Abstract
Assembly of pili in Gram-positive bacteria and their attachment to the cell wall envelope are mediated by sortases. In Bacillus cereus and its close relative Bacillus anthracis, the major pilin protein BcpA is cleaved between the threonine and the glycine of its C-terminal LPXTG motif sorting signal by the pilin-specific sortase D. The resulting acyl enzyme intermediate is relieved by the nucleophilic attack of the side-chain amino group of lysine within the YPKN motif of another BcpA subunit. Cell wall anchoring of assembled BcpA pili requires sortase A, which also cleaves the LPXTG sorting signal of BcpA between its threonine and glycine residues. We show here that sortases A and D require only the C-terminal sorting signal of BcpA for substrate cleavage. Unlike sortase D, which accepts the YPKN motif as a nucleophile, sortase A forms an amide bond between the BcpA C-terminal carboxyl group of threonine and the side-chain amino group of diaminopimelic acid within the cell wall peptidoglycan of bacilli. These results represent the first demonstration of a cell wall anchor structure for pili, which are deposited by sortase A into the envelope of many different microbes.
Many Gram-positive bacteria use pili, protein fibers that project from microbial surfaces, to adhere to and invade host tissues or form biofilms (1). Gram-positive pili are assembled from pilin subunits that are synthesized as precursors with N-terminal signal peptides and C-terminal sorting signals in the bacterial cytoplasm (P1) (2). The signal peptide is cleaved and precursors translocated through the bacterial membrane by the Sec machinery, thereby generating the P2 precursor (3, 4). Assembly of pili involves the polymerization of the major pilin protein, a reaction that is catalyzed by pilin-specific sortase (2, 5), designated sortase D in pathogenic Bacillus sp. (6). Pilin-specific sortases perform a transpeptidation reaction, whereby the C-terminal LPXTG motif sorting signal of the major pilin protein, BcpA in bacilli, is cleaved between the threonine and the glycine residue (4). Intermediary product of this reaction is a thioester-linked acyl-enzyme between the active site cysteine residue of sortase and the carboxyl group of threonine (7, 8). Nucleophilic attack of the ε-amino group of lysine within the YPKN pilin motif of another BcpA subunit resolves the sortase D intermediate and generates the amide bond between the C-terminal carboxyl group of threonine and the side-chain amino group of lysine of adjacent BcpA subunits (4). Conservation of pilin-specific sortase, the YPKN motif, and C-terminal sorting signal in the major subunit of pilin proteins suggests that pilus assembly occurs by a universal mechanism in Gram-positive bacteria (2).
Pili are anchored to the cell wall envelope of Gram-positive bacteria by a mechanism that requires sortase A (6, 9-11). Sortase A is otherwise known to cut the C-terminal LPXTG sorting signal of surface proteins and immobilize anchored products in the cell wall envelope of Gram-positive bacteria (12, 13). In Staphylococcus aureus, sortase A acyl enzyme intermediates are resolved by the nucleophilic attack of lipid II (14, 15), the biosynthetic precursor of peptidoglycan biosynthesis or, more specifically, the amino group of its cross-bridge, pentaglycine in S. aureus (16, 17). In contrast to surface proteins, which are directly incorporated by sortase A into peptidoglycan (18-20), the mechanisms that provide for the deposition of pili in the cell wall envelope are still unknown. Unlike surface proteins, pili must first be assembled by pilin-specific sortase and then be deposited in the envelope (21). The latter requires the aforementioned signal peptide, YPKN pilin motif, and sorting signal, however, the requirements for substrate recognition of the major pilin by sortase A are still unknown. Further, the anchored product of the sortase A-mediated reaction, presumably pilin protein linked to cell wall, has not been characterized. Both of these questions are addressed in this report.
EXPERIMENTAL PROCEDURES
Bacterial Plasmids and Strains—PCR with primer pairs P2 (encompassing an XbaI site) and P101 (NheI) amplified bcpA between residues 26 and 515 (Table 1). Primers P102 (NheI) and P103 (KpnI) amplified a fragment of bcpA beginning at residue 516 with the MH6 tag and srtD. Primer pairs P2/P101 and P102/103 generated PCR products that were digested with XbaI/NheI and NheI/KpnI and ligated into pLM5 digested with XbaI/KpnI to create pJB40. pJB39 (6) encodes wild-type bcpA-srtD under control of the IPTG3-inducible Pspac promoter. To generate pJB173 and pJB169, plasmids lacking srtD, pJB39 and pJB40 were amplified with Pfu DNA polymerase and P88/P89. Amplified plasmids were first incubated for 3 h with DpnI to digest methylated parental plasmid DNA and were then digested with KpnI for 2 h. Digested plasmid DNA was ligated and transformed into Escherichia coli DH5α (22).
TABLE 1.
Primers used in this study
Non-capitalized letters indicate the sequence of the restriction site inserted. Underlined nucleotides encode for the MH6 tag. Bolded residues denote nucleotide changes introduced.
| Primer | Restriction site | Nucleic acid sequence (5′ -3′) |
|---|---|---|
| 2 | XbaI | AAAtctagaGCACACTATTGCTTTTAAGAAGG |
| 17 | PstI | GTTAAATTAACGATAGAActgcagAAAAGTGGATGGATTCTTC |
| 18 | PstI | GAAGAATCCATCCACTTTTctgcagTTCTATCGTTAATTTAAC |
| 19 | PstI | ActcgagATCTTTTGAAGGGTGATGAATATATG |
| 20 | PstI | AAActgcagTTTAATACTGTTCCCATCAAATAC |
| 88 | KpnI | AAAggtaccAATACCTCCCAAAACAAGATTTC |
| 89 | KpnI | AAAggtaccATCCTATTGTTATGTGTGTTTCTA |
| 101 | NheI | AAAgctagcACTTTTATTATTTTCTATCGTTAATTTA |
| 102 | NheI | AAAgctagcatgcatcaccatcaccatcacGGATGGATTCTTCCGGTAACG |
| 103 | KpnI | AAAggtaccTTATCTTTGAATTTCCGGTCCC |
| 173 | None | TTATACAACTTTAATTACGGCTACGCCTTATGGAATAAAC |
| 174 | None | GTTTATTCCATAAGGCGTAGCCGTAATTAAAGTTGTATAA |
pJB12 (6) encodes bcpA-srtD-bcpB under control of the IPTG-inducible Pspac promoter. pJB1 (23) was used for expression of an untagged version of PlyL amidase (24). pJB69 encodes bcpA-gst-srtD driven by the Pspac promoter (4) and was used as a template to generate pSY8. pSY8 encodes for the cell wall sorting signal of bcpA fused to isdX1 at is N terminus and gst at its C terminus. To swap the bcpA coding region preceding its cell wall sorting signal with isdX1 in pJB69, a PstI site was generated 15 nucleotides upstream of the DNA sequence encoding for the LPVTG cell wall sorting signal with quick change mutagenesis and primers P17/18, as described previously (6). The resulting plasmid, pJB69-PstI, was digested with XbaI, blunt-ended, and digested with PstI. The purified plasmid fragment was ligated with PstI-digested isdX1 Pfu PCR product. Plasmid variants lacking srtD or containing srtD with active site cysteine 207 to alanine substitution were generated by quick change. Primers P173/174 generated pSY13 (isdX1SS-gst-srtDC207A). Primers P88/89 generated pSY14 (isdX1SS-GST).
Fractionation of Bacillus Cultures—B. anthracis strain Sterne F32 or its isogenic variant AHG263 (srtA::ermC) (20) harboring pLM5 or plasmids encoding for B. cereus pilus genes were grown overnight at 30 °C on LB agar plates containing kanamycin and IPTG. Bacilli were suspended in 100 mm NaCl, vortexed briefly, and centrifuged for 2 min at 6000 × g. Cells were washed two more times in 0.1 m NaCl. The sediment was analyzed by immunoelectron microscopy with α-BcpA antisera and 10 nm gold anti-rabbit IgG conjugate as previously reported with additional wash steps between secondary antibody labeling and fixation with glutaraldehyde (6). For immunoblotting experiments, cells were extracted for 10 min by boiling with 500 μl of 6 m urea, 1% SDS, 50 mm Tris-HCl, pH 9.5, releasing the cytoplasm and membrane fractions (25). Bacterial samples were washed in water, extracted with 5% trichloroacetic acid, and peptidoglycan-digested with mutanolysin (6). Sortase A was detected by immunoblotting of extracts of bacilli that had been concentrated by precipitation with 8% trichloroacetic acid.
