Abstract
Autocrine granulocyte macrophage-colony stimulating factor (GM-CSF) sequentially activates intracellular components in monocyte/macrophage production of the pro-inflammatory and immunoregulatory prostanoid, prostaglandin E2 (PGE2). GM-CSF first induces STAT5 signaling protein phosphorylation, then prostaglandin synthase 2 (COX2/PGS2) gene expression, and finally IL-10 production, to downregulate the cascade. Without activation, monocytes of at-risk, type 1 diabetic (T1D), and autoimmune thyroid disease (AITD) humans, and macrophages of nonobese diabetic (NOD) mice have aberrantly high GM-CSF, PGS2, and PGE2 expression, but normal levels of IL-10. After GM-CSF stimulation, repressor STAT5A and B isoforms (80–77 kDa) in autoimmune human and NOD monocytes and activator STAT5A (96–94 kDa) and B (94–92 kDa) isoforms in NOD macrophages stay persistently tyrosine phosphorylated. This STAT5 phosphorylation persisted despite treatment in vitro with IL-10, anti-GM-CSF antibody, or the JAK2/3 inhibitor, AG490. Phosphorylated STAT5 repressor isoforms in autoimmune monocytes had diminished DNA binding capacity on GAS sequences found in the PGS2 gene enhancer. In contrast, STAT5 activator isoforms in NOD macrophages retained their DNA binding capacity on these sites much longer than in healthy control strain macrophages. These findings suggest that STAT5 dysfunction may contribute to dysregulation of GM-CSF signaling and gene activation, including PGS2, in autoimmune monocytes and macrophages.
Keywords: Autoimmune, Cytokine, Prostaglandin, Monocyte/macrophage, Signal transduction
1. Introduction
Both overexpression and knock-out deletion of GM-CSF in mice have been shown to lead to dysregulation of myeloid differentiation and autoimmune disease [1]. Though GM-CSF is best known for its role in myeloid differentiation, it is also a factor in mature monocyte and macrophage activation and function [2,3]. Activation stimuli such as LPS exposure, CD40/CD40L engagement, or TNFα stimulation promote autocrine GM-CSF production in monocytes, macrophages, and granulocytes [4–6].
GM-CSF uses Janus kinase 2 (Jak2) activation of STAT5A and STAT5B proteins to regulate gene expression [2,6–13]. STAT5A and B signal transduction proteins are transcribed by two closely related genes on Chromosome 11 in mice and on Chromosome 17 in humans [1,14]. These gene products have both shared and independent functions in development and activation of the immune system [1,4,5,8,10,14–16].
STAT5 proteins are activated in early myeloid differentiation by GM-CSF and by IL-3 [2,5]. These cytokines can induce signaling through both full-length STAT5A (94–96) and B (94–92) isoforms, as well as through truncated isoforms (77 and 80) that lack the transcriptional activator motif [2,11–13,17,18]. Truncated STAT5 isoforms are not derived from splice variations as seen in other STAT proteins, but are produced post-translationally by the actions of a myeloid-specific nuclear serine protease [13,17]. As these cells mature and become activated, they down-regulate the protease so that most signaling involving STAT5 in matured, activated monocytes, macrophages, and granulocytes occurs only through the full-length isoforms [3,6,13]. However, in matured but unactivated monocytes, GM-CSF can activate truncated 80 kDa STAT5 isoforms to selectively temper the expression of specific genes [2,18]. Thus in differentiation and in mature unactivated monocytes, GM-CSF-induced truncated STAT5 isoforms predominately act as repressors of gene transcription in immature/unactivated cells, while full-length STAT5 isoforms induced in mature/activated cells act as gene transcription activators [11–13].
We previously reported that unactivated monocytes from diabetic/at-risk humans aberrantly express the normally inducible cyclooxygenase, prostaglandin synthase 2 (PGS2/COX2) [19,20]. This early monocyte dysfunction seen in at-risk individuals correlates with low first phase insulin responsiveness (FPIR) and risk for diabetes [19,20]. PGS2 overexpression, through its overproduction of prostaglandin E2 (PGE2), may promote the microenvironment favorable for chronic inflammation and immune dysregulation in at-risk and autoimmune individuals. Our analysis of trans acting factors affecting PGS2 expression revealed that autoimmune monocyte/macrophage PGS2 expression is highly resistant to IL-10 suppression, and that these cells have high autocrine GM-CSF production [21].
Yamaoka et al. [5] have suggested that autocrine GM-CSF is a key component of PGS2 expression activation in LPS-stimulated monocytes and macrophages, as well as its subsequent immunosuppression by IL-10. In this study, we used unactivated monocytes from established autoimmune patients and their at-risk relatives, and monocytes and macrophages from NOD mice to examine the potential for GM-CSF-induced STAT5 activation and function to affect IL-10 and GM-CSF regulation of PGS2 expression in unactivated autoimmune myeloid cells.
2. Materials and methods
2.1. Human cell samples
Heparinized peripheral blood monocytes were from at-risk relatives, new onset and established T1D patients, established Autoimmune Thyroid Disease (AITD, both Graves Disease (GD) patients and Hashimotos thyroiditis (HT) patients), and non-autoimmune healthy controls. Autoimmune (AI) subject samples were obtained during their participation in one of three clinical Type 1 diabetes trials: Diabetes Prevention Trial-1 (DPT-1), Gainesville Subcutaneous Injection Trial (SQ), and Natural History (NH) studies (IRB approved protocol #372-96, Table 1) [19–21]. In these trials, ‘at-risk’ for Type 1 diabetes was defined by production of autoantibodies (IAA+, GAD+, and/or IA-2+), family history of the disease, and/or low first phase insulin response (FPIR). We remained blinded to the intervention status to DPT participant intervention status throughout our studies; and therefore, treatment subgroups were not considered in our studies. No attempt was made to match samples for age or gender in this study, as these factors found were not significant in our previous studies of PGS2 expression in these populations [19–21]. Due to limited blood sample volume available, not all assays could be run on any one sample. Table 1 contains a breakdown of the number of samples analyzed in each major assay system.
Table 1.
Summary of human sample populations
| Healthy controls | Non-AI relatives | All AI subjects | DPTa | SQ/NHa | T1Da | AITDc |
||
|---|---|---|---|---|---|---|---|---|
| GDd | HTe | |||||||
| Total samples | 92 | 60 | 179b | 106 | 19 | 19 | 28 | 13 |
| Individuals | 35 | 16 | 126b | 64 | 11 | 14 | 25 | 11 |
| Age range (years) | 8–49 | 8–50 | 4–68 | 4–40 | 12–52 | 4–42 | 12–68 | |
| Males | 18 | 9 | 57 | 36 | 4 | 9 | 6 | 11 |
| Females | 19 | 7 | 69 | 28 | 7 | 5 | 19 | 10 |
| Disease duration | – | – | – | – | – | New Onset–30 years | New onset–50 years | |
| #Samples analyzed by | ||||||||
| RT-PCR | 30 | 23 | 83 | 45 | 2 | 7 | 29 | |
| Flow cytometry | 10 | 5 | 14 | 4 | 0 | 6 | 5 | |
| Deconvolution microscopy | 41 | 21 | 49 | 26 | 4 | 7 | 12 | |
| IP–Western | 8 | 6 | 21 | 11 | 4 | 3 | 3 | |
| DAP–Western | 6 | 3 | 12 | 9 | 1 | 0 | 0 | |
| EMSA | 11 | 6 | 24 | 13 | 7 | 2 | 0 | |
Type 1 Diabetes (T1D), Diabetes Prevention Trial-1 (DPT-1), Gainesville Subcutaneous Injection Trial (SQ), and Natural History (NH) studies. In these trials, ‘at-risk’ for Type 1 diabetes was defined by production of autoantibodies, family history of the disease, and/or low first phase insulin response (FPIR).
Total includes individuals with multiple samples that overlap multiple groups (e.g., some T1D subjects originally enrolled through DPT or SQ/NH trials prior to onset).
Autoimmune thyroid disease (AITD).
Graves disease (GD).
Hashimotos thyroiditis (HT) patients.
2.2. Mouse cell culture
Peripheral blood mononuclear cells and peritoneal macrophages were collected from 6- to 10-week-old NOD and C57BL/6 female or male mice by cardiac puncture and peritoneal lavage, respectively (IACUC-approved protocol #B083). Tissues were pooled from three to six animals per strain for each analysis, though cells from different genders were not mixed in any experiment. Peripheral blood monocytes were purified using Lympholyte Mgradients (Woodland Technologies, Accurate Chem, Westbury, NY) and adherence-to-plastic. Mouse peritoneal macrophages were adherence-purified prior to analysis.
