Abstract
Soluble epoxide hydrolase (sEH) is a bifunctional enzyme with two catalytic domains: a C-terminal epoxide hydrolase domain and an N-terminal phosphatase domain. Epidemiology and animal studies have attributed a variety of cardiovascular and anti-inflammatory effects to the C-terminal epoxide hydrolase domain. The recent association of sEH with cholesterol-related disorders, peroxisome proliferator-activated receptor activity, and the isoprenoid/cholesterol biosynthesis pathway additionally suggest a role of sEH in regulating cholesterol metabolism. Here we used sEH knock-out (sEH-KO) mice and transfected HepG2 cells to evaluate the phosphatase and hydrolase domains in regulating cholesterol levels. In sEH-KO male mice we found a ∼25% decrease in plasma total cholesterol as compared with wild type (sEH-WT) male mice. Consistent with plasma cholesterol levels, liver expression of HMG-CoA reductase was found to be ∼2-fold lower in sEH-KO male mice. Additionally, HepG2 cells stably expressing human sEH with phosphatase only or hydrolase only activity demonstrate independent and opposite roles of the two sEH domains. Whereas the phosphatase domain elevated cholesterol levels, the hydrolase domain lowered cholesterol levels. Hydrolase inhibitor treatment in sEH-WT male and female mice as well as HepG2 cells expressing human sEH resulted in higher cholesterol levels, thus mimicking the effect of expressing the phosphatase domain in HepG2 cells. In conclusion, we show that sEH regulates cholesterol levels in vivo and in vitro, and we propose the phosphatase domain as a potential therapeutic target in hypercholesterolemia-related disorders.
Soluble epoxide hydrolase (sEH)2 is a member of the epoxide hydrolase family (EC 3.3.2.3) with broad distribution in human tissues (1). sEH has now been conclusively shown to metabolize endogenous fatty acid epoxides generated by CYP450 epoxygenases (2, 3). These fatty acid epoxides (such as epoxyeicosatrienoic acids) have been shown to possess a wide variety of biological effects, many of which are related to cardiovascular physiology (4, 5). Epoxyeicosatrienoic acids have been described as major components of the endothelium-derived hyperpolarizing factors and are generally known for their vasodilator effects (6). sEH inhibitors have been shown to lower blood pressure in several animal models and have been proposed as therapeutic options for the management of hypertension (7, 8). Additionally, epidemiological studies link sEH polymorphisms with various human diseases, many of which are related to cardiovascular disorders. The R287Q polymorphism was found to be associated with subclinical atherosclerosis and coronary artery calcification in African-Americans and was described as an emerging risk factor for atherosclerosis (9, 10). Another variant (K55R) was associated with coronary heart disease in Caucasians (11). Interestingly, the R287Q variant was also found to be associated with increased levels of plasma cholesterol and triglycerides in familial hypercholesterolemia patients (12).
Recent studies suggest additional role(s)/mechanism(s) for sEH in cardiovascular diseases and metabolic disorders. Several groups showed that fatty acid epoxide substrates and diol products of sEH activate peroxisome proliferator-activated receptors (PPARs) α and γ (13-16). The exact role of sEH in modulating PPARs is currently unknown; however, the documented significance of PPARs in lipid metabolism and metabolic disorders is consistent with the above described epidemiological studies. Another emerging role of sEH is based on the recently described phosphatase activity of the sEH N-terminal domain (17, 18). Isoprenoid pyro- and monophosphates are substrates for the N-terminal domain (19, 20). These lipid phosphates are metabolic precursors of cholesterol biosynthesis and are also utilized for isoprenylation of small G-proteins involved in multiple cell signaling pathways (21). Interestingly, most current sEH literature attributes the hydrolase domain to the cardiovascular effects seen in humans. However, the human sEH single nucleotide polymorphism most often associated with cardiovascular disease (R287Q) encodes a protein with significantly lower hydrolase activity (22). In addition, the R287Q protein has been recently shown to have an altered subcellular localization phenotype (23). Thus, solely incriminating the hydrolase domain for adverse cardiovascular effects seems premature and highlights the importance of further investigating the independent roles of the hydrolase and phosphatase domains.
In this study we tested the hypothesis that sEH plays a significant role in regulating cholesterol metabolism in vitro and in vivo. First, we evaluated the effect of targeted disruption of the soluble epoxide hydrolase gene (sEH-knock-out (sEH-KO)) on plasma total cholesterol levels and liver expression of HMG-CoA reductase in mice. Second, we used HepG2 cell lines stably expressing modified sEH protein constructs and the human sEH R287Q variant to investigate the potential independent roles of the sEH phosphatase and hydrolase domains. Finally, we evaluated the effect of hydrolase inhibition on cholesterol levels in mice and HepG2 cells.
EXPERIMENTAL PROCEDURES
Animal Experiments—Mice on a 129X1/SvJ × C57BL/6 background with targeted disruption in the Ephx2 gene were obtained from Chris Sinal (Dalhousie University, Halifax, Canada) under National Cancer Institute Material Transfer Agreement 1-16268-04 (24). These mice were backcrossed onto a C57BL6 genetic background five generations prior to use in this study (25). All of the procedures were conducted in accordance with approved IACUC protocols. EPHX2-null or wild type mice (25-30 g) were treated with sEH inhibitor TUPS (laboratory number EHI 1709) (5, 26). TUPS was given via drinking water (10-15 mg/liter in 1% polyethylene glycol 400) for 7 days. Water consumption was monitored daily, and the concentration of TUPS was adjusted accordingly to achieve equal plasma concentrations in the treatment groups. A second set of animals was treated with 1% polyethylene glycol 400 alone (vehicle). Blood was collected by cardiac puncture from the right ventricle in EDTA-rinsed syringes following lethal injection of sodium pentobarbital (100 mg/kg body weight intraperitoneally). TUPS was extracted from blood samples, and the concentration was analyzed by liquid chromatography coupled mass spectroscopy liquid chromatography-tandem mass spectrometry as previously described (27). Plasma was separated (10 min, 400 × g) and stored at -80 °C until analysis. Liver samples were collected, frozen immediately, and kept at -80 °C.