Purification of Cell Wall-anchored Pilin—B. anthracis strain Sterne F32 harboring pJB169 was grown for 20 h at 30 °C in 6 liters of LB broth containing 10 μg/ml kanamycin and 1 mm IPTG. For mutanolysin treatment, cells were washed with 100 ml of double distilled water (ddH2O) and extracted by boiling with 100 ml of 6 m urea, 1% SDS, 50 mm Tris-HCl, pH 9.5. Murein sacculi were washed in ddH2O, extracted by boiling in 5% trichloroacetic acid, washed in 50 mm Tris-HCl (pH 6.3)-1.5 mm MgCl2, and peptidoglycan-digested with 20,000 units of mutanolysin (23). For PlyL digestion, sedimented bacilli were washed once in 100 ml of 50 mm Tris-HCl (pH 7.5) and suspended in 50 ml of the same buffer supplemented with 5 mm phenylmethanesulfonyl fluoride (26). Cell walls were broken in a bead beater instrument (Biospec Products Inc.) by 10 pulses of 1 min, followed by 5 min of incubation on ice. The crude lysate was decanted to remove the glass beads and centrifuged at 33,000 × g for 15 min to sediment cell wall sacculi and membranes. Sediment was treated with 1% Triton X-100, 100 mm phosphate (pH 7.5), and 1 mm phenylmethanesulfonyl fluoride, and peptidoglycan was then treated with PlyL as described previously (23). Following cell wall digestion, insoluble material was removed by centrifugation at 33,000 × g. The pH of the samples was adjusted to 7.5 with 2 m sodium phosphate dibasic, and BcpAMH6 was purified by Ni-NTA affinity chromatography (26).
Purification of Pilin Anchor Peptides—Purified BcpAMH6 was methanol-chloroform-precipitated and cleaved with CNBr (26). Anchor peptides were purified by a second round of Ni-NTA affinity chromatography under denaturing conditions and separated by reverse-phase high-performance liquid chromatography (RP-HPLC) with uv detection using a C18 column with a linear gradient from 1% to 99% acetonitrile (CH3CN) in 0.1% formic acid in 100 min, as previously reported for BcpA pilin peptides (4).
MALDI-MS—Aliquots of RP-HPLC fractions (0.5 μl) were co-spotted with matrix (0.5 μl of α-cyano-4-hydroxycinnamic acid) prepared at 10 mg/ml in CH3CN-water-trifluoroacetic acid (30:40:0.1), 4 mm (NH4)2HPO4. Matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) spectra were obtained in a reflectron time-of-flight instrument (ABI Biosystems MALDI 4700) in reflectron mode. Spectra were acquired using external calibration with bovine insulin. Theoretical parent ion and fragmentation ion monoisotopic m/z values were produced with ProteinProspector version 4.27.2 MS-Product web-based program (University of California-San Francisco, prospector.ucsf.edu).
Edman Degradation—RP-HPLC samples of anchor peptides were dried under vacuum and submitted for Edman sequencing at the University of Illinois at Urbana-Champaign Biotechnology Center Protein Sciences Facility.
RESULTS
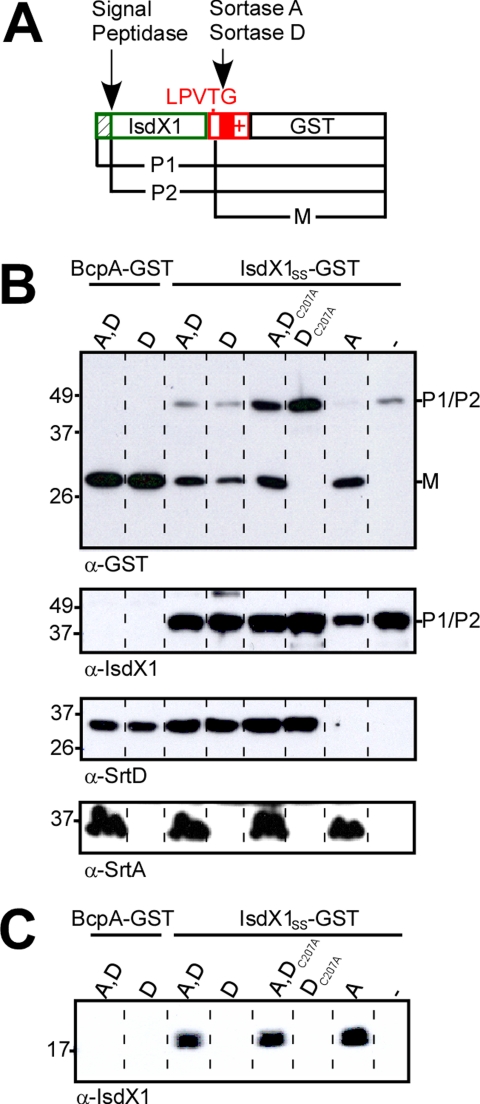
Substrate Properties Control the Fate of Pilin Precursors—Previous work demonstrated that sortase A and sortase D both cleave the major pilin protein precursor BcpA, but left unresolved the substrate requirements of these enzymes. To address this, we used pJB69, encoding bcpA-gst srtD, and transformed this plasmid into B. anthracis Sterne (expressing sortase A) or its isogenic srtA variant (Fig. 1A). Processing of BcpA-GST was monitored by separating whole bacterial extracts on SDS-PAGE followed by immunoblotting with antibodies specific for GST (Fig. 1B). Bacilli expressing both enzymes (A and D) or only sortase D both cut BcpA-GST precursor between the threonine and the glycine of its LPVTG sorting signal and generated GST cleavage products (Fig. 1B). Bacilli lacking sortase A and D cannot cleave this hybrid substrate (4). In the presence of sortase D, BcpA is polymerized into pili, whereas sortase A is thought to immobilize pili in the cell wall envelope (4). Pilus assembly requires the YPKN pilin motif and CNA B domain of BcpA (4). It is not known, however, whether these elements are also required for substrate recognition, defined here as sortase cleavage of the pilin sorting signal (SS). To test this, we generated plasmid pSY8, encoding isdX1SS-gst srtD. Under physiological conditions, bacilli secrete IsdX1 via its N-terminal signal peptide into the extracellular medium, where the soluble protein binds hemoglobin and scavenges heme with its NEAT domain (27). Fusion of IsdX1 to the sorting signal of BcpA (SS) and GST generated a hybrid that was cleaved by bacilli expressing both sortase D and sortase A, or only one of these enzymes, but not by bacilli that lacked sortase A and D (Fig. 1, A and B). To verify that the active site cysteine of sortase D is involved in substrate cleavage, we generated an alanine substitution mutant (DC207A). Although sortase DC207A was expressed at the same level as the wild-type enzyme, the mutant was unable to cleave IsdX1SS-GST substrate (Fig. 1, B and C). Of note, expression of sortase DC207A in wild-type bacilli appeared to reduce the ability of sortase A to cleave IsdX1SS-GST, suggesting that the mutant sortase may interact with the substrate in a manner that interferes with its recognition by the other sortase (Fig. 1, B and C). As a control, expression of sortase DC207A did not affect the abundance of sortase A in bacilli (Fig. 1B). Sortase A, but not sortase D, cleavage of IsdX1SS-GST led to the deposition of IsdX1 in the cell wall envelope of bacilli (Fig. 1C). Together these findings support a model, whereby sortase A acyl intermediates can only be resolved by the nucleophilic attack of lipid II, whereas sortase D intermediates must be resolved by the nucleophilic attack of BcpA unless hydrolysis at the active site thioester precipitates release of product into the extracellular medium (21). Finally, the pilin sorting signal alone is sufficient for sortase A cleavage of BcpA at the LPVTG motif and, following completion of the transpeptidation reaction, for transfer of the cleaved product into the cell wall envelope.
FIGURE 1.
IsdX1SS-GST is anchored to the cell wall by sortase A. A, IsdX1SS-GST is a translational hybrid between IsdX1 (colored green), the cell-wall sorting signal of BcpA (colored red), and glutathione S-transferase. The diagram displays the precursor (P1) of IsdX1SS-GST, and the signal peptidase (P2) and sortase-cleaved products (M). B, sortase cleavage products were detected in urea -SDS-release cytoplasmic fractions by immunoblot with α-GST antisera. Antisera raised against B. cereus sortase D and B. anthracis sortase A allowed for their detection by immunoblot. Labels indicate the sortase (srtA, srtD, srtDC207A, or none) and substrate (bcpA-gst or isdX1SS-gst) expressed in each strain. C, bacilli cell wall extracts were digested with mutanolysin. IsdX1 anchoring was analyzed by immunoblot with α-IsdX1 antibodies.
