Abstract
G-protein-coupled receptors (GPCRs), one of the most versatile groups of cell surface receptors, can recognize specific ligands from neural, hormonal, and paracrine organs and regulate cell growth, proliferation, and differentiation. Gpr48/LGR4 is a recently identified orphan GPCR with unknown functions. To reveal the functions of Gpr48 in vivo, we generated Gpr48-/- mice and found that Gpr48-/- fetuses displayed transient anemia during midgestation and abnormal definitive erythropoiesis. The dramatic decrease of definitive erythroid precursors (Ter119pos population) in Gpr48-/- fetal liver at E13.5 was confirmed by histological analysis and blood smear assays. Real-time PCR analyses showed that in Gpr48-/- mice both adult hemoglobin α and β chains were decreased while embryonic hemoglobin chains (ζ, βH1, and εy) were increased, providing another evidence for the impairment of definitive erythropoiesis. Furthermore, proliferation was suppressed in Gpr48-/- fetal liver with decreased c-Myc and cyclin D1 expression, whereas apoptosis was unaffected. ATF4, a key transcription factor in erythropoiesis, was down-regulated in Gpr48-/- fetal livers during midgestation stage through the cAMP-PKA-CREB pathway, suggesting that Gpr48 regulated definitive erythropoiesis through ATF4-mediated definitive erythropoiesis.
Erythropoiesis occurs sequentially in distinct anatomical locations in two different phases during embryogenesis. The earlier phase is defined as primitive erythropoiesis, which, in the mouse, originates from the yolk sac at embryonic day 7.5 (E7.5),3 and the later phase is definitive erythropoiesis, which was carried out in the fetal liver during midgestation after E12.5. Non-nucleated adult-type red blood cells are first generated in the fetal liver, the primary organ for erythropoiesis during midgestation from E12 to E16 (1). Erythropoiesis then transfers to bone marrow and spleen in the adult (2, 3). However, the molecular mechanism of regulating erythropoiesis has not been completely delineated. Most of the studies were focused on the transcription factors to explore the mechanisms of erythropoiesis (4, 5). In recent years, several transcription factors were also identified to regulate definitive erythropoiesis in fetal liver during midgestation (6-11). One of the transcription factors is ATF4, which has been shown to regulate cell proliferation in response to a broad spectrum of cell stresses and can be either an activator or a repressor in response to different extracellular signals (12). ATF4-/- mice have been reported to cause defective definitive erythropoiesis, and severe anemia at midgestation (13). Although receptors, such as c-Kit and EPOR, have been well studied in erythropoiesis (14), little is known on the function of G-protein-coupled receptors (GPCRs) in erythropoiesis during development (14, 15).
The GPCR family represents the largest and most versatile group of cell surface receptors (16-18). GPCRs can recognize their ligands, a diverse array of extracellular signals, then transmit these signals to intracellular responses by the ligand-receptor interaction. For its versatile roles, the GPCR family is one of the most promising and attractive targets to develop pharmaceutical drugs for human diseases ranging from allergic rhinitis to pain, hypertension, and schizophrenia (16). The glycoprotein hormone receptors represent a subgroup of GPCRs that have a large N-terminal extracellular (ecto-) domain containing leucine-rich repeats, a versatile structural domain that is important for glycoprotein hormone ligands recognition (19-21). Based on the comparison of peptide hormones and glycoprotein hormone receptors, a sub-group of GPCRs, leucine-rich repeat-containing GPCRs (LGRs), was identified (22). Many studies suggest that the expanding family of LGRs, including three known glycoprotein hormone receptors (LH, FSH, and TSH), the orphan receptors LGR4 (also termed Gpr48), LGR5, and LGR6 are homologous and conservative (22-24). The mammalian glycoprotein hormone receptors have diverse structural features and mainly couple through the cAMP-dependent pathway for signal transduction (25).
Gpr48 (LGR4) has a putative horseshoe-like structure composed of 17 leucine-rich repeats, which is proposed to be the ligand-binding site for this family of receptors (21, 26). The molecular structure and evolutionary features of Gpr48 have been well studied; however, the ligands and physiological functions remain unclear (19, 20, 25, 27, 28). Gpr48 is widely expressed in multiple organs at both embryonic and adult stage (19, 27, 29), which suggests that Gpr48 might play a vital role in development and adult physiological functions. Recent studies from our and other laboratories have indicated that Gpr48 plays an important role in renal, eye, and reproductive system development (30-33). However, little is known thus far about the function and molecular mechanism of Gpr48 in erythropoiesis. In this study, we demonstrated that Gpr48 is expressed in both embryonic and adult liver, and that the deletion of Gpr48 in mouse impairs definitive erythropoiesis at midgestation through down-regulation of c-Myc, cyclin D1, and ATF4 pathways.
EXPERIMENTAL PROCEDURES
Generation of GPR48 Knockout Mice—Gpr48 gene trap ES clone (LST020) was obtained from Bay Genomics (34, 35). The Gpr48 ES clones were injected into C57BL/6 blastocysts and transferred to ICR females. Male chimera mice were mated with C57BL/6 females, resulting in transmission of the inserted allele to the germ line. Positive mice were interbred and maintained on a mixed 129×C57BL/6 background.
Gpr48 knockout mice were generated based on the secretory-trap approach as previously described (33, 36) by disrupting the endogenous Gpr48 gene. This vector (Pgt0,1,2tm-pfs, 11.98 kb) includes the mouse En-2 SA sequence, a fragment of CD4 containing the transmembrane domain inserted in-frame with β-geo together with a downstream internal ribosome entry site, the PLAP gene, and the simian virus 40 polyadenylation signal in the Pgt1tm vector. The vector was inserted into intron 1 of Gpr48 to produce a transcript containing exon 1 fused to the β-geo-PLAP cassette. A male chimeric mouse line was intercrossed to B6/129 wild-type mice so as to obtain heterozygous mice with Gpr48 mutant. Homozygosity was obscured by intercrossing these heterozygous mice. Mice were housed under controlled humidity, temperature, and light regimen and fed ad libitum. Animal care was consistent with institutional and National Institutes of Health guidelines.
DNA-based Genotyping—Genotype of mice was determined by using four primers, for wild-type, forward (5′-TGT TTC AAC CTT TTA AAG ACT GTA GCG AGA-3′), reverse (5′-TAA AGG ACT TAA TGC CAA ATG TGA TTG GTT-3′); for mutant, forward (5′-CCA ATC ACC ACT CTT ACA CAA TGG CTA AC-3′), reverse (5′-GGT CTT TGA GCA CCA GAG GAC-3′). PCR was carried out by using standard techniques. PCR conditions were 35 cycles at 94 °C for 40 s, 53 °C for 40 s, and 72 °C for 1 min. DNA was prepared from 0.5 cm of clipped tail of embryos and extracted by using DirecPCR kit (Viagen Biotech, Los Angeles, CA) protocols. Genomic DNA was extracted by ethanol precipitation after proteinase K digestion in a tissue lysis buffer (50 mm Tris (pH 7.5), 50 mm EDTA (pH 8.0), 100 mm NaCl, 0.5 mm spermidine, 1% SDS, 5 mm dithiothreitol). Reaction products were electrophoresed on 1% agarose gels stained with ethidium bromide. Two expected products were 450 bp for wild-type and 750 bp for mutant. Due to the insertion of the large 11.98-kb trap vector between primer A and primer B, the A/B PCR product could not be amplified in the mutant allele under the present PCR conditions.
