Summary
Objective
To provide a more complete picture of the effect of IL-1β on adult human articular chondrocyte gene expression, in contrast to the candidate gene approach.
Design
Chondrocytes from human knee cartilage were cultured in medium containing IL-1β. Changes in gene expression were analyzed by microarray and RT-PCR analysis. The ability of TGF-β1, FGF-18, and BMP-2 to reverse the effects of IL-1β was analyzed. Computational analysis of the promoter regions of differentially expressed genes for transcription factor binding motifs was performed.
Results
IL-1β-treated human chondrocytes showed significant increases in the expression of Granulocyte Stimulating Factor-3, Endothelial Leukocyte Adhesion Molecule 1 and Leukocyte Inhibitor Factor as well as for a large group of chemokines that include CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL8, CCL2, CCL3, CCL4, CCL5, CCL8, CCL20, CCL3L1, CX3CL1 and the cytokine IL-6. As expected, the mRNA for MMP-13 and BMP-2 also increased while mRNA for the matrix genes COL2A1 and aggrecan was down regulated. A subset of chemokines increased rapidly at very low levels of IL-1β. The phenotype induced by IL-1β was partially reversed by TGF-β1, but not by BMP-2. In the presence of IL-1β, FGF-18 increased expression of ADAMTS-4, aggrecan, BMP-2, COL2A1, CCL3, CCL4, CCL20, CXCL1, CXCL3, CXCL6, IL-1β, IL-6, and IL-8 and decreased ADAMTS-5, MMP-13, CCL2, CCL8. Computational analysis revealed a high likelihood that the most up-regulated chemokines are regulated by the transcription factors MEF-3, C/EBP and NF-κB.
Conclusion
IL-1β has a diverse effect on gene expression profile in human chondrocytes affecting matrix genes as well as chemokines and cytokines. TGF-β1 has the ability to antagonize some of the phenotype induced by IL-1β.
Keywords: Chondrocytes, IL-1β, Chemokines, TGF-β1, Transcription Factor, Binding Motifs
INTRODUCTION
IL-1β is an important cytokine in rheumatoid and osteoarthritic (OA) joint diseases. Generally, IL-1β is viewed as a catabolic factor for cartilage, inducing enzymes that degrade the extracellular matrix 1, 2 and reducing synthesis of the primary cartilage components type II collagen (COL2A1) and aggrecan 3. On the other hand, it has recently been shown that IL-1β can also induce the growth and morphogenetic factor BMP-2 4 potentially helping to balance its catabolic effects. In joint diseases, IL-1β is synthesized by synovial cells 5 and cartilage chondrocytes 6, therefore its effect on chondrocytes is highly relevant to the fate of cartilage. In order to obtain a global picture of IL-1β effects on human adult articular chondrocytes, we analyzed the changes in gene expression induced by IL-1β by gene array analysis. A dramatic response was observed in a specific set of chemokine genes.
Chemokines are potent mediators of inflammation and are known to be important in inflammatory diseases such as rheumatoid arthritis, inflammatory bowel disease, multiple sclerosis and transplant rejection 7. Certain chemokines mediate infiltration of leukocytes in synovial tissue and fluid 8. Initially discovered as co-receptors for HIV entry into lymphocytes, they are now known to be involved in chemoattraction, cell adhesion and migration. Extracellular gradients of chemokines are established by binding to glycosaminoglycan chains in the ECM 9 and at the cell surface chemokines can modulate integrin integrity 10. The chemokine CXCL12/SDF-1 increases MMP-3 activity 11.
There exist over 50 chemokine ligands and 18 chemokine receptors 12. The largest families of chemokines have similar protein structure being 8–10 kDa with conserved cysteine residues either adjacent (CC) or separated by 1 amino acid (CXC) 13. Very little is known about their regulation at the gene transcription or protein levels, however specific chemokines have been shown to be up-regulated by NF-κB and factors that enhance NF-κB 14. As IL-1β stimulates anabolic as well as catabolic events, we determined whether treatment with anabolic agents FGF-18, BMP-2 or TGF-β1 could reverse aspects of the IL-1β-induced chemokine phenotype.
METHODS
Materials
PRONASE, Streptomyces griseus Protease, was from CALBIOCHEM (La Jolla, CA). Collagenase P was from Roche (Indianapolis, IN). Recombinant Human IL-1β, TGF-β1 and BMP-2 were from R&D Systems (Minneapolis, MN). Recombinant Human Fibroblast Growth Factor (FGF-18) was from Leinco Technologies (St. Louis, MO). TRIZOL reagent, amplification grade DNase I, SuperScript II Reverse Transcriptase and Platinum Taq DNA Polymerase were from Invitrogen (Carlsbad, CA). RNeasy Mini kit was from Qiagen (Valencia, CA).
Cell and Tissue Culture
Cartilage was obtained with approval of the Washington University Human Studies Review Board and permission of the patient. Normal chondrocytes were obtained from normal articular knee cartilage from tissue donors (N = 4) with above the knee amputations due to chondrosarcoma or traumatic injury or from autopsy. Chondrocytes were also obtained from osteoarthritic (OA) cartilage from donors (N = 11) undergoing total joint replacement surgery. For the latter, chondrocytes from macroscopically normal looking cartilage were used; patients were of both sexes and greater than 60 years of age. Chondrocytes were isolated following previously published procedures 4 and plated at a density of 2.5 × 105 cells/cm2 in DMEM/F12 media plus 10% FBS, 50 μg/ml ascorbate and antibiotics (50 U/ml penicillin and 50 μg/ml streptomycin). Cells were allowed to recover for 24 h and cytokines or growth factors (IL-1β/TGF- β1/FGF-18/BMP-2) were added at the concentrations and times indicated. To study the response of cartilage explants to IL-1β, explants from a total joint replacement surgery were used. Cartilage was cultured with and without IL-1β prior to isolation of RNA directly from the tissue. IL-1β was reconstituted in sterile phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin; FGF-18 was reconstituted in 5 mM Tris, pH 8.0; BMP-2 and TGF-β1 were reconstituted in 4 mM HCl containing 0.1% bovine serum albumin. The time course, concentration gradient and effects of TGF-β1, FGF-18 and BMP-2 were repeated on a minimum of three biological replicates (one normal and two from joint replacement surgery). All experiments were repeated three times with cells from the same patient. Each figure shows data from a single donor. At no point were cells pooled from different donors. The experiments shown in the figures are representative of all of the data.
Total RNA Isolation
Total RNA was isolated from primary chondrocytes or cartilage explants by homogenizing these directly into TRIZOL reagent (Invitrogen) and following the protocol recommended by the manufacturer. For microarray analysis, RNA was further purified by Qiagen RNeasy Minikit. For RT-PCR analysis the isolated RNA was treated by DNase I to remove traces of contaminating DNA.
Microarray Analysis
For microarray analysis, first strand cDNA was generated from RNA and labeled with the Cy3 and Cy5 fluorescent dyes using the 3DNA Array 900 kit (Genisphere), without any prior amplification of the RNA. The hybridization was performed on the Human Operon V3.0 Oligo Expression Array (Whitehead Institute, Cambridge) which consists of 34,580 human 70-mer probes representing 24,650 genes and 37,123 gene transcripts. The analysis was repeated with the Cy3 and Cy5 dyes exchanged between the control and experimental RNAs. The arrays were scanned on a Perkin Elmer ScanArray ExpressHT scanner to detect Cy3/Cy5 fluorescence. Analysis of images was performed by ScanArray v.3.0 (Perkin Elmer). To generate a stringent list of candidates demonstrating differential expression only those candidates that scored a local signal-to-background differential intensity of ≥2 in two biological and two technical replicates (due to reversal of dyes) and had a p value of less than 0.05 were considered.
Semi-Quantitative RT-PCR Analysis
The RT-PCR reaction was performed using total RNA isolated from primary chondrocytes or fresh cartilage tissue by SuperScript II Reverse Transcriptase as recommended by Invitrogen. Primers used for PCR were optimized for each gene (Table 1). PCR was performed in a total volume of 20 μl by Platinum Taq DNA Polymerase as recommended by Invitrogen. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a reference for gel loading. cDNA samples from different time points and treatments for each gene were simultaneously amplified in PCR and repeated at least thrice to avoid potential variation between experiments. Results were analyzed using NIH IMAGE J software, tested using standards with known concentrations, to determine the pixel intensity for each concentration. The amplification cycle number used to yield product in the linear range was determined for each set of primers. A standard curve was established for each set of PCR reactions.
Table 1.