2.3. GM-CSF activation timecourse and inhibition studies: IL-10, anti-GM-CSF, and Jak Inhibitor, AG490
Monocytes and macrophages were cultured with IL-10 (500 ng/ml, Pierce-Endogen, Rockford, IL), 2 μg/ml anti-GM-CSF blocking antibodies (Pierce-Endogen), 100 μM AG490 (Calbiochem, San Diego, CA) in DMSO (Sigma, St. Louis, MO), 2 μl/ml DMSO or in media alone (RPMI with 10% fetal bovine serum, 1% penicillin, streptomycin, amphiterin or neomycin, Mediatech-Cellgro, Herndon, VA) for 1 h. The media and non-adherent cells were then washed out, and the cultures activated with medium containing GM-CSF (4 nM for human, 1000 U/ml for mouse; Pierce-Endogen) for timecourses of 0–30 min at 37 °C/5%CO2. The cultures were washed with media again and re-cultured for 24 h at 37 °C/5%CO2, with or without the re-addition of the inhibitory agents (IL-10, anti-GM-CSF, AG490). Cell-free culture supernatants were collected for GM-CSF and PGE2 analysis by ELISA (GM-CSF, BD Biosciences-Pharmingen, San Diego, CA and PGE2, Amersham, Piscataway, NJ). Treated cells were collected for immunohistochemical analysis by flow cytometry and deconvolution microscopy, or extracted for RNA, protein, and/or protein–DNA interaction analyses.
2.4. RT-PCR of PGS2 expression
PGS2 mRNA from human monocytes and NOD macrophages was extracted with Ambion (Austin, TX) RNAqueous Isolation kits, converted cDNA using Ambion RT reagents, and amplified by PCR with Ambion Relative Quantitative RT-PCR Primer sets. Primers for 18S RNA or β-actin RNA (Ambion) were used as internal standards, following the manufacturer’s protocols. PCR products were analyzed on 2% agarose gels (SeaKem, Cambrex, Fisher Scientific, Hampton, NH or Gibco, Invitrogen, Carlsbad, CA) by electrophoresis in 0.5× TBE with 10 μg/ml EtBr (Sigma). Visible band pixel densities were compared using a Stratagene (La Jolla, CA) EagleEye imaging system.
2.5. Flow cytometric analysis of PGS2 expression and STAT5 phosphorylation
Intracellular flow cytometric analysis of human CD14+ monocyte PGS2 expression and STAT5 phosphorylation was done as previously described [19–21]. For human monocyte analyses, PE- and FITC-conjugated anti-PGS2 monoclonal antibodies (Cayman Chemical, Ann Arbor, MI) were used in conjunction with FITC-conjugated anti-STAT5 polyclonal antibodies (Santa Cruz Biotech, Santa Cruz, CA) or PE-conjugated anti-STAT5 tyrosine phosphorylated specific monoclonal antibodies [anti-STAT5Ptyr, Upstate Biotech (Charlottesville, VA) conjugated with Prozyme (San Leandro, CA) PE-modification kit]. Matched isotype conjugates (BD Biosciences-Pharmingen, Cal-Tag, Burlingame, CA) were used to set the background fluorescence for percentage of cells and mean fluorescence data collected on BD Biosciences FACS Calibur flow cytometers.
For similar flow cytometric analysis of STAT5 and phosphorylated STAT5 in mouse cells, anti-CD11b conjugates (BD Biosciences-Pharmingen) were used for identification of monocytes and macrophages. Mouse monocyte and macrophage PGS2 was detected in whole cell extracts by Western blot analysis as described below.
2.6. Immunoprecipitation (IP), subcellular fractionation (SF), and Western blot analysis for PGS2 expression, STAT5 phosphorylation, and STAT5 subcellular localization
Protein extracts were made from whole cell (WC) or nuclear (N)/cytoplasmic (C) subcellular fractionation, as indicated in the figure legends. For WC extract, PBS-washed adherent cells were lysed in situ using STAT5 Lysis Buffer [10 mM HEPES pH 7.3 (Gibco), 1 mM dithiothreitol (Sigma), 2 mM EDTA (Sigma), 400 mM KCl (Sigma), 0.1% Triton X100 (Sigma), 10% glycerol (Sigma); with 5 μg/ml each of aprotinin, leupeptin, pepstatin, (Sigma) and pefabloc (Roche Molecular Biochemicals/Boehringer-Mannheim, Indianapolis, IN) added immediately before use]. For subcellular fractionation, cells were washed in situ with Cytoplasmic Lysis Buffer (2 mM HEPES pH 7.6, 10 mM KCl, 1 mM MgCl2, 0.5 mM DTT, 0.1% Triton X100, 20% glycerol; with 5 μg/ml of each aprotinin, pepstatin, leupeptin, and pefabloc added just prior to use). Lysates were then centrifuged at 5000×g, 4 °C for 5 min to pellet nuclei. The supernatant was clarified by an additional centrifugation at 15 000×g, 4 °C for 15 min, to make the cytoplasmic fraction. Nuclear pellets were extracted with STAT5 Lysis Buffer. All protein extracts were stored at −80 °C and thawed only once for analysis.
Ten micrograms of extract protein was used in each IP or directly separated on 7.5% SDS PAGE gels (BioRad Criterion system, Hercules, CA) and transferred to Hybond P membranes (Amersham) for Western blot analysis. PGS2 expression was detected using anti-PGS2 specific mono- or polyclonal antibodies (Cayman) in Western blot analysis. STAT5 proteins in the extracts were immunoprecipitated with anti-STAT5 polyclonal antibodies (Santa Cruz) coupled to Protein G agarose beads (Sigma) prior to Western blot analysis with anti-STAT5Ptyr (Upstate Biotech) or anti-phosphotyrosine (Upstate Biotech) specific antibodies.
Specific antibody binding was detected using an HRP-conjugated secondary antibody (mouse, rabbit, goat; Upstate Biotech) followed by ECL plus chemiluminescent substrate reagents (Amersham). Bands were visualized on ECL Plus Biofilm (Amersham) and/or using a STORM phosphoimaging system (Amersham). Blots were stripped of antibody using the ECL Plus manufacturer’s recommended protocol, and re-probed with anti-STAT5 antibodies (Santa Cruz) and antibodies to PGS2 (Cayman).
2.7. Deconvolution microscopy for STAT5 phosphorylation and subcellular localization
For immunohistochemical analysis, fresh or cultured cells were stained with fluorescent dye-conjugated antibodies to STAT5 or STAT5Ptyr, or with unlabeled antibody followed by anti-mouse Ig antibodies conjugated with Fluorescein or Cy5, using the same staining protocols as for intracellular flow cytometry [18–21]. Anti-CD14+fluorescent conjugates (-FITC, -PE, or -APC, Beckman-Coulter, Miami, FL or BD Biosciences- Pharmingen) surface staining was for identification of human monocytes, and anti-CD11b fluorescent conjugates (-FITC, -PE, or -APC, BD Biosciences-Pharmingen) for mouse monocytes and macrophages. Suspension cells were transferred to slides by cytocentrifugation (Statspin Cytofuge 2, Norwood, MA). Nuclei were then counter-stained with DAPI (Sigma) and the stained slides were coverslip-mounted with anti-fade Gel Mount (Biomeda, Fisher).
Delta Vision Deconvolution microscopy system with an Olympus OMT inverted microscope (Applied Precision, Issaquah, WA) were used for image analysis. Images were processed through with 15 reiterations of the deconvolution algorithm and presented as 3-dimensional (3D) reconstructions of 10–36 optical slices at 0.01 to 0.2 μm thicknesses. Images in figures are representative of experiments with paired control and subject/NOD samples (2–5 random fields viewed/slide).
2.8. Electrophoretic mobility shift analysis (EMSA) for STAT5 DNA binding
STAT5 DNA binding capacity was analyzed in conventional EMSA using DNA sequences labeled with 32P-dATP (Amersham) added by T4 kinase (Promega, Madison, WI) or FITC-UTP (Amersham) added by TdT transferase or Klenow (Amersham) target. GAS target sequences used were from the PGS2 enhancer (mouse and human, 5′-TTCCAAGAA-3′ (sp3), and human, 5′-TTCTGTTGAA-3′ (sp4) [5]) and the CIS enhancer/promoter region IV (5′-TTCCGGGAA-3′ [22]). Commercially available binding sequences for Akt and NFκB proteins (Promega) and poly(dI·dC) (Sigma) were used as non-specific binding controls. EMSA reaction mixtures contained 10 μg of lysate protein, 16 fmol of 32P-dATP labeled oligos, 50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT, 250 mM NaCl, 0.25 μg/μl poly(dI·dC) (Promega, Sigma, Amersham). Reactions were incubated for 30 min at room temperature (rt) before being run on 6% non-denaturing PAGE gels in 0.5× TBE at 200 V for 4–5 h, 4 °C. Radioisotope-labeled DNA bands in Shift and Supershift complexes were visualized by autoradiography, and fluorescently labeled DNA by STORM Phospho Imager after UV crosslinking (short-wave transilluminator, 15 min, rt). Specific binding of STAT5 to fluorescent DNA was detected on Western blots using anti-FITC antibodies (Amersham), followed by stripping and re-probing with anti-STAT5 and/or anti-STAT5Ptyr antibodies as described above for IP–Westerns.