RNA Extraction and RT-PCR Amplification—Total RNA was extracted from fetal livers by using TRIzol RNA extraction kit (Invitrogen). First-strand cDNA synthesis was performed using Moloney murine leukemia virus reverse transcriptase and oligo(dT) (Promega, Madison, WI) according to the manufacturer's protocol. The primers used for Gpr48 wild-type and mutant transcripts analysis were: forward (5′-GGA ATT CGC CGC CAT GCC GGG CCC G-3′) and reverse (5′-AGC TCT TTA AGG CTG GGA AGG GCT TT-3′). The conditions for reaction were 30 cycles at 94 °C for 40 s, 60 °C for 40 s, and 72 °C for 1 min. The primers used for ATF4 were: forward (5′-CCC ACA ACA TGA CCG AGA T-3′) and reverse (5′-CTC ATC TGG CAT GGT TTC C-3′) under the amplified reaction conditions of 35 cycles at 94 °C for 30 s, 53 °C for 30 s, 72 °C for 1 min, glyceraldehyde-3-phosphate dehydrogenase primers were used as controls. The primers used for glyceraldehyde-3-phosphate dehydrogenase were forward (5′-GGT GAA GGT CGG TGT GAA CG-3′) and reverse (5′-CTT CTG GGT GGC AGT GAT GG-3′).
Sample Preparation and LacZ Histochemistry—Sample preparation and LacZ staining followed the protocol from Dr. James Martin's laboratory (Texas A&M Health Science Center, Institute of Bioscience and Technology, Houston). The embryos were fixed for 1 h at room temperature in the fixation buffer containing 25% glutaraldehyde, 100 mm EGTA (pH 8.0), 500 mm NaH2PO4 (pH 7.3), 1 m MgCl2, and 37% formaldehyde. Samples were then washed three times with rinse buffer containing deoxycholic acid, Nonidet P-40, and 1 m MgCl2. Then these samples were stained overnight at room temperature with LacZ staining buffer containing 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal), 5 mm K3Fe(CN)6, 5 mm K4Fe(CN)6. After LacZ staining, the samples were stored in 70% alcohol overnight, then dehydrated sequentially by 95% alcohol, 100% alcohol, 100% isopropyl alcohol, and embedded in paraffin to be sectioned at 6 and 12 μm. Subsequently these slides were counter-stained with eosin using standard procedures.
Quantitative Real-time PCR—Total RNA extraction and cDNA synthesize from E13.5 blood cells collected by centrifuge of whole blood, and fetal livers were performed as previously described. Primers were used to detect embryonic hemoglobin chains for blood cells: εy, forward (5′-CAA GCT ACA TGT GGA TCC TGA GAA-3′) and reverse (5′-TGCCGAAGTGACTAGCCAAA-3′); ζ, forward (5′-GCGAGCTGCATGCCTACAT-3′), reverse (5′-GCCATTGTGACCAGCAGACA-3′); βh1, forward (5′-AGGCAGCTATCACAAGCATCTG-3′) and reverse (5′-AACTTGTCAAAGAATCTCTGAGTCC-3′); and glycophorin A, forward (5′-GCC GAA TGA CAA AGA AAA GTT CA-3′) and reverse (5′-TCA ATA GAA CTC AAA GGC ACA CTG T-3′). Glycophorin A mRNAs were used as internal controls. Amplification reactions (25 μl) were prepared using SuperArray Real-time SYBR Green/ROX PCR master mix (SuperArray) according to the manufacturer's protocol. Quantitative real-time PCR were carried out using an Mx3000PTM PCR System (Stratagene) and analyzed with Mx3000P software. The reaction conditions were performed at 40 cycles (95 °C for 30 s, 53 °C for 1 min, and 72 °C for 30 s) with glycophorin A as an internal control.
For fetal livers, the following primers were used: adult α-globin forward (5′-AAT ATG GAG CTG AAG CCC TGG-3′) and reverse (5′-AAC ATC AAA GTG AGG GAA GTA GGT CT-3′); adult-β, forward (5′-GTG AGC TCC ACT GTG ACA AGC T-3′) and reverse (5′-GGT GGC CCA GCA CAA TCA CGA TC-3′); and β-actin, forward (5′-GCA CCA GGG TGT GAT GGT G-3′) and reverse (5′-TGG ATG GCT ACG TAC ATG GC-3′). The conditions of amplification reactions (25 μl) were performed at 40 cycles (95 °C for 30 s, 52 °C for 1 min, and 72 °C for 30 s), and β-actin was used as an internal control. ATF4 primers used for real-time PCR were the same as performed in RT-PCR with β-actin used as an internal control. The conditions of amplification reactions (25 μl) were performed at 37 cycles (95 °C for 30 s, 54 °C for 1 min, and 72 °C for 30 s).
Western Immunoblot Analysis—Fetal liver lysates were harvested according to the method described by Mitchell et al. (37). The protein concentration was determined using a BCATM Protein Assay Kit (Pierce) following the manufacturer's procedure. Samples were then diluted into loading buffer at 1 mg/ml. Following heat denaturation, samples containing 8 μg of protein were loaded onto and separated on 12% SDS-PAGE gels as needed. Proteins were then transferred electrophoretically to a 140-μm nitrocellulose membrane (Pall Corp., Ann Arbor, MI). After incubating the membranes in blocking solution, primary antibody and second antibody were used at appropriate dilution. Proteins were detected using the Super-Signal West Pico Chemiluminescent substrate (Pierce) according to the manufacturer's instructions. The following antibodies from Santa Cruz Biotechnology were used: rabbit polyclonal anti-ATF4 (1:300), rabbit polyclonal anti-c-Myc (1:400), and rabbit polyclonal anti-Cyclin D1 (1:400). Goat polyclonal anti-actin (1:500) was used as internal control.
Histology and Immunohistochemistry—Embryos dissected from mice were fixed in 10% formalin and embedded in paraffin according to the standard techniques. 5-μm-thick sections were stained with hematoxylin and eosin. For proliferation assays, the sections were stained with anti-PCNA using a mouse PCNA kit (Zymed Laboratories Inc., South San Francisco, CA) and then counterstained with hematoxylin. For BrdUrd incorporation, mice were injected with 100 mg/kg BrdUrd (Sigma) intraperitoneally 2 h before liver tissue harvesting. BrdUrd incorporation studies were performed using a BrdUrd labeling kit (Zymed Laboratories Inc.) on paraffin-covered slides. The ApopTaq peroxidase in situ apoptosis detection kit (Chemicon, Temecula, CA) was used for apoptosis assay according to the manufacturer's instructions with methyl green as counterstain. Immunohistochemical staining was performed by using ABC staining kit (Santa Cruz Biotechnology). The following antibodies from Santa Cruz Biotechnology were used: rabbit polyclonal anti-ATF4 (C-20) (1:100), rabbit polyclonal anti-c-Myc (1:200), and rabbit polyclonal anti-Cyclin D1(1:200). Hematoxylin was carried out as counterstain.
Blood Smear—Blood was harvested from the carotid arteries of decapitated embryos. Blood smears were prepared by using the wedge technique followed by air drying and Wright-Giemsa staining (Sigma-Aldrich) following the manufacturer's staining protocol. The number of nucleated erythrocytes and enucleated erythrocytes were counted in high power views.
Flow Cytometry—Single-cell suspensions were obtained from E13.5 wild-type and homozygous fetal livers. Briefly, fetal livers were dissected from embryos, then placed in Dulbecco's modified Eagle's medium (HyClone, Logan, UT) with 5% fetal bovine serum. The cells were dissociated by repeated flushing with a 23-gauge needle to obtain the single cell suspension, and the cell number was counted. Cell suspensions with the same concentration were first incubated on ice with rat anti-mouse CD16/CD32 (Pharmingen) to block nonspecific binding to Fc-receptors. Subsequently, cells were incubated with rat anti-mouse phycoerythrin-conjugated anti-c-Kit and anti-CD44, fluorescein isothiocyanate-conjugated anti-CD34 and anti-Ter-119 (all from Pharmingen). Appropriate isotype control antibodies were used. Then the cells were washed and transferred to phosphate-buffered saline 1% paraformaldehyde A. Cell surface expression of different markers was analyzed in a BD Biosciences FACScan using CellQuest software. To measure cell size of erythrocyte in fetal livers, liver cells from E13.5 GPR48-/- fetus and their wild-type littermates were harvested by flushing the femoral medullary cavity with Dulbecco's modified Eagle's medium. Cells are then centrifuged at 4 °C 1500 rpm for 5 min followed by resuspending in phosphate-buffered saline (0.1 m and pH 7.2) and staining with anti-Ter119 (fluorescein isothiocyanate-conjugated) as a marker of mature erythrocyte for flow cytometry assay.