HepG2 Stable Cell Lines—The human hepatoma (HepG2) cell line was obtained from ATCC (Manassas, VA). HepG2 cells were maintained in minimum essential medium supplemented with 2 mm l-glutamine, 0.1 mm nonessential amino acids, 1.0 mm sodium pyruvate, 10% (v/v) fetal bovine serum, and 0.5% antibiotics at 37 °C in a humidified incubator under 95% air and 5% CO2. To prepare HepG2 stable cell lines, human sEH was first cloned into BamHI/XhoI sites of pFastBac and pCR-Script vectors. The fragment was then subcloned into the BglII/SalI site of the pAcGFP-C1 vector (BD Bioscience) to produce green fluorescent protein fusion constructs. Site-directed mutagenesis was used to generate the hsEH R287Q variant, the phosphatase knock-out (D9A), and the hydrolase knock-out (D335S) constructs as previously described (22). All of the PCR products were completely sequenced to verify their identity. For transfections, the cells were grown to 90-100% confluency in T75 flasks then seeded at 7 × 105 cells/well in 6-well plates. After allowing the cells to attach for 24 h, they were transfected with sEH-green fluorescent protein fusion proteins using Effectene (Qiagen) according to the manufacturer's protocol. The cells were then passaged at a 1:20 ratio into selection medium (maintenance medium supplemented with 0.7 mg/ml G418) 24 h after transfection. Green fluorescent protein-positive colonies were subsequently isolated and maintained in selection medium. sEH activity and expression in HepG2 cells were evaluated using t-DPPO (28) and Western blotting (1). Isoprenoid compounds were obtained from Sigma-Aldrich and Cayman Chemicals (Ann Arbor, MI). All treatments for measuring cholesterol in the media were carried out in 1% fetal bovine serum.
HepG2 Stable Cell Line Treatment—Transfected HepG2 cells (7 × 105 cells/well) were treated with the hydrolase inhibitor TUPS (1 μm) (5, 26)y or the phosphatase inhibitor farnesyl thioacetic acid (5 μm) (19) for 24 h. At the end of the incubation, the medium was collected, and total cholesterol was measured.
HMG-CoA Reductase Western Blot Analysis—Liver samples and HepG2 cells were homogenized in lysis buffer containing 10 mm HEPES (pH 7.4), 10 mm NaCl, 1 mm KH2PO4, 5 mm NaHCO3, 5 mm EDTA, 1 mm CaCl2, 0.5 mm MgCl2, and protease inhibitor mixture (Sigma-Aldrich). The homogenates were centrifuged three times at 13,000 × g for 30 min at 4 °C discarding the pellet each time. The collected supernatants were aliquoted and stored at -80 °C. Protein concentration was determined using the bicinchoninic acid assay (Pierce) with bovine serum albumin as a standard. SDS-PAGE was performed using 10% resolving gels. Western transfer using polyvinylidene difluoride membrane (Millipore, Bedford, MA) was performed overnight at 30 V. The membrane was blocked for 30 min in Tris-buffered saline containing 5% (w/v) nonfat dry milk and 0.25% Tween 20 (pH 7.4) and then incubated with either rabbit polyclonal anti-HMG-CoA reductase (1:1000; Santa Cruz Biotechnology) or rabbit polyclonal anti-sEH (1:5000) for 2 h. The secondary antibody, goat anti-rabbit IgG-peroxidase conjugate (1:10,000; Sigma) was incubated for 2 h. After washing, bound secondary antibody was detected with supersignal chemiluminescent substrate (Pierce). The proteins were visualized with a Kodak image station 440CF.
Analysis of Gene Expression—Forward and reverse primers were obtained from Integrated DNA Technologies (IDT). 5′-GGCCTCCATTGAGATCCG-3′ and 5′-CACAATAACTTCCCAGGGGT-3′ for HMG-CoA reductase, and 5′-TTCTTTGCAGCTCCTTCGTT-3′ and 5′-ATGGAGGGGAATACAGCCC-3′ for β-actin. Total RNA was extracted from liver samples using TRIzol reagent (Invitrogen). Reverse transcription was done using SuperScript reverse transcription-PCR kit (Invitrogen), and the concentration of cDNA was analyzed by real time PCR using SYBR green Supermix (Invitrogen). The experimental steps were essentially following those provided by the manufacturer. PCR was carried out for 40 cycles, and a final melting curve verified the authenticity of the target product.
Amplex Red Cholesterol Assay—Aliquots of growing medium or plasma were collected for measuring total cholesterol with the Amplex Red Cholesterol Assay (Molecular Probes, Eugene, OR) as described by the manufacturer. HepG2 cells expressing inactive sEH with mutations at both active sites (sEH-D9A/D335S) were used as control cells. Furthermore, sEH-D9A/D335S was compared with normal nontransfected HepG2 cells and found to exhibit similar cholesterol levels (p < 0.05).