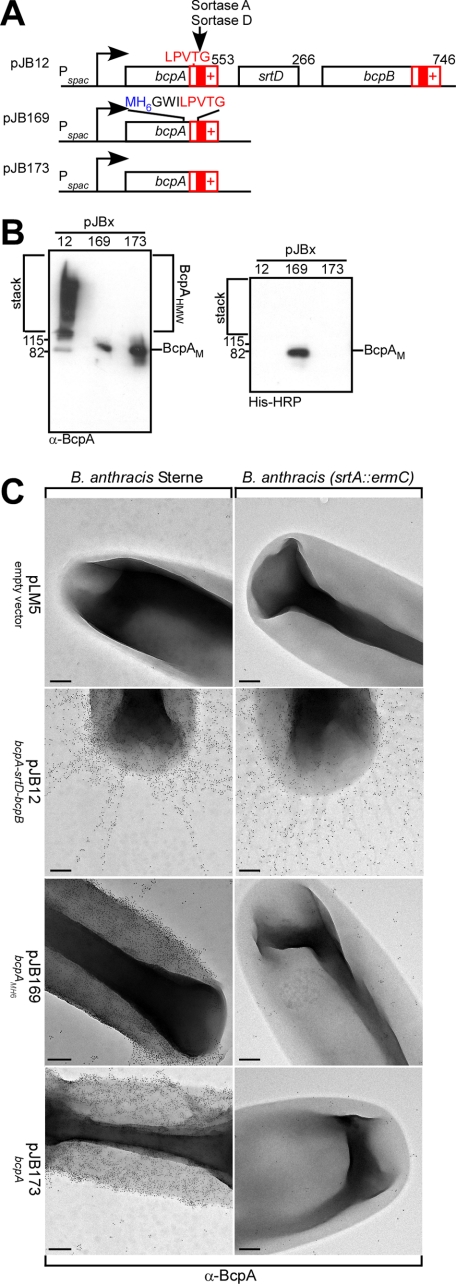
Purification of Anchored Pilin from the Cell Wall Envelope of Bacilli—B. anthracis was transformed with pJB12, a plasmid that encodes bcpA-srtD-bcpB under control of the IPTG-inducible Pspac promoter (6) (Fig. 2A). In the presence of sortase A and D, BcpA was assembled into cell wall-anchored pili as demonstrated by immunogold-electron microscopy (Fig. 2C). Pilus formation was also confirmed by immunoblotting of covalently cross-linked high molecular weight polymers in the stack of SDS-PAGE that had been released from murein sacculi by treatment with mutanolysin (Fig. 2B). In the absence of sortase D (pJB173), sortase A deposited BcpA into the cell wall envelope (Fig. 2B). Sortase A did not form pili from BcpA and instead caused the near uniform distribution of anchored product in the cell wall envelope of bacilli (Fig. 2C). In the absence of both sortase A and sortase D, BcpA was not found in the envelope of bacilli, similar to a B. anthracis control harboring the empty vector plasmid pLM5 (Fig. 2C). To analyze the cell wall anchor structure of BcpA, we engineered BcpAMH6 with an insertion of methionyl-six histidyl (MH6) upstream of the LPVTG motif sorting signal of BcpA (Fig. 2A). Cell wall-anchored BcpAMH6 was detected in mutanolysin extracts of B. anthracis (pJB169) by immunoblot with antibodies specific for BcpA (α-BcpA) or staining with His-HRP (Fig. 2B). Electron microscopy and immunogold labeling with BcpA-specific antibodies revealed gold particle deposition on the surface of bacilli expressing BcpAMH6 (pJB169), but not for B. anthracis harboring the empty vector (pLM5) control (Fig. 2C). In the absence of srtA, BcpA- and BcpAMH6-specific gold conjugate deposits were not detected in the envelope of bacilli (Fig. 2C). As BcpAMH6 is anchored to the cell wall of bacilli in a manner similar to BcpA, we employed this reporter to examine the anchoring of pilin protein to the peptidoglycan.
FIGURE 2.
Purification of BcpAMH6 from the cell wall envelope of bacilli. A, diagram displays plasmids with pilin genes under control of the IPTG inducible Pspac promoter. pJB12 expressed wild-type bcpA-srtD-bcpB. Plasmids pJB173 and pJB169 expressed bcpA. pJB169 contained the MH6 peptide three residues upstream from the LPVTG sorting signal. B, B. anthracis cell wall extracts were digested with mutanolysin. BcpA anchoring was examined by immunoblotting with α-BcpA and by binding to His-HRP in cells harboring pJB12, pJB169, and pJB173. C, bacilli were analyzed by immunogold labeling with α-BcpA antisera and viewed by transmission electron microscopy at a magnification of 35,000 × (scale bars, 200 nm).
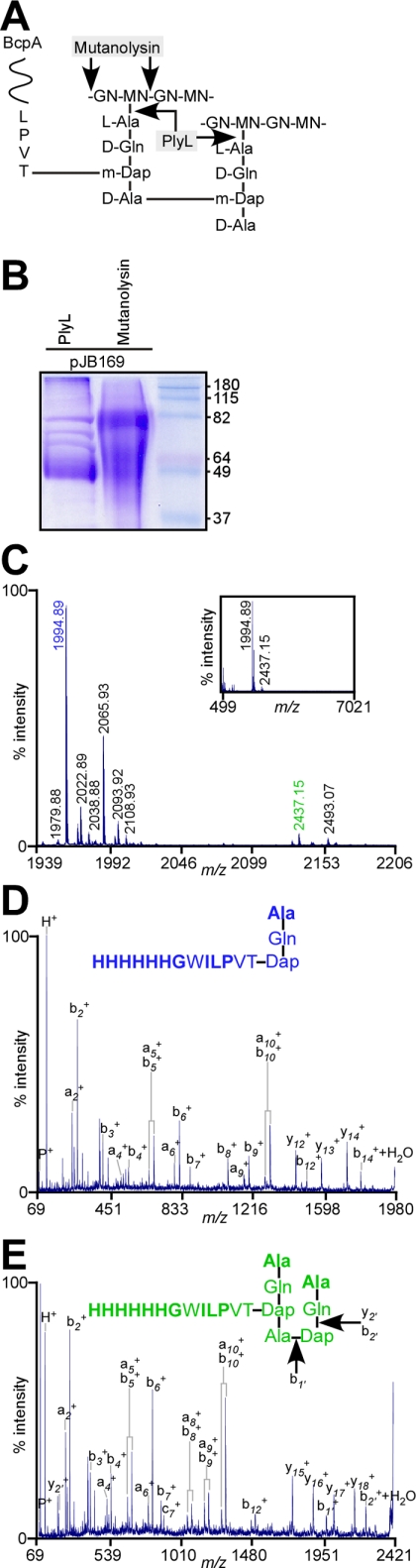
BcpAMH6 was solubilized from purified cell wall of B. anthracis (pJB169) with two different murein hydrolases and released protein was subjected to affinity chromatography on Ni-NTA. PlyL amidase cleaves the amide bond between MurNAc and l-Ala (24) (Fig. 3A). In agreement with the C-terminal cell wall anchor structures proposed in Fig. 3A, PlyL-solubilized BcpAMH6 migrated as a spectrum of fragments on Coomassie-stained SDS-PAGE (49-79 kDa), as ∼19% of wall peptides in B. anthracis peptidoglycan are cross-linked (28)(Fig. 3B). Purified peptidoglycan of B. anthracis was also digested with mutanolysin, a muramidase that cleaves the repeating disaccharides MurNAc-GlcNAc or MurNAc-GlcNH2 (Fig. 3A) (29, 30). Muramidase treatment released protein that also migrated as a spectrum fragments on SDS-PAGE (49-79 kDa), albeit that the mobility of these species differed from those of PlyL-solubilized BcpAMH6 (Fig. 3B).
FIGURE 3.
