Summary
Protein domain movement of the Rieske iron-sulfur protein has been speculated to play an essential role in the bifurcated oxidation of ubiquinol catalyzed by the cytochrome bc1 complex. To better understand the electron transfer mechanism of the bifurcated ubiquinol oxidation at Qp site, we fixed the head domain of ISP at the cyt c1 position by creating an intersubunit disulfide bond between two genetically engineered cysteine residues: one at position 141 of ISP and the other at position 180 of the cyt c1 [S141C(ISP)/G180C(cyt c1)]. The formation of a disulfide bond between ISP and cyt c1 in this mutant complex is confirmed by SDS-PAGE and Western blot. In this mutant complex, the disulfide bond formation is concurrent with the loss of the electron transfer activity of the complex. When the disulfide bond is released by treatment with β-mercaptoethanol, the activity is restored. These results further support the hypothesis that the mobility of the head domain of ISP is functionally important in the cytochrome bc1 complex. Formation of the disulfide bond between ISP and cyt c1 shortens the distance between the [2Fe-2S] cluster and heme c1, hence the rate of intersubunit electron transfer between these two redox prosthetic groups induced by pH change is increased. The intersubunit disulfide bond formation also decreases the rate of stigmatellin induced reduction of ISP in the fully oxidized complex, suggesting that an endogenous electron donor comes from the vicinity of the b position in the cytochrome b.
Keywords: bacterial cytochrome bc1 complex, formation of a disulfide bond between ISP and cyt c1, head domain of ISP, Rhodobacter sphaeroides
1. Introduction
The cytochrome bc1 complex (also known as ubiquinol-cytochrome c oxidoreductase or complex III) is a multi-subunit dimeric integral membrane protein complex. It is an essential segment of cellular energy-conserving electron transport chains in animals, plants, and bacteria [1, 2]. This complex catalyzes electron transfer from ubiquinol to cytochrome c (or cytochrome c2 in bacteria) and concomitantly translocates protons across the membrane to generate a membrane potential and pH gradient for ATP synthesis. Although the cytochrome bc1 complexes from different sources vary in their polypeptide compositions, they all contain four redox prosthetic groups: two b-type cytochromes (bL and bH), one c-type cytochrome (c1), and one high potential Rieske iron-sulfur cluster [2Fe-2S]. Structural information obtained from X-ray crystallographic studies [3–5] has revealed the arrangement of the redox centers, transmembrane helices, inhibitor binding sites and the intertwined dimeric structure. The longer than expected distance between [2Fe-2S] and heme c1 and the less defined structure of the head domain of the iron-sulfur protein suggest a domain movement of ISP during the electron transfer in bc1 complex. The domain movement hypothesis was further supported by the observation of various positions of the [2Fe-2S] cluster in different crystal forms [4, 5] or in complexes loaded with different inhibitors [5, 6].
Structurally, the iron-sulfur protein (ISP) in cytochrome bc1 complex can be divided into three domains: the soluble C-terminal extramembrane domain (head), which houses the [2Fe-2S] cluster at its tip, the membrane-spanning N-terminal domain (tail), and the flexible linking domain (neck), which links the head and tail domains [7, 8]. Bending of the neck is required for movement of the ISP head domain between the two positions (heme b and heme c1). This movement is essential for electron transfer [9–12].
The study of the neck region of ISP using Rhodobacter sphaeroides cytochrome bc1 complex [9, 12] provided the first evidence for movement of the head domain of ISP during cytochrome bc1 complex catalysis. Since the head and tail domains are rather rigid, a flexible neck region is necessary for head domain movement. Mutants with increased neck rigidity, generated by deletion or double- or triple-proline substitution, have greatly reduced electron transfer activity with an increase in activation energy [12]. Formation of a disulfide bond between two engineered cysteines with only one amino acid residue between them in the neck region near the transmembrane helix also drastically reduces electron transfer activity [9], presumably because of increased neck rigidity. As expected, cleavage of the disulfide bond by reduction or alkylation restores activity to that of the wild type enzyme [9]. Elongation of the neck region by insertion of multiple alanine residues significantly slows down the movement of the head domain of ISP and affects the bc1 complex activity [13]. All of these results clearly demonstrate a need for neck flexibility in catalysis.
To further establish that the movement of the head domain of ISP is essential for the bc1 complex, the mutant expressing His6-tagged bc1 complex with a pair of cysteine substituted residues at the interface between cytochrome b (A185) and the head domain of ISP (K70) [11] was generated and characterized. The disulfide bond between the engineered cysteine pair arrests the mobility of ISP to the fixed state (b position) and thus decreases electron transfer activity.
While the requirement of head domain movement of ISP is well established, the bifurcated ubiquinol oxidation at Qp site is not yet fully understood. It is generally believed that ISP is reduced by ubiquinol when its head domain is located at the b position. In other words, if the head domain is at c1 position the ISP will not be able to accept electron from the substrate. To test this hypothesis, we recently generated the mutant [S141C(ISP)/G180C(cyt c1)] with head domain of ISP fixed at the c1 position by mutating residues S141 of ISP and G180 of cyt c1 to cysteines to form an intersubunit disulfide bond (Fig. 1). Mutants with single cysteine substitution at the indicated positions were also generated and characterized to confirm that the residues are not at a critical for the activity or other properties of the bc1 complex. The photosynthetic growth behavior, electron transfer activity, SDS-PAGE patterns, Western blot, pH-induced reduction and oxidation of ISP and cytochrome c1 in the partially reduced complex, redox potential titration and EPR characteristics of the [2Fe-2S] cluster in purified complexes from wild type and mutant strains were examined and compared.
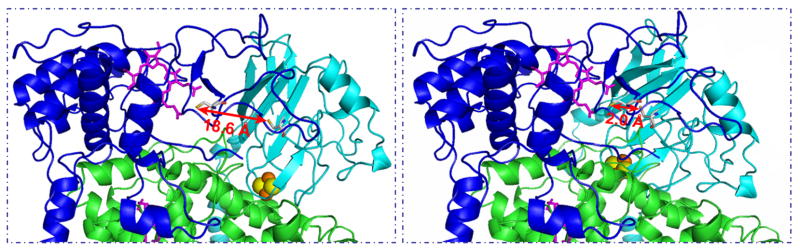
Fig. 1.
Relative distances between engineered cysteines in ISP and cytochrome c1 in the cytochrome bc1 complex from R. sphaeroides mutant S141C(ISP)/G180C(cyt c1). Pictures show the distances between two engineered cysteines when the head domain of ISP is at the b position (left) and the ‘c1’ position (right). Cytochrome c1 is shown by the blue ribbon, cytochrome b is in green, and ISP (from the other monomer) is in cyan. The bH and c1 heme groups are shown as pink stick models, and the [2Fe-2S] cluster is depicted in gold and yellow spheres.
To study whether the formation of an intersubunit disulfide bond between ISP and cytochrome c1 fixes the [2Fe-2S] cluster close to heme c1, the rate of electron transfer between these two redox prosthetic groups, induced by pH change, is tested. Stigmatellin is also used to induce the reduction of ISP, in the absence of exogenous electron donor, in the fully oxidized complex to study whether the formation of the disulfide bond between cyt c1 and ISP could pull the [2Fe-2S] cluster away from the Qp site. The EPR signals of ISP mutant complex with or without stigmatellin are examined and compared with wild type complex. All results show that the mutant S141C(ISP)/G180C(cyt c1) complex obtained is indeed the one whose head domain of ISP is fixed at the cyt c1 position when the disulfide bond is formed between the two engineered cysteines.
2. Materials and methods
2.1. Materials
Cytochrome c (horse heart, type III) was purchased from Sigma. N-Dodecyl-β-D-maltoside (LM) and N-octyl-β-D-glucoside (OG) were from Anatrace. 2,3-Dimethoxy-5-methyl-6-(10-bromodecyl)-1,4-benzoquinol (Q0C10BrH2) was prepared in our laboratory as previously reported [14]. Ni-NTA gel, Qiaprep Spin Miniprep kit and PCR purification kit were from Qiagen. All other chemicals were of the highest purity commercially available.
2.2. Growth of Bacteria
Escherichia coli cells were grown at 37 °C in LB medium. R. sphaeroides BC17 cells [15] were grown photosynthetically at 30 °C in an enriched Siström’s medium containing 5 mM glutamate and 0.2% casamino acids. Photosynthetic growth conditions for R. sphaeroides were essentially as described previously [16]. The concentrations and antibiotics used were: ampicillin, 125 μg/ml; kanamycin sulfate, 30 μg/ml; tetracycline, 10 μg/ml for E. coli, and 1 μg/ml for R. sphaeroides; and trimethoprim, 100 μg/ml for E. coli and 30 μg/ml for R. sphaeroides.