Statistical Analysis—All of the experiments were repeated in replicates with appropriate controls as indicated. Statistical significance was evaluated using t test or ANOVA with p < 0.05. The statistics were calculated based on absolute cholesterol levels and then transformed to ratios for presentation.
RESULTS
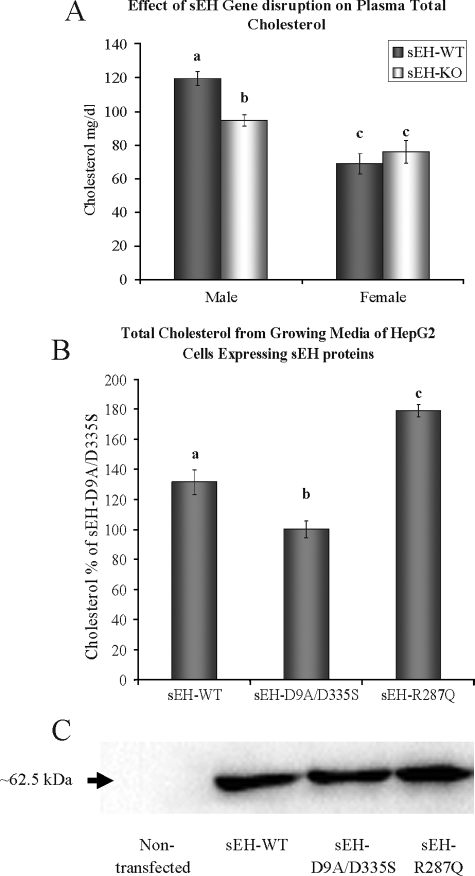
Effect of sEH Disruption on Cholesterol Levels—We first examined plasma cholesterol in response to targeted disruption of the Ephx2 gene in mice. Total plasma cholesterol was ∼25% lower in the 16-week-old male sEH-KO mice as compared with sEH-WT male mice (Fig. 1A). Interestingly, however, disruption of sEH in female mice did not have a significant effect on plasma cholesterol levels (Fig. 1A).
FIGURE 1.
A, sEH gene disruption shows ∼25% decrease in total plasma cholesterol level in male mice but not in females (four animals/group). B, HepG2 cells stably expressing sEH-WT or sEH-R287Q show a significant increase in total cholesterol in the growing medium as compared with HepG2 cells stably expressing sEH with mutated phosphatase and hydrolase active sites (sEH-D9A/D335S). C, representative Western blot analysis demonstrating stable transfection of sEH proteins in HepG2 cells throughout the course of the experiments as compared with nontransfected HepG2 cells. The total protein/lane is 30 μg. The different letters indicate statistical significance (ANOVA, p < 0.05).
Next we used HepG2 cells stably expressing sEH-WT, sEH-R287Q variant or sEH protein with mutated phosphatase and hydrolase active sites (D9A/D335S). The cells were seeded in six-well plates at 7 × 105 cells/well, and total cholesterol in the growing medium was measured after 48 h. Consistent with the above findings in mice, total cholesterol was significantly lower with sEH-D9A/D335S as compared with sEH-WT, whereas R287Q had the highest total cholesterol level (Fig. 1B). The level of sEH protein was similar in each of these three stable cell lines (Fig. 1C).
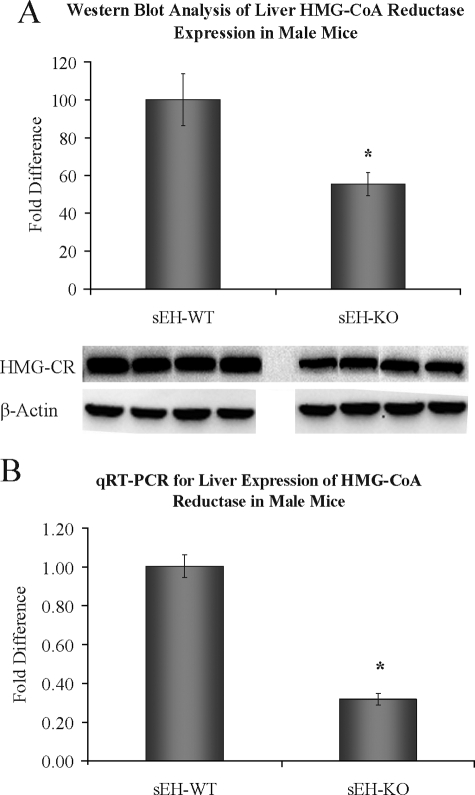
HMG-CoA Reductase Expression in Murine Liver—HMG-CoA reductase is considered the rate-limiting enzyme in cholesterol biosynthesis in vivo and is a common therapeutic target in the management of cholesterol related disorders. We evaluated the correlation of lower plasma total cholesterol in sEH-KO male mice with liver expression of HMG-CoA reductase. Indeed, both quantitative real time PCR and Western blot analysis indicate significantly lower liver expression levels of HMG-CoA reductase in sEH-KO male mice as compared with sEH-WT (Fig. 2).
FIGURE 2.
Liver HMG-CoA reductase expression level in sEH-WT and sEH-KO male mice by Western blot analysis (A) and quantitative real time PCR (B). n = four animals/group; *, significant statistical difference (p < 0.05).