Generation of Constitutive Activated Gpr48—PCR-based point mutagenesis was performed to generate mutant Gpr48 using cDNA encoding human Gpr48 receptors. To introduce the amino acid mutation at T755I into the wild-type cDNA of human Gpr48 gene, overlapping primers containing mutated sequences (forward: 5′-CGC TTG GCT AAT CTT CAT CAA TTG C-3′; reverse: 5′-GCA ATT GAT GAA GAT TAG CCA AGC G-3′) were used to replace the wild-type amino acid in the region. All cDNAs were subcloned into the expression vector pcDNA3.1 (-) (Invitrogen Corp., Carlsbad, CA), and plasmids were purified using a Maxi plasmid preparation kit (Qiagen). Fidelity of the PCR was confirmed by sequencing on both strands of the final constructs before use in expression studies.
Cell Transfection and Luciferase Assays—Human embryonic kidney 293T was maintained in modified Eagle's medium/Ham's F-12 supplemented with 10% fetal bovine serum, 100 μg/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine. Before transfection, cells (5 × 104/culture) were seeded in a 24-well plate (Nalge Nunc International, Naperville, IL). When cells were 70-80% confluent, transient transfection was performed with 0.25 μg of plasmid using the Lipofectamine method following replacement of culture media. After 6-12 h of incubation with the Lipofectamine, media were replaced with Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. 48 h after transfection, cells were washed twice with Dulbecco's phosphate-buffered saline, harvested from the plate. For luciferase transfection experiments, 48 h after transfection, cell lysate was collected, and luminescence assays were performed. Within a given assay, plate wells were set up in triplicate for each transfected construct or control vector. To allow for normalization of firefly luciferase values based on transfection efficiency, a co-reporter vector expressing β-galactosidase (pcDNA3-LacZ) was included at a ratio of 1:20 of co-reporter plasmid to experimental promoter construct (or control vector) in the transfection mixture.
Statistical Analysis—Data are presented as mean ± S.D. and analyzed by one-way analysis of variance or Student t test. For all analyses, p < 0.05 was considered statistically significant.
RESULTS
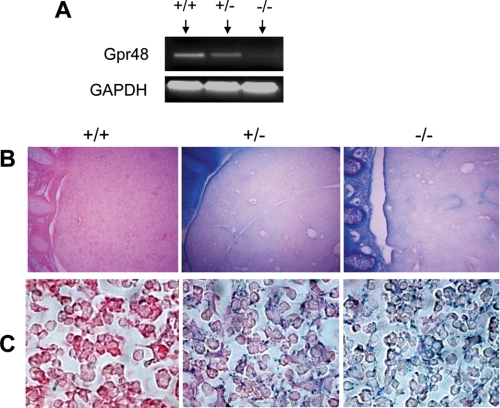
Targeted Inactivation of Gpr48 Gene and the Expression of Gpr48 in Fetal Liver—The murine Gpr48/LGR4 gene was disrupted by inserting a large secretory trap vector (11.98 kb) to the intron 1 (supplemental Fig. S1A). The Gpr48 ES clones were injected into C57BL/6 blastocysts to generate chimeric mice. Heterozygous Gpr48 mutant offspring were obtained by intercross between male chimera mice and C57BL/6 female mice. Heterozygous mice were intercrossed to obtain homozygous mice. The genotypes of offspring of mutant mice were confirmed by PCR analysis (supplemental Fig. S1B). The mRNA level of Gpr48 in the liver of homozygous, heterozygous, and wild-type mice was detected by RT-PCR analysis. The results showed that Gpr48 is expressed in the fetal liver of wild-type fetus, and that there was no expression of Gpr48 in homozygous mutant (Fig. 1A), confirming the successful deletion of Gpr48 in homozygous mutant. Because the mutant Gpr48 gene generates a chimeric protein containing the N-terminal leucine-rich repeat (LRRNT) domain and β-galactosidase, the expression patterns of Gpr48 in both heterozygous and homozygous mutant mice can be examined with the activity of β-galactosidase (LacZ staining) in fetal liver. As shown in Fig. 1B, LacZ staining was performed to measure β-galactosidase activity and the expression of Gpr48 in E15.5 fetal liver tissue sections. The expression of Gpr48 was found not only in hepatocytes but also in erythroid precursor cells (Fig. 1C). During midgestation, the erythroid precursors are mainly derived from liver cells, homologous to the hepatocytes. The expression of Gpr48 in erythroid precursor cells suggests a potential role of the receptor in erythropoiesis.
FIGURE 1.
Expression of Gpr48 in fetal liver. A, RT-PCR analysis of Gpr48 mRNA levels from E13.5 Gpr48 wild-type (+/+), heterozygous (+/-), and homozygous (-/-) fetal livers, showing no Gpr48 expression in Gpr48-/- fetal liver. B and C, LacZ staining of E15.5 Gpr48 wild-type (+/+), heterozygous (+/-), and homozygous (-/-) fetal liver sections. Gpr48+/- and Gpr48-/- fetal liver showed positive staining in liver (upper right panel, B, magnification 100×) and at cell surface membrane (lower right panel, C, magnification 1000×). Sections were counterstained with eosin.
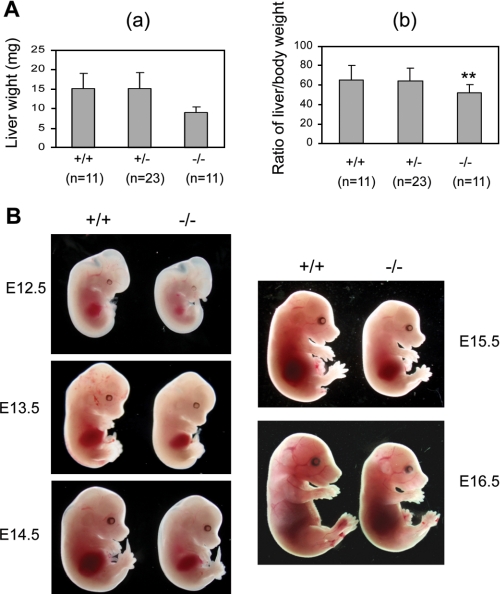
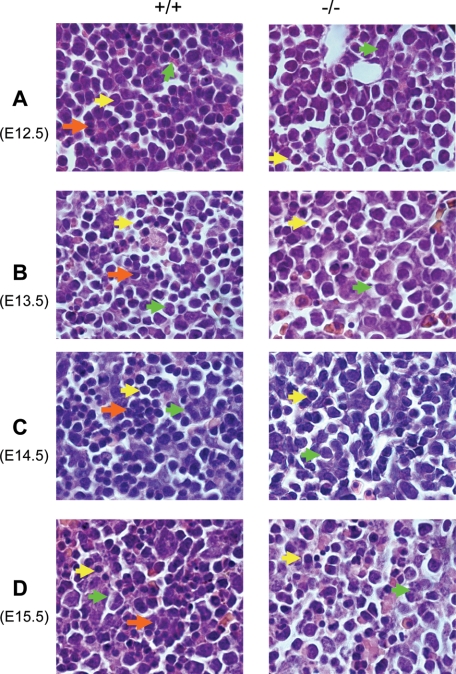
Deletion of Gpr48 Affects Fetal Liver Size and Definitive Erythropoiesis at Midgestation in Gpr48-/- Embryos—To understand the in vivo function of Gpr48 in fetal liver growth and erythropoiesis, we examined fetal liver weight and body weight in Gpr48-/- mice compared with wt mice. As shown in Fig. 2A, at E14.5, inactivation of Gpr48 gene affected liver growth with a 41% weight reduction (Fig. 2A, panel a) and 25% reduction in body weight (data not shown). Furthermore, the ratio of liver to body weight showed significant decrease (Fig. 2A, panel b), and the fetal livers of Gpr48 null embryos showed marked smaller size, indicating hypoplastic development during fetal liver development from E12.5 to E16.5 (Fig. 2B). Because fetal liver is the predominant site for definitive erythropoiesis during embryonic stages, we further examined the morphological features of fetal liver from E12.5 to E16.5. As shown in Fig. 2B, Gpr48-/- embryos and fetal livers from E12.5 to E15.5 appeared considerably paler than their control wt littermates, suggesting less number of erythrocytes with low hemoglobin. However, little difference was found at E16.5 between wt and Gpr48-/- mice (Fig. 2B). In fetal liver, erythropoietic precursor cells can be distinguished from hepatocytes based on their smaller cell size and their more condensed and deeply stained nucleus. Histological examination and hematoxylin and eosin staining show that the gross morphologic features of homozygous fetal livers (green arrows) were normal in cellular architecture (Fig. 3). However, fewer erythroid precursor or definitive progenitors (yellow arrows) and erythroid foci (orange arrows) were visible in the homozygous fetal livers from E12.5-E15.5 embryos compared with that in wild type (Fig. 3). The fetal livers of Gpr48-/- embryos showed marked smaller size, indicating hypoplastic development during E12.5-E16.5.