To confirm specificity of binding, parallel reactions were set with an excess of unlabeled specific (same) or other oligonucleotides (as described in the figure legend) added 10 min prior to the addition of the labeled oligonucleotide, and anti-STAT5A/B (1 μg/reaction, Santa Cruz) or non-specific isotype antibodies (1 μg/ reaction, anti-PGS2, Cayman) were added after the specific oligo for supershift reactions.
2.9. DNA affinity precipitation (DAP)–Western blot analysis for STAT5 DNA binding
Cell protein extracts (10 μg) were reacted with FITC-UTP- labeled mouse and human PGS2 and CIS GAS sequences, as well as a consensus STAT5/STAT6 GAS sequence (5′-GTATTTCCCAGAAAAGGAAC- 3′, Santa Cruz) in binding reactions as described for EMSA. Protein G agarose (Sigma) and anti-FITC antibody (Amersham) were used to precipitate out protein–DNA complexes. Proteins were released from the complexes by boiling in 1× Leammli buffer for analysis as described above for STAT5 IP/Western blot analysis.
2.10. Statistical analyses
Where possible, data were analyzed by one-way ANOVA for multiple timepoint analysis, and using Student t test or nonparametric Mann–Whitney U tests for pairwise analyses. The results of these analyses are listed with appropriate p values and statistical parameters (n, mean, SD and/or SEM) in Table 2, on the figures, in figure legends and/or in Section 3. The designation ‘ns’ is listed where statistical analysis was applied and no statistical significance between the groups was obtained.
Table 2.
Persistent STAT5 tyrosine phosphorylation in unactivated autoimmune human monocytes and mouse macrophages
| Human peripheral blood monocytes
|
Mouse macrophages
|
||||||
|---|---|---|---|---|---|---|---|
| Healthy controls | All AI subjectsa | At-risk subjectsb | T1D patients | AITD patients | NOD | C57BL/6 | |
| Mean/SD % of cells CD14+/STAT5Ptyr+ by flow cytometryc | 7.106 ± 8.235 | 27.07 ± 30.41 | 46.97 ± 47.96 | 23.65 ± 27.38 | 10.51 ± 9.694 | 66.09 ± 11.56 | 33.21± 4.236 |
| n = 10 | n = 12 | n = 3 | n = 5 | n = 4 | n = 4 | n = 4 | |
| *p = 0.0444 | * | ns | ns | ** | **p = 0.0370 | ||
| ***p = 0.0400 | *** | ||||||
| 0/16 | 4/12 | 2/3 | 2/5 | 1/4 | 4/4 | 1/4 | |
| Mean/SD MF of cells CD14+/STAT5Ptyr+ by flow cytometryc | 589 ± 882 | 1278 ± 1593 | 727 ± 688 | 2155 ± 20 | 1155 ± 1582 | 449 ± 154 | 195 ± 107 |
| n = 10 | n = 14 | n = 4 | 82 | n = 5 | n = 4 | n = 4 | |
| *p = 0.009 | * | n = 6 | ** | **p = 0.0347 | |||
| ***p = 0.0140 | *** | 4/4 | 1/4 | ||||
| ****p = 0.0030 | **** | ||||||
| *****p = 0.193 | ***** | ||||||
| 1/10 | 9/14 | 3/4 | 5/6 | 2/5 | |||
| Dysfunctional STAT5 by IP/Western/DAP/EMSA analysesd | 17% (5/29) | 69% (43/62) | 74% (35/47) | 75% (6/8) | 44% (4/9) | 95% (20/21) | 14% (3/21) |
| High STAT5Ptyr+ detectable by immunohistochemical analysis of unstimulated monocytes and macrophages | 0% (0/23) | 92% (35/38) | 95% (20/21) | 100% (7/7) | 80% (8/10) | 100% (9/9) | 22% (2/9) |
| % Positive samples of total STAT5Ptyr+ | 7% (6/85) | 70% (88/126) | 80% (60/75) | 77% (20/26) | 54% (15/28) | 97% (37/38) | 18% (7/38) |
Some subjects are more than one group (e.g., some DPT subjects developed T1D and some subjects had multiple autoimmune diseases (Hashimotos and T1D)).
At-risk subjects include autoimmune subjects from DPT and SQ/NH trials that did not progress to T1D or have AITD during the study period.
p Values within assay data represent comparison of mean of control versus group indicated in nonparametric pairwise comparison using Mann–Whitney U test; ns = not significant; the # of stars (*) indicate the groups compared to the controls for each p value listed under the control data. SD = standard deviation; MF = mean fluorescence.
The criteria for ‘dysfunctional STAT5’ was defined as detectable binding with anti-STAT5Ptyr in IP/western blot analysis, either without stimulation or 24 h after brief exposure to GM-CSF. The criteria for ‘dysfunctional STAT5’ was defined as a lack of binding to PGS2 GAS DNA sequences by truncated STAT5 in DAP and EMSA analyses for human monocytes. In NOD macrophages, persistent binding 24 h after GM-CSF exposure by full-length STAT5 was used as the criteria for ‘dysfunctional STAT5’ in DAP and EMSA analyses.
3. Results
3.1. Persistent STAT5 phosphorylation in autoimmune human monocytes
STAT5A and STAT5B proteins in unactivated, freshly isolated peripheral blood human monocytes were analyzed by flow cytometric, Western blot, and immunohistochemical analyses (Fig. 1). There was an overall marked increase in the basal level of STAT5 tyrosine phosphorylation in 70% (88/126) of T1D, AITD, and at-risk subjects (collectively called ‘autoimmune’ (AI) subjects) compared with 7% (6/85) of healthy controls (Table 2).
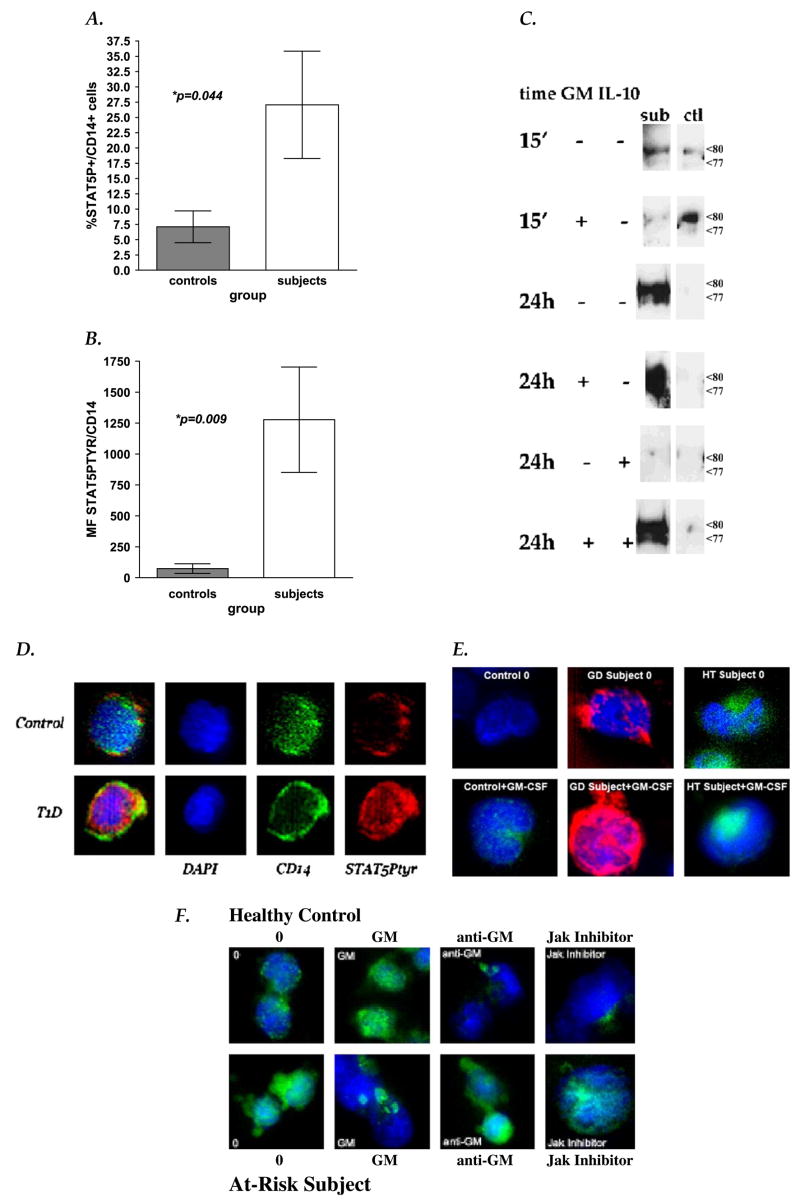
Fig. 1.