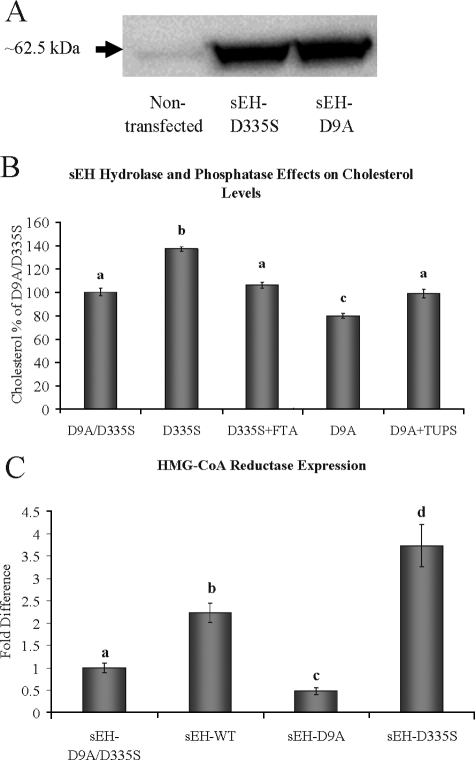
Independent Roles of the sEH Hydrolase and Phosphatase Domains—The results above indicate a significant role of sEH in modulating cholesterol levels in mice and in our HepG2 stable cell lines. However, these data do not differentiate phosphatase from hydrolase effects. Therefore, we measured total cholesterol levels from the growing medium of transfected HepG2 cells stably expressing sEH with inactive phosphatase or hydrolase domains (sEH-D9A or sEH-D335S, respectively). Protein expression levels for the two constructs were similar (Fig. 3A). Surprisingly, both domains had significant but opposite effects on total cholesterol levels (Fig. 3B). The sEH D9A mutation (possessing hydrolase activity only) resulted in a modest but statistically significant lowering of total cholesterol concentration, whereas the sEH D335S (possessing phosphatase activity only) resulted in a 40% increase in total cholesterol. The independent effects of the sEH phosphatase and hydrolase domains on cholesterol levels were further verified by reversing their effects using the corresponding inhibitors farnesyl thioacetic acid (19) and TUPS (26), respectively (Fig. 3B). These results are consistent with the results shown for the sEH-R287Q variant (Fig. 1B), which possess lower hydrolase (22) and higher phosphatase (19) activities as compared with sEH-WT. Next we evaluated the correlation between the media cholesterol levels and HMG-CoA reductase expression levels in HepG2 cells expressing different sEH proteins (Fig. 3C). Quantitative real time PCR displays a ∼2-fold higher expression level of HMG-CoA reductase in sEH-WT cells as compared with sEH-D9A/D335S control cells. However, sEH-D9A (hydrolase only) and sEH-D335S (phosphatase only) cells exhibited ∼0.5- and ∼4-fold differences, respectively.
FIGURE 3.
A, representative Western blot analysis demonstrating stable transfection of sEH proteins with mutated hydrolase (sEH-D335S) or phosphatase (sEH-D9A) active sites in HepG2 cells as compared with nontransfected cells. B, HepG2 cells stably expressing sEH-D335S or sEH-D9A show the opposite effects on total cholesterol level in the growing medium as compared with HepG2 cells stably expressing sEH-D9A/D335S. The cholesterol effects of sEH-D9A (active hydrolase) and sEH-D335S (active phosphatase) were reversed by the corresponding inhibitors TUPS (1 μm) and farnesyl thioacetic acid (5 μm), respectively. C, quantitative real time PCR demonstrates the correlation between HMG-CoA reductase expression and media cholesterol levels. The different letters indicate statistical significance (ANOVA, p < 0.05).
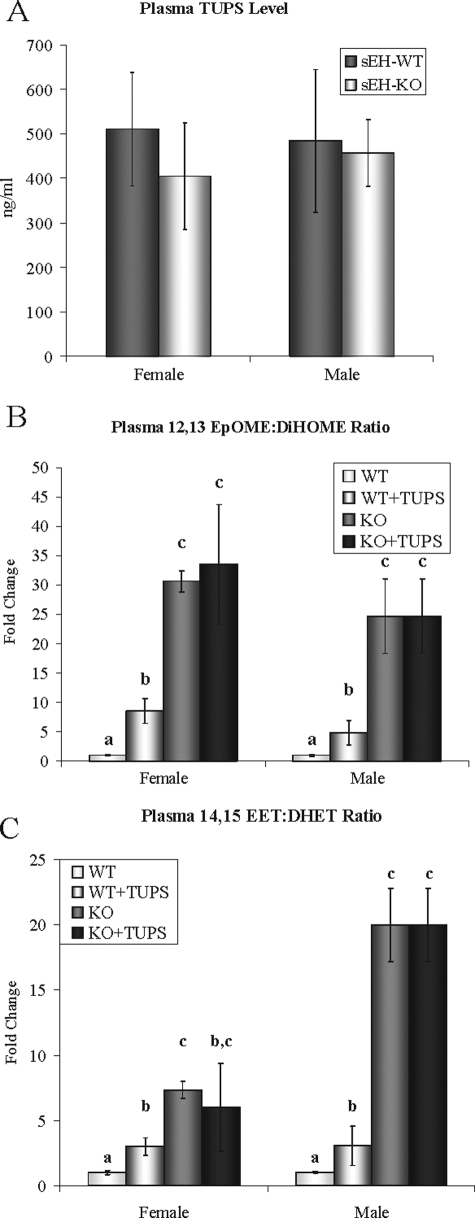
sEH Hydrolase Inhibition in Vivo—Sixteen-week-old male and female sEH-WT and sEH-KO mice were treated with TUPS (10-15 mg/liter) in drinking water for 1 week. Because of observed variation in water consumption between the different animal groups, the concentration of TUPS was adjusted to achieve comparable blood drug levels (Fig. 4A). This compound is a potent inhibitor of sEH (IC50 of 5 and 3 nm for murine and human sEH, respectively) and has higher bioavailability/water solubility (13.9 ± 1.5 μg/ml) than previously used inhibitors such as AUDA and AEPU (26). TUPS was formulated in water using 1% polyethylene glycol 400 to increase solubility. After 7 days of treatment, TUPS blood levels were 1.3 μm (Fig. 4A), which is significantly above the IC50 value.