FIGURE 2.
Defects of Gpr48-/- mice in body size and fetal liver. A, Gpr48-/- fetal mice at E14.5 showed significantly decreased liver weight and the ratio of liver weight/body weight. a, Gpr48-/- fetal liver weight was decreased ∼1.68-fold compared with their wild-type littermates (p < 0.001). b, the ratio of liver weight/body weight was decreased ∼1.25-fold in Gpr48-/- mice compared with wild-type littermates (p < 0.02). B, Gpr48-/- mice embryos and livers (right) during E12.5-E15.5 are paler and smaller than wild-type. E16.5 Gpr48-/- embryos show smaller in size but similar in color compared with wild-type fetuses.
FIGURE 3.
Significant decrease of erythroid precursor cells and erythroblast islands in Gpr48-/- fetal liver by histological analysis. Gpr48-/- fetal liver showed significant decrease of definitive erythroid precursor cells and erythroblast islands from E12.5 to E15.5 (A-D). Erythropoietic precursor cells are indicated by yellow arrows, hepatocytes by green arrows, and erythroblast islands by orange arrows (magnification, 1000×).
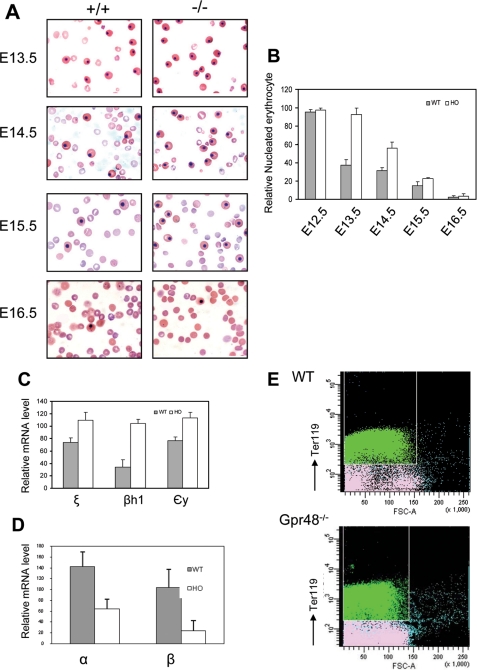
Primitive erythrocytes are nucleated red blood cells in peripheral blood. From a blood smear, the nucleated erythrocytes showed the same size and shape in homozygous mice compared with that in wild-type, but significant less enucleated erythrocytes appeared in homozygous mice from E13.5 to E15.5 days (Fig. 4A). To quantify the nucleated erythrocytes, the number of both nucleated and enucleated erythrocyte was counted in the same high powder views. In Gpr48-/- embryos from E13.5 to E15.5, the percentage of nucleated erythrocytes was elevated 2.5-, 1.8-, and 1.5-fold compared with that in wild-type (Fig. 4B, p < 0.05, 0.01, and 0.01, respectively). However, no marked difference between homozygous and wild-type controls was found at E12.5 and E16.5 (p > 0.05). These nucleated erythrocytes in homozygous appeared the same morphologic characteristics as from wild-type controls, suggesting that nucleated erythrocytes in homozygous blood of these early embryos represented primitive erythrocytes produced by blood islands in yolk sac rather than the premature definitive erythrocytes releasing to blood stream from fetal liver.
FIGURE 4.
Increased nucleated erythrocytes at midgestation of Gpr48-/- mice. A, blood smears from E13.5-E16.5 embryos were stained with Wright-Giemsa stain. Gpr48-/- embryos (right) of E13.5-E15.5 showed increased percentage of nucleated erythrocytes compared with the samples from their wild-type littermates (left), but no significant difference from E16.5 samples between Gpr48-/- and their wild-type littermates (magnification, 1000×). B, relative number in percentage of nucleated erythrocytes from blood smear slides. E13.5 and E14.5 Gpr48-/- embryos showed ∼2.5-fold and 1.8-fold increase in nucleated erythrocytes percentage compared with wild-type, respectively (p < 0.03). E12.5 and E16.5 littermates showed little differences in percentage of nucleated erythrocytes between wild-type and null embryos (p > 0.05). C and D, real-time PCR of embryonic hemoglobin chains (ζ, βh1, and εy) in E13.5 blood cell samples (C) and adult hemoglobin chains (α and β) in E13.5 livers (D). E, the cell size of erythrocytes was reduced by Gpr48 inactivation in E13.5 fetal livers. Ter119pos cells in Gpr48-/- mice bone marrow displayed a reduction in cell size compared with those in their wt littermates (x-axis represents the cell size). Green in the histogram represents Ter119-positive cells (mature red blood cells). Similar reduction of red blood cell size was observed from adult Gpr48-/- cells. However, the cell number did not show significant decrease in Gpr48-/- null mice.
The discrepancy of hemoglobin chain provides another way to identify these two kinds of erythrocytes. Primitive erythrocytes express embryonic globins (ζ, βH1, and εy) in contrast to that definitive erythrocytes express adult forms of hemoglobins (β and α) (2, 38-40). Real-time PCR analyses were performed to examine the mRNA levels of different kinds of globins. In E13.5 embryos, the mRNA expression levels of embryonic globins ζ, βH1, and εy from blood samples showed marked increase in Gpr48-/- mice compared with their wt littermates (p < 0.05) (Fig. 4C). This result was consistent with the elevated ratio of primitive erythrocytes in the blood of homozygous mice (Fig. 4, A and B). On the other hand, the fetal liver is the primary site to produce adult forms of hemoglobins, the expression levels of α and β globin mRNAs in the liver of Gpr48-/- mice at E13.5 showed significantly decreased (p < 0.01 and p < 0.05, respectively) (Fig. 4D), indicating that deletion of Gpr48 affected definitive erythropoiesis in fatal liver rather than primitive erythropoiesis. The decreased α and β globin expression also implies a smaller erythrocytes size in Gpr48-/- fetal livers. Indeed, the red blood cell size of the Ter-119-positive population, representing committed erythroid cells in definitive erythropoiesis (5, 10), was decreased in E13.5 Gpr48-/- fetal livers (Fig. 4E, x-axis represents cell size), indicating deletion of Gpr48 reduced red blood cell size by regulating the expression levels of mature hemoglobins. Furthermore, the less amounts of adult forms of globins lead to the paler appearance of the homozygous Gpr48 livers compared with the wt livers as shown in Fig. 2B.