STAT5 phosphorylation in human monocytes. Flow cytometric analysis of STAT5 phosphorylation in unactivated at-risk/T1D subject PBMC monocytes showed that at-risk/T1D subjects had (A) higher number of cells with detectable STAT5 expression (%CD14+/STAT5Ptyr+ cells, p = 0.0444; Mann–Whitney U test) and (B) higher STAT5 phosphorylation levels per cell (mean fluorescence (MF) CD14+/STAT5Ptyr+ cells, p = 0.009; Mann–Whitney U tests) than seen in healthy controls. Means of controls (grey bars) and subjects (white bars) are shown with SEM error bars. (C) Anti-STAT5 IP/phosphotyrosine Western blot analyses of whole cell lysates show high phosphorylated truncated STAT5 present in control monocytes only after 15 min (15′) exposure to 4 nM GM-CSF (GM), while at-risk subject monocytes have detectable phosphorylated truncated STAT5 without stimulation and 24 h (24 h) after GM-CSF exposure. STAT5 phosphorylation in at-risk monocytes appears sensitive to downregulation by IL-10 (100 ng/ml), unless previously stimulated by GM-CSF. Arrows indicate the position of truncated STAT5 isoforms (77, 80 kDa) on the same blots re-probed with anti-STAT5 A/B (data not shown). (D) Immunohistochemical analysis of STAT5 phosphorylation in unactivated PBMC cells from a healthy control (top panels) and from an established T1D patient (bottom panels). The 3-dimensional composites of 20 deconvolved 0.2-μm optical slices depict DAPI (blue), anti-CD14-APC (pseudo-colored green)), and anti-STAT5Ptyr-PE staining (red). (E) Intense anti-STAT5Ptyr staining (shown as red or green staining over blue DAPI-stained chromatin) is also seen in both Graves Disease (GD) and Hashimotos thyroiditis (HD) patient monocytes, before and after GM-CSF exposure (4 nM, 15 min). (F) Deconvolution composite images (Anti-STAT5Ptyr-anti-mouse IgG-FITC, green; DAPI, blue) of monocytes after 1 h culture in media with DMSO (‘0’), 2 μg/ml anti-GM-CSF blocking antibody (‘anti-GM’) or with 100 μM Jak2/3 kinase inhibitor, AG490 (‘Jak Inhibitor’), or 15 min activation with 4 nM GM-CSF (‘GM’). GM-CSF stimulation appears to cause rapid aggregation of STAT5Ptyr staining in cytoplasm of AI monocytes.
Flow cytometric analysis showed a higher percentage of unactivated monocytes with phosphorylated STAT5 (Table 2 and Fig. 1a, p = 0.0444, Mann–Whitney U test) in AI subjects and an overall higher mean fluorescence (MF) of STAT5Ptyr staining compared with controls (p = 0.009, Mann–Whitney U test, Table 2 and Fig. 1b). The highest percentage of STAT5Ptyr+ monocytes within the AI subject population was contributed by the T1D subgroup. Whereas, all subgroups (at-risk, T1D, and AITD) showed significantly higher STAT5Ptyr per cell (MF of STAT5Ptyr, Fig. 1b and Table 2). In IP–Western blot analyses, the majority of this persistently phosphorylated STAT5 was found as truncated isoforms, though there was variability between individuals as to which isoform was most prevalent (77 kDa or 80 kDa) (Fig. 1c). Deconvolution analysis suggests that the majority of the phosphorylated STAT5 found in AI monocytes is cytoplasmic and aggregated in discrete areas after GM-CSF (Fig. 1d–f).
STAT5 phosphorylation was enhanced when human AI monocytes were activated with GM-CSF for 15 min; a timeframe in which GM-CSF activation of STAT5Ptyr could be also detected in control cells (Fig. 1c, e, f). Removal of exogenous GM-CSF after 15 min caused a loss of STAT5 phosphorylation in healthy control monocytes within 24 h, but little to no diminution of STAT5 phosphorylation in human AI monocytes (Fig. 1c). These data suggest that human AI monocytes have an abundance of persistently activated STAT5 protein without exogenous stimulation. In contrast, control monocytes do not activate STAT5 until treated with GM-CSF and deactivate it quickly once the stimulus is removed.
3.2. IL-10 resistance and GM-CSF independence of PGS2 expression and STAT5 activation in AI human monocytes
We had previously reported that autoimmune human monocytes have significantly higher GM-CSF production without stimulation than that seen in control monocytes [21]. However, we found no correlation between subject GM-CSF production levels and their PGS2 expression levels or PGS2 resistance to IL-10 suppression. High levels of IL-10 added to human AI monocyte cultures could completely block their GM-CSF production while still allowing their PGS2 expression to go unchecked [21].
To determine if the high endogenous GM-CSF in autoimmune monocytes was promoting their persistent STAT5 activation, we blocked GM-CSF production in vitro with IL-10 (Fig. 1c), blocked its ability to bind its receptor with anti-GM-CSF antibodies (Fig. 1f), or blocked Jak2 phosphorylation of STAT5 with AG490, a potent inhibitor of both Jak2 and Jak3 kinase activities (Fig. 1f). Like PGS2 expression [21], STAT5 phosphorylation was resistant to IL-10 suppression in autoimmune human monocytes, while both were sensitive in control cells. However, unlike PGS2 expression, IL-10 resistance of autoimmune monocyte STAT5 phosphorylation appears dependent on at least a brief GM-CSF exposure (Fig. 1c). Anti-GM-CSF antibodies or AG490 Jak inhibition had little to no effect on STAT5 phosphorylation in AI monocytes (Fig. 1f), suggesting that STAT5 phosphorylation in autoimmune monocytes can become GM-CSF and kinase independent.
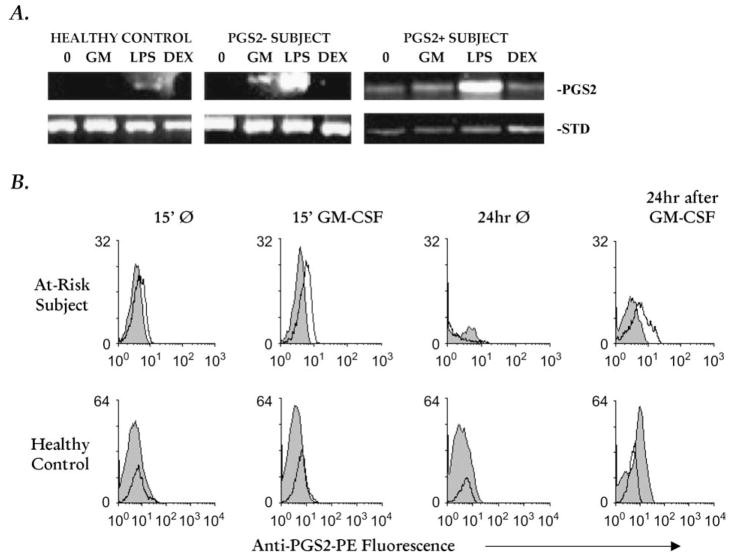
In 5/7 (71%) autoimmune human subjects without high levels of PGS2 in their freshly isolated monocytes (i.e., PGS2 negative [19]), GM-CSF could directly activate modest amounts of PGS2 mRNA and protein expression (Fig. 2a and 2b, respectively). However, in monocytes from human AI subjects who had high PGS2 expression ex vivo (i.e., PGS2 positive [19]), GM-CSF did not increase their PGS2 mRNA or protein expression (Fig. 2a and data not shown).
Fig. 2.
PGS2 mRNA and protein expression in human monocytes. (A) Adherence-isolated monocytes from PGS2+[19] at-risk subject, PGS2− [19] at-risk subject, and control human subjects were cultured without (‘0’, 15 min) or with GM-CSF (‘GM’, 4 nM, 15 min), LPS (‘LPS’, 5 μg/ml, 90 min) or dexamethasone (‘Dex’, 1 μmol/ml, 90 min), then extracted for mRNA extracts and analyzed for PGS2 expression by relative quantitative RT-PCR. 18S RNA and/or β-actin were used as experimental controls (‘Std’). The 18S RNA and/or β-actin internal and external standards were used for the calculation of densitometric ratios (PGS2/standard (Std) RNA RT-PCR product band) which showed that GM-CSF stimulation of PGS2 mRNA expression in both PGS2 positive and PGS2 negative at-risk/T1D subject monocytes 15 min after the stimulus was removed was significantly higher than detected at the same time point in healthy control monocytes [(p = 0.0136, ANOVA, data from timecourse analysis of nine control and 15 subject samples (nine PGS2 negative and six PGS2 positive)]. (B) At-risk/T1D and healthy control monocytes were stimulated with 4 nM GM-CSF for 15 min, washed, and held 24 h without GM-CSF prior to PGS2 protein expression flow cytometric analysis. Grey peaks represent isotype control and unfilled peaks anti-PGS2 conjugate fluorescence; representative of six independent assay runs with nine at-risk/T1D and six control samples.