FIGURE 4.
A, the concentration of TUPS in blood samples from sEH-WT and sEH-KO female and male mice after 7 days of treatment with TUPS in drinking water. The water consumption was monitored, and the TUPS concentration in drinking water (10-15 mg/liter in 1% polyethylene glycol 400) was adjusted accordingly (as described under “Experimental Procedures”). B and C, the epoxide/diol ratios of 12,13-linoleic acid and 14,15-arachidonic acid (epoxyocta-decenoic acid:dihydroxyoctadecenoic acid and epoxyeicosatrienoic acid:dihydroxy-eicosatrienoic acid, respectively) in plasma from hydrolase inhibitor-treated male and female sEH-WT mice as compared with sEH-KO. n = four animals/group. The letters indicate statistical significance (ANOVA, p < 0.05).
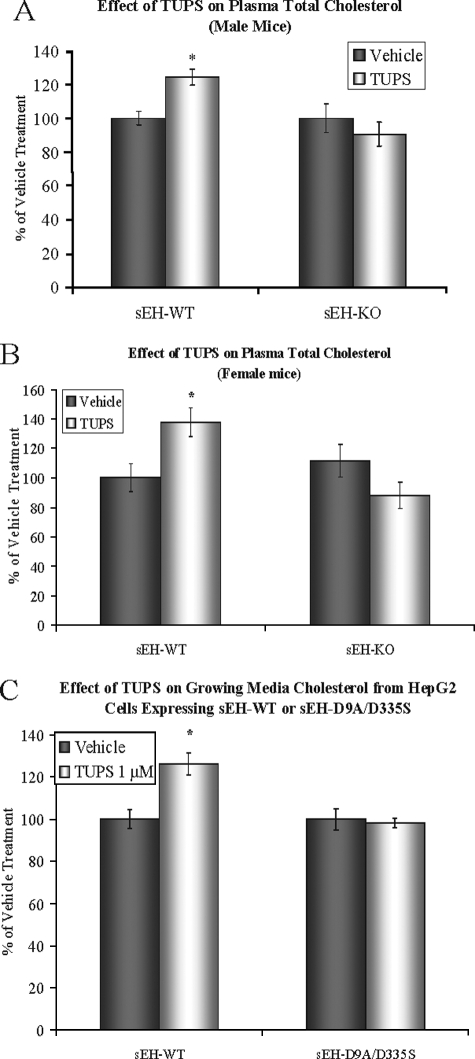
The in vivo effects of TUPS on epoxide hydrolase activity were estimated by measuring the ratios of linoleic acid and arachidonic acid epoxides to their corresponding diols (supplemental Figs. S1 and S2). TUPS treatment was effective in both male and female mice as demonstrated by similar increase of epoxide/diol ratios; however, the fold change was less than that of sEH gene disruption (Fig. 4, B and C). In agreement with the HepG2 results for the hydrolase and phosphatase effects (Fig. 3B), the total plasma cholesterol level showed a ∼25% increase following TUPS treatment in both male and female mice (Fig. 5, A and B). Similarly, HepG2 cells expressing sEH-WT showed ∼25% higher cholesterol levels in the growing medium following TUPS treatment (Fig. 5C). TUPS produced no significant effects on cholesterol levels in sEH-KO mice or HepG2 cells expressing inactive hsEH (Fig. 5), indicating that the effects of TUPS were caused by the inhibition of sEH.
FIGURE 5.
Increased plasma total cholesterol level in both male (A) and female (B) sEH-WT mice following 7 days of treatment with TUPS as compared with vehicle alone, as described in Fig. 3. TUPS treatment had no statistically significant effect on plasma cholesterol in sEH-KO mice. Four animals were included in each treatment or vehicle group. TUPS treatment of HepG2 cells stably expressing sEH-WT resulted in increased total cholesterol in the growing medium (C). * = statistically significant difference as compared with vehicle treatment (ANOVA, p < 0.05).
DISCUSSION
Over the past five years there has been mounting evidence suggesting a role for sEH in lipid metabolism and lipid related disorders. First, several epidemiological studies link human sEH polymorphisms with dyslipidemia (12) and related disorders such as atherosclerosis and coronary heart disease (9-11). Second, fatty acid epoxide substrates and diol products of the sEH hydrolase domain were found to activate PPARs (13-15, 29). PPARs regulate lipid metabolism (30, 31) and are known to modulate plasma lipid profiles in rodents (32) and humans (33-35). Our findings suggest that the hydrolase domain of sEH may exert its effect on lowering cholesterol by reducing the expression level of HMG-CoA reductase consistent with the effects of PPARγ agonists in HepG2 cells (30). Third, the recently identified phosphatase domain of sEH (17, 18) was shown to hydrolyze intermediates of the cholesterol biosynthesis pathway (19, 20). These isoprenoid intermediates are known to regulate various nuclear receptors involved in cholesterol metabolism, such as liver X receptor and PPARα (36-38). We show here that the effect of the phosphatase domain on cholesterol in HepG2 cells may also involve altering the expression level of HMG-CoA reductase. However, it is plausible that the effect of the phosphatase domain could be indirect through modulating liver X receptor activity by isoprenoid metabolites (36) and/or downstream interactions with PPARγ (39).