Primers for PCR Analysis
| IL-6: F (5′-GCCACTCACCTCTTCAGAA-3′), R (5′-GTACTCATCTGCACAGCTCT-3′); |
| CXCL8 (IL-8): F (5′-GACTTCCAAGCTGGCCGT-3′), R (5′-GAATTCTCAGCCCTCT TCA-3′); |
| Nitric Oxide Synthase IIB (NOS2B): F (5′-GAGTGCAGGGAGGCCGCAT-3′), R (5′-CTTGGGCTTGCCAGGCAG-3′); |
| NF kappa B inhibitor alpha (NFKBIA): F (5′-GCTGAAGAAGGAGCGGCTA-3′), R (5′-CTGGCTGGTTGGTGATCA-3′); |
| CCL2 (MCP-1): F (5′-GCATGAAAGTCTCTGCCG-3′), R (5′-GAGTGTTCAAGTCTTCGGA-3′); |
| CCL3 (MIP-1α): F (5′-GCTGCCCTTGCTGTCCTCCTC-3′), R (5′-GGTCAGCACAGACCTGCCGG-3′); |
| CCL4 (MIP-1β): F (5′-GCGCTCTCAGCACCAATGGGC-3′), R (5′-GCATCCGGGCT CAGGTGACC-3′); |
| CCL5 (RANTES): F (5′-GAAGGTCTCCGCGGCAGCC-3′), R (5′-CTGGGCCCTTCAAGGAGCGG-3′); |
| CCL8 (MCP-2): F (5′-GCCTGCTGCTCATGGCAGCC-3′), R (5′-GCACAGACCTCCTTGCCCCG-3′); |
| CCL20 (MIP-3α): F (5′-GCTGTACCAAGAGTTTGCT-3′), R (5′GGCTATGTCCAATTCCATT-3′); |
| CCL3L1 (LD78β): F (5′-GTCTCCACTGCTGCCCTTGCC-3′), R (5′-CTGAGGTCGCTGGGCCTCGA-3′); |
| CXCL1 (Gro-α): F (5′-GCTCCTGCGAGTGGCACTGC-3′), R (5′-GGGCCTCCTTCAGGAACAGCC-3′); |
| CXCL2 (Gro-β): F (5′-CACAGCCGCTCGAACCGCCT-3′), R (5′-GCGCAAGCCAGGTGGCCTCT-3′); |
| CXCL3 (Gro-γ): F (5′-GCAGGAGCGTCCGTGGTCAC-3′), R (5′-GCTCTGGTAAGG GCAGGGACC-3′); |
| CXCL5 (ENA-78): F (5′-GAGCCTCCTGTCCAGCCGC-3′), R (5′-CACCTTGGAGCACTGTGGGCC-3′); |
| CXCL6 (GCP-2): F (5′-GCGAACCCTCTCTTGACCA-3′), R (5′-CTTGTTTCCACTGTCCAA-3′); |
| CX3CL1 (Fractalkine): F (5′-GCTTGGCCACCTTCTGCCAT-3′), R (5′-GTCGGCTCCAGGCTACTGCT-3′); |
| BMP-2: F (5′-GGACACGCCAACCATGGATT-3′), R (5′-CAACCATGTCCTGATAGTTCT-3′); |
| ADAMTS-4: F (5′-CAGATGTGGTACTGCCTGGG-3′), R (5′-GCCGCCGAAGGATCTCCAGA-3′); |
| IL-1β; COL2A1; MMP-13; ADAMTS-5; Aggrecan and TNF-α 58 GAPDH 59 |
Computational Analysis
Potential regulatory DNA surrounding the chemokine genes was analyzed by the Promoter Analysis Pipeline 15, 16. Briefly, the Promoter Analysis Pipeline has two components: a set of algorithms to generate the results of a genome-wide promoter analysis and a user interface to query and process the stored data according to specific parameters. Promoters (10 kb upstream and 5 kb downstream of the transcription start site) were acquired from three species human, mouse and rat, and repetitive elements in the promoters were masked. Promoters of orthologous genes were aligned and transcription factor binding sites were identified using the TRANSFAC 7.2 database, a curated database of transcription factor profiles 17. Probability scores for each promoter and each transcription factor were calculated and a distribution of probability scores was generated for each transcription factor. R-scores were then computed using these distributions 15. This system was used to predict the transcription factors that are most likely to bind to and regulate the set of genes. For each transcription factor binding site motif (identified by the TRANSFAC accession number) and each promoter in the genome, the probability score of the transcription factor binding to the promoter was computed by summing the exponential of the score of each individual site predicted in the promoter on either strand 18. This score is set to a minimum value of 1 for a promoter with no sites exceeding the cutoff. The rank of this score is converted to the R-score which is related to the fraction of promoters with a higher rank, by R-score = lnN-ln(rank). Promoters ranked in the top half have R-score > or equal to ln2, those in the top 10% have R-score > ln10, those in the top 1% have R-score > ln100. The R-score for a set of n promoters, the average R-score, <R-score>, is calculated by <R-score> = (1/n)Σ R-score.
RESULTS
Microarray Analysis
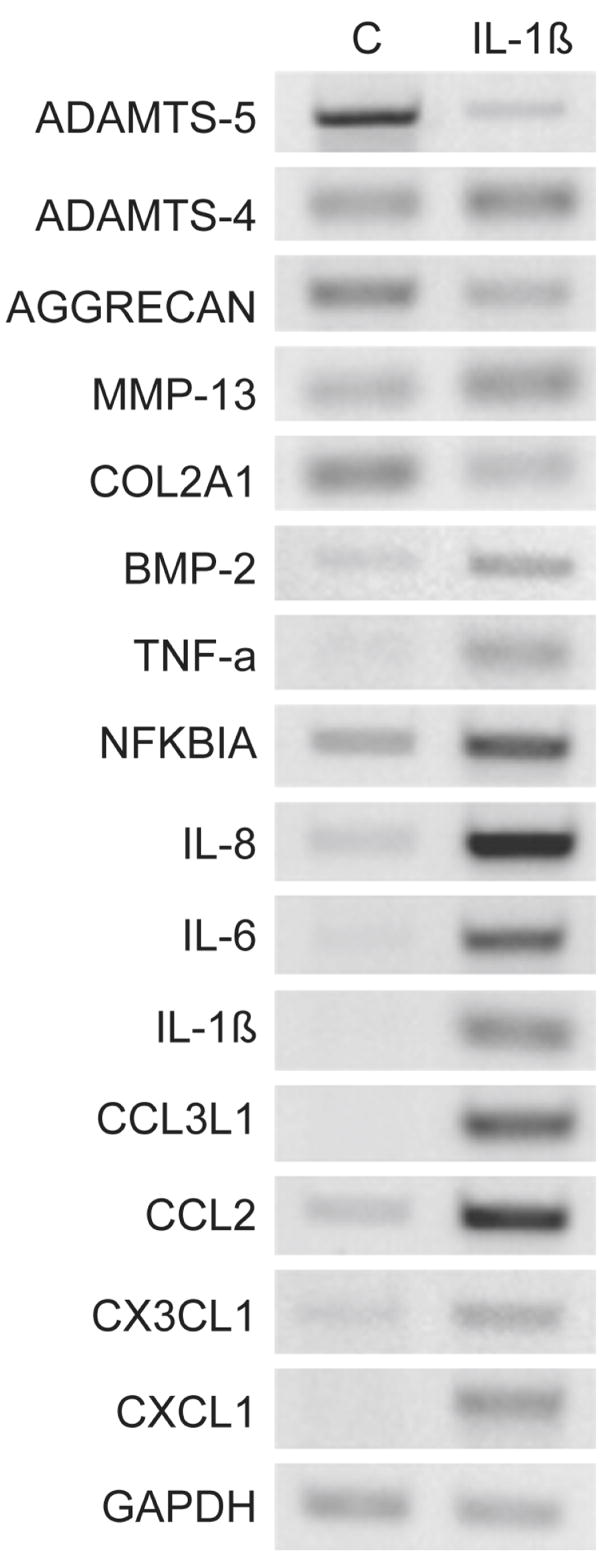
Changes in mRNA levels induced by IL-1β were assessed by microarray analysis (Table 2), several of which were confirmed by PCR analysis of mRNA (Fig 1. and Table 2). Themost highly regulated genes are listed in Table 2. Among the most increased were CSF-3 (Granulocyte colony stimulating factor-3), SELE (Endothelial leukocyte adhesion molecule 1/E-selectin) and LIF (Leukocyte Inhibitor Factor), 76 fold, 65 fold and 49 fold, respectively. As a group of related genes, the chemokines were the most highly up regulated: CXCL1, CXCL2, CXCL3, CXCL6, CXCL8, CCL3, CCL3L1, CCL4 and CCL20. Genes that have been previously reported to respond to IL-1β and thus can be considered as controls were regulated as expected: MMP-13 was up-regulated 6 fold 19 and type II collagen (COL2A1) was down-regulated by 60% 3. The nitric oxide synthases, NOS2B (nitric oxide synthase IIB), and NOS2A (inducible NOS, type IIA) were increased 23 fold and 20 fold, respectively. NFKBIA (NFκB inhibitor alpha) was increased 15 fold. The cytokines IL-8 (also known as the chemokine CXCL8), IL-6 and IL-1β were also increased over 20 fold. Interestingly, the enzyme, ADAMTS-5 (aggrecanase-2) was present in the cultured chondrocytes at high levels and reproducibly down-regulated by exposure to IL-1β (Table 2 and Fig. 1, 2). In some cases of verification of the gene array results by PCR, increases in mRNA level were difficult to quantify as the normal chondrocyte control used for the gene array contains so little chemokine mRNA. However, the increases are in the order of magnitude revealed by the gene array data.
Table 2.
Microarray and RT-PCR analysis.