Structure of PlyL-released BcpAMH6 anchor peptides. A, cell wall anchor structure of BcpA and cleavage sites of the muralytic enzymes mutanolysin and PlyL amidase. B, Coomassie Blue-stained SDS-PAGE of Ni-NTA-purified BcpAMH6 solubilized from peptidoglycan by treatment with mutanolysin or PlyL amidase. C, MALDI-MS spectrum of BcpAMH6 anchor peptides released from B. anthracis peptidoglycan. Purified cell wall was treated with an E. coli extract containing PlyL amidase, and cell wall extracts were subjected to Ni-NTA affinity chromatography. Purified BcpAMH6 was cleaved with CNBr, and anchor peptides were purified by a second round of Ni-NTA affinity chromatography. Purified anchor peptides were separated by RP-HPLC. Inset displays the mass spectrum (m/z) from 499 to 7021 Da of RP-HPLC fraction 45 (35% acetonitrile-0.1% formic acid) determined by MALDI-MS. Parent ions 1994.89 m/z (colored blue) and 2437.15 m/z (colored green) were subjected to tandem mass spectrometry. D, CAD fragmentation spectrum (m/z) of parent ion 1994.89 m/z and the structural assignment of the anchor peptide. Residues in bold were identified by Edman degradation. E, CAD fragmentation spectrum (m/z) of parent ion 2437.15 m/z and the structure assignment of the anchor peptide. Arrows denote names assigned to fragment ions produced during fragmentation. Residues in bold were identified by Edman degradation.
Cell Wall Anchor Structure of PlyL-solubilized BcpAMH6—PlyL-solubilized and Ni-NTA-purified BcpAMH6 was cleaved at methionyl residues with CNBr. C-terminal peptides harboring the six-histidyl tag and cell wall anchor structure were purified by a second round of affinity purification on Ni-NTA. Peptides were separated by RP-HPLC with a C18 column. PlyL-released anchor peptides eluted at 35% CH3CN-0.1% formic acid and were analyzed by MALDI-MS and Edman degradation (Fig. 3C and Table 2, and see Table 5). The predominant ions in this spectrum could be explained as BcpAMH6 C-terminal anchor peptides linked to the side-chain amino group of diaminopimelic acid of wall peptides (Table 2). meso-Diaminopimelic acid (m-Dap) is a diamino acid at position 3 of the Bacillus cell wall pentapeptide precursor (l-Ala-d-iGln-m-Dap-d-Ala-d-Ala), and its side-chain amino group is involved in peptidoglycan cross-linking by forming an amide bond with d-Ala at position four of neighboring wall peptides [l-Ala-d-iGln-(l-Ala-d-iGln-m-Dap-d-Ala-)m-Dap-d-Ala-d-Ala] (28). The predicted m/z of the anchor peptide [l-Ala-d-iGln-(H6GWILPVT-)m-Dap] is 1978.99. Isoforms containing tryptophan oxidation products hydroxytryptophan or N-formylkynurenine are expected at a mass of 1994.98 and 2010.98, respectively (Table 2) (31). Formylated, diformylated, and carbamylated anchor peptide species were detected at m/z 2021.9, 2022.8, 2037.9, 2038.88, 2050.89, and 2053.89 (Table 2). Formylation and carbamylation of α-NH2 groups in cell wall anchor peptides are known to occur following CNBr cleavage (70% formic acid) and purification under denaturing conditions in urea buffer. The predominant ion at m/z 1994.89 was fragmented by collision-activated dissociation (CAD) (Fig. 3D). Table 3 summarizes the observed fragment ions and their structural interpretation, which is in agreement with the peptide structure l-Ala-d-iGln-(H6GWILPVT-)m-Dap harboring the tryptophan oxidation product hydroxytryptophan (Table 3 and Fig. 3D). The second most abundant ion at m/z 2065.93 was explained as l-Ala-d-iGln-(H6GWILPVT-)m-Dap-d-Ala, a C-terminal anchor peptide containing d-Ala at position 4 of the cell wall pentapeptide precursor (calculated m/z 2066.02) (Table 2 and Fig. 3D). The structure of ion 2065.93 m/z was also confirmed by tandem mass spectrometry (data not shown).
TABLE 2.
Summary of ions observed in the mass spectrum of PlyL-released BcpAMH6 anchor peptides
|
m/z
|
Δcalc-obsa
|
Proposed structure
|
Modificationb
|
|
|---|---|---|---|---|
| Observed | Calculated | |||
| 1979.88 | 1978.99 | −0.89 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap] | none |
| 1994.89 | 1994.98 | 0.09 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap] | HTRP |
| 2010.90 | 2010.98 | 0.08 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap] | NFK |
| 2021.90 | 2022.00 | 0.10 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap] | NH2-C(0)- |
| 2022.80 | 2022.98 | 0.18 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap] | HTRP; HC(O)- |
| 2037.90 | 2037.99 | 0.09 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap] | HTRP; NH2-C(O)- |
| 2038.88 | 2038.97 | 0.09 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap] | NFK; HC(O)- |
| 2050.89 | 2050.99 | 0.10 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap] | HTRP; 2 X HC(O)- |
| 2053.89 | 2053.99 | 0.10 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap] | NFK; NH2-C(O)- |
| 2065.93 | 2066.02 | 0.09 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap-d-Ala] | HTRP |
| 2081.92 | 2082.02 | 0.10 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap-d-Ala] | NFK |
| 2093.92 | 2094.02 | 0.10 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap-d-Ala] | HTRP; HC(O)- |
| 2108.93 | 2109.03 | 0.10 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap-d-Ala] | HTRP; NH2-C(O)- |
| 2122.93 | 2122.03 | 0.10 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap-d-Ala] | HTRP; 2 × HC(O)- |
| 2124.95 | 2125.02 | 0.07 | [l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap-d-Ala] | NFK; NH2-C(O)- |
| 2437.08 | 2437.20 | 0.12 | l-Ala-d-iGln-[l-Ala-d-iGln(HHHHHHGWIPLVT -)m-Dap-d-Ala]m-Dap | HTRP |
| 2465.09 | 2465.20 | 0.11 | l-Ala-d-iGln-[l-Ala-d-iGln(HHHHHHGWIPLVT -)m-Dap-d-Ala]m-Dap | HTRP; HC(O)- |
| 2480.08 | 2480.21 | 0.13 | l-Ala-d-iGln-[l-Ala-d-iGln(HHHHHHGWIPLVT -)m-Dap-d-Ala]m-Dap | HTRP; NH2-C(O)- |
| 2493.07 | 2493.21 | 0.14 | l-Ala-d-iGln-[l-Ala-d-iGln(HHHHHHGWIPLVT -)m-Dap-d-Ala]m-Dap | HTRP; 2 × HC(O)- |
The difference between the calculated and observed ion masses.
Modification of αNH2 groups: formylation, HC(O)-, carbamylation, NH2-C(O)-; modification of tryptophan: hydroxytryptophan (HTRP), N-formylkynurenine (NFK).
TABLE 5.
Edman degradation of PlyL-released anchor peptides
A slash (/) indicates that no amino acid could be assigned in this cycle.
| Cycle | Amino acid |
|---|---|
| pM | |
| 1 | H (32.73), A (61.62) |
| 2 | H (23.96) |
| 3 | H (27.18) |
| 4 | H (24.46) |
| 5 | H (22.56) |
| 6 | H (21.86) |
| 7 | G (4.49) |
| 8 | / |
| 9 | I (2.94) |
| 10 | L (2.29) |
| 11 | P (1.49) |
| 12 | / |
TABLE 3.
Summary of ions produced during MS/MS of the PlyL-released m/z 1994.89 parent ion
|
m/z
|
Δcalc-obsa
|
Ion
typeb
|
Proposed
structurec
|
|
|---|---|---|---|---|
| Observed | Calculated | |||
| 70.07 | 70.13 | 0.07 | Immonium | P |
| 110.07 | 110.15 | 0.08 | Immonium | H |
| 247.13 | 247.24 | 0.11 | a2 | HH |
| 260.10 | 260.19 | 0.08 | i | GW |
| 275.13 | 275.21 | 0.09 | b2 | HH |
| 332.15 | 332.23 | 0.08 | i | HHG |
| 397.16 | 397.27 | 0.11 | i | HGW |
| 412.18 | 412.27 | 0.09 | b3 | HHH |
| 510.25 | 510.32 | 0.08 | i | HGWI |
| 521.25 | 521.31 | 0.06 | a4 | HHHH |
| 534.22 | 534.29 | 0.07 | i | HHGW |
| 549.24 | 549.36 | 0.12 | b4 | HHHH |
| 623.33 | 623.39 | 0.06 | i | HGWIL |
| 658.31 | 658.40 | 0.09 | a5 | HHHHH |
| 686.30 | 686.37 | 0.07 | b5 | HHHHH |
| 687.33 | 688.42 | 1.09 | y6 | l-Ala-d-iGln-(PVT-)m-Dap |
| 760.39 | 760.45 | 0.06 | i | HHGWIL |
| 795.37 | 795.47 | 0.11 | a6 | HHHHHH |
| 823.36 | 823.44 | 0.08 | b6 | HHHHHH |
| 880.38 | 880.49 | 0.11 | b7 | HHHHHHG |
| 1034.51 | 1034.68 | 0.17 | i | HHHHGWIL |
| 1054.46 | 1054.59 | 0.13 | a8 | HHHHHHGW |
| 1082.46 | 1082.61 | 0.15 | b8 | HHHHHHGW |
| 1167.55 | 1167.78 | 0.23 | a9 | HHHHHHGWI |
| 1171.57 | 1171.80 | 0.23 | i | HHHHHGWILP |
| 1195.54 | 1195.71 | 0.17 | b9 | HHHHHHGWI |
| 1280.63 | 1280.79 | 0.16 | a10 | HHHHHHGWIL |
| 1308.62 | 1308.82 | 0.19 | b10 | HHHHHHGWIL |
| 1446.71 | 1446.95 | 0.24 | y12 | l-Ala-d-iGln-(HHGWILPVT-)m-Dap |
| 1504.75 | 1505.06 | 0.32 | b12 | HHHHHHGWILPV |
| 1583.77 | 1584.10 | 0.33 | y13 | l-Ala-d-iGln-(HHHGWILPVT-)m-Dap |
| 1703.80 | 1704.17 | 0.36 | y14-NH3 | l-Ala-d-iGln-(HHHHGWILPVT-)m-Dap |
| 1720.83 | 1721.16 | 0.33 | y14 | l-Ala-d-iGln-(HHHHGWILPVT-)m-Dap |
| 1795.85 | 1795.27 | −0.58 | b14+H2O | HHHHHHGWILPVT-m-Dap |
The difference between the monoisotopic calculated and observed m/z of fragment ions.