These findings taken together suggest that GM-CSF induces persistence of IL-10 resistant STAT5 activation that can become GM-CSF/Jak independent and, may influence the IL-10 resistance of PGS2 expression in autoimmune monocytes and macrophages.
3.3. Aberrant DNA binding capacities of STAT5 in AI human monocytes
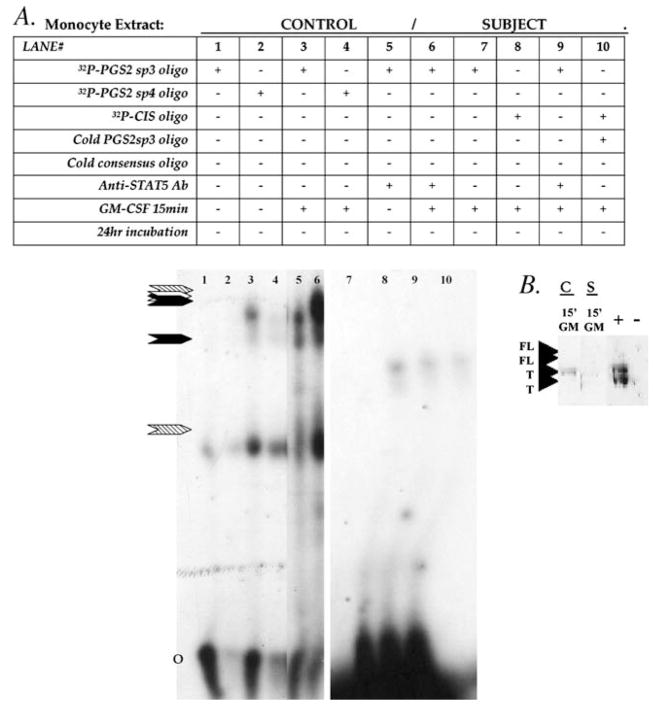
To begin to look at the function of persistently phosphorylated STAT5 isoforms in human AI monocytes, we examined their ability to bind GAS recognition sequences derived from the PGS2 gene enhancer region [5] and from the promoter/enhancer region of STAT5 activated gene, CIS [22] (Fig. 3). Truncated STAT5 isoforms from healthy control monocytes had specific and strong DNA binding, with their binding to CIS sequences being the strongest and PGS2 sequences the weakest signals seen. However, we were unable to detect any DNA binding of STAT5 truncated isoforms from autoimmune monocytes on PGS2 enhancer sequences and only very low levels on CIS sequences.
Fig. 3.
STAT5 DNA binding capacities in human monocytes. (A) Monocytes were analyzed after exposure to 4 nMGM-CSF for 15 min or 24 h after GM-CSF exposure. EMSA analysis of human whole cell using 32P labeled sp3 or sp4 STAT5 [5] or CIS IV [22]GAS binding sequences alone or with excess unlabeled (‘cold oligo’) specific sp3 or consensus STAT5 GAS oligonucleotide sequence show specificity of bind. Supershift reactions were run using 2 μg/reaction of anti- STAT5A/B antibody to specifically identify STAT5 in the binding complex. Arrows indicate the position of STAT5 GAS specific bind (black arrows for PGS2, white arrow for CIS IV) and supershift bands (hatched arrows). O = unbound oligo position. (B) The presence and isoform size of STAT5 available for binding DNA in these extracts was confirmed by DNA affinity precipitation analysis (DAP) using a commercially available (Santa Cruz) consensus GAS sequence and 10 μg protein extracted from control (C) and at-risk subject (S) monocytes after 15 min exposure to 4 nM GM-CSF (‘15′GM’). Protein bound to DNA was identified by Western blot using anti- STAT5A/B or anti-STAT5Ptyr antibodies. ‘T’ = molecular size of truncated STAT5, ‘FL’ = full-length STAT5, and ‘+’ = extract from Jurkat cell as a positive control. ‘−’ = sham DNA binding/precipitation reaction run without extract added.
3.4. STAT5 dysfunction in NOD mouse monocytes
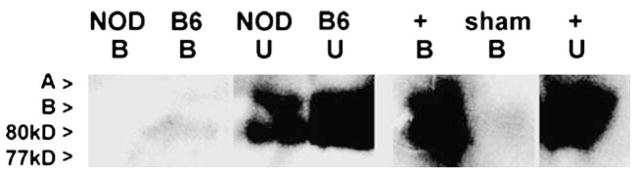
We similarly examined STAT5 dysfunction in peripheral blood monocytes isolated from NOD and C57BL/6 mice. As seen in AI human monocytes, unactivated NOD mouse monocytes show prolonged truncated STAT5 isoform phosphorylation, but no STAT5 DNA binding capacity in vitro on GAS sequences derived from PGS2 gene enhancer (Fig. 4 and data not shown).
Fig. 4.
Phosphorylated STAT5 DNA binding in mouse monocytes. DAP analysis of unactivated NOD and C57BL/6 (B6) peripheral blood monocytes. B = DNA bound fraction of DAP; U = unbound fraction of DAP. No specific phosphorylated STAT5 GAS DNA binding was seen in NOD nuclear extracts despite an abundance of phosphorylated STAT5 (seen in unbound fraction). Nuclear extracts from C57BL/6 gave low but detectable phosphorylated truncated STAT5 isoform DNA binding. DAP with activated Jurkat cell extracts (+ control) were included as positive control for STAT5 isoform band localization (A, B, 77 kDa, 80 kDa). Sham (no extract) DAP were run in parallel as negative control. Gels are representative of data obtained in two separate DAP runs.
These data indicate that in both autoimmune human and NOD mouse monocytes, truncated repressor STAT5 isoforms can bind DNA in vitro compared to controls. If a similar lack of STAT5 DNA binding capacity is true in vivo, STAT5 dysfunction might allow inappropriate gene transcription in GM-CSF activation of autoimmune monocytes.
3.5. Prolonged STAT5 phosphorylation and function in NOD mouse macrophages
In contrast to monocytes, GM-CSF activation of mature macrophages primarily elicits phosphorylation full-length transcriptional activator STAT5 isoforms [2,3,6,13,18]. Thus, similar lack of STAT5 DNA binding capacity in autoimmune macrophages might cause a decrease in GM-CSF-induced gene expression. To test this possibility, we repeated our analyses of STAT5 phosphorylation, activation by GM-CSF, and DNA binding capacity in unelicited peritoneal macrophages of NOD and C57BL/6 mice.
As seen in human and NOD monocytes, we found that unelicited NOD mouse peritoneal macrophages had a higher overall STAT5 phosphorylation (97% (37/38), when compared with macrophages of age-gender matched C57BL/6 mice in flow cytometric and deconvolution analyses (18% (7/38); Table 2 and Fig. 5).
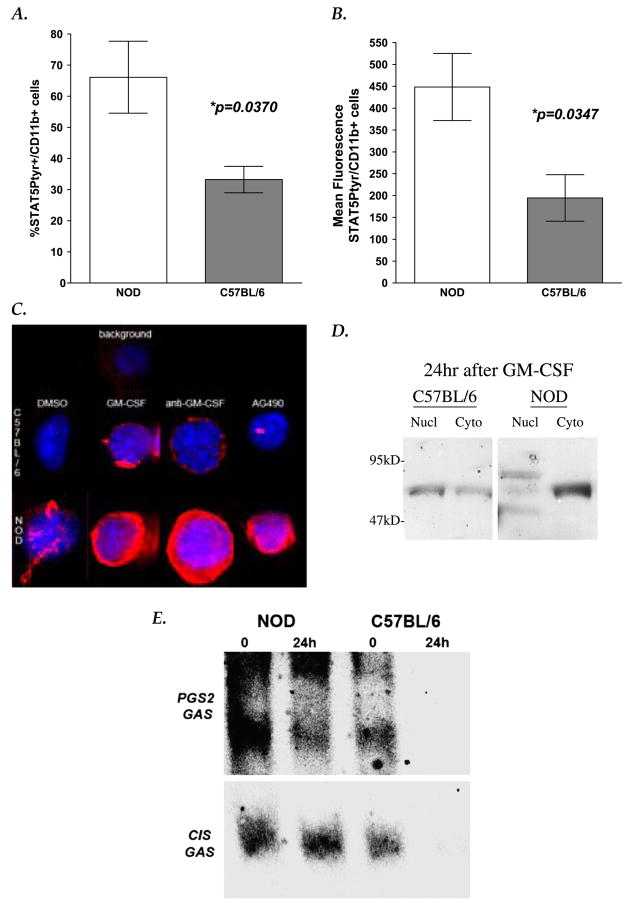
Fig. 5.