| Gene Transcripts Highest Upregulated (20–76 fold) | Microarray (relative intensity) | RT-PCR (pixel intensity) | ||||
|---|---|---|---|---|---|---|
| control | IL-1β 10ng/ml | Fold change | control | IL-1β 10ng/ml | Fold Change | |
| CSF3 (Granulocyte colony stimulating factor 3) | 45.99 | 2530.75 | 76 | ND | ||
| SELE (Endothelial leukocyte adhesion molecule1/E-selectin) | 80.88 | 3683.50 | 65 | ND | ||
| LIF (Leukemia inhibitory factor) | 332.04 | 16253.50 | 49 | ND | ||
| ADORA2A (Adenosine A2A receptor) | 68.51 | 1699.00 | 31 | ND | ||
| PIM2 (Serine threonine protein kinase) | 251.93 | 6564.50 | 26 | ND | ||
| PTGS2 (Cox-2) | 1042.71 | 24325.25 | 23 | ND | ||
| IL-6 | 2508.05 | 65050.50 | 26 | 0 | 8923 | ≫ |
| NOS2B (Nitric Oxide Synthase IIB) | 491.39 | 11247.50 | 23 | 0 | 8576 | ≫ |
| IL-1β | 219.19 | 4123.00 | 20 | 0 | 8232 | ≫ |
| RAC1 (RAS-like protein TC25) | 574.16 | 12403.50 | 20 | ND | ||
| NOS2A (Inducible NOS, type II) | 324.74 | 6186.00 | 20 | ND | ||
| Chemokine Transcripts Group 1 (25–75 fold) | ||||||
| CXCL2 (Gro-β) | 434.41 | 32632.25 | 75 | ND | ||
| CXCL8 (IL8) | 1159.28 | 65126.50 | 57 | 0 | 8607 | ≫ |
| CXCL1 (Gro-α) | 1648.54 | 65214.75 | 40 | 0 | 7337 | ≫ |
| CXCL3 (Gro-γ) | 485.25 | 18605.75 | 40 | ND | ||
| CCL4 (MIP-1β) | 221.41 | 6616.00 | 36 | ND | ||
| CCL20 (MIP-3α) | 1902.24 | 65190.25 | 35 | 0 | 8926 | ≫ |
| CCL3L1 (LD78β) | 346.80 | 12004.50 | 34 | ND | ||
| CXCL6 (GCP-2) | 2040.93 | 56281.75 | 28 | 0 | 8532 | ≫ |
| CCL3 (MIP-1α) | 343.49 | 8495.75 | 25 | ND | ||
| Group 2 (3–12 fold) | ||||||
| CCL5 (RANTES) | 149.47 | 1686.00 | 12 | ND | ||
| CXCL5 (ENA-78) | 663.20 | 4155.00 | 6 | ND | ||
| CX3CL1 (Fractalkine) | 237.90 | 1414.25 | 6 | 0 | 8080 | ≫ |
| CCL2 (MCP-1) | 3151.44 | 13374.75 | 4 | 2345 | 8996 | 3.84 |
| CCL8 (MCP-2) | 47.77 | 181.00 | 3 | ND | ||
| Other Transcripts of Interest | ||||||
| NOS2C | 825.84 | 12485.75 | 15 | ND | ||
| IL-24 | 79.43 | 906.75 | 15 | ND | ||
| NFKBIA (NF kappa B inhibitor alpha) | 1424.45 | 20898.5 | 15 | 0 | 8834 | ≫ |
| MMP-13 | 6784.55 | 42234.75 | 6 | 2097 | 8232 | 3.93 |
| COL2A1 | 11152.09 | 4873.00 | 0.44 | 8524 | 4341 | 0.51 |
| ADAMTS-5 | 1566.57 | 205.75 | 0.13 | 8473 | 4211 | 0.50 |
| Aggrecan | NDT | NDT | - | 8718 | 4771 | 0.55 |
| BMP-2 | NDT | NDT | - | 6742 | 8482 | 1.26 |
Gene changes in mRNA levels induced by IL-1β were assessed by microarray analysis, several of which were confirmed by RT-PCR analysis. NDT: not detected. ND: not determined. ≫: IL-1β-induced gene change is radically higher than in control group without IL-1β treatment.
Figure 1. RT-PCR validation of microarray.
Chondrocytes from normal human knee cartilage were treated with 10 ng/ml of IL-1β for 24 h. RNA extracted from these cells was used for microarray analysis as well for RT-PCR validation. Control cells (C) received vehicle only. Total RNA samples from treated and untreated cells were analyzed by RT-PCR with the primers indicated in Table 1. GAPDH was used as reference for gel loading. Quantification of individual bands was done by NIH IMAGE J software analysis with PCR in the linear range of product (Table 2).
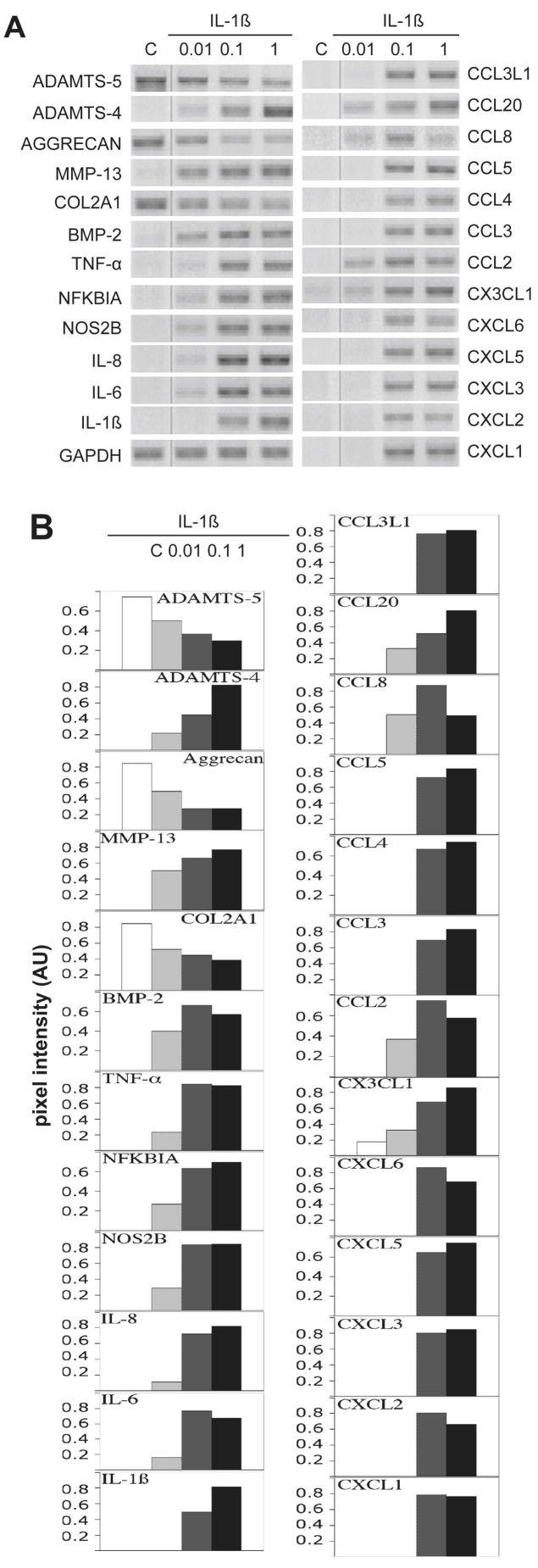
Figure 2. Dose response of chemokines and matrix molecules to IL-1β.
(A) Chondrocytes from normal human knee cartilage were treated with 0.01, 0.1 and 1 ng/ml of IL-1β for 24 h. Control cells (C) received vehicle only. RT-PCR was done with indicated primers and GAPDH was used as reference. (B) Quantitation of RT-PCR analysis with gene expression normalized to GAPDH expression using NIH IMAGE J software. Pixel intensity is given in arbitrary units (AU).
Dose Response of IL-1β-Induced Changes in Phenotype
As the response to exposure to 10 ng/ml was reproducibly strong, we determined the effect of physiological concentrations of IL-1β on the increase in chemokine mRNA by chondrocytes. Normal chondrocytes (from individuals with no history of OA) were exposed to IL-1β at 0.01, 0.1 and 1 ng/ml. At even the lowest concentration of 0.01 ng/ml of IL-1β, many of the chemokine mRNAs were dramatically increased (Fig. 2A). The down-regulation of COL2A1 gene showed a dose response beginning at 0.01ng/ml. MMP-13 was strongly induced by 0.01 ng/ml while ADAMTS-4 was weakly induced at 0.01 ng/ml, but strongly induced by 0.1 ng/ml IL-1β. In summary, the genes induced by 0.01 ng/ml of IL-1β were ADAMTS-4, MMP13, NFKBIA, BMP-2, NOS2B, IL8, IL6, CCL2, CCL8, CCL20 and CX3CL1. The quantification of these results is shown in Fig. 2B. The response of chondrocytes to IL-1β was found to be very reproducible between donors and experiments. Note that some of the chemokine PCR products seem to be at maximum levels with 0.1 ng/ml, however, these were tested at lower rounds of amplification and found to be indeed at maximum levels as shown.
Time Course of IL-1β-Induced Changes in Phenotype
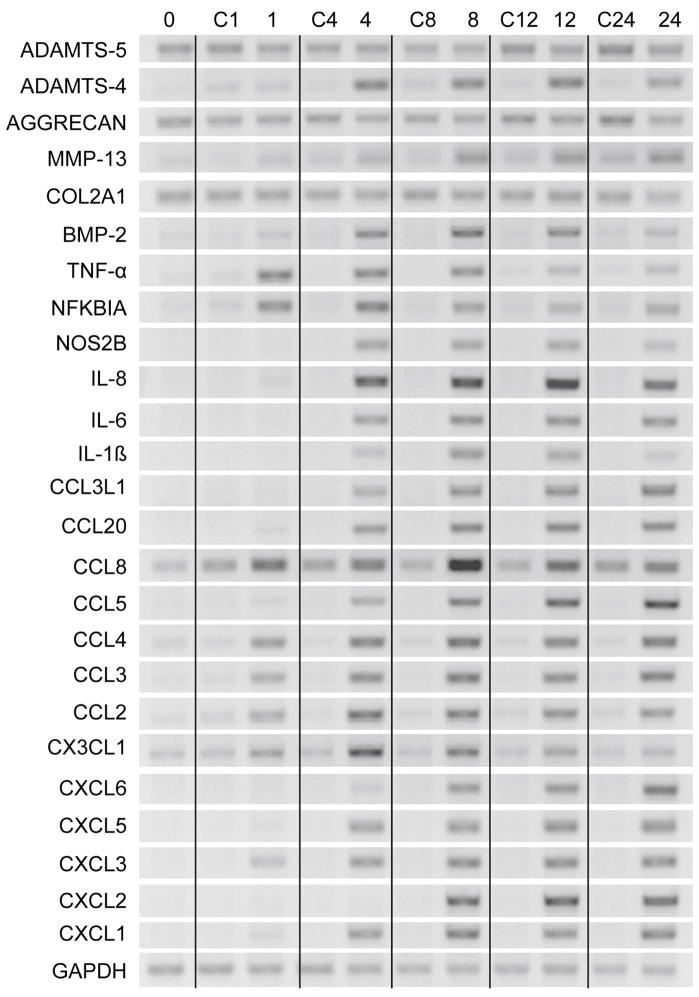
In order to begin to ascertain which genes are coordinately regulated by IL-1β, RNA was isolated at 1, 4, 8, 12 and 24 h after treatment. Three patterns of regulation were observed (Fig. 3). The first pattern was molecules up regulated by 1 h: TNF-α, NFKBIA, CCL2, CCL3, CCL4, CCL8, CX3CL1 and CXCL3. Of these genes up regulated at 1 h, TNF-α and NFKBIA decreased throughout the remaining time period. CCL4, CCL3, CCL2 and CXCL3 remained high throughout the time period. At 4 h, ADAMTS-4, MMP-13, BMP-2, NOS2B, IL-6, CXCL8/IL-8, IL-1β, CCL3L1, CCL20, CCL5, CXCL5 and CXCL1 were up-regulated. ADAMTS-4, BMP-2, IL-1β and NOS2B remained up-regulated at 8 and 12 h, but were reduced by 24 h. At 8 h, CXCL6 and CXCL2 appeared and remained high even at 24 h. The down-regulation of the matrix molecules, COL2A1 and aggrecan, occurred at 24 h. Thus, there appear to be a number of signal transduction pathways leading to the phenotypic changes induced by IL-1β. A consistent observation is that up-regulation of cytokines, chemokines and degradative enzymes appear to be more rapid than the down regulation of matrix gene expression. In addition to providing insight on the different pathways of regulation, these results indicate that it is very important to consider the time point at which effects of IL-1β are analyzed.
