Abstract
A highly specific and sensitive mass assay for inositol hexakisphosphate (InsP6) was characterized. This centres around phosphorylating InsP6 with [32P]ATP using a recombinant InsP6 kinase from Giardia lambia, followed by HPLC of the 32P-labelled products with an internal [3H]InsP7 standard. This assay was used to quantify InsP6 levels in a variety of biological samples. Concentrations of InsP6 in rat tissues varied from 10–20 μM (assuming 64% of wet weight of tissue is cytosol water), whereas using the same assumption axenic Dictyostelium discoideum cells contained 352±11 μM InsP6. HeLa cells were seeded at low density and grown to confluence, at which point they contained InsP6 levels per mg of protein similar to rat tissues. This amounted to 1.952±0.117 nmol InsP6 per culture dish, despite the cells being grown in serum shown to contain no detectable (less than 20 pmol per dish) InsP6. These results demonstrate that mammalian cells synthesize all their own InsP6. Human blood was analysed, and although the white cell fraction contained InsP6 at a concentration comparable with other tissues, in serum and platelet-free plasma no InsP6 was detected (<1 nM InsP6). Human urine was also examined, and also contained no detectable (<5 nM) InsP6. These results suggest that dietary studies purporting to measure InsP6 at micromolar concentrations in human plasma or urine may not have been quantifying this inositol phosphate. Therefore claims that administrating InsP6 in the diet or applying it topically can produce health benefits by increasing extracellular InsP6 levels may require reassessment.
Keywords: dietary phytate, inositol hexakisphosphate (InsP6), mass assay, phytate, phytic acid
Abbreviations: d.p.m., disintegrations per min; Ipk2, inositol polyphosphate multikinase; TCA, trichloroacetic acid
INTRODUCTION
InsP6 was originally discovered as the major plant phosphate storage compound [1,2], hence its alternative name, phytic acid. It was generally assumed to be plant specific until the rekindled interest in inositol phosphates that followed the discovery of Ins(1,4,5)P3 as a second messenger [3] led to the realization that InsP6 is also found in animal cells (e.g. [4,5] and see [6,7] for reviews).
In the intervening years, the route by which animals synthesize InsP6 was controversial [6], but further studies have led to a likely consensus. Unlike slime moulds [8] and plants [9], which can synthesize InsP6 by stepwise phosphorylation directly from inositol, animals apparently incorporate the first three phosphates while the inositol is in its lipid form. That is, Ins(1,4,5)P3 is formed by phospholipase C action on PtdIns(4,5)P2, and Ins(1,4,5)P3 is then phosphorylated up to InsP6. The synthesis of Ins(1,3,4,5,6)P5 from Ins(1,4,5)P3 is most likely to be mediated by a two-step conversion catalysed by a single enzyme, Ipk2 (inositol polyphosphate multikinase) [10–12]; this enzyme is essential for mammalian embryonic survival [13], though it is not certain yet whether that is because of its activity in converting Ins(1,4,5)P3 into Ins(1,3,4,5,6)P5. The alternative route from Ins(1,4,5)P3 to Ins(1,3,4,5,6)P5 via Ins(1,3,4,5)P4, Ins(1,3,4)P3 and Ins-(1,3,4,6)P4 is probably quantitatively minor, at least in fibroblasts, as suggested by the results of Leyman et al. [14], who showed that mouse embryonic fibroblasts devoid of any Ins(1,4,5)P3 3-kinase isoforms synthesized InsP6 at a rate indistinguishable from wild-type fibroblasts; in contrast, fibroblasts derived from Ipk2 knockout mice show a greatly compromised InsP6 synthesis [13]. The final step of InsP6 synthesis (as in plants and slime moulds [8,9]) is the 2-phosphorylation of Ins(1,3,4,5,6)P5 by InsP5 2-kinase [11,15], an enzyme which is also essential to viable mammalian development [16].
However, although we now know that mammalian cells can synthesize InsP6, how they probably do so, and that it is essential that they do synthesize InsP6, the contribution of exogenous InsP6 to mammalian InsP6 homoeostasis is not so clear. A number of studies have reported increases in InsP6 levels in tissue and body fluids resulting from the supply of exogenous InsP6 [17–20], and other work has suggested that InsP6 added to cell cultures or tissues can impinge on their physiology or pathology [21–23]. As a result of these and other studies there are many claims of therapeutic or other beneficial effects of InsP6 (e.g. [23,24] and many of these are predicated on the idea that exogenous InsP6 can directly and significantly influence both extra- and intracellular InsP6 levels in the body.
In the present study, we have designed and characterized an unambiguous mass assay for InsP6. This assay is more time-consuming and complex than others (e.g. [18,25]), but its particular strength is a very high sensitivity and applicability to most sample types. Our results provide some new insights into InsP6 levels in a number of tissues and address whether mammalian cells can make all their own InsP6. Our results also cast doubts on the idea that InsP6 is directly taken up into human tissues and cells.
EXPERIMENTAL
Reagents
All chemicals and biochemicals used were of AnalaR grade unless specified otherwise. [3H]InsP6 was from PerkinElmer. A [3H]InsP7 standard (probably 5-[PP]InsP5, see the Results section) was prepared by incubating [3H]InsP6 with Giardia lambia InsP6 kinase under the normal assay conditions (see below for details) such that approx. 50% of the InsP6 was phosphorylated. The correct nomenclature for the product of InsP6 phosphorylation by InsP6 kinase is [PP]InsP5, and we have used this nomenclature when necessary for unambiguity (e.g. when specifying isomers). However, for most of the present paper we have used InsP7 as a simpler alternative. Sources of InsP5 isomers and scyllo-inositol hexakisphosphate were as described previously [26].
InsP6 kinase
This is a recombinant enzyme from G. lambia. The sequence of Giardia InsP6 kinase (Genbank® Nucleotide Sequence Database accession number AY227443) was amplified from Giardia genomic DNA (the gene contains no introns), and then cloned into the hexahistidine bacterial vector pET43A, which fuses the solubility-enhancing bacterial protein NusA to the N-terminus of the fusion protein. The enzyme was purifed by talon-bead absorption and imidazole elution as described previously [27]. As discussed previously [27], there is no obvious reason why a recombinant mammalian InsP6 kinase should not be substituted for the Giardia enzyme in this assay, although the high affinity and specificity of the Giardia InsP6 kinase (see the Results section) does make it particularly suitable in this context.