STAT5 phosphorylation and function in mouse macrophages. Flow cytometric analysis of STAT5 phosphorylation in unactivated NOD and C57BL/6 peritoneal macrophages showed that NOD macrophages had (A) two-fold higher % of cells with detectable STAT5 expression (%CD14+/ STAT5Ptyr+ cells; p = 0.0370, Mann–Whitney U test), and (B) higher levels of STAT5 phosphorylation per cell (mean fluorescence (MF) CD14+/ STAT5+cells; p = 0.0347, Mann–Whitney U tests) than seen in C57BL/6macrophages. Graphs depictmean and SEM of four independent experiments (four sets of pooled samples from three mice each per stain, statistics are listed in Table 2). (C) Immunohistochemical staining of STAT5 phosphorylation in unactivated macrophages fromC57BL/6 (left panels) and from NOD(right panels) mice are shown as 3-dimensional composite images derived from 20 deconvolved 0.2 μmoptical slices. DAPI (blue), anti-STAT5A/B(pan)-FITC (green)), and anti-STAT5Ptyr-PE staining (red) shown are representative of nine separate experiments. (D) Subcellular fractionation/anti-STAT5 IP-Western blot analyses indicate that phosphorylated truncated (80kDa) and fulllength STAT5A and B isoforms (92–96kDa) are detectable in the nuclei of both NOD and C57BL/6 macrophages after 15minGM-CSF exposure (data not shown). However, 24 h after GM-CSF was removed, full-length STAT5 isoforms (96–94 kDa) remain phosphorylated and in the nuclear fractions only in NOD macrophages. Truncated isoforms (80 kDa) were still visible in all 24 h control and AI fractions. Lane key: Nucl = Nuclear fraction IP; Cyto = Cytoplasmic fraction IP; IP were done using anti-STAT5A/B, and Western blots were probed with anti-STAT5Ptyr. Blots shown are representative of 12 independently run IP–Western blot analyses. (E) Nuclear extracts were made from GM-CSF-activated (1000 U GM-CSF/ml, 15 min), and 24 h post GM-CSF exposure cultures of adherence-isolated peritoneal macrophages from NOD and C57BL/6 mice. EMSA was performed using fluorescently labeled DNA for mouse PGS2 [5] and CIS IV [22] GAS sequences, followed by Western blot analyses with anti-FITC-DNA (shown) and then with anti-STAT5A/B antibodies (not shown). Data presented are representative of nine independent analyses.
STAT5 phosphorylation was markedly enhanced in both control and NOD macrophages after 15 min GM-CSF exposure (Fig. 5c). As seen in human monocytes, inhibition of GM-CSF binding with anti-GM-CSF antibodies or Jak kinase activity with AG490 diminished STAT5 phosphorylation in C57BL/6 macrophage but not NOD macrophages (Fig. 5c and data not shown).
As predicted, IP–Western blot analyses showed that full-length STAT5 (96–92 kDa) and 80 kDa truncated proteins were the predominant isoforms found phosphorylated in GM-CSF-stimulated mouse macrophages. However, anti-STAT5 positive bands of size predicted for full-length, phosphorylated STAT5B (94 kDa) isoforms persisted for 24 h after stimulation only in GM-CSF-activated NOD macrophage nuclei, not in C57BL/6 nuclei (Fig. 6d).
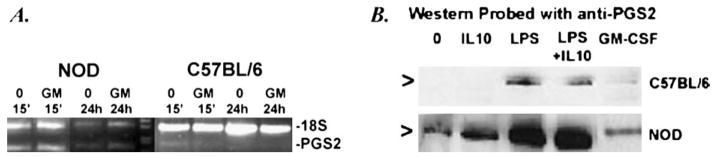
Fig. 6.
PGS2 mRNA and protein expression in mouse macrophages. (A) Adherence-isolated peritoneal macrophages from NOD and C57BL/6 mice were cultured without (0, 15 min, 24 h) or with GM-CSF (GM, 1000 U/ml, 15 min) followed by 24 h culture without GM-CSF (GM 24 h). mRNA extracts from these cultures were analyzed for PGS2 expression (PGS2) by relative quantitative RT-PCR. Primers for 18SRNAwere used as the internal experimental control (18S). Little to no PGS2 mRNA was detectable in the C57BL/6 cultures compared to NOD, but no detectable change in PGS2 mRNA expression was seen between the treatments in the NOD. Data shown are representative of two independent experiments. (D)NODand C57BL/ 6 peritoneal macrophages were also cultured with and without 500 ng/ml IL-10, 1 mg/ml LPS, IL-10+LPS, or 1000 U/ml GM-CSF for 24 h, prior to Western blot analysis with anti-PGS2 anti-sera. NOD macrophages had higher PGS2 protein detectable in all treatments than C57BL/6. Arrows indicate the location of PGS2 protein band in purified standard (lane not shown). Data shown are representative of three independent experiments.
When we analyzed STAT5 DNA binding in NOD and C57BL/6 mouse control strain peritoneal macrophages, we found opposite results as to that seen in AI human and NOD monocytes. STAT5 from NOD macrophages bound avidly to the PGS2 enhancer GAS sequence even 24 h in culture after GM-CSF was removed (Fig. 5e).
These data suggest that NOD mouse macrophages have persistent full-length activator STAT5 isoforms DNA binding after GM-CSF stimulation. Similar DNA binding in vivo could lead to aberrantly persistent gene expression in macrophages.
3.6. GM-CSF effects on NOD macrophage PGS2 expression
Direct GM-CSF treatment of NOD mouse macrophages did not affect their PGS2 mRNA or protein expression (Fig. 6). However, we find that like PGS2+ human monocytes, the PGS2 expression of unactivated NOD macrophages is very resistant to IL-10 suppression, despite the complete suppression of GM-CSF with IL-10 (Fig. 6b and data not shown).
These data suggest that the persistence of IL-10-resistant STAT5 activation can become GM-CSF/Jak independent after a brief activation sometime in its past, and may be influencing the persistence and stability of PGS2 expression in both autoimmune monocytes and macrophages.
4. Discussion
We have found that unactivated monocytes from autoimmune T1D, AITD, and at-risk human subjects, and monocytes and macrophages of the NOD mouse have three potential interrelated phenotypes in cytokine induced activation: 1)PGS2 overexpression, generating high levels of the pro-inflammatory prostanoid, PGE2 [19,20]; 2) high autocrine GM-CSF production in vitro [21], and 3) STAT5 dysfunction. We hypothesize that these phenotypes may contribute to a pro-inflammatory milieu underlying immunopathogenesis of autoimmune disease.
Davoodi-Semiromi et al. [23] recently reported a unique single base pair polymorphism within NOD DNA binding domain of STAT5B DNA. However, no similar defect has been reported for T1D or AITD human STAT5. Due in part to limitations associated with the lack of useful truncated and specific isoform specific antibodies, we cannot definitively detect differences between STAT5A and STAT5B dysfunction in our macrophage phenotypic studies. However, we did detect differentiation stage (i.e., monocyte versus macrophage), and activation state differences in truncated and full-length STAT5 isoform activation and DNA binding capacities in our analyses.
4.1. Persistence of STAT5 phosphorylation
STAT5 proteins in unactivated autoimmune human and mouse monocytes are found primarily in the cytoplasm as persistently phosphorylated dominant negative truncated isoforms that are unable to bind GAS recognition sequences. In more mature myeloid cells, represented in our studies by unactivated NOD peritoneal macrophages, we see persistent phosphorylation of the full-length activator isoforms of STAT5A and B. These isoforms remain in the nucleus of NOD macrophages longer and have prolonged DNA binding capacities compared with C57BL/6 control macrophages.
We noted that STAT5 phosphorylation in autoimmune cells could be or become GM-CSF independent and Jak2/3 independent after activation. However, some component of STAT5 dephosphorylation/deactivation in these cells remains IL-10 sensitive without GM-CSF stimulation. Our data suggest that STAT5 persistent phosphorylation in autoimmune monocytes and macrophages is not dependent on persistent kinase activity; thus, leaving dephosphorylation and recycling/degradation as potential mechanisms for its dysregulation.
In angiotensin II signaling, calcineurin dephosphorylates STAT5 in the nucleus, but whether this is the mechanism used in GM-CSF-stimulated myeloid cells is unknown [24]. In the cytoplasm, STAT5 can be dephosphorylated by PTP1B, SHP-1, and SHP-2, with the choice of phosphatase varying with cell type and in response to different hormones and cytokines [25–33]. SHP-2 is activated by GM-CSF, IFNα, prolactin, IL-2, and IL-6, but not G-CSF or M-CSF [28–33]. SHP-2 can bind STAT5 in conjunction with the Jak inhibitory protein SOCS-3 [30,33]. This interaction dephosphorylates and re-activates Jak 2 in response to IL-6 and possibly prolactin [29,30,33]. SHP-1 activity, which can be activated by M-CSF [27], appears to dephosphorylate STAT5 after activation by any of the three CSF cytokines [26–28]. In light of the GM-CSF-induced STAT5 activation IL-10 resistance we observe in autoimmune monocytes, it is interesting to note that IL-10 can also activate SOCS-3 [34]. Thus, the role of GM-CSF in the persistence of STAT5 phosphorylation in autoimmune monocytes and macrophages may encompass whether or not it can be dephosphorylated by SHP-1 or re-activated by SHP-2/SOCS-3 mechanisms as seen in IL-6.