Figure 3. Time course of response of chemokines and matrix molecules to IL-1β.
Chondrocytes from preserved areas of cartilage from knees of OA patients were treated with 0.1 ng/ml of IL-1β for 0, 1, 4, 8, 12, and 24 h. Control cells (C) received vehicle only. RT-PCR was done with indicated primers with GAPDH used as reference.
Reversal of IL-1β-Induced Phenotype by BMP-2 and TGF-β1
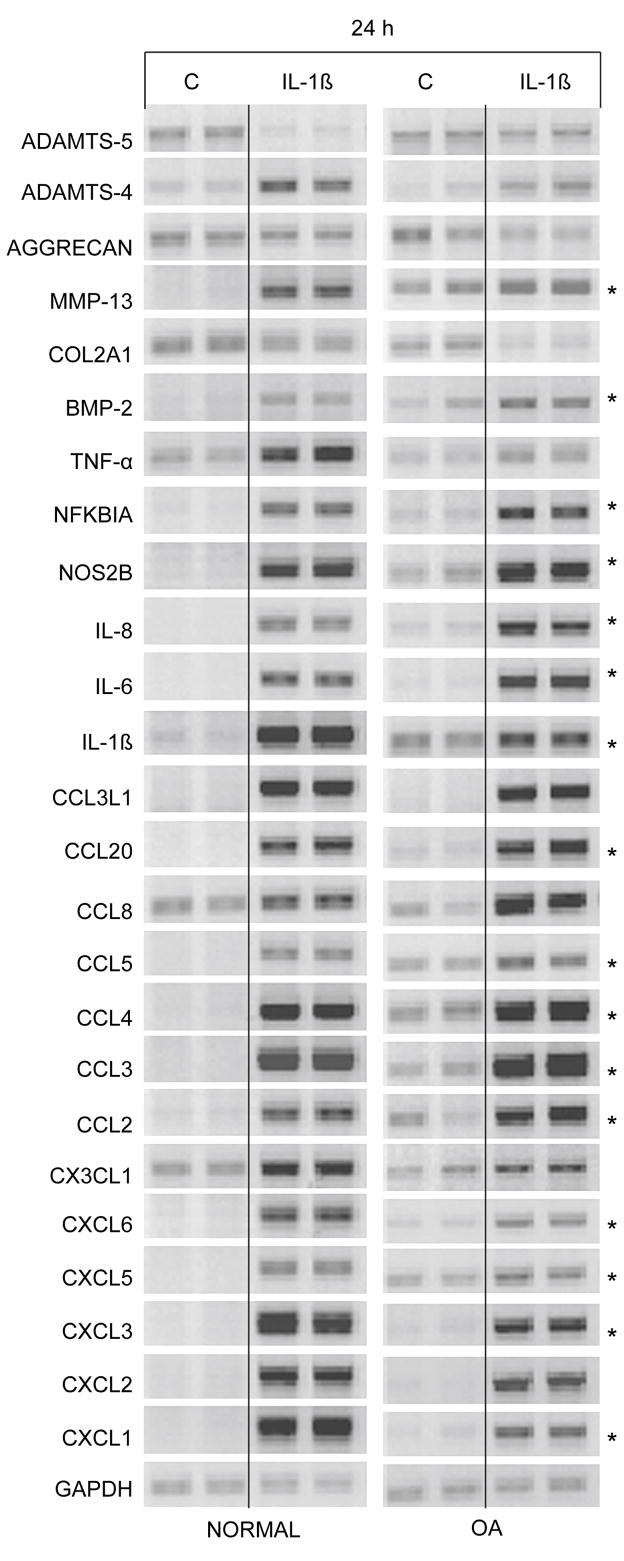
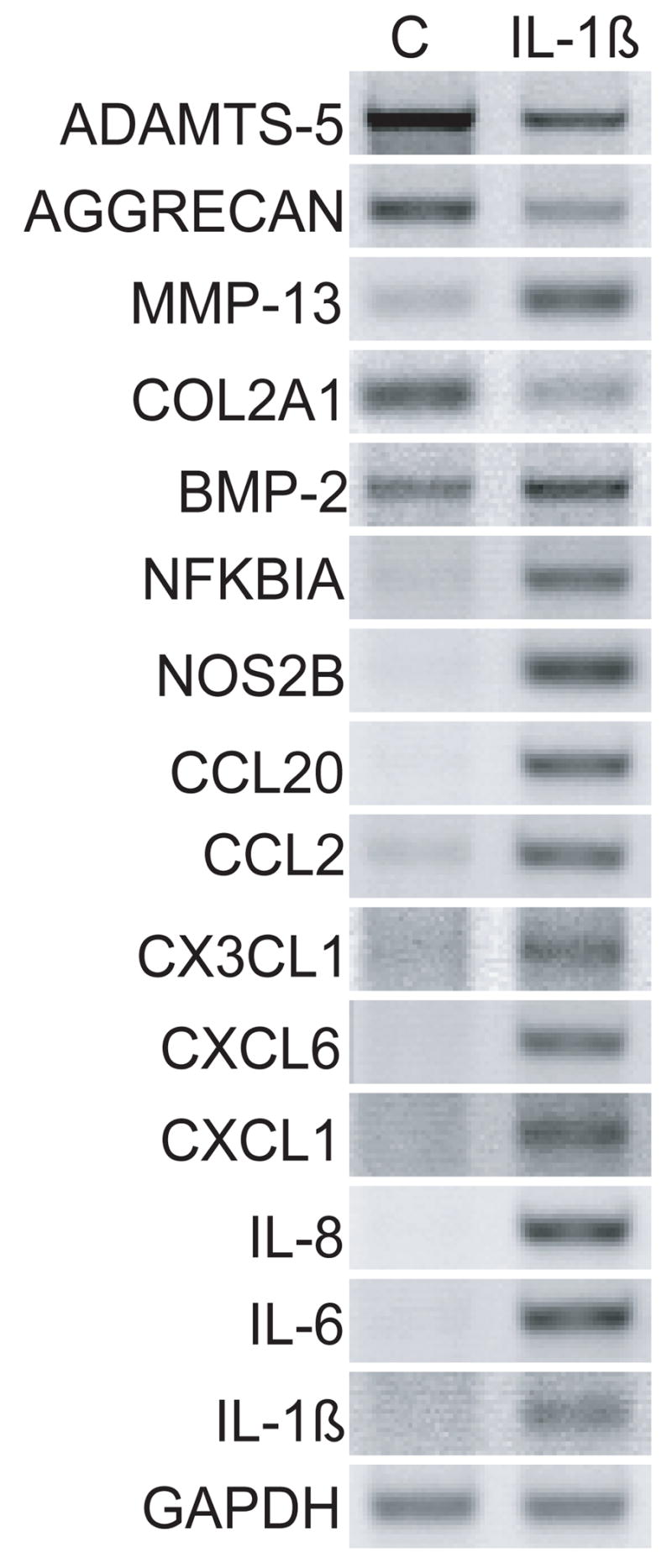
We tested the potential for these growth and differentiation factors to reverse the IL-1β-induced increase in chemokine expression (Fig. 4). In these set of experiments, chondrocytes were cultured with IL-1β for 24 h, the medium changed and the growth factors TGF-β1 and BMP-2 were added to cell cultures for an additional 48 h. 0.1 ng/ml IL-1β was used as it is sufficient to easily see the effects of the cytokine on gene expression. Chondrocytes from normal cartilage and cartilage removed from patients undergoing total joint replacement for OA were used. In cells from both normal cartilage and OA cartilage, all genes were regulated by IL-1β as predicted from the gene array results. Interestingly, cells from OA cartilage without IL-1β treatment demonstrated changes in gene expression very similar to normal chondrocytes exposed to lower concentration (0.1 ng/ml) of IL-1β (Fig. 4, denoted by asterisk). Many of these genes are also up-regulated at the lowest concentration (0.01 ng/ml) of IL-1β (Fig. 2A) in normal chondrocytes. This suggests that our in vitro experiments are duplicating a potential in vivo affect taking place during OA. Under the reversal conditions used in this set of experiments, TGF-β1 was able to reduce the up-regulatory effects of IL-1β on CCL2, CCL4, CCL8, CCL20, and MMP-13 and reversed the down regulation of COL2A1 (Fig. 5). In some cases such as that seen with CXCL1, CXCL3, and CCL3L1 the reversal was more pronounced with chondrocytes from normal patients and in case of CCL3, CXCL2, and IL-1β the reversal was more pronounced with chondrocytes from OA patients. BMP-2 somewhat reversed the down regulation of COL2A1, but had little effect on chemokine levels (data not shown). Surprisingly, while TGF-β1 was able to reverse much of the IL-1β-induced phenotype, TGF-β1 also induced a dramatic increase in ADAMTS-4 in both normal and OA cartilage.
Figure 4. Response of normal and OA chondrocytes to IL-1β.
Chondrocytes from normal human knee cartilage and preserved areas of OA knee cartilage were exposed to 0.1 ng/ml of IL-1β for 24 h. Control cells (C) received vehicle alone. Genes which showed an increase in expression from OA chondrocytes without addition of IL-1β are denoted by *.
Figure 5. Reversal of IL-1β-induced phenotype by TGF-β1.