Extraction protocol
For solid tissue samples, 20–50 mg of tissue was weighed before homogenizing with a ground glass pestle in 180 μl of water. This was rinsed with 70 μl of water and then transferred to a 1.5 ml Eppendorf tube, and 50% (w/v) TCA (trichloroacetic acid) added to give a final concentration of 8.3% (v/v). After vortexing and standing on ice for 30 min, samples were centrifuged at 13000 g for 15 min in a Microfuge, the supernatant was removed and the pellet was kept for protein determination (see below). The supernatant containing inositol phosphates was washed ten times with water-saturated diethyl ether to remove TCA [28] and finally neutralized with 15 μl of 1:10 (v/v) saturated ammonia. Tubes were spun in a centrifugal vacuum dryer for 5 min at 35 °C and then for 30 min at 45 °C to remove the ether, before being adjusted to 300 μl with water and the pH checked to ensure it was approx. pH 6.0. For most assays, we took as an aliquot the equivalent of 1 mg of wet weight of starting tissue for the InsP6 assay.
InsP6 mass assay
Samples of tissue extract were incubated in a final volume of 300 μl containing the following: 30 μl of 10× assay buffer [500 mM Hepes/NaOH (pH 7.5), 1 M KCl, 50 mM MgCl2, 10 mM EGTA and 5 mg/ml BSA], 6 μl of 600 μM 2-mercaptoethanol and 10 μl of [γ-32P]ATP (10 μCi; PerkinElmer). After 60 min incubation at 37 °C, 45 μl of 0.1 M glucose and 6 μl of hexokinase (20 units, Sigma) were added, and the incubation was continued at 37 °C for another 90 min to convert all the remaining radioactivity into [32P]glucose 6-phosphate.
The reaction was stopped by the addition of 0.5 ml of 0.4 M ammonium formate/0.1 M formic acid, and the samples were loaded on to 0.5 ml of Dowex Formate in a BioRad Econo Column (with a disc of Whatman Filter paper number 1 on top of the Dowex resin) [29]. Samples were rinsed on to the column with 4 ml of 0.4 M ammonium formate/0.1 M formic acid, and the column was washed with 10 ml of 0.8 M ammonium formate/0.1 M formic acid to remove any remaining [32P]glucose 6-phosphate. The higher inositol phosphates were eluted with 2×1 ml of 2.5 M ammonium formate/0.1 M formic acid. They were spiked with 600–2000 d.p.m. (disintegrations per min) of [3H]InsP7 and loaded on to a Whatman Patisil 10 SAX WCS analytical HPLC column (4.6×250 mm, catalogue number 4226-001) or a similar-sized column packed with the same material by Phenomenex.
HPLC
The column was eluted at 1 ml/min flow rate with the following programme: solution A, water and solution B, 1.2 M (NH4)2HPO4 (pH 3.8) adjusted with phosphoric acid. The profile gives the percentage of solution B (v/v) added: 0 min, 0%; 5 min, 0%; 30 min, 78%; 75 min, 78%; 76 min, 100%; 81 min, 100%; 82 min, 0%; 102 min, 0%.
Fractions (1 ml) were usually collected between 40 and 75 min (InsP7 elutes at approx. 60 min) and were counted in a scintillation counter after adding 10 ml of Ultima-Flo AP ammonium phosphate-tolerant scintillation fluid (PerkinElmer). The [3H] and [32P] windows were calibrated using standards to ensure each isotope was counted independently (spill over between channels was <1%).
Protein assay
Pellets from the TCA tissue extracts were dissolved in 4 ml of 8 M urea by sonication (in a Decon ultrasonic bath) and vortexing. The majority of material dissolved, but in samples with a high fat content some fat could remain in suspension and may interfere with the assay (this did not happen in any sample analysed in the present study). Samples were adjusted to a final concentration of 6 M urea, and assayed for protein using the Bio-Rad protein assay, based on a method published previously by Bradford [30], using BSA in 6 M urea as a protein standard.
Blood fractionation
Blood was taken from unfasted human volunteers who denied having taken aspirin in the preceding 48 h, mixed with acid citrate dextrose (97 mM sodium citrate, 78 mM citric acid and 110 mM D-glucose) and centrifuged at 200 g for 20 min (all centrifugations were performed at 4 °C). The supernatant from this centrifugation (platelet-rich plasma) was further centrifuged at 2500 g for 20 min, and the resulting supernatant (platelet-free plasma) was removed and frozen. The pellet from the first centrifugation was resuspended in 25 ml of resuspension buffer [10 mM Hepes/NaOH (pH 7.4), 10 mM glucose, 1 mM MgSO4, 145 mM NaCl and 5 mM KCl], and then centrifuged (2500 g for 20 min). The ‘buffy coat’ (mostly white cells) on top of the pellet was removed, along with the supernatant. The pellet (mostly erythrocytes) was resuspended in 25 ml of resuspension buffer and re-centrifuged (2500 g for 20 min). The small amount of buffy coat on this pellet was removed, along with the supernatant, and the remaining pellet (erythrocytes) was then resuspended in 10 ml of resuspension buffer and frozen. The combined supernatants from the two preceding centrifugations were re-centrifuged and resuspended several times as above (2500 g for 20 min) until no erythrocytes were evident at the bottom of the pellet, at which point the final pellet of mostly white cells was resuspended and frozen.
For preparation of serum, fresh blood was left to stand for 4 h, then centrifuged at 3000 g for 20 min, and the supernatant (serum) was decanted and frozen.
Human experimentation
All human fluid samples were taken with informed consent of the donors in accordance with the Declaration of Helsinki (2000) of the World Medical Association and with the relevant ethical guidelines of the University of Cambridge. The urine sample was taken from an adult male at 10:30 h, 3 h after a meal consisting of a bowl of breakfast cereal and a cup of coffee, with the subject subsequently having free access to water before the sample was taken.
RESULTS
Characterization of the enzyme and the assay
Using [3H]InsP6, we established that the Giardia kinase has a Km of approx. 60 nM (results not shown). We used this enzyme to make our own [3H]InsP7 standard for HPLC. Because the Giardia gene is closely related to InsP6 kinases from other eukaryotes, we assumed that its product is 5-[PP]InsP5 (see [31] for discussion), though we have not established its structure. Draskovic et al. [32] have shown that the mammalian InsP6 kinases are remarkably specific for the 5-position such that even on prolonged incubation they prefer to add a third phosphate there rather than use another position on the inositol ring. We exposed bona fide 5-[PP]InsP5 synthesized from InsP6 by a mammalian InsP6 kinase [a gift from Dr S.B. Shears, Inositol Signaling Group, NIEHS (National Institute of Environmental Health Sciences), Research Triangle Park, NC, U.S.A.] to the Giardia enzyme for several hours, but detected no formation of any InsP8, which suggests that Giardia InsP6 kinase is a 5-kinase too, and that extra phosphorylation of 5-[PP]InsP5 is not a complicating factor in the InsP6 assay.