In this study we found significantly lower plasma total cholesterol levels in sEH-KO male mice as compared with their sEH-WT counterparts. Interestingly, the decrease in plasma cholesterol level as a result of sEH gene disruption described here is equal to or greater than what has been previously described in mice following statin treatment (40). This suggests that sEH may be a novel therapeutic target in the management of dyslipidemia. However, it should be noted that the effect of sEH gene disruption on plasma cholesterol was detected in male but not female mice. Intriguingly, the gender-specific effect of sEH gene disruption on cholesterol described here is in line with the previously described effect on blood pressure in male but not female mice (24). Previous studies have shown higher activity and expression levels of sEH in male mice as compared with females, and this could explain the gender dimorphism (41-43). Because the hydrolase and phosphatase domains of sEH are shown here to exhibit opposite effects on cholesterol levels, we suggest that the balance between the two domains may vary with the overall expression level of sEH. Therefore, targeted disruption of sEH in male mice results in a plasma cholesterol level similar to that of females. On the other hand and despite the lack of effect of sEH disruption on plasma cholesterol levels in females, hydrolase inhibition by TUPS in females results in a response similar to that found in males.
Even though there are significant differences in cholesterol metabolism between mice and human cells, our results suggest that sEH exerts its effects through conserved mechanisms. For example, the effect of the hydrolase inhibitor TUPs on cholesterol levels had a similar direction and magnitude in both cells and mice. Similarly, knocking out sEH had the same effect in both models. However, the effect of the N-terminal domain in HepG2 cells was addressed by genetically eliminating (mutating) the C-terminal domain. Genetic ablation of the C terminus could not be performed in mice, and hence the effects of the N terminus were examined by chemically inhibiting the C-terminal hydrolase domain using TUPs. It is conceivable that significant differences may exist between the mouse model and the human cells based on differences in cholesterol metabolism and experimental conditions; however, based on our findings from this study, sEH effects on cholesterol levels appear to be consistent and therefore mediated via conserved mechanisms.
sEH hydrolase inhibitors are currently being developed as potential therapeutic agents for inflammatory disorders (44) and cardiovascular diseases such as hypertension (45). This is based on a growing body of research demonstrating potent anti-inflammatory and vasodilator properties of the arachidonic acid epoxide substrates of the hydrolase domain. Here we showed that treatment with the TUPS hydrolase inhibitor resulted in a significantly higher cholesterol level in sEH-WT but not in sEH-KO mice. A similar effect was found in HepG2 cells stably expressing sEH-WT. The lack of a significant effect of TUPS on cholesterol levels in gene disrupted mice and sEH-D9A/D335S HepG2 cells argues against nonspecific drug effects. The effectiveness of hydrolase inhibition was monitored by measuring plasma epoxide/diol ratios. Despite the previously demonstrated potency of TUPS as an epoxide hydrolase inhibitor (26) and the significant change of epoxide/diol ratios with TUPS treatment in both male and female mice, it should be noted that genetic disruption of sEH had a much greater effect on epoxide/diol ratios. This might be expected with a small molecule enzyme inhibitor because of drug exclusion from some tissues or cellular compartments in an animal model. Also higher levels of epoxides are likely stored as esters in the genetically disrupted mice.
Surprisingly, the higher plasma cholesterol levels following hydrolase inhibition by TUPS are not in agreement with the lower plasma cholesterol levels detected in sEH-KO mice. However, a major difference is that sEH-KO mice lack both hydrolase and phosphatase activities, whereas hydrolase inhibitor treatment inhibits only the hydrolase activity. This suggests potential roles for both the hydrolase and phosphatase domains in regulating cholesterol metabolism. This is further supported by our findings in HepG2 cells stably expressing hydrolase only (sEH-D9A) or phosphatase only (sEH-D335S) sEH. This is also supported by our findings for the R287Q polymorphism, which we have previously shown to exhibit lower hydrolase (22) and higher phosphatase (19) activity. The effect of the hydrolase inhibitor on cholesterol as well as the analogous R287Q model and its association with several cardiovascular-related disorders suggests a cautionary note for developing hydrolase inhibition based therapeutic strategies in cardiovascular diseases. Additionally, further understanding the effects of the phosphatase domain on increasing cholesterol levels may provide novel therapeutic approaches for dyslipidemia-related disorders. The phosphatase domain of sEH metabolizes intermediates in cholesterol biosynthesis, which would be expected to reduce plasma cholesterol. Because we observe the opposite effect, we hypothesize that the phosphatase domain influences cholesterol metabolism at the regulatory level rather than the biosynthetic level. This is further supported by the effect of the phosphatase domain on up-regulating the expression levels of the rate-limiting enzyme HMG-CoA reductase in HepG2 cells.
sEH polymorphisms have a high rate of occurrence in humans (∼50% of samples tested), among which the R287Q was found to be one of the more common (14%) (22). Recent epidemiological studies finding an association of the R287Q polymorphism with human disorders such as atherosclerosis and higher plasma cholesterol and triglycerides in familial hypercholesterolemia patients (9, 10, 12) are in agreement with our findings regarding its effect on the total cholesterol level.
In conclusion, our results suggest a role for sEH in regulating cholesterol metabolism in mice and human liver cells and that both the hydrolase and phosphatase domains are involved. Whereas the effect of hydrolase inhibition on cholesterol levels warrants caution with the currently proposed therapeutic targeting in cardiovascular disease, further investigation of the phosphatase domain effect on cholesterol levels may provide a novel therapeutic approach in the management of dyslipidemia and related disorders. Finally, we suggest that the association of the sEH-R287Q polymorphism with cardiovascular disease is related to its effects on cholesterol levels and that further investigation of the functional significance of the phenotypic properties associated with sEH polymorphisms is warranted.