Chondrocytes from normal knee cartilage and preserved areas of OA knee cartilage were exposed to 0.1 ng/ml of IL-1β for 24 h. Control cells (C) received vehicle alone. After 24 h, media containing IL-1β was removed and fresh media containing 10 ng/ml of TGF-β1 was added for 48 h. RNA samples were analyzed by RT-PCR with indicated primers. GAPDH was used as reference for gel loading. Lanes denoted as (IL-1β) indicate that cells were treated with IL-1β for 24 h and were not treated with TGF-β1; lanes denoted as TGF-β1 (IL-1β) indicate cells received TGF-β1 after removal of IL-1β; ^ indicates that the bands were too low to quantify; ~ indicates no change.
FGF-18 Effect on IL-1β-Induced Phenotype
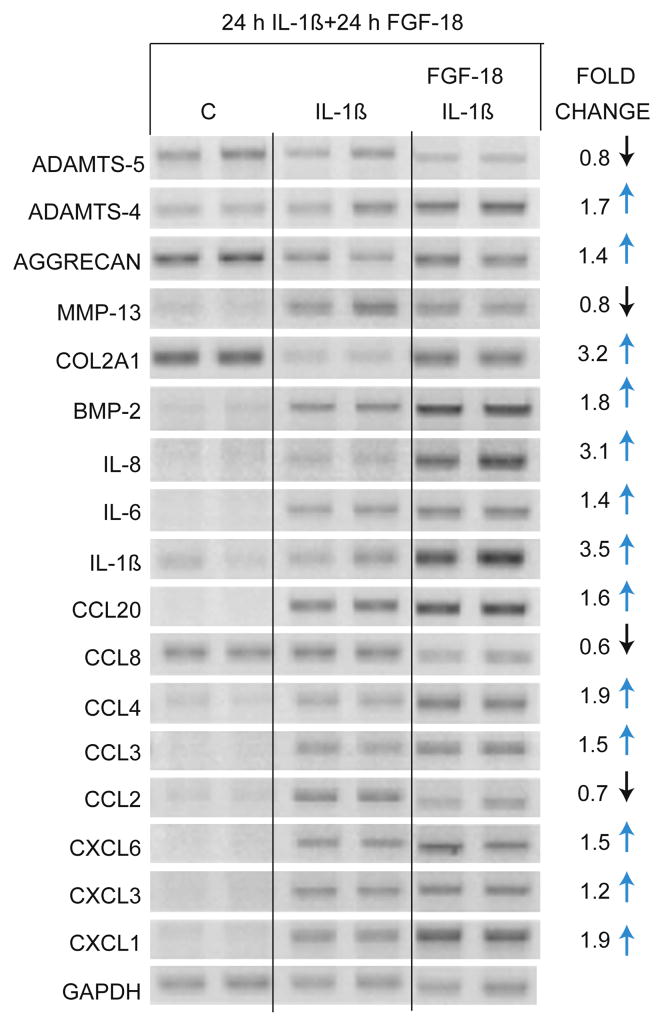
In an independent set of experiments, we asked whether there would be more effect of growth factors when the lowest concentration of IL-1β was used (0.01 ng/ml). In these experiments, IL-1β remained in the culture and TGF-β1, BMP-2 and FGF-18 were added after 24 h. Under these conditions, the BMP-2 and TGF-β1 demonstrated effects similar to the previous set of experiments, where, in general, TGF-β1 was able to reverse much of the IL-1β-induced change in phenotype, but BMP-2 did not (data not shown). However, FGF-18 (200 ng/ml) showed a novel pattern. FGF-18 increased ADAMTS-4, COL2A1, aggrecan and BMP-2, but also increased IL-8/CXCL8, IL-6, IL-1β, CCL3, CCL4, CCL20, CXCL3, CXCL6 and CXCL1 expression (Fig. 6). Increasing the concentration to 400 ng/ml of FGF-18 did not demonstrate any additional affect (data not shown). ADAMTS-5 and MMP-13 expressions were reduced slightly by FGF-18 as well as CCL2 and CCL8.
Figure 6. Effect of FGF-18 on IL-1β-induced phenotype.
Normal and OA chondrocytes were treated with 0.01 ng/ml of IL-1β for 24 h. At 24 h, 200 ng/ml of FGF-18 was added and cultured for another 24 h. Control cells (C) received vehicle alone. RNA samples were analyzed by RT-PCR with indicated primers. GAPDH was used as reference for gel loading.
Effects of IL-1β on Cartilage
Although cell culture experiments can provide valuable information on the potential for cells to respond in a particular manner, the isolation of chondrocytes from the matrix, as well as the stimulation of cells by culture conditions may not reflect the response of cells when they are resident in the extensive extracellular matrix of cartilage. In order to test whether these cell culture experiments simulate in vivo effects, IL-1β was cultured with freshly isolated cartilage explants (Fig. 7). RNA was isolated directly from the cartilage without prior isolation of cells. Genes representing the different classes of changes in gene expression were tested and all demonstrated the same results as the cell culture experiments. For example, ADAMTS-5, COL2A1 and aggrecan were reduced; ADAMTS-4, MMP-13, BMP-2, TNF-α, NFKBIA, IL8, IL6, IL-1β, CCL3L1, CCL2, CX3CL1 and CXCL1 were increased.
Figure 7. Effect of IL-1β on cartilage explants in culture.
Cartilage explants from the preserved areas of knees of OA patients were treated with 10 ng/ml of IL-1β for 24 h. Control tissue (C) received vehicle alone. RNA samples were analyzed by RT-PCR with indicated primers. GAPDH was used as reference for gel loading.
Computational Analysis to Predict Regulatory Domains
Genes that are transcriptionally co-expressed may contain common regulatory motifs in their DNA flanking domains. To begin to analyze the regulatory mechanism of the chemokine genes, the up-regulated chemokines (Table 2) were subdivided into two categories: Group 1 mRNAs were increased 25–75 fold when exposed to 10 ng/ml IL-1β and consisted of CCL3, CCL4, CCL20, CXCL1, CXCL2, CXCL3, CXCL6, CXCL8, CCL3L1, and the cytokine IL-6. Group 2 mRNAs were increased 3–12 fold and consisted of CCL2, CCL5, CCL8, CXCL5, and CX3CL1. The promoters of all chemokine genes identified were either analyzed as a single group (Table 3), or those belonging to Group 1 were analyzed separately (Table 4). We have previously used this methodology to analyze transcription factor binding motifs in a group of cartilage genes 16. The R-score indicates the probability that the transcription factor corresponding to this motif will bind to the promoter of these genes; the higher the R-score, the more likely it is to bind [> ln10 (ln10 = 2.3) is in the top 10%]. Although the binding must be verified experimentally, R-scores over two have been demonstrated to have a high likelihood of functional significance 15. Overall, many transcription factor binding motifs known to be involved in expression of pro-inflammatory cytokine-induced genes were identified: NF-κB, AML1, MEF-3, IRF-7, C/EBPβ, AP-1, ORF and TCF11 20–23. However, when only the Group 1 genes were analyzed the overall R-scores of the transcription factor binding motifs increased indicating more similarity in gene regulatory domains (Table 4). The predominant transcription factor binding motifs identified in the Group 1 genes were MEF-3, C/EBPβ, IRF-7, Pax-4, AML-1 and those related to NF-κB, c-Rel, and p65.
Table 3.
Transcription Factor Binding Motif Prevalence in Genes of Groups 1 and 2
| TRANSFAC Motif Accession Number | Transcription Factor | R-score |
|---|---|---|
| M00774 | NF-κB | 2.1467 |
| M00751 | AML1 | 2.0688 |
| M00271 | AML-1a | 2.0656 |
| Generalized motif (Gary Stormo, personal communication) | NF-κB | 2.0468 |
| M00319 | MEF-3 | 1.906 |
| M00453 | IRF-7 | 1.8815 |
| M00109 | C/EBPβ | 1.8484 |
| M00517 | AP-1 | 1.8354 |
| M00772 | ORF | 1.7779 |
| M0089 | TCF11-Maf | 1.7505 |
Table 4.
Transcription Factor Binding Motif Prevalence in Group 1 Chemokine Genes
| TRANSFAC Motif Accession Number | Transcription Factor | R-score |
|---|---|---|
| M00319 | MEF-3 | 3.59 |
| M00109 | C/EBPβ | 3.1041 |
| M0074 | NF-κB | 2.6676 |
| Generalized motif (Gary Stormo, personal communication) | NF-κB | 2.6286 |
| M00453 | IRF-7 | 2.426 |
| M0068 | Pax-4 | 2.316 |
| M0053 | c-Rel | 2.154 |
| MA0107 | P65 | 2.148 |
| M00052 | NF-κB | 2.145 |
| M00751 | AML1 | 2.130 |
The final group of IL-1β-induced genes examined for common transcription factor binding sites was a group that was highly responsive to IL-1β (increased with only 0.01 ng/ml) and rapidly up regulated in 1–4 h. These genes include IL-6, CXCL8/IL-8, CX3CL1, CCL2, CCL8, CCL20, NOS2B, NFKBIA, TNF-α, and BMP-2. Interestingly, the analysis suggests that in these genes, the highly representated transcription factor binding sites are almost solely related to NF-κB (Table 5).
Table 5.