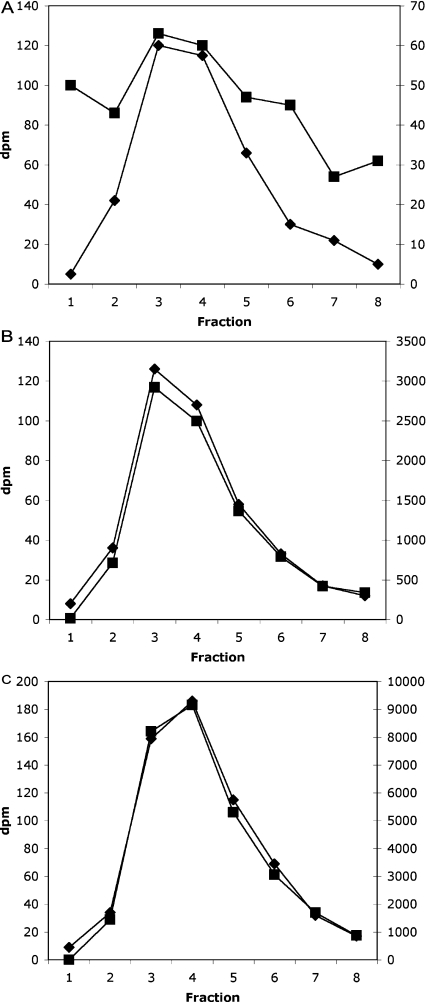
In the context of this assay, the crucial point is that we always included a [3H]InsP7 standard made by the Giardia enzyme in every HPLC run, and collected and counted 1 min samples individually for both isotopes. This leads to a precise co-chromatography of the 3H-labelled standard and the 32P-labelled product formed from InsP6 in the cell extract (e.g. Figure 1); any sample in which this co-chromatography was not absolutely precise was discarded (except for some blanks, where low levels of radioactivity meant that the correlation was less evident: see below). Note that in the Figures, the left-hand axis (♦) is always the 3H-labelled InsP7 standard, and the right-hand axis (■), whose scale varies greatly between Figures (which serves to illustrate the precise co-chromatography of the two isotopes), is the 32P counts for the same fractions.
Figure 1. HPLC profile of a liver InsP6 assay.
Three typical profiles from three assay tubes are illustrated, with the radioactivity in the InsP7 region (indicated by the peak in [3H]InsP7 elution) shown. Left-hand axis (◆), [3H]; right-hand axis (■), [32P]. Note the differing scales on the [32P] axes [dpm (d.p.m.)]. (A) Blank incubation (control), (B) 5 μl of liver extract, and (C) 5 μl of liver extract with 10 pmol InsP6 added just before the assay was performed.
We have discussed elsewhere previously [27], and discounted, the possibility of InsP5 in extracts interfering significantly with the assay. Briefly, InsP5s are poor substrates for the Giardia enzyme (see below), and any PPInsP4 products formed elute well before InsP7 under our HPLC protocol. Any tri-phosphorylated InsP5 [32] would be formed in minute amounts during our 60 min incubations and also would not precisely co-elute with 5-[PP]InsP5. High background levels in the region of the HPLC profile where PPInsP4 elutes prevented us from extensive quantitative analysis of InsP5s as substrates for the Giardia InsP6 kinase. However, in one set of experiments with a fresh batch of [γ-32P]ATP (and con-sequently lower background) we were able to gain some results on the approximate efficacy of the six InsP5 isomers (four of which are presented as racemic pairs) as substrates. Using 300 nM substrate, the rate of phosphorylation of InsP5 isomers relative to InsP6 (InsP6=100%) was 2-OH-InsP5, 0.5%; 1/3-OH-InsP5, 3.2%; 4/6-OH-InsP5, 3.0%, and 5-OH-InsP5 showed no detectable phosphorylation over background levels. In parallel experiments, scyllo-inositol hexakisphosphate was phosphorylated at 4.7% of the rate of InsP6.
We explored whether other compounds present in rat tissue extracts might interfere with the assay by ‘spiking’ samples with known quantities of InsP6. We found significant levels of interference caused by factors present in the extracts, which varied considerably between tissues (from a 2–10-fold underestimate of the InsP6 added), and we attribute this to endogenous ATP diluting our [γ-32P]ATP, plus other unknown compounds inhibiting the enzyme. We tried a number of ‘purification’ protocols to separate InsP6 from these interfering factors, but this only led to losses of InsP6, so, in the end, we decided to adopt a protocol where we performed six assays for any single tissue sample. Three are aliquots of the tissue extract, and three are identical aliquots spiked just before the assay with a known amount (usually 10 pmol) of InsP6. From the difference between these latter spiked samples and the unspiked samples, we can calculate the d.p.m. of InsP7 formed from the known InsP6, and comparing this with a 10 pmol standard assayed at the same time quantifies the ‘quenching’ present in each individual tissue sample. This can then be used to correct the InsP7 formed in the unquenched samples. This is a rather laborious process (as we also included blank incubated controls in all experiments), but it does allow us to be confident that each sample is quantitatively assayed.
This approach assumes that the quenching is linear with InsP6 concentration (doing a complete standard curve within each sample would be the only rigorous way of ensuring that). Although we believe such linearity is indeed likely, some non-linearity would not seriously compromise the conclusions drawn below other than to lead to an underestimate of InsP6 in some tissues listed in Table 1. Whenever possible, we used quantities of cell extract that contained approximately the same amount of InsP6 as the added spike, which self-evidently helps to ensure linearity and also simplifies the quantification of the degree of quenching. We tested our extraction protocol by spiking several samples of rat tissues with [3H]InsP6 before extraction, and found that we recovered 80–85% up to the point of adding the enzyme to the assay. We have corrected all our results for tissue extracts by assuming 80% recovery.
Table 1. InsP6 content of tissues and fluids.
