color of excavated soil: cryptic coloration in the plains pocket gopher (Geomys bursarius) J. Mammal. 2000;81:86–96. doi:10.1644/1545-1542(2000)081<0086:MTCOES>2.0.CO;2 [Google Scholar]
- Lacey E. Spatial and social systems of subterranean rodents. In: Lacey E, Patton J, Cameron G, editors. Life underground: the biology of subterranean rodents. University of Chicago Press; Chicago, IL: 2000. pp. 257–296. [Google Scholar]
- Lessa E, Cook J. The molecular phylogenetics of tuco-tucos (genus Ctenomys, Rodentia: Octodontidae) suggests an early burst of speciation. Mol. Phylogenet. Evol. 1998;9:88–99. doi: 10.1006/mpev.1997.0445. doi:10.1006/mpev.1997.0445 [DOI] [PubMed] [Google Scholar]
- Lessa E, Thaeler C. A reassessment of morphological specializations for digging in pocket gophers. J. Mammal. 1989;70:689–700. doi:10.2307/1381704 [Google Scholar]
- Massarini A, de Freitas T. Morphological and cytogenetics comparison in species of the mendocinus group (genus Ctenomys) with emphasis in C. australis and C. flamarioni (Rodentia: Ctenomyidae) Caryologia. 2005;58:21–27. [Google Scholar]
- Matocq M, Patton J, da Silva M. Population genetic structure of two ecologically distinct Amazonian spiny rats: separating history and current ecology. Evolution. 2000;54:1423–1432. doi: 10.1111/j.0014-3820.2000.tb00574.x. [DOI] [PubMed] [Google Scholar]
- McRae B.H. Isolation by resistance. Evolution. 2006;60:1551–1561. [PubMed] [Google Scholar]
- Mora M.S, Lessa E.P, Kittlein M.J, Vassallo A.I. Phylogeography of the subterranean rodent Ctenomys australis in sand-dune habitats: evidence of population expansion. J. Mammal. 2006;87:1192–1203. doi:10.1644/05-MAMM-A-399R1.1 [Google Scholar]
- Reig O, Busch C, Ortells M, Contreras J. An overview of evolution, systematics, population biology, cytogenetics, molecular biology and speciation in Ctenomys. Prog. Clin. Biol. Res. 1990;335:71–96. [PubMed] [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. doi:10.2307/2410134 [DOI] [PubMed] [Google Scholar]
- Spear S, Peterson C, Matocq M, Storfer A. Landscape genetics of the blotched tiger salamander (Ambystoma tigrinum melanostictum) Mol. Ecol. 2005;14:2553–2564. doi: 10.1111/j.1365-294X.2005.02573.x. doi:10.1111/j.1365-294X.2005.02573.x [DOI] [PubMed] [Google Scholar]
- Sprechmann P. The paleoecology and paleogeography of the Uruguayan coastal area during the neogene and quaternary. Zitteliana. 1978;4:3–72. [Google Scholar]
- Vuilleumier S, Fontanillas P. Landscape structure affects dispersal in the greater white-toothed shrew: inference between genetic and simulated ecological distances. Ecol. Mod. 2007;201:369–376. doi:10.1016/j.ecolmodel.2006.10.002 [Google Scholar]
- Wlasiuk G, Garza J.C, Lessa E.P. Genetic and geographic differentiation in the Rio Negro tuco-tuco (Ctenomys rionegrensis): inferring the roles of migration and drift from multiple genetic markers. Evolution. 2003;57:913–926. doi: 10.1111/j.0014-3820.2003.tb00302.x. doi:10.1554/0014-3820(2003)057[0913:GAGDIT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]