Transcription Factor Binding Motif Prevalence in Genes Induced by Low IL-1β/1–4 h
| TRANSFAC Motif Accession Number | Transcription Factor | R-score |
|---|---|---|
| M00774 | NF-κB | 3.40 |
| M00208 | NF-κB | 3.01 |
| MA0061 | NF-κB | 2.88 |
| M00054 | NF-κB | 2.82 |
| Generalized motif (Gary Stormo, personal communication) | NF-κB | 2.60 |
| M00052 | NF-κB | 2.57 |
| MA0107 | p65 | 2.54 |
| M00261 | Olf-1 | 2.43 |
| M00053 | c-REL | 2.42 |
| M00194 | NF-κB | 2.39 |
| MA0101 | c-REL | 2.36 |
| M00977 | EBF | 2.26 |
DISCUSSION
The large scale screening procedure where 34,580 probes representing 24,650 genes and 37,123 gene transcripts are analyzed at once provided an overall view of the primary response of normal adult articular chondrocytes to IL-1β. We show that one of the predominant responses of human adult articular chondrocytes to exposure to IL-1β is a dramatic increase in a large set of chemokines and other genes related to the inflammatory cascade. Chemokines are produced in inflamed synovial tissue by the synovium, macrophages and fibroblast-like synoviocytes 24–26 and are thought to be key regulators of the inflammatory process 27 where they function in the recruitment of neutrophils, monocytes, immature dendritic cells, B cells and activated T cells 28. Therefore, the production of a large array of chemokines and other pro-inflammatory molecules under the influence of IL-1β could significantly alter the metabolism of chondrocytes in addition to the well-accepted increase in certain degradative enzymes. Furthermore, specific chemokines, CXCL8/IL8 and CXCL1 have been shown to alter the chondrocyte phenotype by inducing hypertrophic differentiation 29.
The most highly up regulated genes by IL-1β, E-selectin (Endothelial Leukocyte Adhesion Molecule, SELE), Leukocyte Inhibitory Factor (LIF) and Colony Stimulating Factor (CSF-3) have been shown to be regulated by IL-1β in other tissues. CSF has not been previously demonstrated in chondrocytes induced by IL-1β, but has been identified in synovial cells in rheumatoid arthritis 30, 31 and shown to exacerbate collagen-induced arthritis in mice 32. E-selectin is an adhesion molecule that is thought to be responsible for the accumulation of blood leukocytes at sites of inflammation by mediating the adhesion of cells to the vascular lining. To our knowledge, E-selectin has not been identified in cartilage, but is known to be up regulated by IL-1β in endothelial cells 33. The expression of SELE was not further analyzed in these studies, but it could be an important player in the pathogenesis of OA due to the ability to recruit and retain additional inflammatory players. LIF has been demonstrated in cartilage from OA patients and up regulated by IL-1β 25, 34–36.
In this study, we further analyzed the increased production of the chemokines CCL2, CCL3, CCL4, CCL5, CCL8, CCL20, CCL3L1, CX3CL1, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, and CXCL8 (IL-8). Their expression at different concentrations of IL-1β and over a 24 h time course demonstrated that there are specific subsets of genes that are co-regulated. The expression of these genes was compared with the expression of the proteases ADAMTS-4, ADAMTS-5, MMP-13; the matrix genes COL2A1 and aggrecan; the growth and differentiation factor BMP-2; the cytokines IL-1β, IL-6 and TNF-α; the nitric oxide producing enzyme NOS2B; and the NFκB inhibitor NFKBIA. Interestingly, the chemokine SDF-1 (CXCL12), a chemokine associated with chemoattraction of stem cells, was not detected in cartilage stimulated by IL-1β. This result is not unexpected as SDF-1 is considered a homeostatic regulator of tissue remodeling rather than an inflammatory mediator. SDF-1 is normally expressed by stromal cells of numerous tissues and actually down regulated by IL-1β and TNF-α in dermal wound healing 37, thus providing additional support for the validity of the chondrocyte response.
The production of a limited group of chemokines has been reported in cartilage 9, 20, 38. Loeser and colleagues reported that NF-κB mediates the stimulation of cytokine and chemokine expression in response to fibronectin fragments. In particular, they examined the expression of chemokines GRO-α (CXCL1), GRO-β (CXCL2), GRO-γ (CXCL3), MCP-1 (CCL-2), IL-8 (CXCL8) and the cytokine IL-6 and showed that IL-1β could mimic the effect of the fibronectin fragments 38. These authors reported low levels of chemokines in the cells from OA patients and moderate increases with IL-1β (2–5 fold). This study differs from ours in that all of their chondrocytes were isolated from cartilage from OA patients while a majority of our studies were done on normal chondrocytes. This is an important difference because we find that normal chondrocytes have very low levels of chemokines and we observed a trend to an increase in our compliment of chemokines when tissue from OA patients was used. In fact, in our experiments, when tissues from patients with OA are probed for this set of chemokines, the result resembles the chemokine pattern established here for IL-1β induction and may reflect the exposure of the osteoarthritic cartilage to the cytokine.
We consistently observed an increase in the chemokine CCL5/RANTES with 0.1 ng/ml of IL-1β beginning at 4 h and continuing through 24 h. It is present in chondrocytes from OA tissue, but not in normal chondrocytes. CCL5/RANTES has been associated with rheumatoid arthritis where it is synthesized by synovial fibroblasts, T cells and mononuclear cells in synovial fluid, and is thought to serve as a chemoattractant for T cells and monocytes In fact, inhibition of CCL5/RANTES by Met-RANTES, which blocks the receptors CCR1 and CCR5, caused amelioration of adjuvant-induced arthritis in rats 39. CCL5 has previously been shown to be made by chondrocytes 20.
ADAMTS-4 and ADAMTS-5 are the two enzymes that have been isolated from articular cartilage and identified as aggrecanase-1 and -2, respectively. In our studies, ADAMTS-4 is present at very low levels in cartilage from both normal and OA patients, however, it is induced with IL-1β, consistent with published studies 40, 41. We investigated this induction in more detail and found that ADAMTS-4 can be detected with exposure to as little as 0.01 ng/ml of IL-1β. When compared to MMP-13 gene expression, the concentration dependent rate of increase was greater, and the time course of increase was different: ADAMSTS-4 increased by 4 h and decreased by 24 h, whereas MMP13 increased at 8 h and remained high at 24 h. Clearly, these two enzymes are regulated through independent signal transduction pathways.
For ADAMTS-5, our studies clearly demonstrate high expression in both chondrocyte cell and cartilage explant cultures of human cartilage, again consistent with previous findings 41, 42. Whereas previous studies showed no effect of IL-1β on the expression of ADAMTS-5, we showed that IL-1β consistently and significantly decreased the mRNA expression level of ADAMTS-5 in cultured chondrocytes and in explant tissue. However, it is becoming apparent that ADAMTS-5 is highly expressed in normal articular 43, 44 and growth plate cartilage tissues 45 in which it is thought to participate in normal proteoglycan turnover. In the study by Aigner and colleagues 41, ADAMTS-5 mRNA levels were higher when cells were cultured with serum. In our studies, serum is used in cell cultures, and our time-course experiments indicate that over 24 h, ADAMTS-5 mRNA increases; this increase is either susceptible to down-regulation by IL-1β or IL-1β inhibits the serum-induced increase (see Fig. 3).
To explore potential transcriptional regulation of the chemokines, they were sub-classified into two sets based on the extent of up-regulation and subjected to a computational analysis for transcription factor binding sites that are highly represented in each set 15, 16. For each group, a distinct set of transcription factor binding sites were found, many of which have previously been experimentally verified. Actual binding probability would depend on a variety of other factors, including the cooperative binding of transcription factors and the concentration of the transcription factors within the nucleus. Chang and colleagues (2006) 15 have demonstrated that the computed scores are highly correlated with binding probability, such that promoters with higher combined scores were more likely to be bound by the transcription factor than promoters with lower scores. In the genes most highly up regulated, binding sites to factors related to NF-κB and IRF were highly represented. Both these transcription factor families have been documented to up regulate RANTES transcription (12 fold increase) on their own, 38, 46, 47 and in a concerted manner 21. NF-κB has been shown to regulate a specific subset of chemokines 38, however, Amos and colleagues recently demonstrated that inhibition of NF-κB activity did not inhibit all inflammatory mediators 48, therefore there are likely other transcriptional mechanisms involved. Here, we identified two additional potential transcription factors for this set of genes: MEF-3 and C/EBPβ; in fact, binding sites for these two transcription factors were the most highly represented in this set of highly expressed chemokine genes induced by IL-1β. C/EBPβ has been previously associated with IL-1β induced changes in chondrocyte gene expression. C/EBPβ is increased in chondrocytes by IL-1β and down-regulates the cartilage matrix genes COL2A1 and MIA/CD-RAP 23, 49; in addition, we have shown that C/EBPβ plays an important role in repressing cartilage gene expression in non-cartilaginous tissues 50. Neither MEF-3 nor C/EBPβ has been shown to regulate chemokine genes, but future experiments will be designed to test their role.
This computational method was also applied to the group of co-expressed genes that are very sensitive to low doses of IL-1β and are expressed at early time points. This group of genes was predicted to be predominantly regulated by proteins that bind at NF-κB binding sites on the gene. However, two other related factors, Olf-1 and EBF scored greater than 2.0. These two proteins are closely related members of the helix-loop-helix transcription factor family and have been shown to function in olfactory gene regulation, neuronal differentiation 51, B-cell development 52, adipogenesis 53 and are expressed in the connective tissues surrounding chondrogenic condensations and developing tendons54.
The chemokine receptors CCR-1, CCR-2, CCR-3, CCR-5, CXCR-1 and CXCR-2 are present in cartilage and up-regulated by IL-1β 28, 29. The presence of receptors, as well as production of chemokines, strongly suggests that chemokines may regulate cellular responses that are indirectly or directly related to inflammation and immune responses. These studies also showed that release of the collagenase, MMP-3, was markedly enhanced by stimulation with chemokines, particularly MCP-1, RANTES and GROα, and that this response was receptor mediated 28. In light of the previous finding of the presence of a full complement of receptors, it is reasonable to predict that chondrocytes will be able to respond to the chemokines up regulated by IL-1β.
Reversal of certain aspects of the IL-1β-induced phenotype has been evaluated for TGF-β1 55. We compared the potential to reverse this newly identified phenotype by TGF-β1, BMP-2 and FGF-18. TGF-β1 was best able to reverse the phenotype having effects on reversal of the down-regulation of matrix molecules, and reversal of some of the up-regulation of chemokines. TGF-β1, however, had opposite effects on two of the enzymes considered important for matrix degradation: it lowered the IL-1β-induced increase in MMP-13, but increased the expression of ADAMTS-4. These observations confirm data presented by Hasty and colleagues 56 who demonstrated that TGF-β could decrease MMP-13 and Moulharat and colleagues 57 who have shown that TGF-β1 increases both mRNA levels of ADAMTS-4 and cleavage products of aggrecan. Potential reversal by BMP-2 proved to be disappointing as BMP-2 did not affect matrix, enzyme or chemokine synthesis: higher concentration did not increase the effect (up to 200 ng/ml). Without IL-1β, control experiments showed that BMP-2 could increase COL2A1 gene expression (data not shown).