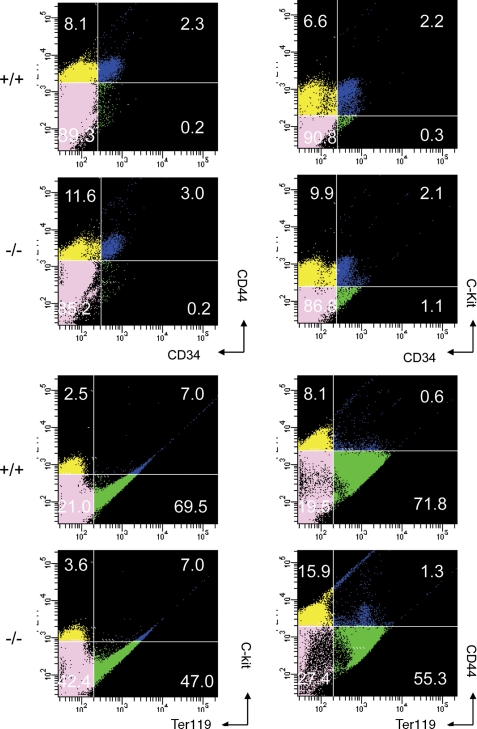
Analysis of Hematopoietic Cell Populations and Their Differentiation in Gpr48-/- Fetal Liver—Hematopoietic cell populations and differentiation stages can be distinguished by the expression of characteristic markers such as CD34, CD44, c-Kit, and Ter-119 (5, 10). Flow cytometry was performed to verify the expression of these cell surface markers (Fig. 5). Single cell suspensions of fetal livers at E13.5 isolated from embryos were prepared for cell sorting. Ter-119pos cells were significantly reduced in Gpr48-/- fetal liver. In addition, the population harboring the long term reconstitution hematopoietic stem cells (CD34posc-Kitpos) was also at similar levels in both wild-type and homozygous fetal liver (Fig. 5). On the other hand, Gpr48-/- fetal liver contained a relative higher level of CD44posTer-119neg hematopoietic progenitors compared with the wt littermates. These data further confirmed that definitive erythropoiesis, the late stage of erythroid differentiation, is affected by the inactivation of Gpr48.
FIGURE 5.
Flow cytometry analysis of fetal liver hematopoietic cells from E13. 5 Gpr48+/+ and Gpr48-/- embryos. Cell suspensions from E13.5 fetal livers were labeled with anti-c-kit, anti-CD34, anti-CD44, or anti-Ter119-specific antibody. The populations of Ter119-positive cells, which represent definitive erythrocytes in Gpr48-/- embryos, were markedly decreased.
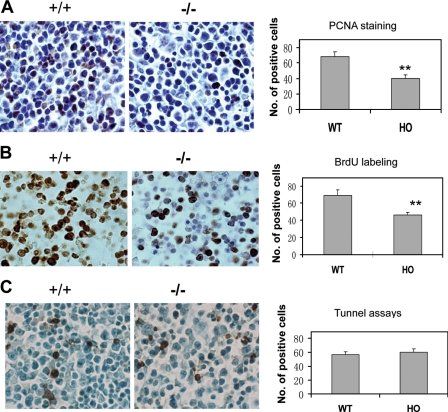
Deletion of Gpr48 Reduces Cell Proliferation in Fetal Liver—As described above, both the number of liver cells and definitive erythroid precursors in Gpr48-/- fetal liver actually decreased. The reduction of definitive erythroid progenitors and liver cells in Gpr48-/- fetal liver might be associated with either decreased cell proliferation or increased apoptosis. Immunohistochemical assays of PCNA and BrdUrd incorporation assays indicated that the number of proliferating cells was markedly reduced in the fetal liver of homozygous Gpr48-/- mice compared with their control littermates (Fig. 6A for PCNA and Fig. 6B for BrdUrd labeling). However, no marked differences were observed in the apoptosis of fetal liver cells using TUNEL assays at E13.5 (Fig. 6C).
FIGURE 6.
Reduced cell proliferation in Gpr48-/- fetal liver. A, E13.5 Gpr48-/- fetal liver showed dramatically decreased number of positive cells (dark brown) compared with wild-type sections using PCNA by immunohistochemistry. Hematoxylin stain was performed as counterstain (magnification, 1000×). B, proliferation assays with BrdUrd labeling demonstrated that Gpr48-/- E13.5 fatal liver have marked decrease in the number of proliferating cells (BrdUrd-positive sells, brown color). Sections were counterstained by methylene green (magnification, 1000×). C, TUNEL assays of E13.5 fetal liver showed no significant differences in the number of positive cells (brown) between Gpr48-/- (left panel) and Gpr48+/+ fetal livers. Sections were counterstained by methylene green (magnification, 1000×).
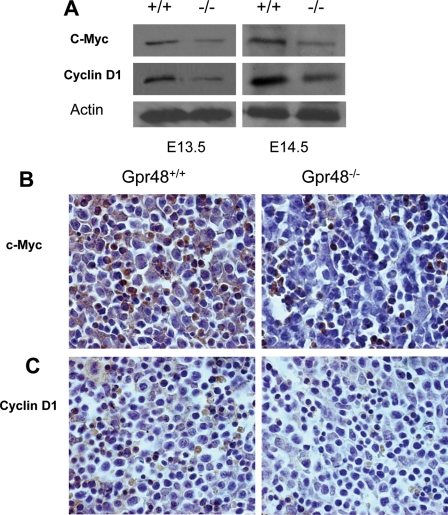
To understand the mechanism of how Gpr48 regulates cell proliferation, we examined the expression levels of key proteins involved in cell proliferation in Gpr48-/- fetal liver. As shown in Fig. 7A, both c-Myc and cyclin D1 were markedly reduced in Gpr48-/- mouse liver at E13.5 and E14.5 using Western blot assays (Fig. 7A). This result was confirmed by immunohistochemistry staining with specific antibodies for c-Myc and cyclin D1, respectively (Fig. 7, B and C). Both cyclin D1 and c-Myc play critical roles in cell proliferation and the cell cycle. Cyclin D1, expressed at the stage of G1, is the downstream gene of c-Myc. The proliferation of mammalian cells is governed by cyclins and their associated cyclin-dependent kinases (CDKs) (41). These cyclin-CDK complexes phosphorylate critical cellular substrates, thereby allowing the ordered progression of the cell cycle (42).
FIGURE 7.
Down-regulation of c-Myc and Cyclin D1 in Gpr48-/- fetal liver. A, Western blot analysis showed that protein levels of c-Myc and Cyclin D1 were both decreased in Gpr48-/- fetal livers compared with that in the wild type. Actin was used as an internal control. B, fetal liver sections from E13.5 Gpr48+/+ and Gpr48-/- embryos were stained with anti-c-Myc antibody. Significant decrease of c-Myc protein level was observed in Gpr48-/- fetal liver (brown color, magnification, 1000×). C, decreased protein levels of Cyclin D1 in Gpr48-/- fetal liver compared with wild-type. Fetal liver sections from E13.5 Gpr48+/+ and Gpr48-/- embryos were stained with anti-Cyclin D1 antibody (brown color, magnification, 1000×).
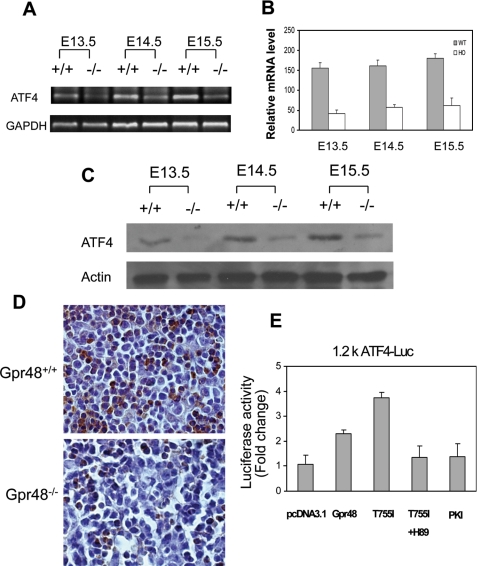
ATF4 Is a Potential Target Gene of Gpr48 in Regulating Definitive Erythropoiesis—ATF4 has been shown to regulate cell proliferation in response to a broad spectrum of cell stresses and can be either an activator or a repressor in response to different extracellular signals (12). ATF4-/- affects the development of multiple organs or systems such as skeleton (43, 44), lens (45), liver (13), and reproductive system (46). ATF4-/- mice have defective definitive erythropoiesis and severe anemia at midgestation (13). Interestingly, all of those implicated tissues in ATF4-/- mice express high levels of Gpr48. Gpr48-/- and ATF4-/- mice have similar phenotypes (29-31). To determine whether Gpr48-mediated anemia at midgestation is caused by the regulation of ATF4 transcription factor, we measured the expression level of ATF4 using different approaches. As shown in Fig. 8, deletion of Gpr48 markedly down-regulated the mRNA expression levels of ATF4 in fetal livers from E13.5 to E15.5 days using RT-PCR and real-time PCR assays (Fig. 8, A and B). To determine whether the ATF4 protein was affected in Gpr48-/- mice, we performed Western blot and immunohistochemistry staining with specific anti-ATF4 antibodies. The protein expression level of ATF4 in Gpr48-/- fetal liver was significantly decreased by both Western blot (Fig. 8C) and by immunohistochemistry analysis (Fig. 8D). These results suggest that ATF4 is a downstream target of Gpr48, and the impairment of definitive erythropoiesis in Gpr48-/- fetal embryos at midgestation was likely caused by the marked reduction of ATF4 expression in Gpr48-/- fetal liver.