Supplementary Material
Acknowledgments
We thank Dr. Christophe Morisseau for critical review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant ES011630. This work was also supported by a grant from the University of Connecticut Foundation. Additional funding was from the National Institute of Environmental Health Sciences R37 ES002710, the National Institute of Environmental Health Sciences Superfund Basic Research Program P42 ES004699 and the University of Connecticut Tobacco-related Disease Research Program New Investigator Award 16KT-0037. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: sEH, soluble epoxide hydrolase; KO, knockout; WT, wild type; PPAR, peroxisome proliferator-activated receptor; TUPS, 1-(1-methylsulfonyl-piperidin-4-yl)-3-(4-trifluoromethoxy-phenyl)-urea; ANOVA, analysis of variance.
References
- 1.Enayetallah, A. E., French, R. A., Thibodeau, M. S., and Grant, D. F. (2004) J. Histochem. Cytochem. 52 447-454 [DOI] [PubMed] [Google Scholar]
- 2.Capdevila, J. H., and Falck, J. R. (2001) Biochem. Biophys. Res. Commun. 285 571-576 [DOI] [PubMed] [Google Scholar]
- 3.Spector, A. A., Fang, X., Snyder, G. D., and Weintraub, N. L. (2004) Prog. Lipid Res. 43 55-90 [DOI] [PubMed] [Google Scholar]
- 4.Larsen, B. T., Campbell, W. B., and Gutterman, D. D. (2007) Trends Pharmacol. Sci. 28 32-38 [DOI] [PubMed] [Google Scholar]
- 5.Chiamvimonvat, N., Ho, C. M., Tsai, H. J., and Hammock, B. D. (2007) J. Cardiovasc. Pharmacol. 50 225-237 [DOI] [PubMed] [Google Scholar]
- 6.Fleming, I., and Busse, R. (2006) Hypertension 47 629-633 [DOI] [PubMed] [Google Scholar]
- 7.Imig, J. D., Zhao, X., Capdevila, J. H., Morisseau, C., and Hammock, B. D. (2002) Hypertension 39 690-694 [DOI] [PubMed] [Google Scholar]
- 8.Imig, J. D., Zhao, X., Zaharis, C. Z., Olearczyk, J. J., Pollock, D. M., Newman, J. W., Kim, I. H., Watanabe, T., and Hammock, B. D. (2005) Hypertension 46 975-981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei, Q., Doris, P. A., Pollizotto, M. V., Boerwinkle, E., Jacobs, D. R., Jr., Siscovick, D. S., and Fornage, M. (2007) Atherosclerosis 190 26-34 [DOI] [PubMed] [Google Scholar]
- 10.Fornage, M., Boerwinkle, E., Doris, P. A., Jacobs, D., Liu, K., and Wong, N. D. (2004) Circulation 109 335-339 [DOI] [PubMed] [Google Scholar]
- 11.Lee, C. R., North, K. E., Bray, M. S., Fornage, M., Seubert, J. M., Newman, J. W., Hammock, B. D., Couper, D. J., Heiss, G., and Zeldin, D. C. (2006) Hum. Mol. Genet 15 1640-1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato, K., Emi, M., Ezura, Y., Fujita, Y., Takada, D., Ishigami, T., Umemura, S., Xin, Y., Wu, L. L., Larrinaga-Shum, S., Stephenson, S. H., Hunt, S. C., and Hopkins, P. N. (2004) J. Hum. Genet. 49 29-34 [DOI] [PubMed] [Google Scholar]
- 13.Fang, X., Hu, S., Xu, B., Snyder, G. D., Harmon, S., Yao, J., Liu, Y., Sangras, B., Falck, J. R., Weintraub, N. L., and Spector, A. A. (2006) Am. J. Physiol. 290 H55-H63 [DOI] [PubMed] [Google Scholar]
- 14.Lecka-Czernik, B., Moerman, E. J., Grant, D. F., Lehmann, J. M., Manolagas, S. C., and Jilka, R. L. (2002) Endocrinology 143 2376-2384 [DOI] [PubMed] [Google Scholar]
- 15.Liu, Y., Zhang, Y., Schmelzer, K., Lee, T. S., Fang, X., Zhu, Y., Spector, A. A., Gill, S., Morisseau, C., Hammock, B. D., and Shyy, J. Y. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 16747-16752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng, V. Y., Huang, Y., Reddy, L. M., Falck, J. R., Lin, E. T., and Kroetz, D. L. (2007) Drug Metab. Dispos. 35 1126-1134 [DOI] [PubMed] [Google Scholar]
- 17.Newman, J. W., Morisseau, C., Harris, T. R., and Hammock, B. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1558-1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cronin, A., Mowbray, S., Durk, H., Homburg, S., Fleming, I., Fisslthaler, B., Oesch, F., and Arand, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1552-1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enayetallah, A. E., and Grant, D. F. (2006) Biochem. Biophys. Res. Commun. 341 254-260 [DOI] [PubMed] [Google Scholar]
- 20.Tran, K. L., Aronov, P. A., Tanaka, H., Newman, J. W., Hammock, B. D., and Morisseau, C. (2005) Biochemistry 44 12179-12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovacs, W. J., Olivier, L. M., and Krisans, S. K. (2002) Prog. Lipid Res. 41 369-391 [DOI] [PubMed] [Google Scholar]