In summary, we have shown a large set of genes that are up regulated by the cytokine IL-1β in adult normal cartilage and from patients with OA. Genes from both types of cartilage are affected similarly by IL-1β. By computational analysis, two new transcription factors are now associated with this set of up regulated genes, MEF-3 and C/EBP; however, the early response of genes to IL-1β is most likely due to the NF-κB pathway. Reversal of the IL-1β induced phenotype is not accomplished by BMP-2, although it is expressed in the tissue. TGF-β1 can partially reverse the phenotype, while FGF-18 can reverse certain aspects and augments others. Given the results from rheumatoid arthritis and other inflammatory diseases, it can be expected that this exuberant increase in a wide range of chemokines will have a significant impact on the cells of cartilage and should be considered in the pathophysiology of OA. The chemokine profile from OA patient tissue may reflect the cytokine history of the tissue, therefore we are profiling a large sample of patient and normal tissue to determine whether there is a “IL-1β footprint” for OA.
Acknowledgments
The authors would like to thank Drs. Douglas McDonald and Joseph Borrelli for normal cartilage and Drs. John Clohisy, and Robert Barrack and Head Nurse, Keith Foreman, for cartilage from patients with osteoarthritis. The authors would also like to thank the Genome Sequencing Center at the Washington University School of Medicine for their assistance with the microarray analysis. These studies were funded by The National Institute for Arthritis, Musculoskeletal and Skin Diseases R01 AR05084, R01 AR045550 and R01 AR036994.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Poole AR, Guilak F, Abramson S. Etiopathogenesis of osteoarthritis. In: Moskowitz RW, Altman RD, Hochberg MC, Buckwalter JA, Goldberg VM, editors. Osteoarthritis: Diagnosis and Medical/Surgical Management. 4. Philadelphia: Kluwer; 2007. pp. 27–49. [Google Scholar]
- 2.Sandell LJ, Heinegard D, Hering TM. Cell Biology, Biochemistry, and Molecular Biology of Articular Cartilage in Osteoarthritis. In: Moskowitz RW, Altman RD, Hochberg MC, Buckwalter JA, Goldberg VM, editors. Osteoarthritis: Diagnosis and Medical/Surgical Management. 4. Philadelphia: Kluwer; 2007. pp. 73–106. [Google Scholar]
- 3.Goldring MB, Birkhead J, Sandell LJ, Kimura T, Krane SM. Interleukin 1 suppresses expression of cartilage-specific types II and IX collagens and increases types I and III collagens in human chondrocytes. J Clin Invest. 1988;82:2026–2037. doi: 10.1172/JCI113823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukui N, Zhu Y, Maloney WJ, Clohisy J, Sandell LJ. Stimulation of BMP-2 expression by pro-inflammatory cytokines IL-1 and TNF-alpha in normal and osteoarthritic chondrocytes. J Bone Joint Surg Am. 2003;85-A(Suppl 3):59–66. doi: 10.2106/00004623-200300003-00011. [DOI] [PubMed] [Google Scholar]
- 5.Martel-Pelletier J, Alaaeddine N, Pelletier JP. Cytokines and their role in the pathophysiology of osteoarthritis. Frontiers in Bioscience. 1999;4:694–703. doi: 10.2741/martel. [DOI] [PubMed] [Google Scholar]
- 6.Tiku K, Thakker-Varia S, Ramachandrula A, Tiku ML. Articular chondrocytes secrete IL-1, express membrane IL-1, and have IL-1 inhibitory activity. Cell Immunol. 1992;140:1–20. doi: 10.1016/0008-8749(92)90172-l. [DOI] [PubMed] [Google Scholar]
- 7.Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2:108–115. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 8.Mobasheri A, Vannucci SJ, Bondy CA, Carter SD, Innes JF, Arteaga MF, et al. Glucose transport and metabolism in chondrocytes: a key to understanding chondrogenesis, skeletal development and cartilage degradation in osteoarthritis. Histol Histopathol. 2002;17:1239–1267. doi: 10.14670/HH-17.1239. [DOI] [PubMed] [Google Scholar]
- 9.Cecil DL, Johnson K, Rediske J, Lotz M, Schmidt AM, Terkeltaub R. Inflammation-induced chondrocyte hypertrophy is driven by receptor for advanced glycation end products. J Immunol. 2005;175:8296–8302. doi: 10.4049/jimmunol.175.12.8296. [DOI] [PubMed] [Google Scholar]
- 10.Constantin G, Majeed M, Giagulli C, Piccio L, Kim JY, Butcher EC, et al. Chemokines trigger immediate beta2 integrin affinity and mobility changes: differential regulation and roles in lymphocyte arrest under flow. Immunity. 2000;13:759–769. doi: 10.1016/s1074-7613(00)00074-1. [DOI] [PubMed] [Google Scholar]
- 11.Kanbe K, Takagishi K, Chen Q. Stimulation of matrix metalloprotease 3 release from human chondrocytes by the interaction of stromal cell-derived factor 1 and CXC chemokine receptor 4. Arthritis Rheum. 2002;46:130–137. doi: 10.1002/1529-0131(200201)46:1<130::aid-art10020>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 12.Mantovani A, Bonecchi R, Locati M. Tuning inflammation and immunity by chemokine sequestration: decoys and more. Nat Rev Immunol. 2006;6:907–918. doi: 10.1038/nri1964. [DOI] [PubMed] [Google Scholar]
- 13.Haringman JJ, Ludikhuize J, Tak PP. Chemokines in joint disease: the key to inflammation? Ann Rheum Dis. 2004;63:1186–1194. doi: 10.1136/ard.2004.020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beinke S, Ley SC. Functions of NF-kappaB1 and NF-kappaB2 in immune cell biology. Biochem J. 2004;382:393–409. doi: 10.1042/BJ20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang LW, Nagarajan R, Magee JA, Milbrandt J, Stormo GD. A systematic model to predict transcriptional regulatory mechanisms based on overrepresentation of transcription factor binding profiles. Genome Res. 2006;16:405–413. doi: 10.1101/gr.4303406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies SR, Chang L-W, Patra D, Xing X, Posey K, Hecht J, et al. Computational identification and functional validation of regulatory motifs in cartilage-expressed genes. Genome Res. 2007;17:1438–1447. doi: 10.1101/gr.6224007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.GuhaThakurta D, Schriefer LA, Waterston RH, Stormo GD. Novel transcription regulatory elements in Caenorhabditis elegans muscle genes. Genome Res. 2004;14:2457–2468. doi: 10.1101/gr.2961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mengshol JA, Vincenti MP, Coon CI, Barchowsky A, Brinckerhoff CE. Interleukin-1 induction of collagenase 3 (matrix metalloproteinase 13) gene expression in chondrocytes requires p38, c-Jun N-terminal kinase, and nuclear factor kappaB: differential regulation of collagenase 1 and collagenase 3. Arthritis Rheum. 2000;43:801–811. doi: 10.1002/1529-0131(200004)43:4<801::AID-ANR10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 20.Alaaeddine N, Olee T, Hashimoto S, Creighton-Achermann L, Lotz M. Production of the chemokine RANTES by articular chondrocytes and role in cartilage degradation. Arthritis Rheum. 2001;44:1633–1643. doi: 10.1002/1529-0131(200107)44:7<1633::AID-ART286>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 21.Genin P, Algarte M, Roof P, Lin R, Hiscott J. Regulation of RANTES chemokine gene expression requires cooperativity between NF-kappa B and IFN-regulatory factor transcription factors. J Immunol. 2000;164:5352–5361. doi: 10.4049/jimmunol.164.10.5352. [DOI] [PubMed] [Google Scholar]
- 22.Goldring MB, Sandell LJ. VIII. Transcriptional Control of Chondrocyte Gene Expression. Osteoarthritis and Cartilage. 2007;70:118–142. [Google Scholar]
- 23.Okazaki K, Li J, Yu H, Fukui N, Sandell LJ. CCAAT/enhancer binding protein beta and delta mediate the repression of gene transcription of cartilage-derived retinoic acid-sensitive protein Induced by interleukin-1b. J Biol Chem. 2002;277:31526–31533. doi: 10.1074/jbc.M202815200. [DOI] [PubMed] [Google Scholar]
- 24.Hosaka N, Nagata N, Miyashima S, Ikehara S. Attenuation of lpr-graft-versus-host disease (GVHD) in MRL/lpr spleen cell-injected SCID mice by in vivo treatment with anti-V beta 8.1,2 monoclonal antibody. Clin Exp Immunol. 1994;96:500–507. doi: 10.1111/j.1365-2249.1994.tb06057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lotz M, Terkeltaub R, Villiger PM. Regulation of IL-8 expression by human articular chondrocytes. J Immunol. 1992;1448:466–473. [PubMed] [Google Scholar]
- 26.Koch AE, Kunkel SL, Harlow LA, Mazarakis DD, Haines GK, Burdick MD, et al. Macrophage inflammatory protein-1 alpha. A novel chemotactic cytokine for macrophages in rheumatoid arthritis. J Clin Invest. 1994;93:921–928. doi: 10.1172/JCI117097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haringman JJ, Smeets TJM, Reinders-Blankert P, Tak PP. Chemokine and chemokine receptor expression in paired peripheral blood mononuclear cells and synovial tissue of patients with rheumatoid arthritis, osteoarthritis, and reactive arthritis. Ann Rheum Dis. 2006;65:294–300. doi: 10.1136/ard.2005.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borzi RM, Mazzetti I, Cattini L, Uguccioni M, Baggiolini M, Facchini A. Human chondrocytes express functional chemokine receptors and release matrix-degrading enzymes in response to C-X-C and C-C chemokines. Arthritis Rheum. 2000;43:1734–1741. doi: 10.1002/1529-0131(200008)43:8<1734::AID-ANR9>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 29.Merz D, Liu R, Johnson K, Terkeltaub R. IL-8/CXCL8 and growth-related oncogene alpha/CXCL1 induce chondrocyte hypertrophic differentiation. J Immunol. 2003;171:4406–4415. doi: 10.4049/jimmunol.171.8.4406. [DOI] [PubMed] [Google Scholar]
