Abstract
Multidrug resistance associated protein MRP3/Mrp3 (ABCC3) is upregulated in cholestasis, an adaptive response that may protect the liver from accumulation of toxic compounds, such as bile salts and bilirubin conjugates. However, the mechanism of this upregulation is poorly understood. We and others have previously reported that fetoprotein transcription factor/liver receptor homolog-1 is an activator of MRP3/Mrp3 expression. In searching for additional regulatory elements in the human MRP3 promoter, we have now identified nuclear receptor retinoic X receptor-α:retinoic acid receptor-α (RXRα:RARα) as a repressor of MRP3 activation by transcription factor Sp1. A luciferase reporter assay demonstrated that cotransfection of transcription factor Sp1 stimulates the MRP3 promoter activity and that additions of RXRα:RARα abrogated this activation in a dose-dependent manner. Site mutations and gel shift assays have identified a Sp1 binding GC box motif at −113 to −108 nts upstream from the MRP3 translation start site, where RXRα:RARα specifically reduced Sp1 binding to this site. Mutation of the GC box also reduced MRP3 promoter activity. The functional role of RXRα:RARα as a repressor of MRP3 expression was further confirmed by RARα small-interfering RNA knock-down in HepG2 cells, which upregulated endogenous MRP3 expression. In summary, our results indicate that activator Sp1 and repressor RXRα:RARα act in concert to regulate MRP3 expression. Since RXRα:RARα expression is diminished by cholestatic liver injury, loss of RXRα:RARα may lead to upregulation of MRP3/Mrp3 expression in these disorders.
Keywords: gene expression, transporter, cholestasis, multidrug resistance-associated protein 3
THE MULTIDRUG RESISTANCE-ASSOCIATED PROTEINS, the MRP (ABCC) family, consist of at least 12 ATP-binding cassette transport proteins that function to export a variety of organic solutes out of cells (3, 14). The MRP3/Mrp3 ortholog (ABCC3) is expressed in a number of cell types and tissues including hepatocytes, cholangiocytes, intestine, and kidney where it is localized to the basolateral membrane (15, 16, 21). Substrates for MRP3/Mrp3 include sulfated and nonsulfated bile salts, bilirubin glucuronides, 17β-glucuronosyl estradiol, leukotrienes, and some anticancer drugs (1, 8, 9, 16). Mrp3 expression in hepatocytes varies between species but is weakly expressed in rat compared with the mouse and human liver (13, 19). However, rat Mrp3 is markedly upregulated in the liver following bile duct obstruction (7, 19, 24), and human MRP3 is upregulated in patients with the Dubin-Johnson syndrome (15) and primary biliary cirrhosis (32). These findings suggest that this transporter functions to extrude bilirubin and bile salt conjugates from the hepatocyte when bile secretion is impaired. Recent studies in bile duct-ligated Mrp3 knockout mice support a role for extrusion of endogenous glucuronide conjugates (18, 30, 31). However, the mechanisms that regulate MRP3/Mrp3 gene expression are not well understood.
The transcription factor Sp1 has been reported to be an activator of the human and rat MRP3/Mrp3 promoters by use of luciferase reporter assays (25, 26). But how this constitutively active transcription factor may contribute to the upregulation of MRP3/Mrp3 expression in cholestasis is unknown. Recently, human MRP3 was shown to be upregulated by bile acids in Caco2 cells via the fetoprotein transcription factor (FTF), also known as liver receptor homolog-1 (Lrh-1) and cholesterol 7α hydroxylase promoter factor (CPF) (12). Previous studies from our laboratory indicate that bile duct obstruction increases Lrh-1 and Mrp3 expression in mouse liver and Lrh-1 binding to the mouse Mrp3 promoter (2). This adaptive response is mediated by TNF-α and functionally diminishes hepatocyte injury compared with bile duct obstruction in TNF-R1-/- mice (2). Nevertheless, it seems unlikely that increases in FTF/Lrh-1 expression and binding to the MRP3 promoter can fully account for the marked upregulation of its protein expression in patients with cholestasis and following bile duct ligation in the rat.
In the present study, the observation that MRP3 promoter activity was suppressed in retinoic X receptor-α:retinoic acid receptor-α (RXRα:RARα)-cotransfected HepG2 and HEK293 cells led us to search for additional nuclear receptors that may regulate MRP3/Mrp3 expression.
MATERIALS AND METHODS
Materials
Chemicals were purchased from Sigma, except when the source is specified. Cell culture medium DMEM, fetal bovine serum (FBS), penicillin and streptomycin, trypsin, and PBS were from Invitrogen (Carlsbad, CA). Enhanced chemiluminescence reagents were from Amersham (Piscataway, NJ). DNA oligos and sequencing were provided by the Keck Biotechnology Center at Yale University. TaqMan probes for real-time PCR were generated by Applied Biosystems (Foster City, CA). Luciferase assay kit was purchased from Promega (Madison, WI). DIG-Gel shift kit was from Roche Boehringer (Indianapolis, IN).