Tissues were extracted and assayed as described in the Experimental section. Each assay was performed in triplicate, and results are means±S.E.M. For all tissues, at least one independent assay was performed on another sample, with similar results (results not shown). Note that the InsP6 concentration for all samples except serum, plasma and urine, is an estimated intracellular concentration.
| Tissue | InsP6 (pmol/mg of protein) | InsP6 concentration |
|---|---|---|
| Rat liver | 39.8±0.43 | 11±0.12 μM |
| Rat brain | 194.7±25.5 | 14.5±1.9 μM |
| Rat kidney | 55.5±7.1 | 10.9±1.4 μM |
| Rat lung | 69.8±12.4 | 15.8±2.8 μM |
| HeLa cells | 175±16.6 | 37.6±3.5 μM |
| Slime mould | 3645±114 | 352±11 μM |
| Calf serum | − | <8 nM |
| Human erythrocytes | 0.037±0.0057 | 26±4.0 nM |
| Human white cells | 43.5±6.1 | − |
| Human serum | − | <0.5 nM |
| Human plasma | − | <0.5 nM |
| Human urine | − | <5 nM |
InsP6 content of rat tissues
Figure 1 shows a typical set of single HPLC profiles from an assay performed on an extract from rat liver. The combination of the triplicate samples leads to a level of 4.75 pmol in the sample, which represents 0.84 mg of wet weight of tissue (0.149 mg of protein). If we assume an 80% recovery and that there is 0.64 g water/g of tissue (see [33,34] for derivation of this latter assumption), then the complete assay leads to an estimated intracellular level of 11±0.12 μM InsP6 in the liver.
In a further set of experiments, we then assayed InsP6 levels in a selection of rat tissues, and the results are shown in Table 1. We observe no obvious pointers to tissue-specific functions in these results, as the concentrations of InsP6 were broadly similar. Perhaps the most relevant observation in that regard is that comparing the two internal tissues in the body exposed to the highest and lowest O2 levels (lung and kidney respectively), there is no large difference. If a principal function of InsP6 in vivo was to act as an antioxidant [26,35], then one might expect there to be much more InsP6 in the lung. In fact, the likelihood of InsP6 interacting with iron ions (the reason for its antioxidant action) inside cells has been questioned in the light of the discovery that InsP6 probably exists mostly as a penta-Mg2+ salt in vivo [36].
Slime moulds
Our calculated concentrations of InsP6 in mammalian tissues are overall consistent with the indirect calculations derived from equilibrium labelling of cell cultures with [3H]inositol (e.g. [6,37]), but nevertheless we sought an independent validation of the assay. One organism for which there is already an accurate mass estimation of InsP6 is D. discoideum, which has an estimated intracellular concentration of InsP6 assayed by inorganic phosphorous determination as 594 μM [8]. This value is significantly higher than the estimated maximum solubility of the penta-Mg2+ salt of InsP6 proposed to exist in vivo (approx. 50 μM [38]), and may be explained by some form of compartmentalization in acidic vesicles (see [38] for discussion).
We therefore assayed pellets of axenal cultures of slime moulds for InsP6. The pellets were weighed before extraction, and the mass levels of InsP6 measured gave an estimated concentration in the cells of 352±11 μM (Table 1). The figure of 594 μM [8] used an assumption of 0.4 g of water/g of pelleted cells, which corrects to 371 μM if we use the assumption of 0.64 g of water as determined above. The close agreement between the two sets of data provides an independent validation of the present assay, and confirms the really remarkably high InsP6 levels in these organisms.
HeLa cells
We next assayed InsP6 in confluent cultures of HeLa cells (Table 1). These have apparently higher levels of InsP6 than rat tissues; however, the difficulty of accurately measuring cellular wet weight, plus the absence of connective tissue in a cell culture, limit the interpretability of such comparisons. Nevertheless, these experiments provided us with an opportunity to investigate the extent to which mammalian cell lines make all their own InsP6. InsP6 is reported to be present in human and rat serum or plasma [17–19], so it is possible that cultured cells derive some or even all of their InsP6 from the calf serum in the medium in which they are grown. In this respect, cell cultures could be argued to be a paradigm for whole animal requirements, as they are with inositol (see [39] and the Discussion section). So we tried to measure InsP6 in the calf serum in which the cells were cultured, but could detect none within the limits of the assay. The arguments for these limits are detailed below when we consider human serum, and, using this logic, we estimate that there could be no more than 16 pmol of InsP6 in the medium of each 10-cm-diameter dish of cells. Yet a dish of HeLa cells grown to confluence in that medium contained 1.952±0.117 nmol of InsP6. The same medium has been used in our laboratory to grow numerous other cell lines [e.g. HEK-293 cells (human embryonic kidney cells), COS cells and CHO cells (Chinese-hamster ovary cells)] for many passages, and so from these results we conclude that cultured mammalian cells make all their own InsP6.
Human blood
The absence of detectable InsP6 in calf serum raised the question of whether this is unique to cows, given that human and rat serum are reported to have levels of InsP6 from 30–460 nM (e.g. [19,25] and see also the Discussion section). So we tested human blood. We fractionated the blood in two ways. As serum would be a direct comparison with the calf serum above, for some samples we let the blood clot and took the supernatant (serum) remaining. For comparison with measurements of plasma InsP6, and also to gain some information about InsP6 in some of the cells in blood, we also collected blood into citrate, and fractionated it into erythrocytes, white (buffy coat) cells and platelet-free plasma (we discarded the platelet fraction).
The results in Table 1 show that we could detect virtually no InsP6 in erythrocytes (the small amount present is probably a result of white-cell contamination), which is in itself an interesting comparison with nucleated erythrocytes of amphibians and reptiles, which make high levels of Ins(1,3,4,5,6)P5 [6,40]. For the white cell (buffy coat) fraction, we only used cellular protein as a standard, and the results suggest that, as one might expect from nucleated mammalian cells, human white cells contain InsP6 at a concentration similar to the rat tissues assayed above (Table 1).