2.3. Generation of R. sphaeroides Strains Expressing the His6-tagged bc1 Complexes with Single or Pair of Cysteine Substitutions on Cytochrome c1 and ISP
The QuickChange™ XL site-directed mutagenesis kit from Stratagene was used for mutagenesis. The oligonucleotides used for mutagenesis were as follows: S141C (ISP), F, 5′ CCGATCGGCGGCGTGT GCGGTGACTTCGGGGGC 3′; R, 5′ GCCCCC GAAGTCACCGCACACGCCGCCGATCGG 3′; G180C (cyt c1), F, 5′ GCCTGCTCGTGGAT CGCCATG 3′; R, 5′ CTAGGCGATCCACGAG CAGGC 3′; H111C (cyt c1), F, 5′ GCGCGGGC GGGCTTCTGCGGCCCGATGGGCACC; R, 5′ GGTGCCCATCGGGCCGCAGAAGCCCGCCCGC GC; G112C (cyt c1), F, 5′ CGGGCGGGCTTCCACTGCCCGATGG GCACCGGC; R, 5′ GCCGGTGCCCATCGGGCAGTGGAAGCCCGCCCG; M114C (cyt c1), F, 5′ GGCTTCCA CGGCCCGTGCGGCACCGGCATGAGC; R, 5′ GCTCATGCCGGTGCCGCACGGGCCGTG GAAGCC.
The double-stranded plasmid pGEM-7Zf(+)-fbcFB was used as the template for mutagenesis. Forward and reverse primers were used for PCR amplification. The pGEM-7Zf(+)-fbcFB plasmid was constructed by ligating the EcoR I-Xba I fragment from pRKD418-fbcFBCHQ [15] into EcoR I and Xba I sites of the pGEM-7Zf(+) plasmid. After the mutagenesis, the fragment containing the mutant from pGEM-7Zf(+)-fbcFmB was inserted into pRKD418-fbcFBKmCHQ by EcoR I and Nsi I double digestion and ligation reaction. Loss of kanamycin resistance was then used to screen for the recombinant cloning containing the mutant ISP gene (pRKD418-fbcFmBCHQ). A similar process was used to get the clones with the mutant cytochrome c1 gene (pRKD418-fbcFBCHmQ) and double mutant pRKD418-fbcFmBCHmQ.
The pRKD418-fbcFmBCHmQ plasmid in Escherichia coli S17-1 cells was mobilized into R. sphaeroides BC17 cells by the plate-mating procedure [15]. The presence of engineered mutations was confirmed by DNA sequencing before and after photosynthetic or semi-aerobic growth of the cells as previously reported [11, 15]. Expression plasmid pRKD418-fbcFmBCHmQ was purified from an aliquot of a photosynthetic or semi-aerobic culture using the Qiagen Plasmid Mini Prep kit. Because R. sphaeroides cells contain more than one type of endogenous plasmids [11], the isolated plasmids lack the purity and concentration needed for direct sequencing. Therefore, the DNA segments containing the mutation sequence in ISP and cytochrome c1 were amplified from the isolated plasmids by the polymerase chain reaction. The PCR products were purified with an extraction kit from Sigma. The DNA sequencing was performed by the Recombinant DNA/Protein Core Facility at the Oklahoma State University.
2.4. Enzyme Preparations and Assay of Cytochromes
Chromatophore membranes were prepared from frozen cell paste and cytochrome bc1 complexes with a His6-tag placed at the C-terminus of cytochrome c1 were purified from chromatophores as described previously [16] and stored at −80 °C in the presence of 10% glycerol. Protein concentrations were determined by the absorbance at 280 nm, using a converting factor of 1 OD280 = 0.56 mg/ml. The concentrations of cytochromes b and c1 were determined spectrophotometrically using published molar extinction coefficients [17–19].
2.5. Activity Assay of the Cytochrome bc1 Complex
To assay ubiquinol-cytochrome c reductase activity, chromatophores or purified cytochrome bc1 complexes were diluted with 50 mM Tris-Cl, pH 8.0, containing 200 mM NaCl and 0.01% N-dodecyl-β-D-maltoside (LM) to a final concentration of cytochrome b of 1 μM unless otherwise specified. Appropriate amounts of the diluted samples were added to 1 ml of assay mixture containing 100 mM Na+/K+ phosphate buffer, pH 7.4, 0.3 mM EDTA, 100 μM cytochrome c, and 25 μM Q0C10BrH2. Activities were determined by measuring the reduction of cytochrome c (the increase of the absorbance at wavelength 550 nm) in a Shimadzu UV-2401 PC spectrophotometer at 23 °C, using a millimolar extinction coefficient of 18.5 for calculation. The non-enzymatic oxidation of Q0C10BrH2, determined under the same conditions, in the absence of enzyme, was subtracted during specific activity calculations. Although the chemical properties of Q0C10BrH2 are comparable with those of Q0C10H2, the former is a better substrate for the cytochrome bc1 complex [14].
2.6. Gel Electrophoresis and Western Blot Preparation
SDS-PAGE was performed according to the method of Laemmli [20] using a Bio-Rad Mini-Protean dual slab vertical cell. Samples were digested with 10 mM Tris-Cl buffer, pH 6.8, containing 1% SDS and 3% glycerol in the presence or absence of 0.4% β-mercaptoethanol (β-ME) for 10 min at room temperature before being subjected to electrophoresis. Western blotting was performed with polyclonal rabbit antibodies against cytochrome c1 or ISP of the R. Sphaeroides bc1 complex. The polypeptides separated by SDS-PAGE gel were transferred to 45 Micron nitrocellulose membrane for immunoblotting. Protein A conjugated to horseradish peroxidase (HRP) was used as the second antibody. Color development was carried out using a HRP color development solution.
2.7. Determination of the Redox Potential of Cytochromes c1 in the Wild Type and the Mutant Cytochrome bc1 Complexes
Redox titrations of cytochromes c1 in the wild type and the mutant bc1 complexes were determined essentially according to the previously published method [21, 22]. 3 ml aliquots of the bc1 complex (7 μM cytochrome c1) in 0.1 M Na+/K+ phosphate buffer, pH 7.0, containing 25 μM 1,4-benzoquinone (Em, 293 mV), 25 μM 2,3,5,6-tetramethyl-p-phenylenediamine (Em, 260 mV), 25 μM 1,2-naphthoquinone (Em, 143 mV), 20 μM phenazine methosulfate (Em, 80 mV), 20 μM phenazine ethosulfate (Em, 55 mV), 25 μM 1,4-naphthoquinone (Em, 36 mV) and 25 μM duroquinone (Em, 5 mV) were used. Reductive titrations were carried out by addition of sodium dithionite solution to the ferricyanide-oxidized sample. Oxidative titrations were carried out by addition of ferricyanide solution to the dithionite-reduced sample. At indicated Eh values during the redox titration, absorption spectra, 600 to 500 nm, were taken with a Shimadzu model UV-2401 PC spectrophotometer. The optical density at 550 nm, minus that at 535 nm, was used for cytochrome c1 measurement. The midpoint potential of cytochrome c1 was calculated by fitting the redox titration data, obtained for cytochrome c1, using the Nernst equation for a one-electron carrier (n = 1) by Kaleidagraph [21].
2.8. Determination of pH-induced Reduction and Oxidation of ISP and Cytochrome c1 in the Partially Reduced Wild Type and Mutant bc1 Complexes
The wild type or mutant bc1 complex was diluted in 3 ml of 20 mM Tris-Cl buffer, pH 8.0, containing 200 mM NaCl and 0.01% LM. The concentration of cytochrome c1 was adjusted to about 10 μM. Different amounts of NaOH or HCl were added to give the indicated pHs. Fully oxidized or reduced cytochrome c1 and ISP were obtained by addition of K3Fe(CN)6 or sodium ascorbate. Reduction of cytochrome c1 was followed by measuring the increase of the α-absorption (550–535 nm) in a Shimadzu UV-2401 PC spectrophotometer. Reduction of ISP was followed by measuring the negative CD peak at 500 nm of partially reduced ISP minus fully oxidized complex in a JASCO J-715 spectropolarimeter [11, 23, 24]. The same samples were used for the absorption and CD measurement. Instrument settings for the spectropolarimeter were: scan speed, 100 nm/min; step resolution, 1 nm; accumulation, 10 traces for averaging; response, 1 s; bandwidth, 2.0 nm; sensitivity, 10 mdeg; and slit width, 500 μm.
2.9. Determination of the pH induced Electron Transfer Rates between the [2Fe-2S] Cluster and Heme c1 in the Wild Type and the Mutant bc1 Complexes
The method used is essentially the same as that previously reported [11, 24]. The cytochrome bc1 was diluted in 20 mM Tris-Cl buffer, pH 8.0, containing 200 mM NaCl and 0.01% LM to a cytochrome c1 concentration around 10 μM. The percentage of cytochrome c1 reduction in the sample was adjusted to around 50% at pH 8.0 by adding sodium ascorbate or ferricyanide. For the electron transfer from the [2Fe-2S] cluster to heme c1, the pH of the sample was decreased to pH 6.9 by adding HCl. This solution was rapidly mixed with an equal volume of the same buffer but containing enough NaOH to cause the pH change from 6.9 to 8.9 in the stopped flow spectrophotometer at room temperature. Increased reduction of cytochrome c1 was monitored spectrophotometrically. The reaction was monitored at 550nm–535nm. Complex mixed with an equal volume of the same buffer at pH 6.9 was used as a baseline.