Plasmid constructs
Human RARα (NR1B1) and RXRα (NR2B1) expression vectors are gifts from Dr. David Mangelsdorf (University of Texas Southwestern Medical Center, Dallas, TX). Human Sp1 and Sp3 expression constructs in pCMV4 vector were kindly provided by Drs. Graciela Krikun and Charles Lockwood at Yale University. Five constructs used for luciferase reporter assay were made by inserting 234 bp, 0.5 kb, 1 kb, 2 kb, and 4 kb of MRP3 gene upstream sequence from the transcription initiation site into a plasmid pGL3-basic (Promega); they were named p-200Luc, p-500Luc, p-1 kbLuc, p-2 kbLuc, p-4 kbLuc, and p-500LucM as previously described (2). Oligo DNA primers made for mutating the putative Sp1 binding site in the p-200Luc plasmid are listed in Table 1 (GC5), and the mutants were generated by using the QuikChange XL site-directed mutagenesis kit from Stratagene (La Jolla, CA). These p-200Luc mutants were confirmed by DNA sequencing. Glutathione-S-transferase (GST)-hRXRα and GST-hRARα expression constructs were made by inserting the full-length PCR fragments of human RXRα and RARα into pGEX-6P-1 vector, respectively. The oligo primers are as follows: hRXRα-forward: GTGGAATTCACCATGGACACCAAACATTTCCTGC; hRXRα-reverse: GTAAGAATGCGGCCGCCTAAGTCATTTGGTGC; hRARα-forward: GTAGGATCCATGGCCAGCAACAGCAGCTCC; hRARα-reverse: GTCGCTCGAGTCACGGGGAGTGGGTGGC.
Table 1.
Oligos and probes for gel shift and mutagenesis
| Name | Sequence (5′ → 3′) |
|---|---|
| WT | -124AGGGCTCGGCAGGGCGGGTCCGGGCGGAGCGCG-92 |
| GC5 | -124AGGGCTCGGCAGttCGGGTCCGGGCGGAGCGCG-92 |
| GCB | -124AGGGCTCGGCAGttCGGGTCCGttCGGAGCGCG-92 |
| Sp1 | ATTCGATCGGGGCGGGGCGAGC |
Antisenses were made at the same length and complementary to the sense sequence.
Cell culture and transfection
HepG2 and HEK293 cells were grown at 37°C in DMEM medium with 10% FBS in 5% CO2 atmosphere. Sp1 deficient Drosophila S2 cells (kindly provided by Dr. Tian Xu’s laboratory at Yale University) were maintained in Schneider’s Drosophila medium supplemented with 10% FBS at 25°C. Cells were transfected by Lipofectamine without serum in the medium for 16 h when they were at 60% confluence and then regular growth medium was added. For nuclear receptor ligand treatment, the transfected cells were cultured in serum-free DMEM with or without 100 nM 9-cis-retinoic acid and/or 100 nM all-trans-retinoic acid, or with 0.1% DMSO vehicle for 24 h.
Quantitative real-time PCR analysis
Total RNA was extracted from the tissues or cultured cells by using Trizol reagent according to the manufacturer’s instructions. Concentration and purity were confirmed by spectrophotometer and formaldehyde denaturing agarose gel electrophoresis. Five micrograms of total RNA from each sample were used to generate cDNA by reverse transcription with the ProSTAR RT-PCR kit (Stratagene, La Jolla, CA). TaqMan real-time quantitative PCR assay was performed on an ABI Prism 7700 sequence detection system, according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA). GAPDH was used as reference for normalizing data. MRP3 primers and probe are forward, CGTGAAGATGCGCGCC; reverse, TCAGCTTGATGCGCGAGTC; probe, Fam-TCCAGGTAAAGCAAATG. Proprietary primers and probe of human GAPDH were purchased from ABI (Applied Biosystems).
Western blot analyses
Total membrane proteins were isolated from HepG2 cells as described previously (2), and nuclear proteins were prepared by using the NE-PER kit (Pierce, Rockford, IL) according to the manufacturer’s protocol. Protein concentration was determined according to Bradford (4), and samples were stored at −70°C. For Western blot analysis, 20 μg nuclear extract or 100 μg membrane protein were separated on a 4 −15% SDS-polyacrylamide gel (Bio-Rad, Hercules, CA). After transfer to polyvinylidene fluoride microporous membranes (Bio-Rad), the membranes were blocked with 5% nonfat milk, followed by primary antibody and horseradish peroxidase-conjugated secondary antibody incubation. The immune complexes were detected with the Enhanced Chemiluminescence reagent kit, and the signals were recorded on X-ray film. Immunoreactive bands were quantified by densitometry (Multi-Analyst, Bio-Rad). SH-PTP1 protein was used to normalize band intensity for nuclear proteins. Polyclonal antibodies against human RARα (C-20), SH-PTP1, and human MRP3 (C-18) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Gene knock-down with small interfering RNA
Four sets of small interfering RNAs (siRNAs) that targeted human RARα were designed and chemically synthesized by Dharmacon (Chicago, IL). The sequences are listed in Table 2. The oligos were annealed to duplex according to manufacturer’s instruction. Control RNA, siCONTROL RISC-Free siRNA (sequence proprietary) was purchased from Dharmacon. HEK293 or HepG2 cells were transfected with mixtures of four sets of RARα siRNA duplexes or control siRNA duplex in the amount of 100 pmol/well for a 6-well plate or 25 pmol/well for a 24-well plate using Lipofectamine reagent according to the manufacturer’s instructions (Invitrogen). After 24 and 48 h of transfection, cells were harvested for RNA isolation or luciferase reporter assay.
Table 2.