FIGURE 8.
Down-regulation of ATF4 in Gpr48-/- fetal liver. A, RT-PCR showed that ATF4 mRNA was dramatically decreased in Gpr48-/- livers during E13.5-E15.5, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. B, real-time PCR showed significantly reduced mRNA levels of ATF4 in Gpr48-/- livers during E13.5-E15.5, and β-actin used as an internal control. C, the protein expression level of ATF4 was significantly decreased at E13.5, E14.5, and E15.5 in Gpr48-/- livers by Western blot. Actin was used as an internal control. D, the expression level of ATF4 was markedly reduced in E13.5 Gpr48-/- fetal livers by immunocytochemistry staining. Sections were stained with anti-ATF4-specific antibody. The number of positive cells (brown) was markedly reduced compared with wild type. Hematoxylin was used as counterstain (1000×). E, Gpr48 regulates ATF4 promoter through the cAMP-PKA-CREB pathway. PKA inhibitor PKI strongly inactivated the ATF4 1.2k promoter activity induced by a constitutively activated mutant Gpr48 (T755I). The activation of ATF4 by the active Gpr48 mutant was also markedly inhibited by PKA-specific inhibitor H89, suggesting Gpr48 activates ATF4 through activation of the PKA pathway.
Gpr48 Regulates the Intracellular Activation through the cAMP-PKA-ATF4 Signaling Cascade—ATF4 (also termed CREB 2) is a member of CREB family of transcription factors, a major downstream transcription factor family of the cAMP-PKA pathway. Because the ligands of Gpr48 remain unclear at present, therefore generation of constitutively active mutant receptor provides a ligand-independent method for the function assay of Gpr48. Based on early studies found in FSH and LH receptors, we generated a number of single mutations using specific site-direct mutagenesis method, and we then examined the intracellular cAMP production and activation of cAMP-downstream transcription factor. One of the mutations, Gpr48 T755I, was found to increase intracellular cAMP significantly compared with the control and wild-type receptor without any ligand stimulation (Weng et al. (60)). The result was independently confirmed by Dr. Kitagawa's group (47), suggesting that G-protein Gαs and cAMP pathways are coupled to Gpr48. To further explore the molecular mechanism how Gpr48 regulates ATF4, we examined the proximal promoter region of ATF4 promoter and found a semi-CRE binding site CGTCA (at position -921 relative to the transcription start site) for potential binding and activation of the ATF4 gene by the CREB transcription factor and the cAMP-PKA signaling pathway. To confirm that Gpr48-mediated cAMP-CREB pathways regulate the promoter activity of ATF4, we cotransfected the constitutively active mutant Gpr48 (T755I) and a 1.2-kb ATF4 promoter-luciferase reporter gene into the cells, and then examined the direct activation of ATF4 by Gpr48 and cAMP pathways. As shown in Fig. 8E, the Gpr48-active mutant receptor (T755I) significantly increased the intracellular cAMP production. The activation of cAMP level by Gpr48 active receptor was significantly inhibited by adding PKI, a competitive PKA inhibitor (Fig. 8E). To further determine whether Gpr48 regulates ATF4 through PKA, we also used another PKA inhibitor, H89, a chemical PKA inhibitor, to measure ATF4 promoter activation mediated by the Gpr48-cAMP-PKA pathway. As shown in Fig. 8E, H89 successfully inhibited the activation of ATF4 promoter activated by Gpr48 active mutant receptor (T755I) (Fig. 8E). These data indicate that ATF4 was the direct downstream target gene of the Gpr48-mediated cAMP-PKA-CREB signaling pathway.
DISCUSSION
The present study demonstrated that inactivation of Gpr48 (LGR4) lead to impairment of definitive erythropoiesis at midgestation in fetal liver. Using a gene-trapped approach described previously (36), we constructed the Gpr48 inactivation model by inserting a secretary-trap vector (11.98 kb) at intron 1 of wild-type Gpr48. Ablation of Gpr48 resulted in the impairment of definitive erythropoiesis at midgestation, which is associated with the down-regulation in the expression levels of c-Myc, cyclin D1, and ATF4 through the cAMP-PKA-CREB signaling pathway.
Gpr48 is expressed in a wide range of tissues, including skeleton, cartilage, ovary, testis, adrenal, thyroid, kidney, heart, and liver (19, 27, 29), suggesting that Gpr48 is involved in the developmental regulation of multiple organs, including fetal liver. The facts that Gpr48 is highly expressed in the fetal liver and in the membrane of premature erythroblast at midgestation indicate that Gpr48 is involved in the development of definitive erythropoiesis. Not only was the body size affected in Gpr48-/- mice, but fetal livers at midgestation during E13.5-E15.5 also displayed much smaller and paler phenotype in Gpr48 mutant mice than those in normal control mice. Fetal liver is the predominant site for development of definitive erythropoiesis during midgestation after E12.5, so the deficient fetal liver in Gpr48-/- mice suggests that Gpr48 play a key role for the development of definitive erythropoiesis. Erythropoiesis includes two sequential process termed primitive erythropoiesis and definitive erythropoiesis, respectively. Our data indicated that the silencing of Gpr48 had no evident effects on primitive erythropoiesis but dramatically reduced definitive erythropoiesis. Based on different characteristics between definitive erythropoiesis and primitive erythropoiesis (2, 16), we counted the nucleated erythrocytes (primitive erythrocyte) and enucleated erythrocytes (definitive erythrocytes) from blood smear and distinguish these two populations by real-time PCR arrays for different globin chains. The elevated level of nucleated erythrocytes indicated that more primitive erythrocytes in the circulation of Gpr48-/- mice, which was consistent with the result of real-time PCR analysis for globin chains (ζ, βH1, and εy). However, adult α and β globin chains was decreased markedly, indicating reduced definitive erythrocytes in Gpr48-/- mice. Examinations of the hemoglobin chain suggest that the relative increased number of nucleated erythrocytes in homozygous mutants resulted from persistence of primitive yolk sac-derived erythrocytes rather than the premature release of nucleated definitive erythrocytes into circulation due to stress erythropoiesis.
ATF4, also termed CREB2, is a member of the ATF/CREB ubiquitous basic leucine zipper (bZip) transcription factors. ATF4 is essential for cell proliferation (13, 43), especially for the processes that require high level proliferation, such as fetal liver hematopoiesis (13), bone development (45), lens development (44), and human cancer cells (48). ATF4-/- mice showed severe anemia due to impairment of definitive erythropoiesis during midgestation, indicating that ATF4 plays a pivotal role for definitive fetal-liver erythropoiesis under stress response (13). Similar to ATF4-/- mice, GPR48-/- fetal livers also have elevated nucleated primitive erythrocytes relative to enucleated definitive erythrocytes without morphologically changes of primitive erythrocytes. Here we provide evidences that Gpr48 regulates the expression level of ATF4 through the cAMP-PKA-CREB signaling pathway. As a critical transcription factor in response to hypoxia, ATF4 activates its downstream gene expression, including CHOP and TRB3 (49, 50). These genes are stress-induced factors and collaborate to modulate a complex regulation network for fulfilling a fine regulation under conditions of integrated stress response or erythropoiesis (50-52). Furthermore, the pathway of signaling transduction for ATF4-CHOP-TRB3 might be independent of HIF-1α (49), a critical factor to mediate hematopoiesis through the inducing EPO pathway.