2.10. Stigmatellin Induced Reduction Rate of ISP in the Absence of Exogenous Electron Donor in the Fully Oxidized Complex
In order to know whether position of [2Fe-2S] cluster in ISP was pulled toward the cytochrome c1, far away from the Qp site by the formation of the disulfide bond between cyt c1 and ISP, stigmatellin is used to induce the reduction of ISP in the absence of exogenous electron donor in the fully oxidized complex. The bc1 complexes from wild type, S141C(ISP)/G180C(cyt c1), and A185C(cyt b)/K70C(ISP) [11] were diluted to 10 μM of cytochrome c1 in 3 ml of 20 mM Tris-Cl buffer, pH 8.0, containing 200 mM NaCl and 0.01% LM. K3Fe(CN)6 was used to fully oxidize the bc1 complexes, and the fully oxidized state of complexes was confirmed by measuring CD from 450 nm to 550nm in a JASCO J-715. Stigmatellin induced reduction of ISP were measured after incubating the fully oxidized bc1 complexes with 50 μM inhibitor for 5 min, 1 hr and 2hr, respectively. Fully reduced ISP spectrum was obtained after the bc1 complex was reduced by sodium ascorbate. The reduction of ISP was calculated by the negative CD peak at 500 nm of reduced ISP minus fully oxidized spectrum.
2.11. Stigmatellin Induced Oxidation Rate of Cytochrome c1 in the Partially Reduced Complex
It is well known that binding of stigmatellin to cytochrome bc1 complex causes substantial elevation of the mid-point potential of ISP [25, 26]. As a result, an intersubunit electron transfer between the [2Fe-2S] cluster and heme c1 is expected to take place after stigmatellin is added into the complex. The bc1 complex was diluted to cytochrome c1 5 μM in the buffer containing 20 mM Tris-Cl buffer, pH 8.0, containing 200 mM NaCl and 0.01% LM. Sodium ascorbate was used to adjust the reduction of cyt c1 to around 50%. Stigmatellin was added to the complex to a final concentration 25 μM. The redox state of cytochrome c1 was monitored (550–535 nm) by a Shimadzu UV-2401 PC spectrophotometer at 23 °C.
2.12. EPR Experiments
EPR spectra of the ISP were recorded by a Bruker EMX Spectrometer [27]. The instrument settings were as follows: microwave frequency, 9.28 Hz; microwave power, 20 milliwatt; modulation amplitude, 20 G; modulation frequency, 100 kHz; time constants, 0.1 s; and scan rate, 20 respectively. 50 μM bc1 complexes were incubated with 200 μM sodium ascorbate on ice for 20 min and frozen in liquid nitrogen. After EPR spectra were recorded, the complexes were thawed on ice and stigmatellin was added to 250 μM. The samples were put into liquid nitrogen again after incubation with stigmatellin on ice for 15 min. EPR spectra were record at −156°C.
3. Results and discussion
3.1. Photosynthetic Growth Behaviors of the Wild Type and Mutants
Since cytochrome bc1 complex is absolutely required for photosynthetic growth of R. sphaeroides, a mutant with cysteine substitution at a critical position will not grow photosynthetically, whereas mutants with substitution at noncritical positions will grow. Thus, observing the photosynthetic growth behavior, one can determine whether the engineered cysteines are at critical positions. In mid-log phase, semiaerobically dark grown wild type and mutant cells were inoculated into enriched Siström’s medium and subjected to anaerobic photosynthetic growth conditions. Mutants grew at rates comparable with that of the wild type (Table I), indicating that the engineered cysteine at these positions are noncritical to the complex. However, mutants H111C(cyt c1), G112C(cyt c1) and S141C(ISP)/H111C(cyt c1), S141C(ISP)/G112C(cyt c1) grew at a retarded rate, suggesting that substitutions of H111C(cyt c1), G112C(cyt c1) affect the activity of cytochrome bc1 complex and thus decrease the growth rate of R. sphaeroides.
3.2. Formation of a Disulfide Bond between the ISP Head Domain and Cytochrome c1 in the S141C(ISP)/G180C(cyt c1) Mutant bc1 Complex
Chromatophores freshly prepared from the S141C(ISP), G180C(cyt c1), and S141C(ISP)/G180C(cyt c1) mutant cells have, respectively, 100, 78, and 26% of the bc1 activity found in the wild type chromatophores (Table 1). The purified His6-tagged bc1 complexes isolated from freshly prepared chromatophores have, 100, 80, and 8% of the wild type bc1 activity respectively (Table 1), based on cytochrome b content. Both of the single mutants, S141C(ISP) and G180C(cyt c1) have good activity, indicating the mutagenesis has little effect. The reason that the double mutant S141C(ISP)/G180C(cyt c1) can grow well photosynthetically yet the activity of both the chromatophore and purified bc1 complex are low is because few disulfide bonds are present in the intact cells. As formation of a disulfide bond is an oxidative process, no disulfide bond can be formed in anaerobic conditions, even if the two cysteines are in favorable positions. Since photosynthetic growth is under strict anaerobic conditions, disulfide bond formation between the two engineered cysteines in mutant S141C(ISP)/G180C(cyt c1) is not possible. Hence, the double mutant cell can grow photosynthetically. However, during preparation of chromatophores and purification of the bc1 complex, both engineered cysteines are exposed to the air (oxygen); formation of the disulfide bond occurs, resulting in a decrease of activity.
TABLE 1.
Characterization of mutants. The cytochrome bc1 complexes were in 50 mM Tris-Cl, pH 8.0, containing 200 mM NaCl, 200 mM histidine, 0.5% octyl glucoside and 10% glycerol.
| Strains | Ps a | Activity b |
|
|---|---|---|---|
| Chromatoxphore | Purified complex | ||
| Wild type | + + + | 2.3 (100%) | 2.5 (100%) |
| S141C(ISP) | + + + | 2.3 (100%) | 2.5 (100%) |
| G180C(cyt c1) | + + + | 1.8 (78%) | 2.0 (80%) |
| S141C(ISP)/G180C(cyt c1) | + + | 0.6 (26%) | 0.2 (8%)* |
| H111C(cyt c1) | + | 0.2 (9%) | 0.6 (24%) |
| S141C(ISP)/H111C(cyt c1) | + | 0.2 (9%) | 0.6 (24%) |
| G112C(cyt c1) | + | 0.8 (35%) | 0.9 (36%) |
| S141C(ISP)/G112C(cyt c1) | + | 0.8 (35%) | 0.9 (36%) |
| M114C(cyt c1) | + + + | 2.3 (100%) | 2.5 (100%) |
| S141C(ISP)/M114(cyt c1) | + + + | 2.3 (100%) | 2.5 (100%) |
Ps=photosynthetic growth.
The enzymatic activity is expressed as μmol of cytochrome c reduced/min/nmol cytochrome b at room temperature.
If purification is carried out in the presence of β-ME, this value will be 2.0.
The specific activity of aerobically purified complex from mutant S141C(ISP)/G180C(cyt c1) is around 8% of the wild type complex. It is of interest to know whether this residual activity is from the disulfide linked complex or due to the incomplete disulfide bond formation. One way to know the nature of this residual activity is to determine the sensitivity of this activity to antimycin. Results show that 90% of activity in the mutant was blocked by antimycin, similar to the sensitivity of activity of the wild type complex (data not shown), indicating that the residual activity of mutant complex is resulted from the presence of small portion bc1 complex without disulfide linkage between ISP and cyt c1 subunits. The presence of a faint band of free ISP and cytochrome c1 in the Western blot analysis with antibodies against R. sphaeroides cytochrome c1 or ISP of mutant S141C(ISP)/G180C(cyt c1) complex in the absence of β-ME supports this conclusion.