Ribonucleotide oligo sequence for siRNA targeting of human RARα
| Duplex No. | Strand | Sequence (5′ → 3′) |
|---|---|---|
| 1 | Sense: | GACAAGAACUGCAUCAUCAUU |
| Antisense: | pUGAUGAUGCAGUUCUUGUCUU | |
| 2 | Sense: | GCAAAUACACUACGAACAAUU |
| Antisense: | pUUGUUCGUAGUGUAUUUGCUU | |
| 3 | Sense: | GAACAACAGCUCAGAACAAUU |
| Antisense: | pUUGUUCUGAGCUGUUGUUCUU | |
| 4 | Sense: | GAGCAGCAGUUCUGAAGAGUU |
| Antisense: | pCUCUUCAGAACUGCUGCUCUU |
siRNA, small interfering RNA.
Luciferase reporter assay
HepG2, HEK293, or S2 cells seeded on 24-well plates were cotransfected with 75 ng of RARα plasmid DNA, 75 ng of RXRα plasmid DNA, 0.3 μg of human MRP3 promoter region reporter construct, and 5 ng of phRL-CMV as internal control except where indicated (Promega). After transfection and ligand treatment, cells were washed twice with PBS and lysed with 100 μl of passive lysis buffer (Promega). A fraction of protein was subject to a dual-luciferase system reporter assay from Promega by following the manufacturer’s protocol. The firefly luciferase activity was normalized by Renilla luciferase activity or protein concentration (28). Since Sp1 can stimulate Renilla luciferase expression in the phRL-CMV vector [there are at least three putative Sp1 binding sites in the CMV promoter upstream of Renilla luciferase gene based on MatInspector (www.genomatix.dez)], firefly luciferase activity was normalized to total cell protein when Sp1 was cotransfected. In both cases, promoter activities are given as means ± SD of triplicate transfections.
EMSA and supershift EMSA
For the gel shift assay, double-strand DNA oligos were made with a biotin label at the 3′ end or by the DIG-end labeling technique (Table 1), according to the manufacturer’s instructions (Roche). Recombinant human Sp1 protein and a canonical Sp1 oligo were from Promega. Antibody against human Sp1 was purchased from Santa Cruz Biotechnology, In vitro translated human RARα and RXRα were made by utilizing the TNT kit (Promega). Recombinant GST, GST-hRARα, and GST-hRXRα protein were expressed in Escherichia coli BL21 and purified by using glutathione beads. Electrophoretic mobility shift assays (EMSA) with biotin-labeled probes were performed using the LightShift chemiluminescent EMSA kit (Pierce). EMSA and supershift EMSA with digoxin-labeled probes were performed using the DIG-Gel shift kit by following manufacturer’s instructions (Roche). In supershift studies for RXRα and RARα, 5 μl (0.2 mg/ml) of the relevant rabbit polyclonal antibody (D-20 and C-20, Santa Cruz) or control rabbit IgG was incubated with 5 μg of nuclear extract on ice for 30 min before addition of the labeled probe and further incubated on ice for 30 min. The entire 20 μl binding reaction was resolved on a 6% polyacrylamide gel and then transferred to positively charged nylon membrane (Bio-Rad) in 0.5 × Tris borate-EDTA buffer.
Statistical analysis
Data are expressed as means ± SD. Differences between experimental groups were assessed for significance by the two-tailed unpaired Student’s t-test. A P value of <0.05 was considered to be statistically significant.
RESULTS
The human MRP3 promoter activity is diminished after cotransfection of RXRα and/or RARα
To assess promoter activity in the MRP3 5′-flanking region, p-200Luc, p-500Luc, p-1 kbLuc, p-2 kbLuc, and p-4 kbLuc constructs were cotransfected with phRL-CMV into HepG2 cells. Luciferase assays demonstrated that most of the promoter activity in the MRP3 gene was in the first 2-kb sequence (data not shown). CPF, farnesoid X receptor (FXR) (NR1H4), pregnane X receptor (PXR) (NR1I2), constitutive androstane receptor (CAR) (NR1I3), RARα, and RXRα expression constructs were then cotransfected with p-2 kbLuc into HepG2 cells and treated with their specific ligands to examine whether any of these nuclear receptors might upregulate MRP3 expression. We did not see any significant stimulation on MRP3 promoter activity from FXR, PXR, or CAR in this construct. However, CPF activated the MRP3 promoter, consistent with our previous report (2). In contrast, we repeatedly observed that luciferase activity was significantly lower in RXRα:RARα-transfected cells, and addition of retinoids did not augment this inhibitory effect. To confirm this finding, we cotransfected RXRα:RARα expression constructs with the p-200Luc, p-500Luc, or p-1 kbLuc into HepG2 and HEK293 cells. As demonstrated in Fig. 1, RXRα:RARα suppressed MRP3 promoter activity by 42 ± 2% with p-200Luc, 40 ± 4% with p-500Luc, and 33 ± 2% with p-1 kbLuc in HepG2 cells, indicating that there is an RXRα:RARα response element in the first 234 bp of the MRP3 5′ flank region that represses MRP3 expression.
Fig. 1.
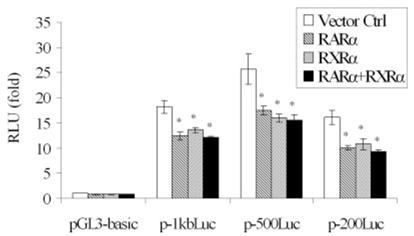
Luciferase reporter assays demonstrate that the first 234-bp sequence of the human multidrug resistance-associated protein MRP3 promoter contain a retinoic X receptor-α:retinoic acid receptor-α (RARα:RXRα) response element. Cotransfection of 75 ng RARα and/or 75 ng RXRα expression plasmid (s) with 300 ng of either of 3 human MRP3 promoter region constructs (p-200Luc, p-500Luc, or p-1 kbLuc) into HepG2 cells significantly inhibit MRP3 promoter activity when compared with empty vector control (Ctrl) (N = 3, *P < 0.01). Firefly luciferase activity was normalized to Renilla luciferase activity. RLU, relative light units.