i denotes internal ions.
W is modified as hydroxytryptophan (HTRP).
Ion signals indicating cross-linking between anchor peptides and neighboring cell wall peptides were detected (Table 2 and Fig. 3C). Several species at m/z 2437.08, 2465.09, 2480.08, and 2493.07 m/z could be explained as the tryptophan-oxidized C-terminal BcpAMH6 peptide anchored to cell wall tetrapeptide that was in turn cross-linked to diaminopimelic acid within another wall tripeptide: l-Ala-d-iGln-[l-Ala-d-iGln-(H6GWILPVT-)m-Dap-d-Ala]m-Dap and its formylated, carbamylated, and diformylated forms (Table 2). To test this prediction, the ion at m/z 2437.08 was subjected to tandem mass spectrometry with CAD, which confirmed its predicted structure (calculated m/z 2437.20) (Table 4 and Fig. 3E). PlyL-released BcpAMH6 anchor peptides were subjected to Edman degradation, which released two amino acid residues in the first cycle, histidine and alanine (Table 5). These data corroborate mass spectrometry results, revealing anchor peptides with branched structure and two N-terminal residues accessible to phenylthiohydantoin-mediated cleavage. The first is generated by CNBr cleavage at the methionyl immediately upstream of the MH6 tag, whereas the other is generated by PlyL cleavage of the amide bond between N-acetylmuramic acid and the l-Ala of wall peptides. Subsequent cycles of Edman sequencing identified amino acids HHHHHG/ILP, which confirmed the predicted structure of the PlyL-released anchor peptides (Table 5). No amino acid could be assigned following cycle eight, which is likely caused by degradation of tryptophan during the Edman cycle (Table 5). Taken together, these results demonstrate that the C-terminal threonine of sortase A-cleaved BcpAMH6 is amide-linked to the side-chain amino group of diaminopimelic acid within wall peptides, some of which are cross-linked with neighboring wall peptides.
TABLE 4.
Summary of ions produced during MS/MS of the PlyL-released m/z 2437.2 parent ion
|
m/z
|
Δcalc-obsa
|
Ion
typeb
|
Proposed
structurec
|
|
|---|---|---|---|---|
| Observed | Calculated | |||
| 70.07 | 70.14 | 0.07 | Immonium | P |
| 110.07 | 110.15 | 0.08 | Immonium | H |
| 195.09 | 195.21 | 0.12 | i | HG |
| 201.09 | 202.20 | 1.11 | y2′-NH3 | L-Ala-D-iGln |
| 247.13 | 247.23 | 0.10 | a2 | HH |
| 260.10 | 260.23 | 0.13 | i | GW |
| 275.13 | 275.23 | 0.10 | b2 | HH |
| 373.21 | 373.29 | 0.08 | i | VT-m-Dap |
| 384.19 | 384.25 | 0.06 | i | D-iGln-(T-)m-Dap |
| 397.16 | 397.26 | 0.10 | i | HGW |
| 412.18 | 412.28 | 0.10 | b3 | HHH |
| 510.25 | 510.37 | 0.13 | i | HGWI |
| 521.25 | 521.38 | 0.13 | a4 | HHHH |
| 534.22 | 534.34 | 0.12 | i | HHGW |
| 549.24 | 549.32 | 0.08 | b4 | HHHH |
| 651.35 | 651.52 | 0.17 | i-H2O | L-Ala-D-iGln-(PVT-)m-Dap |
| 658.31 | 658.41 | 0.10 | a5 | HHHHH |
| 671.28 | 671.33 | 0.05 | i | HHHGW |
| 686.30 | 686.38 | 0.08 | b5 | HHHHH |
| 760.39 | 760.44 | 0.06 | i | HHGWIL |
| 795.37 | 795.49 | 0.13 | a6 | HHHHHH |
| 797.42 | 797.36 | −0.06 | i-H2O | L-Ala-D-iGln-(VT-)m-Dap-D-Ala |
| 823.36 | 823.43 | 0.07 | b6 | HHHHHH |
| 853.48 | 853.42 | −0.06 | i | L-Ala-D-iGln-(LPVT-)m-Dap-D-Ala |
| 880.38 | 880.50 | 0.11 | b7 | HHHHHHG |
| 897.41 | 897.46 | 0.05 | c7 | HHHHHHG |
| 898.50 | 898.45 | −0.06 | i | (WILPVT-)m-Dap |
| 1034.51 | 1034.62 | 0.12 | i | HHHHGWIL |
| 1054.46 | 1054.83 | 0.37 | a8 | HHHHHHGW |
| 1082.46 | 1082.65 | 0.19 | b8 | HHHHHHGW |
| 1167.55 | 1167.73 | 0.18 | a9 | HHHHHHGWI |
| 1168.64 | 1169.08 | 0.45 | i | L-Ala-D-iGln-(WILPVT-)m-Dap-D-Ala |
| 1194.62 | 1194.56 | −0.06 | i | HHHGWILPVT |
| 1195.54 | 1195.63 | 0.09 | b9 | HHHHHHGWI |
| 1280.63 | 1280.83 | 0.20 | a10 | HHHHHHGWIL |
| 1308.62 | 1308.76 | 0.13 | b10 | HHHHHHGWIL |
| 1504.75 | 1504.99 | 0.24 | b12 | HHHHHHGWILPV |
| 1751.91 | 1752.16 | 0.25 | y15 | L-Ala-D-iGln-[L-Ala-D-iGln-(HGWILPVT-)m-Dap-D-Ala]m-Dap |
| 1888.97 | 1889.25 | 0.28 | y16 | L-Ala-D-iGln-[L-Ala-D-iGln-(HHGWILPVT-)m-Dap-D-Ala]m-Dap |
| 1976.97 | 1977.23 | 0.26 | b1′ | [L-Ala-D-iGln-(HHHHHHGWILPVT-)m-Dap-D-Ala] |
| 2026.03 | 2026.42 | 0.40 | y17 | L-Ala-D-iGln-[L-Ala-D-iGln-(HHHGWILPVT-)m-Dap-D-Ala]m-Dap |
| 2163.08 | 2163.44 | 0.36 | y18 | L-Ala-D-iGln-[L-Ala-D-iGln-(HHHHGWILPVT-)m-Dap-D-Ala]m-Dap |
| 2237.12 | 2237.71 | 0.58 | c2′ | [L-Ala-D-iGln-(HHHHHHGWILPVT-)m-Dap-D-Ala]m-Dap |
| 2238.11 | 2238.53 | 0.43 | b2′ + H2O | [L-Ala-D-iGln-(HHHHHHGWILPVT-)m-Dap-D-Ala]m-Dap |
The difference between the monoisotopic calculated and observed masses of fragment ions.
i denotes internal ions. Fig. 3C indicates the proposed site of fragmentation in ions labeled with subscripts 1′ and 2′.
W is modified as hydroxytryptophan (HTRP).