To see whether or not the loss of the bc1 complex activity observed in the mutant S141C(ISP)/G180C(cyt c1) results from disulfide bond formation during the purification, SDS-PAGE patterns of the purified mutant complexes, with and without β-ME treatment, were examined (Fig. 2). The purified complexes were treated with SDS in the absence or presence of β-ME (Fig. 2) at room temperature for 10 min and subjected to electrophoresis. The same process was done with bc1 of the wild type, S141C(ISP), and G180C(cyt c1) mutants. When the β-ME was present, all bc1 complexes showed the same pattern in SDS-PAGE: 4 protein bands, cyt b (41 kDa), cyt c1 (30.4 kDa), ISP (21 kDa), and subunit IV (14 kDa). However, in the absence of β-ME, the two single mutants had the same electrophoretic pattern as the wild type but the mutant S141C(ISP)/G180C(cyt c1) showed two differences: (i) An adduct protein band of the cross-link protein was present and its molecular weight was around 51 kDa, the sum of cyt c1 (30.4 kDa) and ISP (21 kDa); (ii) The density of cyt c1 and ISP bands showed a sharp decrease, indicating that most cyt c1 and ISP are present in the cross-link protein. The intersubunit disulfide bond linked adduct protein from the mutant S141C(ISP)/G180C(cyt c1) was identified as a smear protein band in SDS-PAGE (Fig. 2), consistent with the characteristic of cytochrome c1 which also shows as a smear band in SDS-PAGE in the absence of β-ME. It has been reported that cyt c1 of R. sphaeroides shows as a smear band in SDS-PAGE patterns under non-reducing conditions [28]. It was recently reported that [28], under reducing conditions (in the presence of β-ME), protein with covalently bound heme (such as c-type cytochrome) shows as a sharp band in SDS-PAGE, whereas the protein band of c-type cytochromes become smears in SDS-PAGE when β-ME is absent.
Fig. 2.
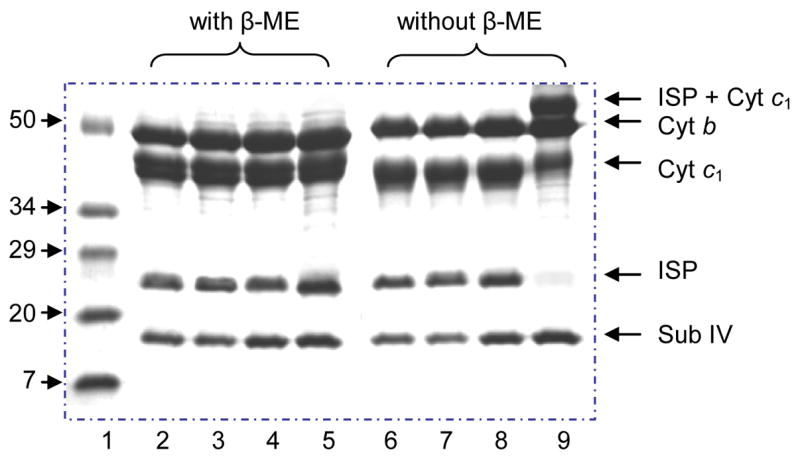
SDS-PAGE of the wild type and the cysteine mutant cytochrome bc1 complexes. Lane 1, protein standard (Std); Lane 2 & 6, the wild type; Lane 3 & 7, S141C(ISP); Lane 4 & 8, G180C(cyt c1); Lane 5 & 9, S141C(ISP)/G180C(cyt c1). Adduct cross-link protein bond (ISP + Cyt c1, lane 9) is disappeared when treated with β-ME (lane 5). Moreover, the intensity of ISP and cyt c1 bands are obviously decreased compared to the corresponding bonds in the lane 5.
Western blot analysis with antibodies against R. sphaeroides cytochrome c1 or ISP was done to characterize the adduct protein band (Fig. 3). The results show that the adduct protein band can be blotted by both anti- cyt c1 and anti-ISP antibodies, indicating the adduct protein contains both cyt c1 and ISP. Trace amount of cytochrome c1 and ISP was also observed in the Western blot analysis of the double mutant complex.
Fig. 3.
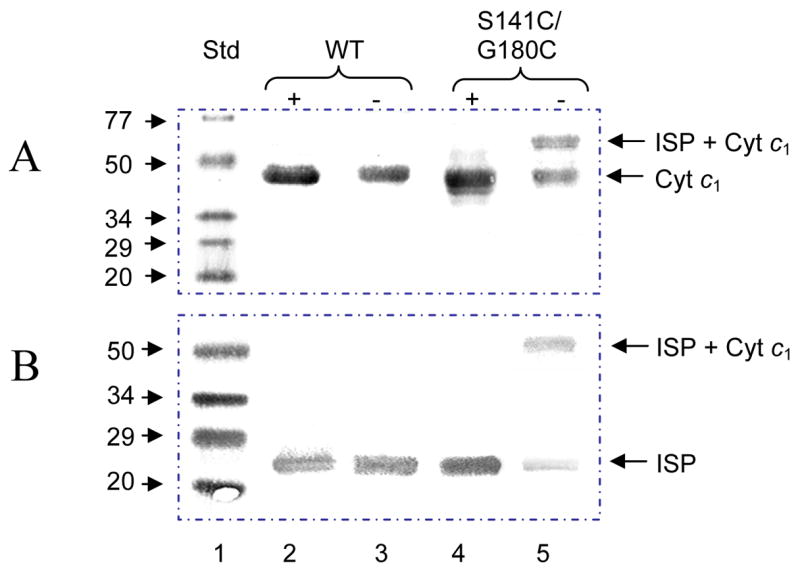
Western blot analysis of ISP or cytochrome c1 in cytochrome bc1 complexes from the wild type and S141C(ISP)/G180C(cyt c1). Aliquots of the wild type and S141C(ISP)/G180C(cyt c1) were incubated in the loading buffer containing 1% SDS with (+) or without (−) 0.4% β-ME at room temperature for 10 min. Digested samples containing 200 pmol of cytochrome b were subjected to electrophoresis. Samples in the gel were transferred to 45 micron nitrocellulose membrane and treated with antibody against R. sphaeroides cytochrome c1 (A) or ISP (B). Protein A-horseradish peroxide conjugate was used as a second antibody. Std, protein standard. WT, the wild type.
From gene sequence, there are nine cysteine residues in the R. sphaeroides cytochrome bc1 complex: one in cytochrome b, four in cytochrome c1, and four in ISP. Two cysteines (Cys129 and Cys149) in ISP serve as ligands for the [2Fe-2S] cluster [29] and two cysteines (Cys37 and Cys40) in cytochrome c1 are covalently bonded to heme c1 [30]. Thus, there are five cysteines in the complex: one in cytochrome b (Cys54), two in ISP (Cys134 and Cys151) and two in cytochrome c1 (Cys145 and Cys169). In the reported stigmatellin loading in cytochrome bc1 complex structure [10], Cys134 is on loop 4–5 and Cys151 is on loop 6–7 of ISP. The distance between these two cysteines is 2.0 Å and they form an intrasubunit disulfide bond that brings the two loops together and stabilizes this region [29]. The distances between the engineered cysteine S141C (ISP) to Cys129 (ISP), Cys134 (ISP), Cys 149(ISP), and Cys 151(ISP) are 13.3 Å, 11.2 Å, 14.4 Å, and 13.7 Å, respectively. Therefore, they are not available for intrasubunit disulfide bond formation between the engineered and endogenous cysteines in ISP. Since it has been established that the pair of cysteines, Cys145 and Cys169 in the head domain of cytochrome c1 form a disulfide bond to maintain the structural integrity [28], their participation in forming a disulfide bond with the engineered cysteine in cytochrome c1 (G180C) is unlikely. Also, the distances between Cys 180 (cyt c1) and Cys145 (cyt c1) and Cys169 (cyt c1) are 10.0 Å and 8.6 Å, too far apart for intrasubunit disulfide bond formation, as the head domain of cytochrome c1 is rather rigid. Although the distance between two engineered residues, S141 (ISP) and G180 (cyt c1) is 18.6 Å when the head domain of ISP is at the b position (Fig. 1), this distance is expected to be much less when the head domain moves ~23 Å to c1 position to deliver the electron [31]. Therefore, the formation of an intersubunit disulfide bond is likely.
The low bc1 complexes activities (Table I) and the retarded rate of photosynthetic growth in mutants H111C(cyt c1), G112C(cyt c1) and S141C(ISP)/H111C(cytc1), S141C(ISP)/G112C (cyt c1), indicate that the mutation on these two amino acid residues have considerable effect on the bc1 complexes. Double mutant S141C(ISP)/M114C (cyt c1), however, has good bc1 complex activity and normal growth rate, suggesting that no significant change in the mutant complexes. SDS-PAGE results (data not shown) show there is no adduct protein band in either S141C(ISP)/H111C(cyt c1), S141C(ISP)/G112C (cyt c1) or S141C(ISP)/M114C(cyt c1), indicating that no disulfide bond is formed between ISP and cytochrome bc1 in these double mutant complexes.