RARα siRNAs knock down RARα nuclear protein and increase expression of endogenous MRP3
To assess directly the potential inhibitory effects of RARα on MRP3 expression, we used siRNA to knock down the endogenous expression of this nuclear receptor. As illustrated in Fig. 2A, transfection of RARα siRNAs resulted in reduction of RARα nuclear protein to 53 ± 5% of control levels by 48 h in HepG2 cells. In contrast, both MRP3 mRNA and protein were significantly increased by 211 ± 7 and 154 ± 15%, respectively (Fig. 2, B and C). This result confirms the findings from the previous promoter reporter assay that RXRα:RARα suppresses MRP3 expression and further suggests that there is an RXRα:RARα response element in this 234-bp sequence.
Fig. 2.
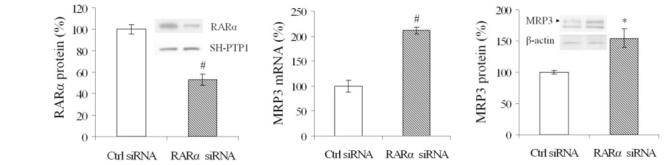
Endogenous MRP3 is upregulated in RARα small-interfering RNA (siRNA) transfected HepG2 cells at 48 h. Left: Western blot and densitometry demonstrate that endogenous RARα expression is knocked down by its specific siRNA, #P < 0.01, N = 3. There was no change in the nuclear protein SH-PTP1. Middle: quantitative PCR demonstrates that MRP3 mRNA is upregulated when RARα is knocked down, #P < 0.01. Right: Western blot and densitometry demonstrate an increase in expression of MRP3 protein when RARα is knocked down, *P < 0.05. There was no change in β-actin.
RXRα:RARα does not directly bind to the proximal promoter of the human MRP3 gene
Because RXRα:RARα repressed the p-200Luc construct, we searched the Genomatix website (http://www.genomatix.de) for nuclear receptor and transcription factor binding sites in the first 234-bp sequence of the MRP3 promoter region to identify the putative retinoic acid receptor response elements (RARE) in the MRP3 gene. However, a typical RARE was not detected. In contrast, Sp1 and FTF/CPF binding sites were detected in this region as previously reported (Fig. 3). Furthermore, to search whether there was an atypical RARE, we constructed seven pairs of complementary oligonucleotides to cover the full 234-bp sequence of the MRP3 promoter in the p-200Luc construct. Each oligonucleotide pair was 50 bp long with 20 bp overlapping with adjacent regions and was labeled with biotin. By using in vitro translated RARα and RXRα, we found that two pairs of oligos (the −152 to −102 nts and −122 to −72 nts) specifically bound to RXRα:RARα in a gel-shift assay. Because there is a 20-bp overlap in these two pairs of oligos, we were able to confine the potential RARE to this 20-bp region. We then used this 20-bp sequence to test whether it binds to in vitro translated RXRα:RARα. This result also appeared to be positive. However, we could not confirm the findings when purified recombinant human RARα and RXRα protein were used (Fig. 5C). Therefore, a specific RXRα:RARα binding site does not seem to be present in this region of the human MRP3 promoter.
Fig. 3.
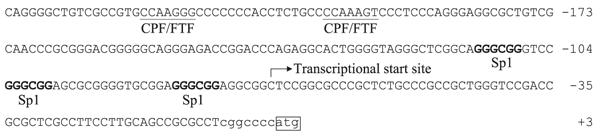
The 234-bp DNA sequence of the MRP3 gene 5′-flank region in p-200Luc construct. Fetoprotein transcription factor (FTF)/cholesterol 7α hydroxylase promoter factor (CPF)/liver receptor homolog-1 (Lrh-1) and putative Sp1 binding GC box motifs are indicated.
Fig. 5.
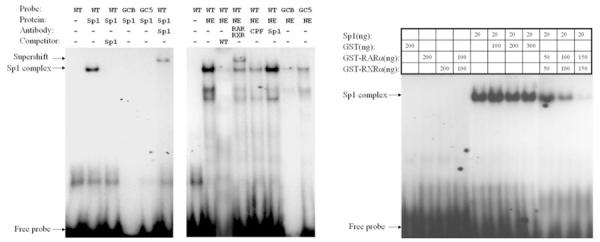
EMSA indicates that RXRα:RARα and Sp1 are in the complex of MRP3 promoter. Sp1 (left) and nuclear extract from HepG2 cell (middle) bind to the GC box motif, and mutation on the GC box abrogates these bindings. Right: recombinant RXRα and RARα specifically reduced Sp1 binding to the GC box motif. NE, nuclear extract. WT, wild type; GCB, both guanine and cytosine box are mutated; GC5, 5′ end of guanine and cytosine box are mutated; GST, glutathione-S-transferase.