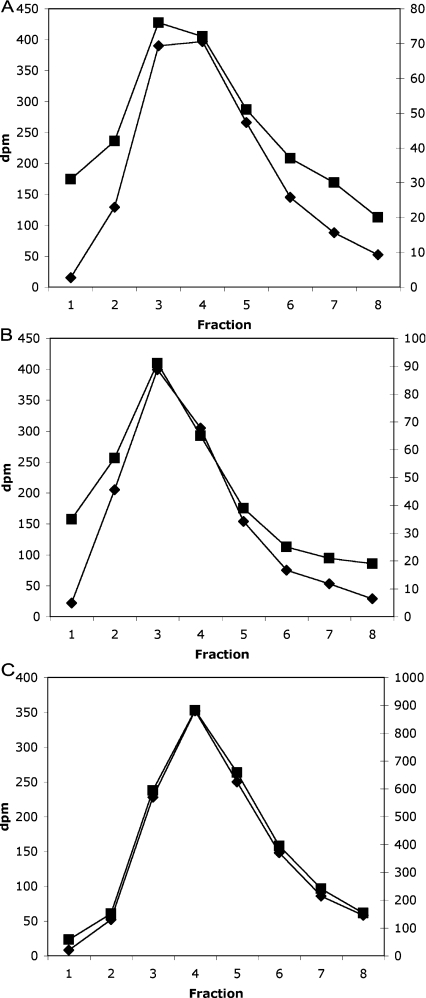
However, in both serum and plasma, we again found no significant quantities of InsP6. To illustrate this point, Figure 2 shows typical HPLC profiles from assays on human serum samples which were (Figure 2C) or were not (Figure 2B) ‘spiked’ with 2.5 pmol of InsP6 before extraction; this extra part of the protocol was introduced to ensure that there was no extraction problem with this particular type of preparation (which has no cells, but a high protein content). Similar experiments were performed on blood taken from three individuals and all gave similar results. From these profiles (Figure 2), it is difficult to be accurate as to how much InsP6 we would have been able to detect. The background in the 32P channel where [3H]InsP7 elutes varied between experiments, in general this was lower if the [γ-32P]ATP was fresh [27], as this 32P background in the HPLC eluate is the ultimate limitation to the sensitivity of the assay. A further complication is that, in that background, there is frequently an apparent minor peak of 32P that approximately co-chromatographs with [3H]InsP7, which is present in both blank and human serum samples (Figures 2A and 2B). Nevertheless, if we suggest that 100 d.p.m. above backgound in triplicate samples would have shown up as a distinct and detectable 32P peak co-incident with [3H]InsP7 in Figure 2(B), we can calculate that this would represent no more than 64 fmol of InsP6, which leads us to suggest that the plasma sample typified by Figure 2 probably contains less than 0.5 nM InsP6. Moreover, during plasma preparation, some damage may have occurred to white cells (or platelets) to release traces of InsP6, and, with this in mind, we suggest that in vivo blood probably contains no extracellular InsP6 at all.
Figure 2. HPLC profile of a human plasma InsP6 assay.
Three typical HPLC profiles from three assay tubes are illustrated, with the radioactivity in the InsP7 region (indicated by the peak in [3H]InsP7 elution) shown. Left-hand axis (♦), [3H]; right-hand axis (■), [32P]. Note the differing scales on the [32P] axes [dpm (d.p.m.)]. (A) Blank incubation (control), (B) extract of 100 μl of human plasma, and (C) extract of 100 μl of human plasma with 2.5 pmol (25 nM final concentration) InsP6 added before extraction.
Human urine
The other human fluid that has been analysed for InsP6 is urine, which is reported to contain 1–3 μM InsP6 (e.g. [20] and see the Discussion section). We therefore measured InsP6 in a urine sample from one of our blood donors, again spiking some samples before extraction as well as afterwards to confirm the recovery of InsP6. The results revealed that there is no detectable InsP6 present. There is more ‘interference’ with the assay from unknown factors present in urine than in plasma or serum, which made the assay less sensitive. But the assays included 50 μl of samples of urine spiked with 2.5 pmol InsP6 (final concentration of 50 nM) before extraction, and these showed a clear peak of InsP7 product; from a similar semi-quantitative logic to that stated above, we can set a lower limit of InsP6 detection in the samples analysed of approx. 250 fmol InsP6, the equivalent of 5 nM InsP6 in human urine.
DISCUSSION
The levels of InsP6 that we have measured in mammalian tissues are of a similar order to those deduced from equilibrium labelling of cultured cells with [3H]inositol (e.g. [37], and see [6] for discussion) and we can conclude that mammalian cells contain tens of μM InsP6. If anything, these may be underestimates compromised by a potential non-linearity of the assay when performed on tissue extracts. Moreover, these calculations assume a uniform cytosolic localization, but obviously local concentrations may be higher. This may be most relevant in the context of well defined nuclear functions for this inositol phosphate and its more highly phosphorylated derivatives [11,41–43]. Slime moulds, which synthesize very high levels of InsP6 very quickly [8], remain an enigma only deepened by our results.
Crucially, we have shown, as far as we know, for the first time, that HeLa cells in culture (and by implication, all mammalian cell lines in perpetuity) synthesize all their own InsP6. This depends, of course, on the cells having sufficient inositol to make PtdIns(4,5)P2 ([39] and also see the Introduction and Discussion sections), and we suggest that they probably serve as a paradigm for humans. The key to this observation is our discovery that cultured cells have no significant external source of InsP6 because there is none detectable in the calf serum in which we grow them. This was the most unexpected finding of our study, and because of its implications for human diet and health, we went on to demonstrate, as thoroughly as the assay allows, that there is also no detectable InsP6 in human serum, plasma (Figure 2) or urine.
These results have important implications for the apparent uptake of exogenous InsP6 into cells or tissues, and for the proposed extracellular actions of InsP6 in health and disease. To consider cells first, when radiolabelled InsP6 is added at micromolar concentrations to mammalian cells in culture it has been reported to be taken up, but the radiolabel is then found as lower inositol phosphates or inositol [23,44]. InsP6 binds strongly to Ca2+, and its solubility limit in the presence of millimolar levels of Ca2+ is less than 1 μM [36,38]. However, the (insoluble) Ca2+ salt of InsP6 has been found to exist naturally, for example, as a component of the extracellular coat around the parasitic cestode Echinococcus granulosus [45] to which it can be transported as nanometre diameter granules [46]. From these observations, it seems that the most plausible mechanism for cellular InsP6 uptake is endocytotic absorption (possibly as a Ca2+–InsP6 precipitate), accompanied by dephosphorylation within the cell.
Consistent with this argument are results from the classic studies of Eagle et al. [39], in which mammalian cell lines in culture were shown to require approx. 0.3 μM inositol for growth, and this requirement could be fully substituted for by inositol monophosphates, or, with a much lower efficacy, by 1 μM InsP6. In this context, we should note that adding InsP6 at much higher (up to millimolar) concentrations to cell cultures will reduce extracellular Ca2+ to 1 μM or lower [36], so any subsequent cell death caused by such high concentrations (e.g. [21,22], and see [23] for a review) would be simply explained by the very well established requirement for extracellular Ca2+ in order to maintain mammalian cell function [47]; in short, the cells die because InsP6 removes all the multivalent cations from the culture medium.