The residence time and half-life of truncated versus full-length isoforms of phosphorylated STAT5 in healthy cells is still debated [35]. Dephosphorylated truncated isoforms maybe rapidly degraded or recycled [36,37]; or as proposed by Wang et al. [35], the loss of the COOH terminus may prohibit ubiquitination of these short forms, causing them to collect in the cytoplasm over time.
4.2. Dysregulation of STAT5–DNA interactions
Changes in STAT5 DNA binding capacities in autoimmune monocytes and macrophages could result from defects in its translocation or its inhibition by trans acting regulatory proteins. Serine/threonine phosphorylation of nuclear translocated STAT5 dimers may support its binding of DNA or facilitate its dephosphorylation and export [36,38]. STAT5 interactions with CRM-1, the nuclear export protein that binds the NES sequence embedded in STAT5’s DNA binding motif, allows for normal recycling and translocation of STAT5 proteins [36]. However, our preliminary data do not support defects in either of these mechanisms. We observed no changes in STAT5 serine/threonine phosphorylation or in its interactions with CRM-1. Additionally, we see STAT5 proteins return to the cytoplasm after activation and translocation to the nucleus in autoimmune monocytes [Litherland, unpublished data].
PIAS3, a STAT regulatory protein that has or associates with a SUMO-1 ligase protein modification activity [39,40], may be a better candidate for STAT5 dysregulation in autoimmune monocytes and macrophages. STAT5 interactions with PIAS3 can prohibit phosphorylated STAT5 binding to DNA as well as target unphosphorylated STAT5 for degradation [39]. Our preliminary IP studies have indicated that NOD macrophage and autoimmune human monocyte STAT5 has decreased interactions with this important regulatory protein [Belkin et al., manuscript in preparation].
4.3. Relating STAT5 dysfunction with GM-CSF, PGS2, and dysfunction of autoimmune monocytes and macrophages
Piazza et al. [13] have shown that GM-CSF-induced STAT5 isoform changes can act as key regulatory ‘switches’ for myeloid differentiation and activation, with truncated STAT5 isoforms acting as repressors of gene transcription in immature/unactivated cells and full-length STAT5 isoforms induced in mature/activated cells act as gene transcription activators [11,13,17]. Previous studies in the NOD have suggested that myeloid APC differentiation and function are defective at a point or points during the immunopathogenesis of its autoimmune diabetes where the determination of specific cell lineage decisions and activation state are made based on the cytokine microenvironment [41–44]. Our data suggest that dysregulation of GM-CSF signaling through STAT5 may be a prime candidate for the cause of these defects, due to its pivotal role at such divergent decision junctions.
Bringing our studies full circle, we have sought to find if there is a link between immunopathology and the GM-CSF, PGS2, and STAT5 autoimmune phenotypes in the monocytes of at-risk/T1D/AITD subjects and in the monocytes and macrophages of the NOD mouse. Overexpression of PGS2 by these cells helps to establish the pro-inflammatory milieu that can promote auto-reactive T cell escape from tolerance and their activation to damage self tissues. GM-CSF overproduction can alter the responsiveness of unactivated autoimmune monocytes and macrophages to itself and other differentiation signals, and thus interfere with their normal progression through differentiation and activation, including regulation of their PGS2 expression. STAT5 dysfunction may be the link between the GM-CSF and PGS2 phenotypes and a mechanism for the unique resistance of PGS2 to IL-10 suppression in autoimmune monocytes and macrophages.
The prolonged STAT5 phosphorylation and cell type/isoform-specific alterations in DNA binding we see in vitro may be indicative of STAT5 dysregulation and/or dysfunction in autoimmune myeloid cells. These data suggest that GM-CSF- and STAT5-induced dysfunction in myeloid differentiation and monocyte/ macrophage activation has the potential to hinder the development and function of myeloid cells as antigen presenting cells (APC) and their role in maintaining immune self-tolerance.
Acknowledgments
The authors would like to thank the Diabetes Prevention Trial, the UF Diabetes Research Group, and the Shands at UF Pediatric and Adult Endocrinology clinics for their support of this project. We gratefully acknowledge the help of J.M. Crawford, M.D., Ph.D., C. Ketcham, Ph.D., K. Madsen, Ph.D., M.D., N. Reich, Ph.D., P. Small, M.D., L. Morel, Ph.D., M. McDuffie, M.D., and A.B. Peck, Ph.D. for their editorial reviews and intellectual support. These studies were supported by grants from the NIH (SAL, NIDDK KO1 DK02947 & MCS, AI PO1 AI42288), the Howard Hughes Research Institute (SAL), and the Children’s Miracle Network (SAL). The authors have no conflicting financial interests.
References
- 1.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–5. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Lehtonen A, Matikainen S, Miettinen M, Julkunen I. Granulocyte- macrophage colony-stimulating factor (GM-CSF)-induced STAT5 activation and target-gene expression during human monocyte/macrophage differentiation. J Leukoc Biol. 2002;71:511–9. [PubMed] [Google Scholar]
- 3.Biethahn S, Alves F, Wilde S, Hiddeman W, Spiekermann K. Expression of granulocyte colony-stimulating factor- and granulocyte- macrophage colony-stimulating factor-associated signal transduction proteins of the JAK/STAT pathway in normal granulopoiesis and in blast cells of acute myelogeneous leukemia. Exp Hematol. 1999;27:885–94. doi: 10.1016/s0301-472x(99)00017-x. [DOI] [PubMed] [Google Scholar]
- 4.Schindler C, Darnell JE., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–51. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 5.Yamaoka K, Otsuka T, Niiro H, Arinobu Y, Nihho Y, Hamasaki N, et al. Activation of STAT5 by lipopolysaccharide through granulocyte-macrophage colony-stimulating factor production in human monocytes. J Immunol. 1998;160:838–45. [PubMed] [Google Scholar]
- 6.Novak U, Mui A, Miyajima A, Paradiso L. Formation of STAT5-containing DNA binding complexes in response to colony-stimulating factor-1 and platelet-derived growth factor. J Biol Chem. 1996;271(31):18250–354. doi: 10.1074/jbc.271.31.18350. [DOI] [PubMed] [Google Scholar]
- 7.Bunting KD, Bradley HL, Hawley TS, Moriggl R, Sorrentino BP, Ihle JN. Reduced lympho-myeloid repopulating activity form adult bone marrow and fetal liver of mice lacking expression of STAT5. Blood. 2002;99(2):479–87. doi: 10.1182/blood.v99.2.479. [DOI] [PubMed] [Google Scholar]
- 8.Kieslinger M, Woldman I, Moriggl R, Hofmann J, Marine J-C, Ihle JN, et al. Antiapoptotic activity of Stat5 required during terminal stages of myeloid differentiation. Gene Dev. 1999;14:232–44. [PMC free article] [PubMed] [Google Scholar]
- 9.Valledor AF, Borras FE, Cullell-Young M, Celada A. Transcription factors that regulate monocyte/macrophage differentiation. J Leukoc Biol. 1998;63:405–17. doi: 10.1002/jlb.63.4.405. [DOI] [PubMed] [Google Scholar]
- 10.Al-Shami A, Mahanna W, Naccache PH. Granulocyte-macrophage colony-stimulating factor-activated signaling pathways in human neutrophils. J Biol Chem. 1998;273(2):1058–63. doi: 10.1074/jbc.273.2.1058. [DOI] [PubMed] [Google Scholar]
- 11.Azam M, Lee C, Strehlow I, Schindler C. Functionally distinct isoforms of STAT5 are generated by protein processing. Immunity. 1997;6:691–701. doi: 10.1016/s1074-7613(00)80445-8. [DOI] [PubMed] [Google Scholar]
- 12.Ilaria RL, Jr, Hawley RG, van Etten RA. Dominant negative mutants implicate STAT5 in myeloid cell proliferation and neutrophil differentiation. Blood. 1999;93(12):4154–66. [PubMed] [Google Scholar]