RXRα:RARα suppresses Sp1 activity in the MRP3 promoter
Previous studies indicate that nuclear receptors can modulate Sp1 activity (a transcriptional activator) in GC-rich regions (22). Because GC content is 78% of the proximal promoter of the human MRP3 gene (Fig. 3), we next examined whether RXRα:RARα might suppress MRP3 expression through Sp1. To test this hypothesis, p-200Luc was cotransfected with a human Sp1 expression construct into S2 cells, an Sp1-deficient insect cell line. As predicted, when 50 and 100 ng of this expression vector were cotransfected into S2 cells, Sp1 greatly stimulated MRP3 promoter activity in a dose-dependent manner by a factor of 5- and 12-fold, respectively. Qualitatively similar results were obtained in transfected HEK293 and HepG2 cells. In contrast, human Sp3 did not show any stimulation (Fig. 4A), indicating that this promoter activity is Sp1 specific. When the Sp1 expression construct was cotransfected with RARα and RXRα expression plasmids into S2 cells, this stimulatory effect was abrogated in a dose-dependent manner (Fig. 4B). Similar results were also observed in transfected HepG2 and HEK293 cells, consistent with our original observation that RXRα:RARα inhibits MRP3 promoter expression in these cell lines.
Fig. 4.
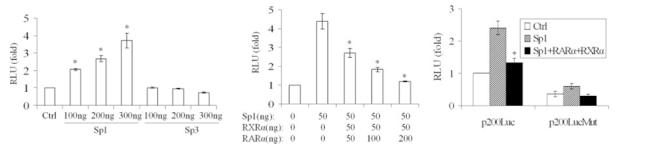
Luciferase reporter assays demonstrate that RXRα:RARα and Sp1 coregulate the MRP3 promoter. Left: Sp1 specifically activates the MRP3 promoter in a dose-dependent manner in transfected HEK293 cells. p-200Luc (200 ng) was cotransfected with the indicated amount of either the Sp1 vector, the Sp3 vector, or the empty vector. Middle: RARα:RXRα abrogated Sp1 activation on MRP3 promoter in a dose-dependent manner in transfected S2 cells. p-200Luc (200 ng) was cotransfected with the indicated amount of Sp1, RAR, and RXR. Right: mutations of the Sp1 binding GC box motif reduced MRP3 promoter activity. p-200Luc (200 ng) wild-type and mutants were cotransfected with or without 100 ng Sp1 and 50 ng RARα and 50 ng RXRα expression vectors in HepG2 cells. N = 3, *P < 0.01. Firefly luciferase activity was normalized to total cell protein.
To assess whether the putative Sp1 binding sites are responsible for Sp1 activation, we mutated the major Sp1 activation site (−113 to −108 nts) in the p-200Luc construct. As shown in Fig. 4C, p-200LucMut demonstrated significantly lower basal promoter activity and Sp1 inducibility compared with the wild type, indicating that Sp1 is a functional activator of MRP3 gene expression.
RXRα:RARα reduced Sp1 binding to its response element in the human MRP3 promoter
Finally to determine whether RXRα:RARα and Sp1 bind as a complex to the GC box motif in the MRP3 promoter, we performed EMSA experiments.
First, as shown in Fig. 5A, purified recombinant Sp1 protein binds to the wild-type sequence, but not when either of the Sp1 putative binding sites was mutated or when it is competed by a canonical Sp1 oligo. Specificity of the binding was confirmed since addition of Sp1 antibody leads to a supershift. These results indicate that the putative Sp1 binding site in the MRP3 promoter is a functional Sp1 site.
Second (Fig. 5B), when the probe was incubated with nuclear extract from HepG2 cells, a significant gel shift was observed. The shifted band was competed when cold (unlabeled) wild-type probe was added. Mutations of the Sp1 binding site reduced nuclear protein binding activity, and addition of antibodies against RXRα and RARα, but not CPF or Sp1, specifically supershifted the band, indicating that RXRα:RARα is part of the transcription factor complex of the MRP3 promoter.
Third, to further explore how RXRα:RARα may suppress Sp1 activity, we tested whether RXRα:RARα interferes with Sp1 binding to its response element. In this experiment, purified recombinant human RARα and RXRα protein was incubated with Sp1 and its response element probe. As shown in Fig. 5C, the EMSA demonstrate that Sp1, but not GST, or RXRα and RARα alone or in combination binds to the wild-type probe. This binding was not affected by GST protein. In contrast, RARα together with RXRα reduced the Sp1 binding in a dose-dependent manner, suggesting that RXRα:RARα represses Sp1 activity on MRP3 expression through a protein-protein interaction.
DISCUSSION
MRP3/Mrp3 is a major export pump for conjugated bile salt and organic solutes on the basolateral membrane of hepatocytes and is adaptively upregulated during cholestatic liver injury (2, 7, 13, 19, 24). However, the mechanisms by which this adaptive response in MRP3/Mrp3 expression occurs have not been fully explained. Takada et al. (25) reported that human MRP3 is under the control of TATA-less proximal promoter and suggested that Sp1 may be involved in its transcription. Similar results were also reported for rat Mrp3 regulation (26). But neither of these reports determined whether Sp1 could upregulate MRP3/Mrp3 in cholestasis. We and others have previously identified FTF/Lrh-1/CPF as a nuclear receptor that positively regulates MRP3/Mrp3 gene expression (2, 12) and that is mediated by the TNF-α signaling pathway (2). However, the magnitude of this response seemed insufficient to be fully explained by the induction of FTF alone. In the process of searching for additional regulators for MRP3/Mrp3 expression, we observed that RXRα:RARα suppressed MRP3 promoter activity in a luciferase reporter assay. Therefore we speculated that the RXRα:RARα heterodimeric complex might be a repressor of MRP3/Mrp3 expression under normal physiological conditions and that loss of these nuclear receptors under cholestatic conditions, as we previously described (7), might contribute to MRP3/Mrp3 upregulation.