Our results have an impact most directly on the issue of the uptake of InsP6 from the human diet. In a number of studies, Grases et al. [17–19] have reported that adding InsP6 to the diet, or the topical application of InsP6-containing creams [20], increases the levels of InsP6 in plasma, urine and tissues. We have not explored different dietary regimes here, but we would argue that any such difference is not likely to be crucial. Typical levels of InsP6 in urine from human subjects kept on an ‘InsP6-free diet’ are 2–3 μM [20], and in plasma approx. 0.1 μM [19], whereas reverting to an ‘InsP6-normal diet’ leads to approx. 4-fold increases in these parameters. Yet our estimates of maximal levels of InsP6 in human subjects consuming a normal, carnivorous, cereal-containing diet are respectively (at least) 200 times (plasma) and 500 times (urine) lower than those reported for human subjects free of exogenous InsP6 [17–19]. These maxima are only estimates, and perhaps a more direct demonstration of the inconsistency between results published previously [17–20 and ours lies the results for human serum in Figure 2(C) [and the corresponding results for urine (results not shown)], where spiking serum or urine with InsP6 concentrations that are still respectively 4-fold and 40-fold lower than those claimed to be found in these fluids in InsP6-deprived individuals [17–19] gives robust signals in our assay that are very much higher than those from the unspiked samples.
For these reasons, we suggest it is our assay that differs from other studies, not the diet of the donors. Experiments feeding radioactive InsP6 to rats showed that it is dephosphorylated within the gut, and that the radiolabel is subsequently found in plasma and urine only as inositol and InsP [48], and it has been shown that microbes in the gut contribute the phytase activity responsible for this hydrolysis [49]. As the measurement of InsP6 has so far been largely performed by (Dowex) ion-exchange chromatography (e.g. [18]) or HPLC [25] of acid extracts, followed by dephosphorylation of the fractions with phytase and then determination of myo-inositol (with a scyllo-inositol standard) by MS, it is conceivable that, as also suggested by Shears [7], the presence of high concentrations of anions (including multivalent anions such as ATP) in the tissue extracts might compromise inositol phosphate separation protocols characterized in their absence.
Thus although our assay is not designed for the quantification of lower inositol phosphates, we can combine our results with the literature published previously [48,49] to suggest that InsP6 is not absorbed directly from the diet, but rather is dephosphorylated before it is taken up; note that the results of Wise and Gilburt [49] confirm that in the absence of any hydrolysis by gut flora, there is no significant InsP6 absorption. The inositol and InsP generated by dephosphorylation will of course not be without effect: inositol is synthesized de novo by animals (from glucose 6-phosphate via Ins3P, see [6]), but most mammalian tissues do not produce enough of it, and so inositol is a vitamin (vitamin Bh) for which a significant dietary source is InsP6 from cereals. Free levels of inositol are high in many mammalian cells (e.g. [50]), but the Km for PtdIns synthetase is also high [51], so if raising levels of dietary InsP6 leads to increases in the availability of inositol and InsP to tissues, a mass-action effect might be expected on PtdIns synthetase, perhaps leading to increases in steady-state inositol lipid and phosphate levels.
So, if the conclusion of the present study is that we (humans) do indeed make all our InsP6 (by the route described in the Introduction), and that we derive none directly from our diet, what effect(s) will exogenous InsP6, something claimed to be of significant medical benefit [23,24], cause? From the arguments in the preceding paragraph, increases may occur in cellular inositide levels (including InsP6), but this effect would be indirect, and high dietary inositol or InsP would be expected to do the same thing. Any specific and direct actions of exogenous InsP6 will be confined to its place of application. Such direct actions would stem from its ability to bind all multivalent cations with high affinity [36,38]. Binding Fe3+ gives InsP6 unique and potent antioxidant properties [26,35], so it is likely that dietary InsP6 will alter oxidative processes within the gut. This, and its general chelation of cations (e.g. Fe3+, Zn2+ and Ca2+) to restrict their uptake, probably underlie the reported effects of InsP6 in vivo (e.g. [23,24]). These ion-binding actions of InsP6 can have undesirable consequences, either environmental, such as depleting Fe3+ or Zn2+ from some soils [52], or physiological, by InsP6 removing these same essential cations from the human diet to a degree that can be deleterious, depending on the nutritional status of the individual [53–56].
However, a major thrust of many of the arguments for the benefits of InsP6 to humans [23,24] stems from a combination of two factors: (a) the killing of tumour-cell cultures by applying to them high extracellular levels of InsP6, which, we argue above, is of questionable physiological relevance; and (b) an assumption that exogenous InsP6 is taken up directly to form a significant extracellular InsP6 pool, which bathes some or all our cells and which, because of its unique chemistry, can in turn influence their physiology or pathology. It is this latter assumption that the results in the present study suggest requires reconsideration.
Acknowledgments
We are grateful to Dr Charles Brearley (Department of Biological Sciences, University of East Anglia, Norwich, U.K.) for help with some experiments, to Dr Rob Kay (MRC Laboratory of Molecular Biology, Cambridge, U.K.) for the gift of pelleted axenic D. discoideum cells, and to Professor Kevin Brindle (Department of Biochemistry, University of Cambridge, Cambridge, U.K.) for helpful discussions about cell water.
FUNDING
This work was supported by a Programme Grant from the Wellcome Trust, and by the Royal Society.