Based on the crystal structure of R. sphaeroide bc1 complex determined in the presence of stigmatellin [10], the distances from S141C(ISP) to H111(cyt c1), G112(cyt c1) and M114(cyt c1) are 20.0, 23.5 and 24.1 Å, respectively, farther than the distance between S141C(ISP) and G180C(cyt c1) (18.6 Å). The reason of designing these 3 mutants was to test whether they are on the path of the head domain of ISP moving to cytochrome c1 position. The results showed that none of them can form disulfide bond with S141C of ISP, suggesting they are not in the proposed path of ISP head domain movement. However, since disulfide bond can be formed between G180C(cyt c1) and S141C(ISP), G180C is located in the proposed path of the ISP head domain movement. The fact, that S141C(ISP)/G180C(cyt c1) can form the disulfide bond between cytochrome c1 and ISP only in the absence and not in the presence of stigmatellin (data not show), suggests that cytochrome bc1 complex structure obtained in the presence of stigmatellin is different from the native one and the head domain of cytochrome c1 may undergo a certain movement in the native structure. A high resolution structure of cytochrome bc1 complex in the absence of inhibitor would resolve these speculations.
3.3. Effect of β-ME on the Activity of Cytochrome bc1 Complex
To confirm that formation of a disulfide bond between ISP and cytochrome c1 in the S141C(ISP)/G180C(cyt c1) mutant causes activity loss, the effect of β-ME on bc1 complex activity and the degree of disulfide bond formation were examined. When purified S141C(ISP)/G180C(cyt c1) mutant complex was incubated at 4°C with 15 mM β-ME, activity increased as incubation time was prolonged (Fig 4), the activity was restored to that of single mutant G180C(cyt c1) after a 24 hour incubation. No adduct protein of ISP and cytochrome c1 was detected in the β-ME treated S141C(ISP)/G180C(cyt c1) mutant complex. When this complex was purified from freshly prepared chromatophores in the presence of 100 mM β-ME, it had the same activity as that found in the G180C(cyt c1) and no ISP- cytochrome c1 adduct was detected. All these results show that the loss of enzymatic activity is caused by the formation of a disulfide bond between S141C(ISP) and G180C(cyt c1), not the presence of cysteine. It must be emphasized that the observed activity restoration is not due to nonenzymatic reduction of cytochrome c by added β-ME [11] nor to long term exposure to 4°C, because the same treatment has little effect on the wild type complex.
Fig. 4.
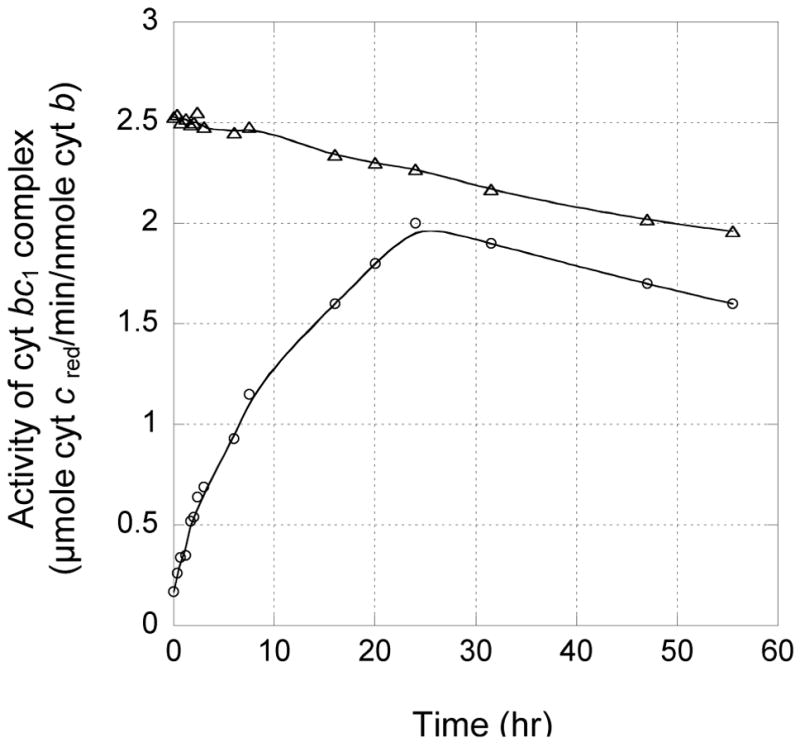
Effect of β-ME on the cytochrome bc1 activity in purified complexes from the wild type (triangle) and the mutant S141C(ISP)/G180C(cyt c1) (circle) at 4°C. 10 μM cytochrome bc1 was incubated at 4°C with 15mM β-ME.
3.4. Effect of the Mutations on Redox Potential of Cytochrome c1 in the bc1 Complex
The redox potential of cyt c1 in the S141C(ISP)/G180C(cyt c1) mutant complex is 220 mV (Fig. 5), slightly lower than 235 mV observed in the wild type bc1 complex [28, 32–34], indicating the formation of a disulfide bond does not affect the redox potential of cyt c1 significantly, and the loss of bc1 activity cannot be attributed to a redox potential change.
Fig. 5.
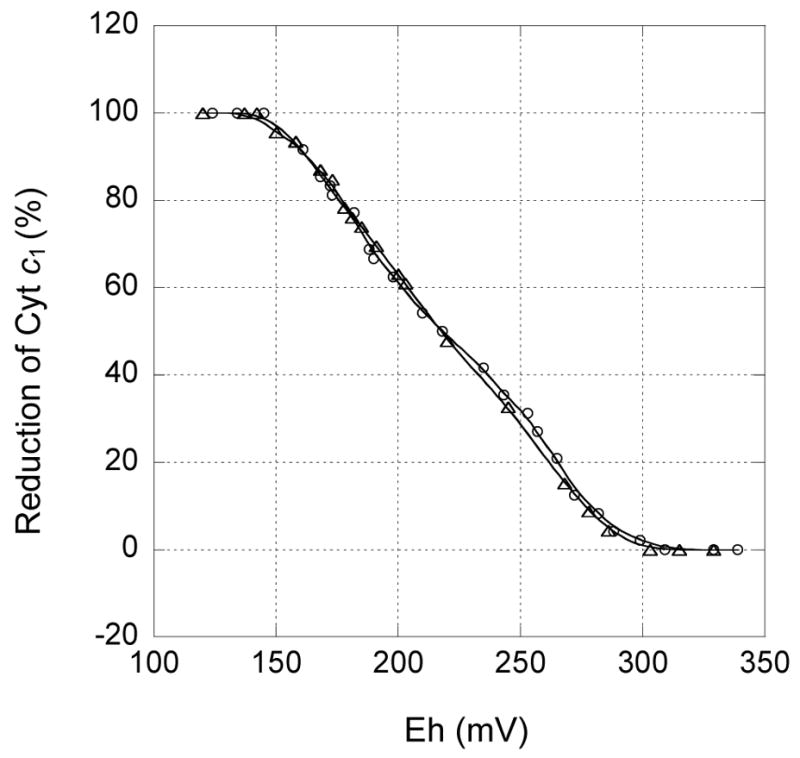
Redox potential titration of cytochrome c1 in S141C(ISP)/G180C(cyt c1) bc1 complex. Oxidative and reductive titrations were performed as described in “Experimental Procedures.” Triangle curve shows oxidative titration by ferricyanide and the circle shows reductive titration by dithionite. The data were fit to the Nernst equation when n = 1
3.5. pH-dependent Oxidation of ISP and Reduction of Cytochrome c1 in the Purified Cytochrome bc1 Complex
It has been reported that intramolecular electron transfer between the [2Fe-2S] cluster and heme c1 in both bovine and R. sphaeroides cytochrome bc1 complexes can be induced by pH change of the enzyme solution [11, 24]. This is based on the fact that the redox potential of heme c1 is independent of pH, whereas the redox potential of the [2Fe-2S] cluster is pH-dependent (negative correlation). A similar phenomenon is observed in the S141C(ISP)/G180C(cyt c1) mutant complex, as shown in Fig. 6. Both ISP and cytochrome c1 are 52% reduced at pH 8.0, indicating that they have the same redox potential at the given pH. However, when the pH is increased to 9.0, cytochrome c1 reduction increases to 78% and ISP reduction decreases to 28%. When the pH is increased, the redox potential of ISP decreases while that of cytochrome c1 remains unchanged. As a result, electrons transfer from ISP to cytochrome c1. Similarly, when the pH is decreased from pH 8.0 to 7.0, cytochrome c1 reduction decreases from 52% to 30%, whereas ISP reduction increases from 52% to 82%. A linear relationship between ISP oxidation and cytochrome c1 reduction is observed (Fig. 6). These results suggest that formation of an intersubunit disulfide bond does not alter the pH dependency of midpoint potentials of either ISP or cytochrome c1.
Fig. 6.