This hypothesis is confirmed in the present study by the following findings: 1) When human MRP3 promoter constructs (p-200Luc, p-500Luc, p-1 kbLuc) are cotransfected with plasmids expressing RXRα and/or RARα in either HepG2 or HEK293 cells, MRP3 promoter activity is significantly suppressed, indicating that RXRα:RARα can repress MRP3 transcription (Fig. 1). 2) When RARα siRNAs knocked down endogenous expression of RARα nuclear protein in human hepatoma cell lines, endogenous MRP3 mRNA and protein reciprocally increased, directly demonstrating that MRP3 expression is enhanced in association with a reduction in expression of this nuclear protein (Fig. 2). Luciferase reporter assays also demonstrate that MRP3 promoter activity is significantly enhanced by these RARα siRNAs (data not shown). 3) We also confirm that Sp1 is a transcriptional activator of MRP3 expression (Fig. 4A) and further show that RXRα:RARα abrogates Sp1 activity in this gene’s promoter (Fig. 4B). 4) EMSAs demonstrate that RXRα and RARα are part of a complex that binds to the Sp1 GC box motif of the MRP3 promoter and that they specifically reduce Sp1 binding to this motif (Fig. 5C). 5) Although a CPF response element is also present distally in the p-200Luc construct of MRP3 promoter, it is unlikely that this region is significant since similar effects exerted by Sp1 and RXRα:RARα were observed with a p-110Luc construct that excluded the CPF site while maintaining the three putative Sp1 response elements (data not shown). Thus, on the basis of these combined experiments, we conclude that RXRα and RARα are suppressors of Sp1’s activation of human MRP3 expression. Because the expression of RAR and RXR is diminished in cholestatic liver injury (6), we speculate that upregulation of MRP3/Mrp3 in cholestasis is in part due to the loss of these nuclear receptors.
RAR is a well-known nuclear receptor that heterodimerizes with RXR and binds to RAREs to regulate gene expression. It is also well documented that as a general transcriptional activator Sp1 binds to a GC box motif in promoters and regulates gene expression. Recent studies indicate that RAR modulates Sp1 transcriptional activity on GC-rich promoters through direct interactions between the two proteins (11, 22). Kojima and colleagues (22) have reported that RXRα:RARα regulate the expression of several genes in an RARE-independent manner, including urokinase, transglutaminase, TGF-β1, and type I and II TGF-β receptors. In these examples, RXRα:RARα strengthened Sp1 binding to their GC box motifs and thus enhanced target gene expression. In contrast, Uruno et al. (27) have observed that RAR repressed Sp1 activity on the expression of the thromboxane receptor and demonstrated that RAR reduced Sp1 binding to its GC box motif in its promoter. This finding is similar to the effect of RAR:RXR on Sp1 activity and MRP3 expression in the present study. It remains to be determined why RAR is an enhancer for Sp1 in some cases but a repressor in others. Differences in adjacent sequence may contribute to this effect.
The regulatory effects on the levels of MRP3 promoter activity that we have observed for RXRα:RARα repression, although highly significant and specific, are not large in magnitude when analyzed in transfected HepG2 and HEK293 cells possibly because of high endogenous expression of Sp1. For example, Sp1 induced higher levels of MRP3 and was more severely repressed by RXRα:RARα when transfected in S2 cells where endogenous Sp1 is deficient (Fig. 4B). It is also possible that other transcriptional factors may be involved in regulating MRP3 gene expression. Previous reports suggest that Mrp3 expression in the liver can be induced by phenobarbital (5, 13, 29), and we have demonstrated similar stimulation of MRP3 mRNA in HepG2 cells (our unpublished observations). These findings suggest that CAR might also regulate MRP3/Mrp3 expression. However, phenobarbital has similar effects in CAR-/- mice (5) and we have been unable to identify a CAR cis-binding element in our constructs of the human MRP3 promoter. Thus the phenobarbital effect must be independent of CAR and CAR may not be a specific regulator of MRP3. Others have reported that nuclear factor E2-related factor 2 may be involved in inducing MRP3/Mrp3 expression in HepG2 cells and in the mouse liver by trans-stilbene oxide (23). Glucocorticoids can stimulate MRP3 expression in A549 cells (non-small-cell lung cancer cell line) but not in Hela cells, findings consistent with additional regulators for MRP3 expression (20).
Human population studies confirm that liver MRP3 mRNA expression is normally low (10). However, the expression is variable and higher levels have been correlated with exposure to the proton pump inhibitor omeprazole, suggesting activation by the aryl hydrocarbon receptor (Ahr) pathway (10). β-Napthoflavone, a ligand for Ahr, also can increase MRP3 mRNA expression (10), and dioxin response elements have been identified in the 5′ flanking region of human MRP3 (25) to which Ahr:Arnt (Aryl hydrocarbon nuclear translocator) heterodimers may bind. Thus MRP3 might also be upregulated by Ahr. Significant correlations in MRP3 expression have been found with a polymorphism in the −211 C>T of the MRP3 promoter (17). This polymorphism is located immediately between the two CPF binding sites and thus might affect FTF regulation of MRP3. Further studies will be necessary to determine the functional significance of these polymorphisms and other candidate receptors as regulators of MRP3 expression.
ACKNOWLEDGMENTS
Present address for W. Chen: Department of Gastroenterology, Southwest Hospital, Third Military Medical University, Chongqing 400038, China.