One of the most important changes in erythroid development is the ongoing maturation and the loss of proliferative capacity in favor of the differentiation program. The balance between erythropoietin (Epo)-mediated erythroid differentiation and stem cell factor-mediated proliferation are strictly regulated during erythropoiesis. In the presence of Epo, Signal transducer and activator of transcription 5 (STAT5) is recruited to the Epo receptor and phosphorylated by the Janus kinase 2 (JAK2). Once phosphorylated, STAT5 is released from EpoR, homodimerizes in the cytosol, and translocates to the nucleus where it activates its target gene expression, including Bcl-X and cyclin D1 (53-58). Phosphorylation of CREB by PKA enhances Epo-stimulated STAT5 transactivation by inducing recruitment of CREB/CBP/p300 to the STAT5 transactivational complex (58, 59). Inactivation of Gpr48 caused dramatic decrease in the proliferation of definitive erythroid progenitors and erythroblast islands in fetal liver. Moreover, analyses of key proliferation genes demonstrate that both c-Myc and cyclin D1 were markedly down-regulated in Gpr48-/- fetal liver. These data suggest that Gpr48 also affect definitive erythropoiesis by regulating Epo-mediated erythroid differentiation through the cAMP-PKA-CREB pathway.
The erythropoiesis of Gpr48-/- embryos was mainly affected during E12.5-E15.5 with E13.5 as the most severe stage. After E15.5, the anemia phenotype in a blood smear of Gpr48-/- mice looked gradually normal, suggesting that the defect of definitive erythropoiesis in Gpr48 homozygous mice is a transient process at midgestation. Other factors might serve as compensatory regulators for Gpr48-/- embryos surviving through midgestation. These compensatory genes might be expressed a bit later than ATF4 during the onset of fetal liver hematopoiesis to coordinate with ATF4 and to regulate hematopoiesis under Gpr48 ablation.
In summary, Gpr48 ablation resulted in impaired fetal-liver definitive erythropoiesis at midgestation. A reduced population of definitive erythrocytes was associated with decreased proliferation and down-regulation of c-Myc and cyclin D1 in Gpr48-/- fetal liver. These results suggest that Gpr48 is a critical GPCR to regulate fetal liver erythropoiesis during midgestation stage through intracellular signaling pathways coupled to the regulation of ATF4.
Supplementary Material
Acknowledgments
The Gpr48 gene trap ES cell clone (LST020) was obtained from Bay Genomics at the Mutant Mouse Regional Resource Center of the University of California, Davis.
This work was supported, in whole or in part, by National Institutes of Health Grants 5R01HL064792 and 1R01CA106479 (to M. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: E, embryonic day(s); GPCR, G-protein-coupled receptor; LGR, leucine-rich repeat-containing GPCR; RT, reverse transcription; BrdUrd, bromodeoxyuridine; wt, wild type; CDK, cyclin-dependent kinase; PKA, protein kinase A; CREB, cAMP-response element-binding protein; Epo, erythropoietin; STAT5, signal transducer and activator of transcription 5; JAK2, Janus kinase 2; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling; PCNA, proliferating cell nuclear antigen.
References
- 1.Socolovsky, M., Fallon, A. E., Wang, S., Brugnara, C., and Lodish, H. F. (1999) Cell 98 181-191 [DOI] [PubMed] [Google Scholar]
- 2.Godin, I., and Cumano, A. (2002) Nat. Rev. Immunol. 2 593-604 [DOI] [PubMed] [Google Scholar]
- 3.Palis, J., Robertson, S., Kennedy, M., Wall, C., and Keller, G. (1999) Development 126 5073-5084 [DOI] [PubMed] [Google Scholar]
- 4.Perry, C., and Soreq, H. (2002) Eur. J. Biochem. 269 3607-3618 [DOI] [PubMed] [Google Scholar]
- 5.Cantor, A. B., and Orkin, S. H. (2002) Oncogene 21 3368-3376 [DOI] [PubMed] [Google Scholar]
- 6.Kawane, K., Fukuyama, H., Kondoh, G., Takeda, J., Ohsawa, Y., Uchiyama, Y., and Nagata, S. (2001) Science 292 1546-1549 [DOI] [PubMed] [Google Scholar]
- 7.Dumitriu, B., Patrick, M. R., Petschek, J. P., Cherukuri, S., Klingmuller, U., Fox, P. L., and Lefebvre, V. (2006) Blood 108 1198-1207 [DOI] [PubMed] [Google Scholar]
- 8.Hodge, D., Coghill, E., Keys, J., Maguire, T., Hartmann, B., McDowall, A., Weiss, M., Grimmond, S., and Perkins, A. (2006) Blood 107 3359-3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iavarone, A., King, E. R., Dai, X. M., Leone, G., Stanley, E. R., and Lasorella, A. (2004) Nature 432 1040-1045 [DOI] [PubMed] [Google Scholar]
- 10.Neubauer, H., Cumano, A., Muller, M., Wu, H., Huffstadt, U., and Pfeffer, K. (1998) Cell 93 397-409 [DOI] [PubMed] [Google Scholar]
- 11.Socolovsky, M., Nam, H., Fleming, M. D., Haase, V. H., Brugnara, C., and Lodish, H. F. (2001) Blood 98 3261-3273 [DOI] [PubMed] [Google Scholar]
- 12.Hai, T., and Hartman, M. G. (2001) Gene (Amst.) 273 1-11 [DOI] [PubMed] [Google Scholar]
- 13.Masuoka, H. C., and Townes, T. M. (2002) Blood 99 736-745 [DOI] [PubMed] [Google Scholar]
- 14.Munugalavadla, V., and Kapur, R. (2005) Crit. Rev. Oncol. Hematol. 54 63-75 [DOI] [PubMed] [Google Scholar]
- 15.Marchese, A., George, S. R., Kolakowski, L. F., Jr., Lynch, K. R., and O'Dowd, B. F. (1999) Trends Pharmacol. Sci. 20 370-375 [DOI] [PubMed] [Google Scholar]
- 16.Hill, S. J. (2006) Br. J. Pharmacol. 147 Suppl. 1, S27-S37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le, T. I., Teller, D. C., Okada, T., Stenkamp, R. E., Yamamoto, M., and Miyano, M. (2000) Science 289 739-745 [DOI] [PubMed] [Google Scholar]
- 18.Iiri, T., Farfel, Z., and Bourne, H. R. (1998) Nature 394 35-38 [DOI] [PubMed] [Google Scholar]
- 19.Hsu, S. Y., Liang, S. G., and Hsueh, A. J. (1998) Mol. Endocrinol. 12 1830-1845 [DOI] [PubMed] [Google Scholar]
- 20.Osuga, Y., Kudo, M., Kaipia, A., Kobilka, B., and Hsueh, A. J. (1997) Mol. Endocrinol. 11 1659-1668 [DOI] [PubMed] [Google Scholar]
- 21.Kobe, B., and Kajava, A. V. (2001) Curr. Opin. Struct. Biol. 11 725-732 [DOI] [PubMed] [Google Scholar]
- 22.Hsu, S. Y. (2003) Trends Endocrinol. Metab. 14 303-309 [DOI] [PubMed] [Google Scholar]
- 23.Fredriksson, R., Lagerstrom, M. C., Lundin, L. G., and Schioth, H. B. (2003) Mol. Pharmacol. 63 1256-1272 [DOI] [PubMed] [Google Scholar]
- 24.Hsu, S. Y., Nakabayashi, K., Nishi, S., Kumagai, J., Kudo, M., Sherwood, O. D., and Hsueh, A. J. (2002) Science 295 671-674 [DOI] [PubMed] [Google Scholar]