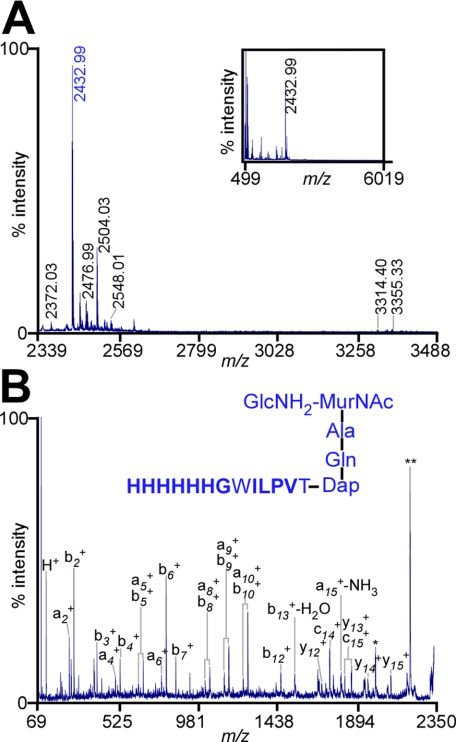
Cell Wall Anchor Structure of Muramidase-solubilized BcpAMH6—Isolated bacterial cell wall was cleaved with mutanolysin at the β(1-4)-glycosidic bond of MurNAc-Glc-NAc or MurNAc-GlcNH2 (Fig. 3A). Following affinity purification by Ni-NTA, BcpAMH6 was cleaved with CNBr and amino sugars of C-terminal anchor peptides were reduced with sodium borohydride (32). Glycopeptides were subjected to RP-HPLC with a C18 column, and anchor peptides were found to elute at 35% CH3CN-0.1% formic acid. MALDI-MS revealed two clusters of peaks (Fig. 4A). The predominant ion in the first cluster, m/z 2432.99, was explained as oxidized C-terminal anchor peptide linked to murein disaccharide tripeptide: GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap], a structure that was confirmed with CAD fragmentation (calculated m/z 2433.17) (Table 6 and Fig. 4B). Other ions in the first cluster were explained as the oxidation product of the tryptophan residue to N-formylkynurenine, formylation, or carbamylation of m/z 2433.17 (m/z 2499.02, 2460.98, 2476.99, and 2489.04) (Table 7 and Fig. 4A). Additional ion signals were explained as the anchor peptides cross-linked to murein disaccharide tetrapeptide GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap-d-Ala] with tryptophan oxidation and/or modification at the α-NH2 amino group (Table 7 and Fig. 4A).
FIGURE 4.
Structure of mutanolysin-released BcpAMH6 anchor peptides. A, MALDI-MS spectrum of BcpAMH6 anchor peptides released from B. anthracis peptidoglycan. Purified cell wall was treated with mutanolysin, and cell wall extracts were subjected to Ni-NTA affinity chromatography. Purified BcpAMH6 was cleaved with CNBr, and anchor peptides were purified by a second round of Ni-NTA affinity chromatography. Purified anchor peptides were separated by RP-HPLC. Inset displays the mass spectrum (m/z) from 499 to 6019 Da of RP-HPLC fraction 45 (35% acetonitrile-0.1% formic acid) determined by MALDI-MS. The parent ion 2432.99 m/z (colored blue) was subjected to tandem mass spectrometry. B, CAD fragmentation spectrum (m/z) of parent ion 2432.99 m/z and the structural assignment of the anchor peptide. Residues in bold were identified by Edman degradation.
TABLE 6.
Summary of fragment ions produced during MS/MS of the mutanolysin-released m/z 2432.99 parent ion
|
m/z
|
Δcalc-obsa
|
Ion
typeb
|
Proposed
structurec
|
|
|---|---|---|---|---|
| Observed | Calculated | |||
| 110.07 | 110.15 | 0.08 | Immonium | |
| 195.09 | 195.17 | 0.09 | i | HG |
| 247.13 | 247.22 | 0.09 | a2 | HH |
| 260.10 | 260.23 | 0.13 | i | GW |
| 275.13 | 275.24 | 0.12 | b2 | HH |
| 332.15 | 332.21 | 0.06 | i | HHG |
| 373.17 | 373.35 | 0.18 | i | l-Ala-d-iGln-m-Dap |
| 384.19 | 384.28 | 0.09 | i-H2O | d-iGln-(T-)m-Dap |
| 395.30 | 395.19 | −0.11 | i-28 | HHH |
| 397.16 | 397.28 | 0.12 | i | HGW |
| 412.18 | 412.32 | 0.14 | b3 | HHH |
| 483.26 | 483.34 | 0.08 | i-H2O | d-iGln-(VT-)m-Dap |
| 506.33 | 506.37 | 0.04 | i-H2O | ILPVT |
| 521.25 | 521.34 | 0.09 | a4 | HHHH |
| 549.24 | 549.36 | 0.12 | b4 | HHHH |
| 623.33 | 623.49 | 0.16 | i | HGWIL |
| 658.31 | 658.39 | 0.08 | a5 | HHHHH |
| 686.30 | 686.44 | 0.14 | b5 | HHHHH |
| 784.36 | 784.42 | 0.06 | i | HHHHGW |
| 795.37 | 795.48 | 0.11 | a6 | HHHHHH |
| 823.36 | 823.45 | 0.09 | b6 | HHHHHH |
| 880.38 | 880.49 | 0.11 | b7 | HHHHHHG |
| 897.41 | 897.59 | 0.18 | c7 | HHHHHHG |
| 1034.51 | 1034.64 | 0.13 | i | HHHHGWIL |
| 1054.46 | 1054.56 | 0.10 | a8 | HHHHHHGW |
| 1082.46 | 1082.61 | 0.15 | b8 | HHHHHHGW |
| 1167.55 | 1167.73 | 0.19 | a9 | HHHHHHGWI |
| 1171.57 | 1171.91 | 0.35 | i | HHHHHGWIL |
| 1195.54 | 1195.69 | 0.15 | b9 | HHHHHHGWI |
| 1280.63 | 1280.96 | 0.33 | a10 | HHHHHHGWIL |
| 1308.62 | 1308.83 | 0.21 | b10 | HHHHHHGWIL |
| 1477.73 | 1477.03 | −0.71 | i-NH3 | HHHGWILPVT-m-Dap-d-iGln |
| 1504.75 | 1504.92 | 0.18 | b12 | HHHHHHGWILPV |
| 1587.78 | 1587.03 | −0.75 | b13-H2O | HHHHHHGWILPVT |
| 1747.87 | 1748.23 | 0.36 | y12 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HGWILPVT-) m-Dap) |
| 1794.90 | 1795.24 | 0.33 | c14 | HHHHHHGWILPVT-m-Dap |
| 1860.92 | 1861.25 | 0.34 | a15-NH3 | d-iGln-(HHHHHHGWILPVT-)m-Dap |
| 1884.93 | 1885.38 | 0.45 | y13 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHGWILPVT-) m-Dap) |
| 1922.96 | 1923.33 | 0.37 | c15 | d-iGln-(HHHHHHGWILPVT-)m-Dap |
| 2021.99 | 2022.43 | 0.43 | y14 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHGWILPV T-)m-Dap) |
| 2066.63 | 2067.41 | 0.78 | -d | lactoyl-[l-Ala-d-iGln-(HHHHHHGWILPVT-)m-Dap] |
| 2159.05 | 2159.41 | 0.36 | y15 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHGWILPV T-)m-Dap) |
| 2271.78 | 2272.80 | 1.02 | -e | MurNAc-[l-Ala-d-iGln-(HHHGWILPVT-)m-Dap) |
The difference between the calculated and observed masses of fragment ions.
i denotes internal ions.
W is modified as HTRP.
The y-type ion originating from cleavage of the ether bond between glucosamine and the lactoyl group of N-acetylmuramitol.
The y-type ion originating from cleavage of the β-1,4-O-glycosidic bond between glucosamien and N-acetylmuramitol.
TABLE 7.