Present address for L. A. Denson: Gastroenterology, Hepatology, and Nutrition, Cincinnati Children’s Hospital Medical Center, MLC 2010, 3333 Burnet Avenue, Cincinnati, OH 45229.
GRANTS This research was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grants DK-25636 (J. L. Boyer) and the Yale Liver Center Cellular and Molecular Physiology and Morphology Cores (P30-34989).
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Akita H, Suzuki H, Hirohashi T, Takikawa H, Sugiyama Y. Transport activity of human MRP3 expressed in Sf9 cells: comparative studies with rat MRP3. Pharm Res. 2002;19:34–41. doi: 10.1023/a:1013699130991. [DOI] [PubMed] [Google Scholar]
- 2.Bohan A, Chen WS, Denson LA, Held MA, Boyer JL. Tumor necrosis factor alpha-dependent up-regulation of Lrh-1 and Mrp3 (Abcc3) reduces liver injury in obstructive cholestasis. J Biol Chem. 2003;278:36688–36698. doi: 10.1074/jbc.M304011200. [DOI] [PubMed] [Google Scholar]
- 3.Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–592. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 4.Bradford MM. A rapid and sensitive method of quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Cherrington NJ, Slitt AL, Maher JM, Zhang XX, Zhang J, Huang W, Wan YJ, Moore DD, Klaassen CD. Induction of multidrug resistance protein 3 (mrp3) in vivo is independent of constitutive androstane receptor. Drug Metab Dispos. 2003;31:1315–1319. doi: 10.1124/dmd.31.11.1315. [DOI] [PubMed] [Google Scholar]
- 6.Denson LA, Bohan A, Held MA, Boyer JL. Organ-specific alterations in RARα:RXRα abundance regulate rat Mrp2 (Abcc2) expression in obstructive cholestasis. Gastroenterology. 2002;123:599–607. doi: 10.1053/gast.2002.34758. [DOI] [PubMed] [Google Scholar]
- 7.Donner MG, Keppler D. Up-regulation of basolateral multidrug resistance protein 3 (Mrp3) in cholestatic rat liver. Hepatology. 2001;34:351–359. doi: 10.1053/jhep.2001.26213. [DOI] [PubMed] [Google Scholar]
- 8.Hirohashi T, Suzuki H, Sugiyama Y. Characterization of the transport properties of cloned rat multidrug resistance-associated protein 3 (MRP3) J Biol Chem. 1999;274:15181–15185. doi: 10.1074/jbc.274.21.15181. [DOI] [PubMed] [Google Scholar]
- 9.Hirohashi T, Suzuki H, Takikawa H, Sugiyama Y. ATP-dependent transport of bile salts by rat multidrug resistance-associated protein 3 (Mrp3) J Biol Chem. 2000;275:2905–2910. doi: 10.1074/jbc.275.4.2905. [DOI] [PubMed] [Google Scholar]
- 10.Hitzl M, Klein K, Zanger UM, Fritz P, Nussler AK, Neuhaus P, Fromm MF. Influence of omeprazole on multidrug resistance protein 3 expression in human liver. J Pharmacol Exp Ther. 2003;304:524–530. doi: 10.1124/jpet.102.043547. [DOI] [PubMed] [Google Scholar]
- 11.Husmann M, Dragneva Y, Romahn E, Jehnichen P. Nuclear receptors modulate the interaction of Sp1 and GC-rich DNA via ternary complex formation. Biochem J. 2000;352:763–772. [PMC free article] [PubMed] [Google Scholar]
- 12.Inokuchi A, Hinoshita E, Iwamoto Y, Kohno K, Kuwano M, Uchiumi T. Enhanced expression of the human multidrug resistance protein 3 by bile salt in human enterocytes. A transcriptional control of a plausible bile acid transporter. J Biol Chem. 2001;276:46822–46829. doi: 10.1074/jbc.M104612200. [DOI] [PubMed] [Google Scholar]
- 13.Kiuchi Y, Suzuki H, Hirohashi T, Tyson CA, Sugiyama Y. cDNA cloning and inducible expression of human multidrug resistance associated protein 3 (MRP3) FEBS Lett. 1998;433:149–152. doi: 10.1016/s0014-5793(98)00899-0. [DOI] [PubMed] [Google Scholar]
- 14.Konig J, Nies AT, Cui Y, Leier I, Keppler D. Conjugate export pumps of the multidrug resistance protein (MRP) family: localization, substrate specificity, and MRP2-mediated drug resistance. Biochim Biophys Acta. 1999;1461:377–394. doi: 10.1016/s0005-2736(99)00169-8. [DOI] [PubMed] [Google Scholar]
- 15.Konig J, Rost D, Cui Y, Keppler D. Characterization of the human multidrug resistance protein isoform MRP3 localized to the basolateral hepatocyte membrane. Hepatology. 1999;29:1156–1163. doi: 10.1002/hep.510290404. [DOI] [PubMed] [Google Scholar]