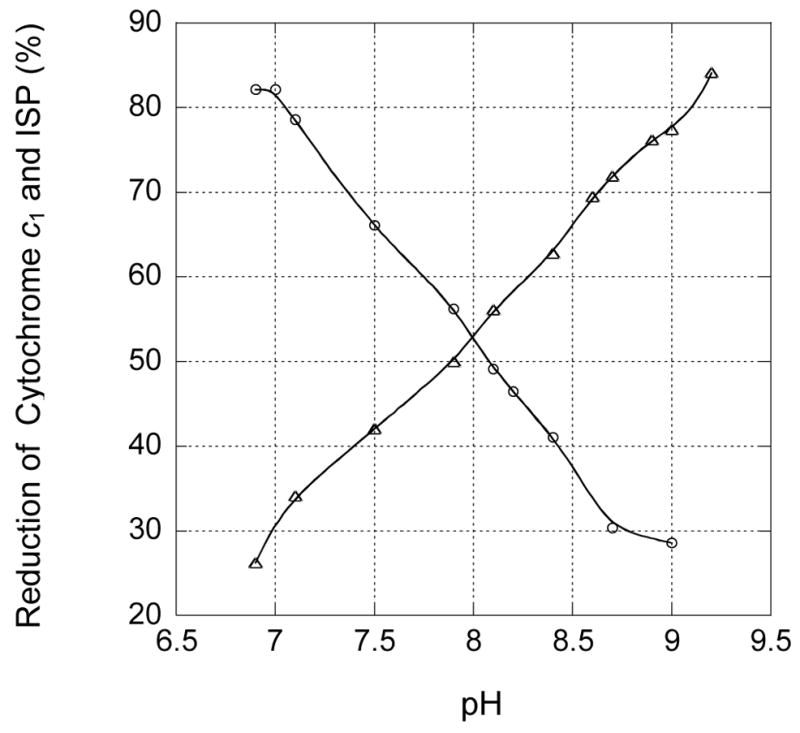
pH induced reduction and oxidation of ISP (circle) and cytochrome c1 (triangle) in the partially reduced cytochrome bc1 complex from S141C(ISP)/G180C(cyt c1) mutant. The experimental conditions and instrument settings are detailed under “Experimental Procedures.”
3.6. Effect of Disulfide Formation, between ISP and Cytochrome c1, on the Rate of Intramolecular Electron Transfer Between the [2Fe-2S] Cluster and Heme c1 Induced by pH Change
The rate of intramolecular electron transfer between the [2Fe-2S] cluster and heme c1 is correlated with the distance between them. Therefore, comparing electron transfer rates in wild type and mutant complexes is a way to prove whether or not intersubunit disulfide bond formation arrests the head domain of ISP at the c1 position. Fig. 7 shows the time trace of alkalization-induced intramolecular electron transfer between [2Fe-2S] cluster and heme c1 in the wild type and the mutant S141C(ISP)/G180C(cyt c1). Reduction of heme c1 in the wild type complex appears to be biphasic, with rate constants of 29 s−1 and 0.6 s−1 (Fig. 7A) respectively. This result is consistent with some (<15%) head domains of ISP molecule being at the b position while others at the c1 position and somewhere in between. Reduction of cytochrome c1 in the mutant S141C(ISP)/G180C(cyt c1) complex is monophasic, with the slower rate constant of 0.7 s−1 (Fig. 7B). We presume that a majority of ISP molecules which are at the c1 position are reduced within the dead-time of the apparatus. In other words, the first phase of cytochrome c1 reduction in the mutant S141C(ISP)/G180C(cyt c1) complex is too fast to be measured by the stopped-flow apparatus. As one can see from Fig. 7B, about 80% of inducible reduction of cytochrome c1 is completed within the dead time of the instrument. These results proved the formation of intersubunit disulfide bond between ISP and cytochrome c1 in the mutant S141C(ISP)/G180C(cyt c1) complex and the head domain of ISP is held at the c1 position by the disulfide bond.
Fig. 7.
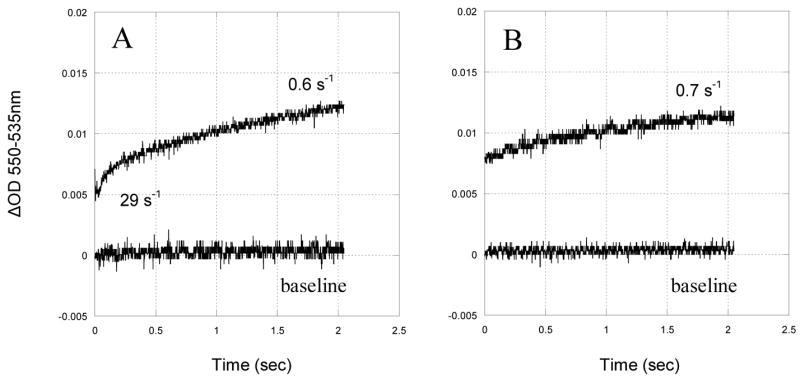
Time trace of the pH induced reduction of cytochrome c1 in the partially reduced cytochrome bc1 complex. 10 μM cytochrome bc1 complex in pH 6.9 phosphate buffer was rapidly mixed with equal volume of the same buffer with enough NaOH to cause the pH change from 6.9 to 8.9 in the stopped flow spectrophotometer at room temperature. A, reduction of cytochrome c1 in the wild type. B, reduction of cytochrome c1 in mutant S141C(ISP)/G180C(cyt c1).
3.7. Stigmatellin Induced Reduction Rate of ISP in the Absence of Exogenous Electron Donor in the Fully Oxidized Complex
Stigmatellin is a tightly bound Qp site inhibitor [35]. The [2Fe-2S] cluster in the mutant S141C(ISP)/G180C(cyt c1) bc1 complex is further from bound stigmatellin than those in the wild type and A185C(cyt b)/K70C(ISP). As a result, its reduction rate induced by stigmatellin is the slowest in these three bc1 complexes (Table 2) with only a small portion of ISP, in the mutant S141C(ISP)/G180C(cyt c1) complex, being reduced. On the contrary, the reduction rate of the [2Fe-2S] cluster in the mutant A185C(cyt b)/K70C(ISP) bc1 complexes, induced by stigmatellin, is the fastest. The slow reduction rate of ISP in mutant S141C(ISP)/G180C(cyt c1) bc1 complex shows that formation of a disulfide bond between S141C(ISP) and G180C(cyt c1) pulls the [2Fe-2S] cluster away from the Qp site, while the fast reduction rate of ISP in A185C(cyt b)/K70C(ISP) complex shows the [2Fe-2S] cluster is pulled close to the Qp site. Since there is no reducing compound present in the ferricyanide oxidized complex, the source of the electron donor is unknown; it might come from amino acid residues located near Qp site. The possibility that the reducing compound is ferricyanide has been ruled out because similar reduction is observed in a complex oxidized by cytochrome c oxidase and cytochrome c.
TABLE 2.
Reduction of ISP induced by stigmatellin in the absence of exogenous electron donor. The bc1 complexes from the wild type, S141C(ISP)/G180C(cyt c1), and A185C(cyt b)/K70C(ISP) were diluted to 10 μM in 3 ml of 20 mM Tris-Cl buffer, pH 8.0, containing 200 mM NaCl and 0.01% LM. After incubated with 50 μM stigmatellin for 5 min, 1 hr, and 2 hr, the bc1 complexes was subjected to the CD to measure the reduction of ISP.
| Reduction percentage of ISP
|
|||
|---|---|---|---|
| 5 min | 1 hr | 2 hr | |
| Wild type | 54% | 81% | 100% |
| S141C(ISP)/G180C(cyt c1) | 43% | 59% | 76% |
| K70C(ISP)/A185C(cyt b) | 91% | 98% | 100% |
3.8. Stigmatellin Induced Oxidation of Cytochrome c1 in the Partially Reduced Cytochrome bc1 Complex
In the partially reduced wild type complex, a rapid oxidation of cytochrome c1 is observed upon addition of stigmatellin. This oxidation is the result of an elevation of the mid-point potential of ISP by stigmatellin bound at the Qp site and the fixation of the head domain of ISP at the b position. In the mutant S141C(ISP)/G180C(cyt c1) complex, the head domain of ISP is linked to the c1 position through formation of the disulfide bond, the binding of stigmatellin is therefore no longer able to fix the head domain of ISP at the b position and no elevation of the mid-point potential of ISP will occur. Thus cytochrome c1 in the mutant S141C(ISP)/G180C(cyt c1) complex will not be oxidized by ISP in spite of the short distance between heme c1 and the [2Fe-2S] cluster. This result confirms the observation that the elevation of ISP mid-point potential caused by stigmatellin binding is due to the fixation of ISP head domain at b position and not due to the binding of the inhibitor to ISP [26]. Fig. 8 compares the stigmatellin induced oxidation of cytochrome c1 in the partially reduced bc1 complexes of wild type and mutant S141C(ISP)/G180C(cyt c1). Only a small portion of cytochrome c1 in the mutant complex is oxidized upon the addition of stigmatellin and the rate of oxidation is much smaller than in the wild type. Although the rate of stigmatellin induced cytochrome c1 oxidation in the wild type complex is much faster than that in the mutant complex, this rate is still much smaller than that induced by pH. The reason is because, in the case of pH induced oxidation, ISP potential is directly affected by pH, regardless of the location of head domain of ISP. In stigmatellin induced reduction, the potential change of iron-sulfur cluster is due to the fixation of the head domain of ISP at the b position. Since in the S141C(ISP)/G180C(cyt c1) mutant complex stigmatellin can no longer fix head domain of ISP at the b position in the mutant complex, little potential elevation is expected. Therefore, little oxidation of cytochrome c1 occurs, in spite of the short distance between the two redox centers.