Summary of ions observed in the mass spectrum of mutanolysin-released BcpAMH6 anchor peptides
|
m/z
|
Δcalc-obsa
|
Proposed structure
|
Modificationb
|
|
|---|---|---|---|---|
| Observed | Calculated | |||
| 2432.99 | 2433.17 | 0.18 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap] | HTRP |
| 2449.02 | 2449.16 | 0.15 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap] | NFK |
| 2460.98 | 2461.16 | 0.18 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap] | HTRP; HC(O)- |
| 2476.99 | 2476.18 | −0.81 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap] | HTRP; NH2-C(O)- |
| 2489.04 | 2489.17 | 0.14 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap] | HTRP; 2 × HC(O)- |
| 2504.03 | 2504.21 | 0.17 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap-d-Ala] | HTRP |
| 2519.03 | 2520.20 | 1.17 | GlcNH2-(β1-4)MurNac-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap-d-Ala] | NFK |
| 2531.06 | 2531.22 | 0.16 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap-d-Ala] | NH2-C(O)- |
| 2533.02 | 2532.20 | −0.82 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap-d-Ala] | HTRP; HC(O)- |
| 2544.04 | 2544.22 | 0.18 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap-d-Ala] | 2 × HC(O)- |
| 2548.02 | 2547.21 | −0.81 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap-d-Ala] | HTRP; NH2-C(O)- |
| 2548.02 | 2548.20 | 0.18 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap-d-Ala] | NFK; HC(O)- |
| 2564.02 | 2563.21 | −0.82 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap-d-Ala] | NFK; NH2-C(O)- |
| 3314.40 | 3313.57 | −0.82 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap-{GlcNH2-(β1-4)-MurNAc- [l-Ala-d-iGln-m-Dap-}d-Ala] | HTRP |
| 3341.38 | 3341.57 | 0.19 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap-{GlcNH2-(β1-4)-MurNAc- [l-Ala-d-iGln-m-Dap-}d-Ala] | HTRP; HC(O)- |
| 3355.33 | 3356.58 | 1.25 | GlcNH2-(β1-4)MurNAc-[l-Ala-d-iGln-(HHHHHH GWILPVT-)m-Dap-{GlcNH2-(β1-4)-MurNAc- [l-Ala-d-iGln-m-Dap-}d-Ala] | HTRP; NH2-C(O)- |
The difference between the monoisotopic calculated and observed ion masses.
Modification of αNH2 groups: formylation, HC(O)-; carbamylation, NH2-C(O)-; modification of tryptophan: hydroxytryptophan (HTRP), N-formylkynurenine (NFK).
The second set of ion signals indicated cross-linking between anchor peptides and neighboring cell wall tripeptide (m/z 3314.40, 3341.38, and 3355.33) in oxidized, formylated, or carbamylated anchor peptide species (Table 7 and Fig. 4A). Anchor peptides were analyzed with twelve cycles of Edman degradation, which yielded the sequence HHHHHHG/ILPV (Table 8). Of note, only one amino acid was released during the first cycle (histidine), as the wall peptides of the anchor structure are engaged in an amide bond with N-acetylmuramic acid. In agreement with previous work on the structure of sortase C-anchored proteins of B. anthracis (23), anchor peptides linked to cell wall peptides with higher degrees of cross-linking could not be detected.
TABLE 8.
Edman degradation of mutanolysin-released anchor peptides
A slash (/) indicates that no amino acid could be assigned in this cycle.
| Cycle | Amino acid |
|---|---|
| pm | |
| 1 | H (7.84) |
| 2 | H (7.15) |
| 3 | H (7.12) |
| 4 | H (7.23) |
| 5 | H (7.37) |
| 6 | H (6.00) |
| 7 | G (3.43) |
| 8 | / |
| 9 | I (3.22) |
| 10 | L (2.39) |
| 11 | P (1.36) |
| 12 | V (1.57) |
DISCUSSION
Sortases assemble proteins in the envelope of Gram-positive bacteria and, on the basis of biochemical criteria, four classes of enzymes can be distinguished (33). Sortase A cuts surface protein precursors at their LPXTG sorting signal and accepts lipid II as a nucleophile, thereby incorporating proteins into the peptidoglycan synthesis pathway (15, 34, 35). Sortase B recognizes surface protein precursors with distinct sorting signals (NPQTN motif in S. aureus) and employs cross-linked cell wall as a nucleophile (26, 36). The product of this reaction is the deposition of NEAT domain proteins in the cell wall envelope to enable heme-iron transport (37). Sortase C cleaves the LPNTA motif sorting signal of proteins in spore-forming bacteria and promotes protein deposition at cross-bridges of mother cell and spore peptidoglycan (23, 38-40). Finally, pilin-specific sortases, sortase D in pathogenic bacilli, polymerize major pilin proteins and assemble pili of Corynebacterium diphtheriae, Bacillus cereus, and many other Gram-positive bacteria (2, 6, 41-45). Pilin-specific sortases recognize the side-chain amino group of lysine within the YPKN motif as nucleophile for their transpeptidation reaction (2, 4, 6).
Although pilin-specific sortase D is clearly responsible for polymerization of the major pilin protein BcpA, a search for additional factors that may be required for the assembly of pili in bacilli identified sortase A, but not sortase B or sortase C (6). Similar results were obtained for Streptococcus agalactiae and Corynebacterium diphtheriae (9, 10). Of note, the B. anthracis sortase A mutant causes release of elongated pili into the extracellular medium (6), and sortase A cleaves the major pilin protein between the threonine and the glycine of the LPXTG motif, i.e. precisely at the same position as sortase D (4). Based on these observations, a model was derived whereby pilus assembly is terminated when sortase A cuts BcpA and accepts its nucleophile, lipid II. As BcpA can still be joined to the pilus via its YPKN motif, incorporation of lipid II at the C-terminal end would cause pili to be immobilized in the cell wall envelope. Thus, deposition of pili into the envelope may occur by a mechanism where sortase A competes with sortase D for its substrates (BcpA) (21). This model was tested here by analyzing the substrate requirements of BcpA for cleavage by sortase A or sortase D and by determining the anchor structure of the pilin protein.
In agreement with the model of competition for the same substrate, sortase A and sortase D displayed similar requirements for substrate recognition and cleavage, i.e. the C-terminal LPVTG motif sorting signal of BcpA. Although both enzymes cleave IsdX1SS-GST substrate, only sortase A can deposit IsdX1 into the cell wall envelope, corroborating the notion that sortases display nucleophile specificity and thereby control the fate of their substrates. The sortase A-anchored pilin protein was linked to the peptidoglycan cross-bridge, diaminopimelic acid, which is part of wall peptides of B. anthracis. These data are in agreement with the model whereby lipid II functions as the nucleophile of sortase A-pilin intermediates, causing biosynthetic incorporation of pili into the cell wall envelope (21). These results also represent the first demonstration of the cell wall anchor structure of pili in Gram-positive bacteria.
Some pili are assembled from two pilin subunits. For example in bacilli, the tip protein, BcpB, is tethered to the BcpAn polymer, whose C-terminal end is immobilized in the cell wall envelope (6, 46). Assembly requires two enzymes that cut BcpA and BcpB (sortase D) or presumably only BcpA (sortase A). Attack of specific nucleophiles, i.e. the YPKN motif of BcpA (sortase D) and lipid II (sortase A), establish assembly order in a manner so that BcpB can only be deposited at the pilus tip and BcpA polymer length is determined by the abundance of pilin substrate and enzyme and by the affinity of substrate for sortases A and D. Most pili in Gram-positive bacteria, however, are assembled from three precursors, one major pilin protein with YPKN motif, and two minor pilins (3). One of the minor pilin proteins appears deposited at the pilus tip (2), whereas in S. agalactiae and C. diphtheriae deletion of the structural gene for the second minor pilin protein causes release of fibers into the extracellular medium (11, 47), similar to mutations in sortase A (9). If so, sortase A may recognize the minor pilin protein, but not the major subunit, whereas the nucleophile within the minor subunit must compete with the YPKN motif of the major subunit and attack intermediates between pilin-specific sortase and the major pilin protein (21, 47). We suspect that the cell wall anchor structures of pili that are assembled from three subunits resemble that of B. anthracis pili. Nevertheless, experimental proof for substrate cleavage sites and nucleophiles is needed to verify the aforementioned models for pili that are derived from three subunits. Of note, Lemieux et al. (48) report that S. pneumoniae does not require sortase A to deposit pili made from three subunits in the cell wall envelope, suggesting that another, thus far unidentified sortase, may be able to fulfill the role of sortase A in pneumococci.
Acknowledgments
We thank the members of our laboratories for discussion and critical comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health, United States Public Health Service Grant AI38897 (to O. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IPTG, isopropyl 1-thio-β-d-galactopyranoside; Ni-NTA, nickel-nitrilotriacetic acid; RP-HPLC, reverse-phase high-performance liquid chromatography; MALDI-MS, matrix-assisted laser desorption ionization-mass spectrometry; GST, glutathione S-transferase; SS, sorting signal; HRP, horseradish peroxidase; m-Dap, meso-diaminopimelic acid; CAD, collision-activated dissociation.