- 22.Przybyla-Zawislak, B. D., Srivastava, P. K., Vazquez-Matias, J., Mohrenweiser, H. W., Maxwell, J. E., Hammock, B. D., Bradbury, J. A., Enayetallah, A. E., Zeldin, D. C., and Grant, D. F. (2003) Mol. Pharmacol. 64 482-490 [DOI] [PubMed] [Google Scholar]
- 23.Luo, B., Norris, C., Bolstad, E. S., Knecht, D. A., and Grant, D. F. (2008) J. Mol. Biol. 380 31-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinal, C. J., Miyata, M., Tohkin, M., Nagata, K., Bend, J. R., and Gonzalez, F. J. (2000) J. Biol. Chem. 275 40504-40510 [DOI] [PubMed] [Google Scholar]
- 25.Luria, A., Weldon, S. M., Kabcenell, A. K., Ingraham, R. H., Matera, D., Jiang, H., Gill, R., Morisseau, C., Newman, J. W., and Hammock, B. D. (2007) J. Biol. Chem. 282 2891-2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, P. D., Tsai, H. J., Do, Z. N., Morisseau, C., and Hammock, B. D. (2006) Bioorg. Med. Chem. Lett 16 5212-5216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe, T., Schulz, D., Morisseau, C., and Hammock, B. D. (2006) Anal. Chim. Acta. 559 37-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borhan, B., Mebrahtu, T., Nazarian, S., Kurth, M. J., and Hammock, B. D. (1995) Anal. Biochem. 231 188-200 [DOI] [PubMed] [Google Scholar]
- 29.Fang, X., Hu, S., Watanabe, T., Weintraub, N. L., Snyder, G. D., Yao, J., Liu, Y., Shyy, J. Y., Hammock, B. D., and Spector, A. A. (2005) J. Pharmacol. Exp. Ther. 314 260-270 [DOI] [PubMed] [Google Scholar]
- 30.Klopotek, A., Hirche, F., and Eder, K. (2006) Exp. Biol. Med. (Maywood) 231 1365-1372 [DOI] [PubMed] [Google Scholar]
- 31.Llaverias, G., Rebollo, A., Pou, J., Vazquez-Carrera, M., Sanchez, R. M., Laguna, J. C., and Alegret, M. (2006) Biochem. Pharmacol. 71 605-614 [DOI] [PubMed] [Google Scholar]
- 32.Ferreira, A. V., Parreira, G. G., Green, A., and Botion, L. M. (2006) Metabolism 55 731-735 [DOI] [PubMed] [Google Scholar]
- 33.Derosa, G., Cicero, A. F., D'Angelo, A., Gaddi, A., Ciccarelli, L., Piccinni, M. N., Salvadeo, S. A., Pricolo, F., Ferrari, I., Gravina, A., and Ragonesi, P. D. (2006) Clin. Ther. 28 679-688 [DOI] [PubMed] [Google Scholar]
- 34.Robins, S. J., and Bloomfield, H. E. (2006) Curr. Opin. Lipidol. 17 431-439 [DOI] [PubMed] [Google Scholar]
- 35.Mimura, K., Umeda, F., Hiramatsu, S., Taniguchi, S., Ono, Y., Nakashima, N., Kobayashi, K., Masakado, M., Sako, Y., and Nawata, H. (1994) Diabet. Med. 11 685-691 [DOI] [PubMed] [Google Scholar]
- 36.Forman, B. M., Ruan, B., Chen, J., Schroepfer, G. J., Jr., and Evans, R. M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 10588-10593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gan, X., Kaplan, R., Menke, J. G., MacNaul, K., Chen, Y., Sparrow, C. P., Zhou, G., Wright, S. D., and Cai, T. Q. (2001) J. Biol. Chem. 276 48702-48708 [DOI] [PubMed] [Google Scholar]
- 38.Martin, G., Duez, H., Blanquart, C., Berezowski, V., Poulain, P., Fruchart, J. C., Najib-Fruchart, J., Glineur, C., and Staels, B. (2001) J. Clin. Investig. 107 1423-1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Argmann, C. A., Edwards, J. Y., Sawyez, C. G., O'Neil, C. H., Hegele, R. A., Pickering, J. G., and Huff, M. W. (2005) J. Biol. Chem. 280 22212-22221 [DOI] [PubMed] [Google Scholar]
- 40.Johnston, T. P., Nguyen, L. B., Chu, W. A., and Shefer, S. (2001) Int. J. Pharm 229 75-86 [DOI] [PubMed] [Google Scholar]
- 41.Inoue, N., Yamada, K., Imai, K., and Aimoto, T. (1993) Biol. Pharm. Bull. 16 1004-1007 [DOI] [PubMed] [Google Scholar]
- 42.Pinot, F., Grant, D. F., Spearow, J. L., Parker, A. G., and Hammock, B. D. (1995) Biochem. Pharmacol. 50 501-508 [DOI] [PubMed] [Google Scholar]
- 43.Gill, S. S., and Hammock, B. D. (1980) Biochem. Pharmacol. 29 389-395 [DOI] [PubMed] [Google Scholar]
- 44.Schmelzer, K. R., Kubala, L., Newman, J. W., Kim, I. H., Eiserich, J. P., and Hammock, B. D. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 9772-9777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin, L., Foss, C. E., Zhao, X., Mills, T. M., Wang, M. H., McCluskey, L. P., Yaddanapud, G. S., Falck, J. R., Imig, J. D., and Webb, R. C. (2006) FASEB J. 20 539-541 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.