Fig. 8.
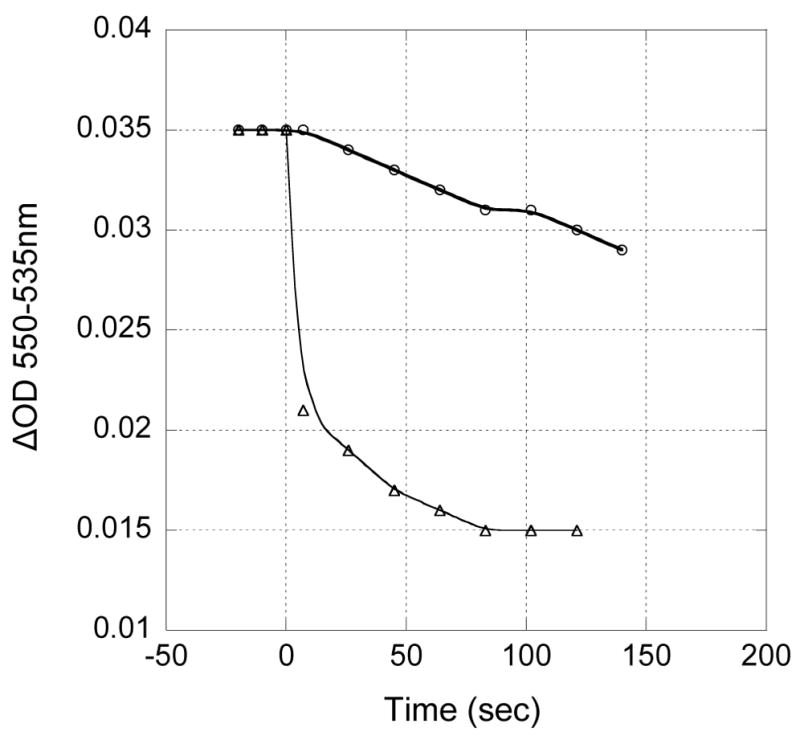
Time trace of stigmatellin induced oxidation of cytochrome c1 in the partially reduced cytochrome bc1 complex. 5 μM of the wild type (triangle) and the mutant S141C(ISP)/G180C(cyt c1) (circle) cytochrome bc1 complexes were treated with 25 μM stigmatellin. The oxidation of cytochrome c1 is followed spectrophotometrically.
3.9. Effect of the Mutations on EPR Characteristics of the [2Fe-2S] Cluster in the bc1 Complex with and without stigmatellin binding
As shown in Fig. 9, the [2Fe-2S] cluster in the S141C(ISP)/G180C(cyt c1) mutant has an EPR spectrum similar to that of the wild type complex, indicating that the microenvironments of the [2Fe-2S] cluster in the mutated bc1 complexes are not affected by disulfide bond formation between the two engineered cysteines. Therefore, the loss of bc1 activity is not due to the change of microenvironments of ISP but to the formation of an intersubunit disulfide. SDS-PAGE results also show that the disulfide bond in the mutant complex remains under the conditions (in the presence of sodium ascorbate) used for the EPR measurement.
Fig. 9.
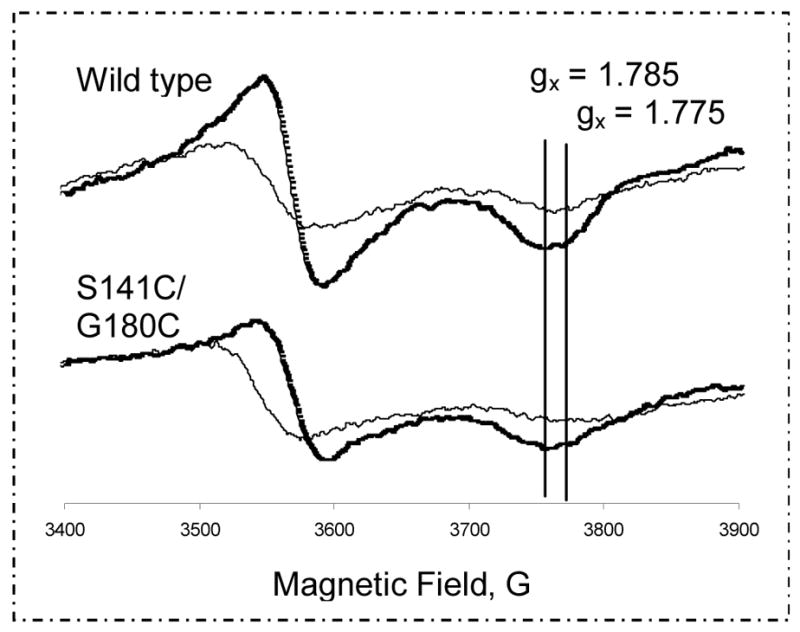
EPR spectra of the [2Fe-2S] cluster in the cytochrome bc1 complexes of the wild type (top) and S141C(ISP)/G180C(cyt c1) (bottom) with (thick) or without (thin) stigmatellin. The experimental conditions and instrument settings are detailed under “Experimental Procedures.”
The EPR signal of ISP increases after the binding Qp site inhibitor, stigmatellin (Fig. 9). However, the amount of increase in the S141C(ISP)/G180C(cyt c1) mutant is much less than that in the wild type complex, indicating the [2Fe-2S] cluster in the mutant is further to the b position in the Qp pocket so the effect of stigmatellin binding is less than that in the wild type complex. It is known that addition of stigmatellin to isolated reduced ISP has little effect on its EPR signals. Stigmatellin also has no effect on EPR signal of ISP if the cytochrome bc1 complex is in the delipidated form. The effect is the result of the fixation of head domain of ISP at b position. In the isolated S141C(ISP)/G180C(cyt c1) mutant complex, the head domain of ISP is fixed at c1 position and it can not be moved to b position upon addition of the inhibitor, therefore no effect on EPR signal is observed. When the mutant complex is pretreated with β-ME to release the disulfide bond, the effect of stigmatellin on EPR signals of ISP is similar to that observed in the wild type complex (data not shown).
Acknowledgments
This work was supported by grants GM30721 (to CAY) from the National Institutes of Health, MCB0077650 (to LY) from National Science Foundation and the Oklahoma Agricultural Experiment Station (Projects # 1819 and #2372), Oklahoma State University. We are grateful to Drs. Jose Soulages and Estela Arrese for their assistance in the CD measurements. We would like to thank Dr. Roger Koeppe for his critical review of this manuscript.