- 16.Kool M, van der Linden M, deHaas M, Scheffer GL, de Vree JML, Smith AJ, Jansen G, Peters GJ, Ponne N, Scheper RJ, Elferink Oude RPJ, Baas F, Borst P. MRP3, an organic anion transporter able to transport anti-cancer drugs. Proc Natl Acad Sci USA. 1999;96:6914–6919. doi: 10.1073/pnas.96.12.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang T, Hitzl M, Burk O, Mornhinweg E, Keil A, Kerb R, Klein K, Zanger UM, Eichelbaum M, Fromm MF. Genetic polymorphisms in the multidrug resistance-associated protein 3 (ABCC3, MRP3) gene and relationship to its mRNA and protein expression in human liver. Pharmacogenetics. 2004;14:155–164. doi: 10.1097/00008571-200403000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Manautou JE, de Waart DR, Kunne C, Zelcer N, Goedken M, Borst P, Elferink RO. Altered disposition of acetaminophen in mice with a disruption of the Mrp3 gene. Hepatology. 2005;42:1091–1098. doi: 10.1002/hep.20898. [DOI] [PubMed] [Google Scholar]
- 19.Ogawa K, Suzuki H, Hirohashi T, Ishikawa T, Meier PJ, Hirose K, Akizawa T, Yoshioka M, Sugiyama Y. Characterization of inducible nature of MRP3 in rat liver. Am J Physiol Gastrointest Liver Physiol. 2000;278:G438–G446. doi: 10.1152/ajpgi.2000.278.3.G438. [DOI] [PubMed] [Google Scholar]
- 20.Pulaski L, Kania K, Ratajewski M, Uchiumi T, Kuwano M, Bartosz G. Differential regulation of the human MRP2 and MRP3 gene expression by glucocorticoids. J Steroid Biochem Mol Biol. 2005;96:229–234. doi: 10.1016/j.jsbmb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Rost D, Mahner S, Sugiyama Y, Stremmel W. Expression and localization of the multidrug resistance-associated protein 3 in rat small and large intestine. Am J Physiol Gastrointest Liver Physiol. 2002;282:G720–G726. doi: 10.1152/ajpgi.00318.2001. [DOI] [PubMed] [Google Scholar]
- 22.Shimada J, Suzuki Y, Kim SJ, Wang PC, Matsumura M, Kojima S. Transactivation via RAR/RXR-Sp1 interaction: characterization of binding between Sp1 and GC box motif. Mol Endocrinol. 2001;15:1677–1692. doi: 10.1210/mend.15.10.0707. [DOI] [PubMed] [Google Scholar]
- 23.Slitt AL, Cherrington NJ, Dieter MZ, Aleksunes LM, Scheffer GL, Huang W, Moore DD, Klaassen CD. trans-Stilbene oxide induces expression of genes involved in metabolism and transport in mouse liver via CAR and Nrf2 transcription factors. Mol Pharmacol. 2006;69:1554–1563. doi: 10.1124/mol.105.014571. [DOI] [PubMed] [Google Scholar]
- 24.Soroka CJ, Lee JM, Azzaroli F, Boyer JL. Cellular localization and up-regulation of multidrug resistance-associated protein 3 in hepatocytes and cholangiocytes during obstructive cholestasis in rat liver. Hepatology. 2001;33:783–791. doi: 10.1053/jhep.2001.23501. [DOI] [PubMed] [Google Scholar]
- 25.Takada T, Suzuki H, Sugiyama Y. Characterization of 5′-flanking region of human MRP3. Biochem Biophys Res Commun. 2000;270:728–732. doi: 10.1006/bbrc.2000.2507. [DOI] [PubMed] [Google Scholar]
- 26.Tzeng SJ, Chang WC, Huang JD. Transcriptional regulation of the rat Mrp3 gene promoter by the specificity protein (Sp) family members and CCAAT/enhancer binding proteins. J Biomed Sci. 2005;12:741–761. doi: 10.1007/s11373-005-9002-5. [DOI] [PubMed] [Google Scholar]
- 27.Uruno A, Sugawara A, Kudo M, Sato M, Sato K, Ito S, Takeuchi K. Transcription suppression of thromboxane receptor gene expression by retinoids in vascular smooth muscle cells. Hypertens Res. 2003;26:815–821. doi: 10.1291/hypres.26.815. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Bannon MJ. Sp1 and Sp3 activate transcription of the human dopamine transporter gene. J Neurochem. 2005;93:474–482. doi: 10.1111/j.1471-4159.2005.03051.x. [DOI] [PubMed] [Google Scholar]
- 29.Xiong H, Yoshinari K, Brouwer KL, Negishi M. Role of constitutive androstane receptor in the in vivo induction of Mrp3 and CYP2B1/2 by phenobarbital. Drug Metab Dispos. 2002;30:918–923. doi: 10.1124/dmd.30.8.918. [DOI] [PubMed] [Google Scholar]
- 30.Zelcer N, van de Wetering K, Hillebrand M, Sarton E, Kuil A, Wielinga PR, Tephly T, Dahan A, Beijnen JH, Borst P. Mice lacking multidrug resistance protein 3 show altered morphine pharmacokinetics and morphine-6-glucuronide antinociception. Proc Natl Acad Sci USA. 2005;102:7274–7279. doi: 10.1073/pnas.0502530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zelcer N, Wetering K, Waart R, Scheffer GL, Marschall HU, Wielinga PR, Kuil A, Kunne C, Smith A, Valk M, Wijnholds J, Elferink RO, Borst P. Mice lacking Mrp3 (Abcc3) have normal bile salt transport, but altered hepatic transport of endogenous glucuronides. J Hepatol. 2006;44:768–775. doi: 10.1016/j.jhep.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 32.Zollner G, Fickert P, Silbert D, Fuchsbichler A, Marschall HU, Zatloukal K, Denk H, Trauner M. Adaptive changes in hepatobiliary transporter expression in primary biliary cirrhosis. J Hepatol. 2003;38:717–727. doi: 10.1016/s0168-8278(03)00096-5. [DOI] [PubMed] [Google Scholar]