- 13.Piazza F, Vlens J, Lagasse E, Schindler C. Myeloid differentiation of FdCP1 cells is dependent on Stat5 processing. Blood. 2000;96(4):1358–65. [PubMed] [Google Scholar]
- 14.Copeland NG, Gilbert DJ, Schindler C, Zhong Z, Wen Z, Darnell JE, Jr, et al. Distribution of the mammalian Stat gene family in mouse chromosomes. Genomics. 1995;29:225–8. doi: 10.1006/geno.1995.1235. [DOI] [PubMed] [Google Scholar]
- 15.Socolovsky M, Fallon AE, Wang S, Brugnara C, Lodish HF. Fetal anemia and apoptosis of red cell progenitors in Stat5a−/−5b−/− mice: a direct role for Stat5 in Bcl-X(L) induction. Cell. 1999;98(2):181–91. doi: 10.1016/s0092-8674(00)81013-2. [DOI] [PubMed] [Google Scholar]
- 16.Itoh T, Liu R, Yokota T, Arai K-I, Watanabe S. Definition of the role of tyrosine residues of the common β subunit regulating signaling pathways of granulocyte-macrophage colony-stimulating factor receptor. Mol Cell Biol. 1998;18(2):742–52. doi: 10.1128/mcb.18.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee C, Piazza F, Brutsaert S, Valens J, Strehlow I, Jarosinski M, et al. Characterization of the Stat5 protease. J Biol Chem. 1999;274(38):26767–75. doi: 10.1074/jbc.274.38.26767. [DOI] [PubMed] [Google Scholar]
- 18.Rosen RL, Winestock KD, Chen G, Liu X, Hennighausen L, Finbloom DS. Granulocyte-macrophage colony-stimulating factor preferentially activates the 94 kDa STAT5A and an 80 kDa STAT5A isoform in human peripheral blood monocytes. Blood. 1996;88(4):1207–14. [PubMed] [Google Scholar]
- 19.Litherland SA, Xie T, Hutson AD, Whittaker DS, Schatz D, Hofig A, et al. Aberrant monocyte prostaglandin synthase 2 (PGS2) expression defines an antigen presenting cell defect and is a novel cellular marker for insulin dependent diabetes mellitus (IDDM) J Clin Invest. 1999;104:515–23. doi: 10.1172/JCI4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Litherland SA, She J-X, Schatz D, Fuller K, Hutson AD, Li Y, et al. Aberrant monocyte prostaglandin synthase 2 (PGS2) in type 1 diabetes before & after disease onset. Pediatr Diabetes. 2003;4:10–8. doi: 10.1034/j.1399-5448.2003.00042.x. [DOI] [PubMed] [Google Scholar]
- 21.Litherland SA, Xie TX, Grebe KM, Li Y, Moldawer LL, Clare-Salzler MJ. IL-10 resistant PGS2 expression in at-risk/type 1 diabetic human monocytes. J Autoimmun. 2004;22:227–33. doi: 10.1016/j.jaut.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, et al. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89(9):3148–54. [PubMed] [Google Scholar]
- 23.Davoodi-Semiromi A, Alorava ML, Kumar GP, Purohit S, Jha RK, She J-X. A mutant STAT5B with weaker DNA binding affinity defines a key defective pathway in nonobese diabetic mice. J Biol Chem. 2004;279(12):11533–61. doi: 10.1074/jbc.M312110200. [DOI] [PubMed] [Google Scholar]
- 24.Liang H, Venema VJ, Wang X, Ju H, Venema RC, Marrero MB. Regulation of angiotensin II-induced phosphorylation of STAT3 in vascular smooth muscle cells. J Biol Chem. 1999;274(28):19846–51. doi: 10.1074/jbc.274.28.19846. [DOI] [PubMed] [Google Scholar]
- 25.Aoki N, Matsuda T. A cytosolic protein-tyrosine phosphatase PTP1B specifically dephosphorylates and deactivates prolactin-activated STAT5a and STAT5b. J Biol Chem. 2000;275(50):39718–26. doi: 10.1074/jbc.M005615200. [DOI] [PubMed] [Google Scholar]
- 26.Jiao H, Yang W, Berrada K, Tabrizi M, Shultz L, Yi T. Macrophages from motheaten and viable motheaten mutant mice show increased proliferative responses to GM-CSF: detection of potential HCP substrates in GM-CSF signal transduction. Exp Hematol. 1997;25:592–600. [PubMed] [Google Scholar]
- 27.Chen HE, Chang S, Trub T, Neel BG. Regulation of colony-stimulating factor 1 receptor signaling by the SH2 domain-containing tyrosine phosphatase SHPTP1. Mol Cell Biol. 1996;16(77):3685–97. doi: 10.1128/mcb.16.7.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timms JF, Carlberg K, Gu H, Chen H, Kamatkar S, Nadler MJS, et al. Identification of major binding proteins and substrates for the SH2-containing protein tyrosine phosphatase SHP-1 in macrophages. Mol Cell Biol. 1998;18(7):3838–50. doi: 10.1128/mcb.18.7.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You M, Yu D-H, Feng G-S. Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Mol Cell Biol. 1999;19(3):2416–24. doi: 10.1128/mcb.19.3.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz J, Weissenbach M, Haan S, Heinrich PC, Schaper R. SOCS3 exerts its inhibitory function on interleukin-6 signal transduction through the SHP2 recruitment site of gp130. J Biol Chem. 2000;276(17):12848–56. doi: 10.1074/jbc.275.17.12848. [DOI] [PubMed] [Google Scholar]
- 31.Berchtold S, Volarevic S, Moriggl R, Mercep M, Groner B. Dominant negative variants of the SHP-2 tyrosine phosphatase inhibit prolactin activation of Jak2 (Janus kinase 2) and induction of Stat5 (signal transducer and activator of transcription 5)-dependent transcription. Mol Endocrinol. 1998;12:556–67. doi: 10.1210/mend.12.4.0086. [DOI] [PubMed] [Google Scholar]
- 32.Yu C-L, Jin Y-J, Burako SJ. Cytosolic tyrosine dephosphorylation of STAT5. J Biol Chem. 2000;275(1):599–604. doi: 10.1074/jbc.275.1.599. [DOI] [PubMed] [Google Scholar]
- 33.Chormarat P, Banchereau J, Davoust J, Palucka AK. IL-6 switches the differentiation of monocytes from dendritic cells to macrophages. Nat Immunol. 2000;1(6):510–3. doi: 10.1038/82763. [DOI] [PubMed] [Google Scholar]
- 34.Berlato C, Cassatella MA, Kinjyo I, Gatto L, Yoshimura A, Bazzoni F. Involvement of suppressor of cytokine signaling-3 as a mediator of the inhibitory effects of IL-10 on lipopolysaccharide- induced macrophage activation. J Immunol. 2002;168:6404–11. doi: 10.4049/jimmunol.168.12.6404. [DOI] [PubMed] [Google Scholar]
- 35.Wang D, Moriggl R, Starvopodis D, Carpino N, Marine J-C, Teglund S, et al. A small amphipathic α-helical region is required for transcriptional activities and proteasome-dependent turnover of the tyrosine-phosphorylated Stat5. EMBO J. 2000;19(3):392–9. doi: 10.1093/emboj/19.3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McBride KM, Reich NC. The ins and outs of STAT1 nuclear transport. Science STKE. 2003;195:1–9. doi: 10.1126/stke.2003.195.re13. re13, http://www.stke.org/cgi/content/full/sigtrans. [DOI] [PubMed]
- 37.Meyer J, Jucker M, Ostertag W, Stocking C. Carboxyl-truncated STAT5b is generated by a nucleus-associated serine protease in early hematopoietic progenitors. Blood. 1998;91(6):1901–8. [PubMed] [Google Scholar]
- 38.Ginger RS, Dalton EC, Ryves WJ, Fukuzawa M, Williams JG, Harwood AJ. Glycogen synthase kinase-3 enhances nuclear export of a dictyostelium STAT protein. EMBO J. 2000;19(20):5483–91. doi: 10.1093/emboj/19.20.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenhalgh CJ, Hilton DJ. Negative regulation of cytokine signaling. J Leukoc Biol. 2001;770:348–56. [PubMed] [Google Scholar]
- 40.Kotaja N, Karvonen U, Janne AO, Palvimo JJ. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol Cell Biol. 2002;22(14):5222–34. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serreze DV, Gaskins HR, Leiter EH. Defects in the differentiation and function of antigen presenting cells in the NOD/Lt Mice. J Immunol. 1993;150:2534–43. [PubMed] [Google Scholar]
- 42.Steptoe RJ, Ritchie JM, Harrison LC. Increased generation of dendritic cells from myeloid progenitors in autoimmune-prone nonobese diabetic mice. J Immunol. 2002;168:5032–41. doi: 10.4049/jimmunol.168.10.5032. [DOI] [PubMed] [Google Scholar]
- 43.Clare-Salzler MJ, Brooks J, Chai A, van Herle K, Anderson C. Prevention of diabetes in nonobese diabetic mice dendritic cell transfer. J Clin Invest. 1992;90(3):741–8. doi: 10.1172/JCI115946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clare-Salzler MJ. The immunopathogenic roles of antigen presenting cells in the NOD mouse. In: Leiter EH, Atkinson MA, editors. NOD mice and related strains: research applications in diabetes, AIDS, cancer and other diseases. Austin, TX: Landes Bioscience Publishers; 1998. pp. 101–20. [Google Scholar]