The abbreviations used are
- cyt
cytochrome
- LM
N-Dodecyl-β-D-maltoside
- EPR
electron paramagnetic resonance
- [2Fe-2S]
Rieske iron-sulfur center
- ISP
Rieske iron-sulfur protein
- PAGE
polyacrylamide gel electrophoresis
- Q0C10BrH2
2,3-Dimethoxy-5-methyl-6-(10-bromodecyl)-1,4-benzoquinol
- β-ME
β-mercaptoethanol
- Em
redox midpoint potential
- CD
circular dichroism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartoschek S, Johansson M, Geierstanger BH, Okun JG, Lancaster CRD, Humpfer E, Yu L, Yu CA, Griesinger C, Brandt U. Three molecules of ubiquinone bind specifically to mitochondrial cytochrome bc1 complex. J Biol Chem. 2001;276:35231–35234. doi: 10.1074/jbc.C100365200. [DOI] [PubMed] [Google Scholar]
- 2.Trumpower BL, Gennis RB. Energy transduction by cytochrome complexes in mitochondrial and bacterial respiration: the enzymology of coupling electron transfer reactions to transmembrane proton translocation. Annual Review of Biochemistry. 1994;63:675–716. doi: 10.1146/annurev.bi.63.070194.003331. [DOI] [PubMed] [Google Scholar]
- 3.Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PubMed] [Google Scholar]
- 4.Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, Crofts AR, Berry EA, Kim SH. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 6.Kim H, Xia D, Yu CA, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J. Inhibitor binding changes domain mobility in the iron-sulfur protein of the mitochondrial bc1 complex from bovine heart. Proc Natl Acad Sci U S A. 1998;95:8026–8033. doi: 10.1073/pnas.95.14.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwata S, Saynovits M, Link TA, Michel H. Structure of a water soluble fragment of the ‘Rieske’ iron-sulfur protein of the bovine heart mitochondrial cytochrome bc1 complex determined by MAD phasing at 1.5 Åresolution. Structure. 1996;4:567–579. doi: 10.1016/s0969-2126(96)00062-7. [DOI] [PubMed] [Google Scholar]
- 8.Link TA, Saynovits M, Assmann C, Iwata S, Ohnishi T, von Jagow G. Isolation, characterisation and crystallisation of a water-soluble fragment of the Rieske iron-sulfur protein of bovine heart mitochondrial bc1 complex. Eur J Biochem. 1996;237:71–75. doi: 10.1111/j.1432-1033.1996.0071n.x. [DOI] [PubMed] [Google Scholar]
- 9.Tian H, White S, Yu L, Yu CA. Evidence for the head domain movement of the rieske iron-sulfur protein in electron transfer reaction of the cytochrome bc1 complex. J Biol Chem. 1999;274:7146–7152. doi: 10.1074/jbc.274.11.7146. [DOI] [PubMed] [Google Scholar]
- 10.Esser L, Gong X, Yang S, Yu L, Yu CA, Xia D. Surface-modulated motion switch: capture and release of iron-sulfur protein in the cytochrome bc1 complex. Proc Natl Acad Sci U S A. 2006;103:13045–13050. doi: 10.1073/pnas.0601149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao K, Yu L, Yu CA. Confirmation of the involvement of protein domain movement during the catalytic cycle of the cytochrome bc1 complex by the formation of an intersubunit disulfide bond between cytochrome b and the iron-sulfur protein. J Biol Chem. 2000;275:38597–38604. doi: 10.1074/jbc.M007444200. [DOI] [PubMed] [Google Scholar]
- 12.Tian H, Yu L, Mather MW, Yu CA. Flexibility of the neck region of the rieske iron-sulfur protein is functionally important in the cytochrome bc1 complex. J Biol Chem. 1998;273:27953–27959. doi: 10.1074/jbc.273.43.27953. [DOI] [PubMed] [Google Scholar]
- 13.Darrouzet E, Valkova-Valchanova M, Daldal F. Probing the role of the Fe-S subunit hinge region during Qo site catalysis in Rhodobacter capsulatus bc1 complex. Biochemistry. 2000;39:15475–15483. doi: 10.1021/bi000750l. [DOI] [PubMed] [Google Scholar]
- 14.Yu CA, Yu L. Syntheses of biologically active ubiquinone derivatives. Biochemistry. 1982;21:4096–4101. doi: 10.1021/bi00260a028. [DOI] [PubMed] [Google Scholar]
- 15.Mather MW, Yu L, Yu CA. The Involvement of threonine 160 of cytochrome b of Rhodobacter sphaeroides cytochrome bc1 complex in quinone binding and interaction with subunit IV. J Biol Chem. 1995;270:28668–28675. doi: 10.1074/jbc.270.48.28668. [DOI] [PubMed] [Google Scholar]
- 16.Tian H, Yu L, Mather MW, Yu CA. The involvement of serine 175 and alanine 185 of cytochrome b of Rhodobacter sphaeroides cytochrome bc1 complex in interaction with iron-sulfur protein. J Biol Chem. 1997;272:23722–23728. doi: 10.1074/jbc.272.38.23722. [DOI] [PubMed] [Google Scholar]
- 17.Berden JA, Slater EC. The reaction of antimycin with a cytochrome b preparation active in reconstitution of the respiratory chain. Biochim Biophys Acta. 1970;216:237–249. doi: 10.1016/0005-2728(70)90215-x. [DOI] [PubMed] [Google Scholar]
- 18.Yu CA, Yu L, King TE. Preparation and properties of cardiac cytochrome c1. J Biol Chem. 1972;247:1012–1019. [PubMed] [Google Scholar]
- 19.Yu L, Dong JH, Yu CA. Characterization of purified cytochrome c1 from Rhodobacter sphaeroides R-26. Biochim Biophys Acta. 1986;852:203–211. doi: 10.1016/0005-2728(86)90225-2. [DOI] [PubMed] [Google Scholar]
- 20.Wilson DF, Erecinska M, Dutton PL, Tsudzuki T. The oxidation-reduction potentials of the iron-sulfur proteins in mitochondria. Biochem Biophys Res Commun. 1970;41:1273–1278. doi: 10.1016/0006-291x(70)90225-1. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Yu CA, Yu L. The role of extra fragment at the C-terminal of cytochrome b (Residues 421–445) in the cytochrome bc1 complex from Rhodobacter sphaeroides. J Biol Chem. 2004;279:47363–47371. doi: 10.1074/jbc.M406497200. [DOI] [PubMed] [Google Scholar]
- 22.Dutton PL. Redox potentiometry: determination of midpoint potentials of oxidation-reduction components of biological electro-transfer systems. Methods in enzymology. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- 23.Ugulava NB, Crofts AR. CD-monitored redox titration of the Rieske Fe-S protein of Rhodobacter sphaeroides: pH dependence of the midpoint potential in isolated bc1 complex and in membranes. FEBS Letters. 1998;440:409–413. doi: 10.1016/s0014-5793(98)01493-8. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Tai CH, Yu L, Yu CA. pH-induced intramolecular electron transfer between the iron-sulfur protein and cytochrome c1 in bovine cytochrome bc1 complex. J Biol Chem. 2000;275:7656–7661. doi: 10.1074/jbc.275.11.7656. [DOI] [PubMed] [Google Scholar]
- 25.von Jagow G, Ohnishi T. The chromone inhibitor stigmatellin - binding to the ubiquinol oxidation center at the C-side of the mitochondrial membrane. FEBS Letters. 1985;185:311–315. doi: 10.1016/0014-5793(85)80929-7. [DOI] [PubMed] [Google Scholar]
- 26.Darrouzet E, Valkova-Valchanova M, Daldal F. The [2Fe-2S] cluster Em as an indicator of the iron-sulfur subunit position in the ubihydroquinone oxidation site of the cytochrome bc1 complex. J Biol Chem. 2002;277:3464–3470. doi: 10.1074/jbc.M107973200. [DOI] [PubMed] [Google Scholar]
- 27.McCurley JP, Miki T, Yu L, Yu CA. EPR characterization of the cytochrome bc1 complex from Rhodobacter sphaeroides. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1990;1020:176–186. doi: 10.1016/0005-2728(90)90049-a. [DOI] [PubMed] [Google Scholar]
- 28.Elberry M, Yu L, Yu CA. The disulfide bridge in the head domain of Rhodobacter sphaeroides cytochrome c1 is needed to maintain its structural integrity. Biochemistry. 2006;45:4991–4997. doi: 10.1021/bi0522246. [DOI] [PubMed] [Google Scholar]
- 29.Davidson E, Ohnishi Tomoko, Atta-Asafo-Adjei E, Daldal F. Potential ligands to the [2Fe-2S] Rieske cluster of the cytochrome bc1 complex of Rhodobacter capsulatus probed by site-directed mutagenesis. Biochemistry. 1992;31:3342–3351. doi: 10.1021/bi00128a006. [DOI] [PubMed] [Google Scholar]
- 30.Gray KA, Davidson E, Daldal F. Mutagenesis of methionine-183 drastically affects the physicochemical properties of cytochrome c1 of the bc1 complex of Rhodobacter capsulatus. Biochemistry. 1992;31:11864–11873. doi: 10.1021/bi00162a027. [DOI] [PubMed] [Google Scholar]
- 31.Crofts AR. The Q-cycle — A Personal perspective. Photosynthesis Research. 2004;80:223–243. doi: 10.1023/B:PRES.0000030444.52579.10. [DOI] [PubMed] [Google Scholar]
- 32.Gabellini N, Bowyer JR, Hurt E, Melandri BA, Hauska G. A cytochrome b/c1 complex with ubiquinol--cytochrome c2 oxidoreductase activity from Rhodopseudomonas sphaeroides GA. Eur J Biochem. 1982;126:105–111. doi: 10.1111/j.1432-1033.1982.tb06753.x. [DOI] [PubMed] [Google Scholar]
- 33.Guergova-Kuras M, Salcedo-Hernandez R, Bechmann G, Kuras R, Gennis RB, Crofts AR. Expression and one-step purification of a fully active polyhistidine-tagged cytochrome bc1 complex from Rhodobacter sphaeroides. Protein Expression and Purification. 1999;15:370–380. doi: 10.1006/prep.1998.1018. [DOI] [PubMed] [Google Scholar]
- 34.Meinhardt SW, Crofts AR. Kinetic and thermodynamic resolution of cytochrome c1 and cytochrome c2 from Rhodopseudomonas sphaeroides. FEBS Letters. 1982;149:223–227. [Google Scholar]
- 35.Covian R, Gutierrez-Cirlos EB, Trumpower BL. Anti-cooperative oxidation of ubiquinol by the yeast cytochrome bc1 complex. J Biol Chem. 2004;279:15040–15049. doi: 10.1074/jbc.M400193200. [DOI] [PubMed] [Google Scholar]