References
- 1.Posternak S. Sur la synthése de l'ether hexaphosphorique de l'inosite avec le principe phospho-organique de réserve des plantes vertes. C. R. Hebd. Seances Acad. Sci. 1919;169:138–140. [Google Scholar]
- 2.Raboy V. Myo-inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry. 2003;64:1033–1043. doi: 10.1016/s0031-9422(03)00446-1. [DOI] [PubMed] [Google Scholar]
- 3.Berridge M. J., Irvine R. F. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature. 1984;312:315–321. doi: 10.1038/312315a0. [DOI] [PubMed] [Google Scholar]
- 4.Heslop J. P., Irvine R. F., Tashjian A. H., Jr, Berridge M. J. Inositol tetrakis- and pentakisphosphates in GH4 cells. J. Exp. Biol. 1985;119:395–401. doi: 10.1242/jeb.119.1.395. [DOI] [PubMed] [Google Scholar]
- 5.Morgan R. O., Chang J. P., Catt K. J. Novel aspects of gonadotropin- releasing hormone action on inositol polyphosphate metabolism in cultured pituitary gonadotrophs. J. Biol. Chem. 1987;262:1166–1171. [PubMed] [Google Scholar]
- 6.Irvine R. F., Schell M. J. Back in the water: the return of the inositol phosphates. Nat. Rev. Mol. Cell Biol. 2001;2:327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- 7.Shears S. B. Assessing the functional omnipotence of inositol hexakisphophosphate. Cell. Signal. 2001;13:151–158. doi: 10.1016/s0898-6568(01)00129-2. [DOI] [PubMed] [Google Scholar]
- 8.Stephens L. R., Irvine R. F. Stepwise phosphorylation of myo-inositol leading to myo-inositol hexakisphosphate in Dictyostelium. Nature. 1990;346:580–583. doi: 10.1038/346580a0. [DOI] [PubMed] [Google Scholar]
- 9.Brearley C. A., Hanke D. E. Metabolic evidence for the order of addition of individual phosphate esters in the myo-inositol moiety of inositol hexakisphosphate in the duckweed Spirodela polyrhiza L. Biochem. J. 1996;314:227–233. doi: 10.1042/bj3140227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saiardi A., Erdjument-Bromage H., Snowman A. M., Tempst P., Snyder S. H. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- 11.York J. D., Odom A. R., Murphy R., Ives E. B., Wente S. R. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 12.Odom A. R., Stahlberg A., Wente S. R., York J. D. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 13.Frederick J. P., Mattiske D., Wofford J. A., Megosh L. C., Drake L. Y., Chiou S. T., Hogan B. L., York J. D. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8454–8459. doi: 10.1073/pnas.0503706102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leyman A., Pouillon V., Bostan A., Schurmans S., Erneux C., Pesesse X. The absence of expression of the three isozymes of the inositol 1,4,5-trisphosphate 3-kinase does not prevent the formation of inositol pentakisphosphate and hexakisphosphate in mouse embryonic fibroblasts. Cell. Signal. 2006;19:1497–1504. doi: 10.1016/j.cellsig.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 15.Verbsky J. W., Wilson M. P., Kisseleva M. V., Majerus P. W., Wente S. R. The synthesis of inositol hexakisphosphate. Characterization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. J. Biol. Chem. 2002;277:31857–31862. doi: 10.1074/jbc.M205682200. [DOI] [PubMed] [Google Scholar]
- 16.Verbsky J., Lavine K., Majerus P. W. Disruption of the mouse inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene, associated lethality, and tissue distribution of 2-kinase expression. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8448–8453. doi: 10.1073/pnas.0503656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grases F., Simonet B. M., March J. G., Prieto R. M. Inositol hexakisphosphate in urine: the relationship between oral intake and urinary excretion. BJU Int. 2000;85:138–142. doi: 10.1046/j.1464-410x.2000.00324.x. [DOI] [PubMed] [Google Scholar]
- 18.Grases F., Simonet B. M., Prieto R. M., March J. G. Variation of InsP4, InsP5 and InsP6 levels in tissues and biological fluids depending on dietary phytate. J. Nutr. Biochem. 2001;12:595–601. doi: 10.1016/s0955-2863(01)00178-4. [DOI] [PubMed] [Google Scholar]
- 19.Grases F., Simonet B. M., Vucenik I., Prieto R. M., Costa-Bauza A., March J. G., Shamsuddin A. M. Absorption and excretion of orally administered inositol hexaphosphate (IP6 or phytate) in humans. Biofactors. 2001;15:53–61. doi: 10.1002/biof.5520150105. [DOI] [PubMed] [Google Scholar]
- 20.Grases F., Isern B., Perello J., Sanchis P., Prieto R. M., Costa-Bauza A. Absorption of myo-inositol hexakisphosphate (InsP6) through the skin in humans. Pharmazie. 2006;61:652. [PubMed] [Google Scholar]
- 21.Rizvi I., Riggs D. R., Jackson B. J., Ng A., Cunningham C., McFadden D. W. Inositol hexaphosphate (IP6) inhibits cellular proliferation in melanoma. J. Surg. Res. 2006;133:3–6. doi: 10.1016/j.jss.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 22.Vucenik I., Passaniti A., Vitolo M. I., Tantivejkul K., Eggleton P., Shamsuddin A. M. Anti-angiogenic activity of inositol hexaphosphate (IP6) Carcinogenesis. 2004;25:2115–2123. doi: 10.1093/carcin/bgh232. [DOI] [PubMed] [Google Scholar]
- 23.Vucenik I., Shamsuddin A. M. Protection against cancer by dietary IP6 and inositol. Nutr. Cancer. 2006;55:109–125. doi: 10.1207/s15327914nc5502_1. [DOI] [PubMed] [Google Scholar]
- 24.Shamsuddin A. M. Demonizing phytate. Nat. Biotechnol. 2008;26:496–497. doi: 10.1038/nbt0508-496b. [DOI] [PubMed] [Google Scholar]
- 25.Grases F., Simonet B. M., Vucenik I., Perello J., Prieto R. M., Shamsuddin A. M. Effects of exogenous inositol hexakisphosphate (InsP6) on the levels of InsP(6) and of inositol trisphosphate (InsP3) in malignant cells, tissues and biological fluids. Life Sci. 2002;71:1535–1546. doi: 10.1016/s0024-3205(02)01927-6. [DOI] [PubMed] [Google Scholar]
- 26.Hawkins P. T., Poyner D. R., Jackson T. R., Letcher A. J., Lander D. A., Irvine R. F. Inhibition of iron-catalysed hydroxyl radical formation by inositol polyphosphates: a possible physiological function for myo-inositol hexakisphosphate. Biochem. J. 1993;294:929–934. doi: 10.1042/bj2940929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letcher A. J., Schell M. J., Irvine R. F. Methods in Molecular Biology: Inositol Phosphates and Lipids. In: Barker C. J., editor. Totowa: Humana Press; 2009. (in the press) [Google Scholar]