- 25.Hsu, S. Y., Kudo, M., Chen, T., Nakabayashi, K., Bhalla, A., van der Spek, P. J., van, D. M., and Hsueh, A. J. (2000) Mol. Endocrinol. 14 1257-1271 [DOI] [PubMed] [Google Scholar]
- 26.Kajava, A. V. (1998) J. Mol. Biol. 277 519-527 [DOI] [PubMed] [Google Scholar]
- 27.Loh, E. D., Broussard, S. R., and Kolakowski, L. F. (2001) Biochem. Biophys. Res. Commun. 282 757-764 [DOI] [PubMed] [Google Scholar]
- 28.Loh, E. D., Broussard, S. R., Liu, Q., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., and Kolakowski, L. F., Jr. (2000) Cytogenet. Cell Genet. 89 2-5 [DOI] [PubMed] [Google Scholar]
- 29.Van, S. G., Mendive, F., Pochet, R., and Vassart, G. (2005) Histochem. Cell Biol. 124 35-50 [DOI] [PubMed] [Google Scholar]
- 30.Hoshii, T., Takeo, T., Nakagata, N., Takeya, M., Araki, K., and Yamamura, K. (2007) Biol. Reprod. 76 303-313 [DOI] [PubMed] [Google Scholar]
- 31.Kato, S., Matsubara, M., Matsuo, T., Mohri, Y., Kazama, I., Hatano, R., Umezawa, A., and Nishimori, K. (2006) Nephron Exp. Nephrol. 104 e63-e75 [DOI] [PubMed] [Google Scholar]
- 32.Mendive, F., Laurent, P., Van, S. G., Skarnes, W., Pochet, R., and Vassart, G. (2006) Dev. Biol. 290 421-434 [DOI] [PubMed] [Google Scholar]
- 33.Mazerbourg, S., Bouley, D. M., Sudo, S., Klein, C. A., Zhang, J. V., Kawamura, K., Goodrich, L. V., Rayburn, H., Tessier-Lavigne, M., and Hsueh, A. J. (2004) Mol. Endocrinol. 18 2241-2254 [DOI] [PubMed] [Google Scholar]
- 34.Stryke, D., Kawamoto, M., Huang, C. C., Johns, S. J., King, L. A., Harper, C. A., Meng, E. C., Lee, R. E., Yee, A., L'Italien, L., Chuang, P. T., Young, S. G., Skarnes, W. C., Babbitt, P. C., and Ferrin, T. E. (2003) Nucleic Acids Res. 31 278-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skarnes, W. C., von, M. H., Wurst, W., Hicks, G., Nord, A. S., Cox, T., Young, S. G., Ruiz, P., Soriano, P., Tessier-Lavigne, M., Conklin, B. R., Stanford, W. L., and Rossant, J. (2004) Nat. Genet. 36 543-544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leighton, P. A., Mitchell, K. J., Goodrich, L. V., Lu, X., Pinson, K., Scherz, P., Skarnes, W. C., and Tessier-Lavigne, M. (2001) Nature 410 174-179 [DOI] [PubMed] [Google Scholar]
- 37.Mitchell, D. C., Abdelrahim, M., Weng, J., Stafford, L. J., Safe, S., Bar-Eli, M., and Liu, M. (2006) J. Biol. Chem. 281 51-58 [DOI] [PubMed] [Google Scholar]
- 38.McGrath, K. E., and Palis, J. (2005) Exp. Hematol. 33 1021-1028 [DOI] [PubMed] [Google Scholar]
- 39.Samakoglu, S., Fattori, E., Lamartina, S., Toniatti, C., Stockholm, D., Heard, J. M., and Bohl, D. (2001) Blood 97 2213-2220 [DOI] [PubMed] [Google Scholar]
- 40.Basu, P., Morris, P. E., Haar, J. L., Wani, M. A., Lingrel, J. B., Gaensler, K. M., and Lloyd, J. A. (2005) Blood 106 2566-2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozar, K., Ciemerych, M. A., Rebel, V. I., Shigematsu, H., Zagozdzon, A., Sicinska, E., Geng, Y., Yu, Q., Bhattacharya, S., Bronson, R. T., Akashi, K., and Sicinski, P. (2004) Cell 118 477-491 [DOI] [PubMed] [Google Scholar]
- 42.Bjorklund, M., Taipale, M., Varjosalo, M., Saharinen, J., Lahdenpera, J., and Taipale, J. (2006) Nature 439 1009-1013 [DOI] [PubMed] [Google Scholar]
- 43.Elefteriou, F., Benson, M. D., Sowa, H., Starbuck, M., Liu, X., Ron, D., Parada, L. F., and Karsenty, G. (2006) Cell Metab. 4 441-451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, X., Matsuda, K., Bialek, P., Jacquot, S., Masuoka, H. C., Schinke, T., Li, L., Brancorsini, S., Sassone-Corsi, P., Townes, T. M., Hanauer, A., and Karsenty, G. (2004) Cell 117 387-398 [DOI] [PubMed] [Google Scholar]
- 45.Tanaka, T., Tsujimura, T., Takeda, K., Sugihara, A., Maekawa, A., Terada, N., Yoshida, N., and Akira, S. (1998) Genes Cells 3 801-810 [DOI] [PubMed] [Google Scholar]
- 46.Fischer, C., Johnson, J., Stillwell, B., Conner, J., Cerovac, Z., Wilson-Rawls, J., and Rawls, A. (2004) Biol. Reprod. 70 371-378 [DOI] [PubMed] [Google Scholar]
- 47.Gao, Y., Kitagawa, K., Shimada, M., Uchida, C., Hattori, T., Oda, T., and Kitagawa, M. (2006) Hokkaido Igaku Zasshi 81 101-105, 107, 109 [PubMed] [Google Scholar]
- 48.Ameri, K., Lewis, C. E., Raida, M., Sowter, H., Hai, T., and Harris, A. L. (2004) Blood 103 1876-1882 [DOI] [PubMed] [Google Scholar]
- 49.Blais, J. D., Filipenko, V., Bi, M., Harding, H. P., Ron, D., Koumenis, C., Wouters, B. G., and Bell, J. C. (2004) Mol. Cell. Biol. 24 7469-7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohoka, N., Yoshii, S., Hattori, T., Onozaki, K., and Hayashi, H. (2005) EMBO J. 24 1243-1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jousse, C., Deval, C., Maurin, A. C., Parry, L., Cherasse, Y., Chaveroux, C., Lefloch, R., Lenormand, P., Bruhat, A., and Fafournoux, P. (2007) J. Biol. Chem. 282 15851-15861 [DOI] [PubMed] [Google Scholar]
- 52.Cui, K., Coutts, M., Stahl, J., and Sytkowski, A. J. (2000) J. Biol. Chem. 275 7591-7596 [DOI] [PubMed] [Google Scholar]
- 53.Grimley, P. M., Dong, F., and Rui, H. (1999) Cytokine Growth Factor Rev. 10 131-157 [DOI] [PubMed] [Google Scholar]
- 54.Damen, J. E., Wakao, H., Miyajima, A., Krosl, J., Humphries, R. K., Cutler, R. L., and Krystal, G. (1995) EMBO J. 14 5557-5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gobert, S., Chretien, S., Gouilleux, F., Muller, O., Pallard, C., Dusanter-Fourt, I., Groner, B., Lacombe, C., Gisselbrecht, S., and Mayeux, P. (1996) EMBO J. 15 2434-2441 [PMC free article] [PubMed] [Google Scholar]
- 56.Quelle, F. W., Wang, D., Nosaka, T., Thierfelder, W. E., Stravopodis, D., Weinstein, Y., and Ihle, J. N. (1996) Mol. Cell. Biol. 16 1622-1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klingmuller, U., Bergelson, S., Hsiao, J. G., and Lodish, H. F. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 8324-8328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boer, A. K., Drayer, A. L., Rui, H., and Vellenga, E. (2002) Blood 100 467-473 [DOI] [PubMed] [Google Scholar]
- 59.Boer, A. K., Drayer, A. L., and Vellenga, E. (2003) Exp. Hematol. 31 512-520 [DOI] [PubMed] [Google Scholar]
- 60.Weng, J., Luo, J., Cheng, X., Jin, C., Zhou, X., Qu, J., Tu, L., Ai, D., Li, D., Wang, J., Martin, J. F., Amendt, B. A., and Liu, M. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 6081-6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.