- 30.Agro A, Jordana M, Chan KH, Cox G, Richards C, Stepien H, et al. Synoviocyte derived granulocyte macrophage colony stimulating factor mediates the survival of human lymphocytes. J Rheumatol. 1992;19:1065–1069. [PubMed] [Google Scholar]
- 31.Haworth C, Brennan FM, Chantry D, Turner M, Maini RN, Feldmann M. Expression of granulocyte-macrophage colony-stimulating factor in rheumatoid arthritis: regulation by tumor necrosis factor-alpha. Eur J Immunol. 1991;21:2575–2579. doi: 10.1002/eji.1830211039. [DOI] [PubMed] [Google Scholar]
- 32.Campbell IK, Bendele A, Smith DA, Hamilton JA. Granulocyte-macrophage colony stimulating factor exacerbates collagen induced arthritis in mice. Ann Rheum Dis. 1997;56:364–368. doi: 10.1136/ard.56.6.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyble CW, Hynes KL, Kuchibhotla J, Marcus BC, Hallahan D, Gewertz BL. TNF-alpha and IL-1 upregulate membrane-bound and soluble E-selectin through a common pathway. J Surg Res. 1997;73:107–112. doi: 10.1006/jsre.1997.5207. [DOI] [PubMed] [Google Scholar]
- 34.Henrotin YE, De Groote DD, Labasse AH, Gaspar SE, Zheng SX, Geenen VG, et al. Effects of exogenous IL-1 beta, TNF alpha, IL-6, IL-8 and LIF on cytokine production by human articular chondrocytes. Osteoarthritis and Cartilage. 1996;4:163–173. doi: 10.1016/s1063-4584(96)80012-4. [DOI] [PubMed] [Google Scholar]
- 35.Fan Z, Bau B, Yang H, Aigner T. IL-1beta induction of IL-6 and LIF in normal articular human chondrocytes involves the ERK, p38 and NFkappaB signaling pathways. Cytokine. 2004;28:17–24. doi: 10.1016/j.cyto.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Campbell IK, Waring P, Novak U, Hamilton JA. Production of leukemia inhibitory factor by human articular chondrocytes and cartilage in response to interleukin-1 and tumor necrosis factor alpha. Arthritis Rheum. 1993;36:790–794. doi: 10.1002/art.1780360608. [DOI] [PubMed] [Google Scholar]
- 37.Fedyk ER, Jones D, Critchley HO, Phipps RP, Blieden TM, Springer TA. Expression of stromal-derived factor-1 is decreased by IL-1 and TNF and in dermal wound healing. J Immunol. 2001;166:5749–5754. doi: 10.4049/jimmunol.166.9.5749. [DOI] [PubMed] [Google Scholar]
- 38.Pulai JI, Chen H, Im HJ, Kumar S, Hanning C, Hegde PS, et al. NF-kappa B mediates the stimulation of cytokine and chemokine expression by human articular chondrocytes in response to fibronectin fragments. J Immunol. 2005;174:5781–5788. doi: 10.4049/jimmunol.174.9.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shahrara S, Proudfoot AE, Woods JM, Ruth JH, Amin MA, Park CC, et al. Amelioration of rat adjuvant-induced arthritis by Met-RANTES. Arthritis Rheum. 2005;52:1907–1919. doi: 10.1002/art.21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tortorella M, Pratta M, Liu RQ, Abbaszade I, Ross H, Burn T, et al. The thrombospondin motif of aggrecanase-1 (ADAMTS-4) is critical for aggrecan substrate recognition and cleavage. J Biol Chem. 2000;275:25791–25797. doi: 10.1074/jbc.M001065200. [DOI] [PubMed] [Google Scholar]
- 41.Bau B, Gebhard PM, Haag J, Knorr T, Bartnik E, Aigner T. Relative messenger RNA expression profiling of collagenases and aggrecanases in human articular chondrocytes in vivo and in vitro. Arthritis Rheum. 2002;46:2648–2657. doi: 10.1002/art.10531. [DOI] [PubMed] [Google Scholar]
- 42.Tortorella MD, Malfait AM, Deccico C, Arner E. The role of ADAM-TS4 (aggrecanase-1) and ADAM-TS5 (aggrecanase-2) in a model of cartilage degradation. Osteoarthritis and Cartilage. 2001;9:539–552. doi: 10.1053/joca.2001.0427. [DOI] [PubMed] [Google Scholar]
- 43.Abbaszade I, Liu RQ, Yang F, Rosenfeld SA, Ross OH, Link JR, et al. Cloning and characterization of ADAMTS11, an aggrecanase from the ADAMTS family. J Biol Chem. 1999;274:23443–23450. doi: 10.1074/jbc.274.33.23443. [DOI] [PubMed] [Google Scholar]
- 44.Stanton H, Rogerson FM, East CJ, Golub SB, Lawlor KE, Meeker CT, et al. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434:648–652. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- 45.Makihira S, Yan W, Murakami H, Furukawa M, Kawai T, Nikawa H, et al. Thyroid hormone enhances aggrecanase-2/ADAM-TS5 expression and proteoglycan degradation in growth plate cartilage. Endocrinology. 2003;144:2480–2488. doi: 10.1210/en.2002-220746. [DOI] [PubMed] [Google Scholar]
- 46.Lin R, Heylbroeck C, Genin P, Pitha PM, Hiscott J. Essential Role of Interferon Regulatory Factor 3 in Direct Activation of RANTES Chemokine Transcription. Mol Cell Biol. 1999;19:959–966. doi: 10.1128/mcb.19.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J, Guan X, Ma X. Interferon Regulatory Factor 1 Is an Essential and Direct Transcriptional Activator for Interferon γ-induced RANTES/CCl5 Expression in Macrophages. J Biol Chem. 2005;280:24347–24355. doi: 10.1074/jbc.M500973200. [DOI] [PubMed] [Google Scholar]
- 48.Amos N, Lauder S, Evans A, Feldmann M, Bondeson J. Adenoviral gene transfer into osteoarthritis synovial cells using the endogenous inhibitor IκBα reveals that most, but not all, inflammatory and destructive mediators are NF-κB dependent. Rheumatology. 2006;45:1201–1209. doi: 10.1093/rheumatology/kel078. [DOI] [PubMed] [Google Scholar]
- 49.Imamura T, Imamura C, Iwamoto Y, Sandell LJ. Transcriptional Co-activators CREB-binding Protein/p300 Increase Chondrocyte Cd-rap Gene Expression by Multiple Mechanisms Including Sequestration of the Repressor CCAAT/Enhancer-binding Protein. J Biol Chem. 2005;280:16625–16634. doi: 10.1074/jbc.M411469200. [DOI] [PubMed] [Google Scholar]
- 50.Okazaki K, Yu H, Davies S, Imamura T, Sandell L. A promoter element of the CD-RAP gene is required for repression of gene expression in non-cartilage tissues in vitro and in vivo. J Cell Biochem. 2006;97:857–868. doi: 10.1002/jcb.20648. [DOI] [PubMed] [Google Scholar]
- 51.Wang SS, Tsai RY, Reed RR. The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J Neurosci. 1997;17:4149–4158. doi: 10.1523/JNEUROSCI.17-11-04149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z, Cotta CV, Stephan RP, deGuzman CG, Klug CA. Enforced expression of EBF in hematopoietic stem cells restricts lymphopoiesis to the B cell lineage. Embo J. 2003;22:4759–4769. doi: 10.1093/emboj/cdg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akerblad P, Lind U, Liberg D, Bamberg K, Sigvardsson M. Early B-cell factor (O/E-1) is a promoter of adipogenesis and involved in control of genes important for terminal adipocyte differentiation. Mol Cell Biol. 2002;22:8015–8025. doi: 10.1128/MCB.22.22.8015-8025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mella S, Soula C, Morello D, Crozatier M, Vincent A. Expression patterns of the coe/ebf transcription factor genes during chicken and mouse limb development. Gene Expr Patterns. 2004;4:537–542. doi: 10.1016/j.modgep.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 55.van den Berg WB, Joosten LA, Kollias G, van De Loo FA. Role of tumour necrosis factor alpha in experimental arthritis: separate activity of interleukin 1beta in chronicity and cartilage destruction. Ann Rheum Dis. 1999;58(Suppl 1):I40–48. doi: 10.1136/ard.58.2008.i40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shlopov BV, Gumanovskaya ML, Hasty KA. Autocrine regulation of collagenase 3 (matrix metalloproteinase 13) during osteoarthritis. Arthritis Rheum. 2000;43:195–205. doi: 10.1002/1529-0131(200001)43:1<195::AID-ANR24>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 57.Moulharat N, Lesur C, Thomas M, Rolland-Valognes G, Pastoureau P, Anract P, et al. Effects of transforming growth factor-beta on aggrecanase production and proteoglycan degradation by human chondrocytes in vitro. Osteoarthritis and Cartilage. 2004;12:296–305. doi: 10.1016/j.joca.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 58.Tchetina EV, Squires G, Poole AR. Increased type II collagen degradation and very early focal cartilage degeneration is associated with upregulation of chondrocyte differentiation related genes in early human articular cartilage lesions. J Rheumatol. 2005;32:876–886. [PubMed] [Google Scholar]
- 59.Legendre F, Dudhia J, Pujol JP, Bogdanowicz P. JAK/STAT but not ERK1/ERK2 pathway mediates interleukin (IL)-6/soluble IL-6R down-regulation of Type II collagen, aggrecan core, and link protein transcription in articular chondrocytes. Association with a down-regulation of SOX9 expression. J Biol Chem. 2003;278:2903–2912. doi: 10.1074/jbc.M110773200. [DOI] [PubMed] [Google Scholar]