- 28.Irvine R. F., Hemington N., Dawson R. M. Phosphatidylinositol-degrading enzymes in liver lysosomes. Biochem. J. 1977;164:277–280. doi: 10.1042/bj1640277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berridge M. J., Dawson R. M., Downes C. P., Heslop J. P., Irvine R. F. Changes in the levels of inositol phosphates after agonist-dependent hydrolysis of membrane phosphoinositides. Biochem. J. 1983;212:473–482. doi: 10.1042/bj2120473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 31.Mulugu S., Bai W., Fridy P. C., Bastidas R. J., Otto J. C., Dollins D. E., Haystead T. A., Ribeiro A. A., York J. D. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science. 2007;316:106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- 32.Draskovic P., Saiardi A., Bhandari R., Burton A., Ilc G., Kovacevic M., Snyder S. H., Podobnik M. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem. Biol. 2008;15:274–286. doi: 10.1016/j.chembiol.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 33.Reitzer L. J., Wice B. M., Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J. Biol. Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- 34.Brindle K. M., Blackledge M. J., Challiss R. A., Radda G. K. 31P NMR magnetization-transfer measurements of ATP turnover during steady-state isometric muscle contraction in the rat hind limb in vivo. Biochemistry. 1989;28:4887–4893. doi: 10.1021/bi00437a054. [DOI] [PubMed] [Google Scholar]
- 35.Graf E., Empson K. L., Eaton J. W. Phytic acid. A natural antioxidant. J. Biol. Chem. 1987;262:11647–11650. [PubMed] [Google Scholar]
- 36.Torres J., Dominguez S., Cerda M. F., Obal G., Mederos A., Irvine R. F., Diaz A., Kremer C. Solution behaviour of myo-inositol hexakisphosphate in the presence of multivalent cations. Prediction of a neutral pentamagnesium species under cytosolic/nuclear conditions. J. Inorg. Biochem. 2005;99:828–840. doi: 10.1016/j.jinorgbio.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 37.Oliver K. G., Putney J. W., Jr, Obie J. F., Shears S. B. The interconversion of inositol 1,3,4,5,6-pentakisphosphate and inositol tetrakisphosphates in AR4-A2J cells. J. Biol. Chem. 1992;267:21528–21534. [PubMed] [Google Scholar]
- 38.Veiga N., Torres J., Dominguez S., Mederos A., Irvine R. F., Diaz A., Kremer C. The behaviour of myo-inositol hexakisphosphate in the presence of magnesium(II) and calcium(II): protein-free soluble InsP6 is limited to 49 μM under cytosolic/nuclear conditions. J. Inorg. Biochem. 2006;100:1800–1810. doi: 10.1016/j.jinorgbio.2006.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eagle H., Oyama V. I., Levy M., Freeman A. E. Myo-inositol as an essential growth factor for normal and malignant human cells in tissue culture. J. Biol. Chem. 1957;226:191–205. [PubMed] [Google Scholar]
- 40.Johnson L. F., Tate M. E. The structure of ‘phytic acids’. Can. J. Chem. 1969;47:63–73. [Google Scholar]
- 41.Alcazar-Roman A. R., Tran E. J., Guo S., Wente S. R. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 42.Weirich C. S., Erzberger J. P., Flick J. S., Berger J. M., Thorner J., Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 43.York J. D. Regulation of nuclear processes by inositol polyphosphates. Biochim. Biophys. Acta. 2006;1761:552–559. doi: 10.1016/j.bbalip.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 44.Vucenik I., Shamsuddin A. M. [3H]inositol hexaphosphate (phytic acid) is rapidly absorbed and metabolized by murine and human malignant cells in vitro. J. Nutr. Biochem. 1994;124:861–868. doi: 10.1093/jn/124.6.861. [DOI] [PubMed] [Google Scholar]
- 45.Irigoin F., Ferreira F., Fernandez C., Sim R. B., Diaz A. myo-Inositol hexakisphosphate is a major component of an extracellular structure in the parasitic cestode Echinococcus granulosus. Biochem. J. 2002;362:297–304. doi: 10.1042/0264-6021:3620297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irigoin F., Casaravilla C., Iborra F., Sim R. B., Ferreira F., Diaz A. Unique precipitation and exocytosis of a calcium salt of myo-inositol hexakisphosphate in larval Echinococcus granulosus. J. Cell. Biochem. 2004;93:1272–1281. doi: 10.1002/jcb.20262. [DOI] [PubMed] [Google Scholar]
- 47.Ringer S. A further contribution regarding the influence of the different constituents of the blood on the contraction of the heart. J. Physiol. 1883;4:29–43. doi: 10.1113/jphysiol.1883.sp000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakamoto K., Vucenik I., Shamsuddin A. M. [3H]Phytic acid (inositol hexaphosphate) is absorbed and distributed to various tissues in rats. J. Nutr. 1993;123:713–720. doi: 10.1093/jn/123.4.713. [DOI] [PubMed] [Google Scholar]
- 49.Wise A., Gilburt D. J. Phytate hydrolysis by germfree and conventional rats. Appl. Environ. Microbiol. 1982;43:753–756. doi: 10.1128/aem.43.4.753-756.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dawson R. M., Freinkel N. The distribution of free mesoinositol in mammalian tissues, including some observations on the lactating rat. Biochem. J. 1961;78:606–610. doi: 10.1042/bj0780606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takenawa T., Egawa K. CDP-diglyceride:inositol transferase from rat liver. Purification and properties. J. Biol. Chem. 1977;252:5419–5423. [PubMed] [Google Scholar]
- 52.Turner B. L., Paphazy M. J., Haygarth P. M., McKelvie I. D. Inositol phosphates in the environment. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2002;357:449–469. doi: 10.1098/rstb.2001.0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manary M. J., Hotz C., Krebs N. F., Gibson R. S., Westcott J. E., Arnold T., Broadhead R. L., Hambidge K. M. Dietary phytate reduction improves zinc absorption in Malawian children recovering from tuberculosis but not in well children. J. Nutr. 2000;130:2959–2964. doi: 10.1093/jn/130.12.2959. [DOI] [PubMed] [Google Scholar]
- 54.Bohn L., Meyer A. S., Rasmussen S. K. Phytate: impact on environment and human nutrition. A challenge for molecular breeding. J. Zhejiang Univ. Sci. B. 2008;9:165–191. doi: 10.1631/jzus.B0710640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raboy V. The ABCs of low-phytate crops. Nat. Biotechnol. 2007;25:874–875. doi: 10.1038/nbt0807-874. [DOI] [PubMed] [Google Scholar]
- 56.Raboy V. Demonizing phytate. Nat. Biotechnol. 2008;26:497–498. doi: 10.1038/nbt0508-496b. [DOI] [PubMed] [Google Scholar]