References
- 1.Telford, J. L., Barocchi, M., Margarit, I., Rappuoli, R., and Grandi, G. (2006) Nat. Rev. Microbiol. 4 509-519 [DOI] [PubMed] [Google Scholar]
- 2.Ton-That, H., and Schneewind, O. (2003) Mol. Microbiol. 50 1429-1438 [DOI] [PubMed] [Google Scholar]
- 3.Ton-That, H., and Schneewind, O. (2004) Trends Microbiol. 12 251-261 [DOI] [PubMed] [Google Scholar]
- 4.Budzik, J. M., Marraffini, L. A., Souda, P., Whitelegge, J. P., Faull, K. F., and Schneewind, O. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 10215-10220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ton-That, H., Marraffini, L., and Schneewind, O. (2004) Mol. Microbiol. 53 1147-1156 [DOI] [PubMed] [Google Scholar]
- 6.Budzik, J. M., Marraffini, L. A., and Schneewind, O. (2007) Mol. Microbiol. 66 495-510 [DOI] [PubMed] [Google Scholar]
- 7.Ton-That, H., Liu, G., Mazmanian, S. K., Faull, K. F., and Schneewind, O. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 12424-12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maresso, A. W., Wu, R., Kern, J. W., Zhang, R., Janik, D., Missiakas, D. M., Duban, M. E., Joachimiak, A., and Schneewind, O. (2007) J. Biol. Chem. 282 23129-23139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dramsi, S., Caliot, E., Bonne, I., Guadagnini, S., Prevost, M. C., Kojadinovic, M., Lalioui, L., Poyart, C., and Trieu-Cuot, P. (2006) Mol. Microbiol. 60 1401-1413 [DOI] [PubMed] [Google Scholar]
- 10.Swaminathan, A., Mandlik, A., Swierczynski, A., Gaspar, A., Das, A., and Ton-That, H. (2007) Mol. Microbiol. 66 961-974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nobbs, A. H., Rosini, R., Rinaudo, C. D., Maione, D., Grandi, G., and Telford, J. L. (2008) Infect. Immun. 76 3550-3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navarre, W. W., and Schneewind, O. (1994) Mol. Microbiol. 14 115-121 [DOI] [PubMed] [Google Scholar]
- 13.Schneewind, O., Fowler, A., and Faull, K. F. (1995) Science 268 103-106 [DOI] [PubMed] [Google Scholar]
- 14.Ton-That, H., Mazmanian, H., Faull, K. F., and Schneewind, O. (2000) J. Biol. Chem. 275 9876-9881 [DOI] [PubMed] [Google Scholar]
- 15.Perry, A. M., Ton-That, H., Mazmanian, S. K., and Schneewind, O. (2002) J. Biol. Chem. 277 16241-16248 [DOI] [PubMed] [Google Scholar]
- 16.Higashi, Y., Strominger, J. L., and Sweeley, C. C. (1967) Proc. Natl. Acad. Sci. U. S. A. 57 1878-1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghuysen, J.-M., Tipper, D. J., Birge, C. H., and Strominger, J. L. (1965) Biochemistry 4 2245-2254 [DOI] [PubMed] [Google Scholar]
- 18.Mazmanian, S. K., Liu, G., Ton-That, H., and Schneewind, O. (1999) Science 285 760-763 [DOI] [PubMed] [Google Scholar]
- 19.Mazmanian, S. K., Liu, G., Jensen, E. R., Lenoy, E., and Schneewind, O. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 5510-5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaspar, A. H., Marraffini, L. A., Glass, E. M., DeBord, K. L., Ton-That, H., and Schneewind, O. (2005) J. Bacteriol. 187 4646-4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh, S.-Y., Budzik, J. M., and Schneewind, O. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 13703-13704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan, D. (1983) J. Mol. Biol. 166 557-572 [DOI] [PubMed] [Google Scholar]
- 23.Marraffini, L. A., and Schneewind, O. (2007) J. Bacteriol. 189 6425-6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Low, L. Y., Yang, C., Perego, M., Osterman, A., and Liddington, R. C. (2005) J. Biol. Chem. 280 35433-35439 [DOI] [PubMed] [Google Scholar]
- 25.Maresso, A. W., Chapa, T. J., and Schneewind, O. (2006) J. Bacteriol. 188 8145-8152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marraffini, L. A., and Schneewind, O. (2005) J. Biol. Chem. 280 16263-16271 [DOI] [PubMed] [Google Scholar]
- 27.Maresso, A. W., Garufi, G., and Schneewind, O. (2008) PLoS Pathogens 4 e1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Severin, A., Tabei, K., and Tomasz, A. (2004) Microb. Drug Resist. 10 77-82 [DOI] [PubMed] [Google Scholar]
- 29.Calandra, G. B., and Cole, R. (1980) Infect. Immun. 28 1033-1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokogawa, K., Kawata, S., Nishimura, S., Ikeda, Y., and Yoshimura, Y. (1974) Antimicrob. Agents Chemother. 6 156-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor, S. W., Fahy, E., Murray, J., Capaldi, R. A., and Ghosh, S. S. (2003) J. Biol. Chem. 278 19587-19590 [DOI] [PubMed] [Google Scholar]
- 32.Arbeloa, A., Hugonnet, J. E., Sentilhes, A. C., Josseaume, N., Dubost, L., Monsempes, C., Blanot, D., Brouard, J. P., and Arthur, M. (2004) J. Biol. Chem. 279 41546-41556 [DOI] [PubMed] [Google Scholar]
- 33.Marraffini, L. A., DeDent, A. C., and Schneewind, O. (2006) Microbiol. Mol. Biol. Rev. 70 192-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneewind, O., Mihaylova-Petkov, D., and Model, P. (1993) EMBO 12 4803-4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ton-That, H., Faull, K. F., and Schneewind, O. (1997) J. Biol. Chem. 272 22285-22292 [DOI] [PubMed] [Google Scholar]
- 36.Mazmanian, S. K., Ton-That, H., Su, K., and Schneewind, O. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2293-2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazmanian, S. K., Skaar, E. P., Gaspar, A. H., Humayun, M., Gornicki, P., Jelenska, J., Joachmiak, A., Missiakas, D. M., and Schneewind, O. (2003) Science 299 906-909 [DOI] [PubMed] [Google Scholar]
- 38.Marraffini, L. A., and Schneewind, O. (2006) Mol. Microbiol. 62 1402-1417 [DOI] [PubMed] [Google Scholar]
- 39.Claessen, D., Rink, R., de Jong, W., Siebring, J., de Vreugd, P., Boersma, F. G., Dijkhuizen, L., and Wosten, H. A. (2003) Genes Dev. 17 1714-1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elliot, M. A., Karoonuthaisiri, N., Huang, J., Bibb, M. J., Cohen, S. N., Kao, C. M., and Buttner, M. J. (2003) Genes Dev. 17 1727-1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosini, R., Rinaudo, C. D., Soriani, M., Lauer, P., Mora, M., Maione, D., Taddei, A., Santi, I., Ghezzo, C., Brettoni, C., Buccato, S., Margarit, I., Grandi, G., and Telford, J. L. (2006) Mol. Microbiol. 61 126-141 [DOI] [PubMed] [Google Scholar]
- 42.Lauer, P., Rinaudo, C. D., Soriani, M., Margarit, I., Maione, D., Rosini, R., Taddei, A. R., Mora, M., Rappuoli, R., Grandi, G., and Telford, J. L. (2005) Science 309 105. [DOI] [PubMed] [Google Scholar]
- 43.Barocchi, M. A., Ries, J., Zogaj, X., Hemsley, C., Albiger, B., Kanth, A., Dahlberg, S., Fernebro, J., Moschioni, M., Masignani, V., Hultenby, K., Taddei, A. R., Beiter, K., Wartha, F., von Euler, A., Covacci, A., Holden, D. W., Normark, S., Rappuoli, R., and Henriques-Normark, B. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 2857-2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nallapareddy, S. R., Singh, K. V., Sillanpää, J., Garsin, D. A., Höök, M., Erlandsen, S. L., and Murray, B. E. (2006) J. Clin. Invest. 116 2799-2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mora, M., Bensi, G., Capo, S., Falugi, F., Zingaretti, C., Manetti, A. G., Maggi, T., Taddei, A. R., Grandi, G., and Telford, J. L. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15641-15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mishra, A., Das, A., Cisar, J. O., and Ton-That, H. (2007) J. Bacteriol. 189 3156-3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mandlik, A., Das, A., and Ton-That, H. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 14147-14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemieux, J., Woody, S., and Camilli, A. (2008) J. Bacteriol. 190 6002-6013 [DOI] [PMC free article] [PubMed] [Google Scholar]