Abstract
SirT1 is an NAD-dependent histone deacetylase that regulates gene expression, differentiation, development, and organism life span. Here we investigate the function of SirT1 in human chondrocytes derived from osteoarthritic patients. Elevation of SirT1 protein levels or activity in these chondrocytes led to a dramatic increase in cartilage-specific gene expression, whereas a reduction in SirT1 levels or activity significantly lowered cartilage gene expression. SirT1 associated with the cartilage-specific transcription factor Sox9, enhancing transcription from the collagen 2(α1) promoter in a Sox9-dependent fashion. Consistent with this association, SirT1 was targeted to the collagen 2(α1) enhancer and promoter, which in turn recruited the coactivators GCN5, PGC1α, and p300. This led to elevated marks of active chromatin within the promoter; that is, acetylated histone K9/K14 and histone H4K5 as well as trimethylated histone H3K4. Finally, alterations in the NAD salvage pathway enzyme nicotinamide phosphoribosyltransferase led to changes in NAD levels, SirT activity, and cartilage-specific gene expression in human chondrocytes. SirT1, nicotinamide phosphoribosyltransferase, and NAD may, therefore, provide a positive function in human cartilage by elevating expression of genes encoding cartilage extracellular matrix.
Transcriptional control over cartilage-specific gene expression plays a critical role in maintenance of the chondrocyte phenotype (1). Much effort has, therefore, gone into the characterization of chondrocyte-specific transcription factors such as Sox9, -5, and -6 (28). However, it is likely that other factors such as chromatin-modifying enzymes play important roles in controlling cartilage-specific gene expression. Chromatin-modifying enzymes, which include the histone acetyltransferases (HATs)2 and the histone deacetylases (HDACs) can act as potent transcriptional coactivators and corepressors, respectively, for a variety of genes (2). HATs modify the core histones through acetylation of lysine residues, thereby relaxing chromatin for transcription initiation and elongation (3). HDACs remove the acetyl groups, leading to chromatin condensation and transcriptional repression (4). Additionally, acetylation and deacetylation of transcription factors provide another level of regulation over gene expression (2).
In some contexts HDACs are more sensitive to environmental or developmental cues than the HATs and can provide a regulatory role in transcription. In this regard, HDACs have been demonstrated to control both cell proliferation and differentiation through the deacetylation of transcription factors, cytoplasmic proteins, and histones (5, 6). Although HDACs are critically involved in diverse biological processes, their function in chondrocyte biology and cartilage diseases have only recently been explored. Recent work indicates that inhibition of HDACs reduces the expression of matrix metalloproteinase in chondrocytes and fibroblasts (7) and, therefore, inhibits arthritis progression in animal models (8, 9). Additionally, it has been demonstrated that HDAC4, a Class II enzyme, plays a critical role in the onset of chondrocyte hypertrophy during endochondral ossification (10). It would appear then that HDACs regulate genes involved in both inflammation and chondrocyte differentiation. Given that there are more than 18 HDAC genes distributed in at least four different classes, it is likely that multiple HDACs will affect the growth, differentiation, and survival of chondrocytes via transcriptional regulation of cartilage-specific genes.
Of particular interest with regard to cartilage biology are the NAD-dependent Class III HDACs comprised of SirT1–7. Within this group SirT1 has been extensively studied and is known to play a crucial part in regulating cell differentiation, proliferation, survival, and organism longevity (11, 12). Class III HDACs yield nicotinamide (NAM) and O-acetyl-ADP-ribose from NAD in the process of deacetylation. NAM thereby becomes a potent feedback inhibitor of SirT1 by binding to a unique site within the enzyme (11, 12). NAD can be generated from NADH via intracellular dehydrogenase reactions as well as by oxidative phosphorylation. Interestingly, the NAD salvage pathway also plays a role in de novo production of NAD (13). Through this pathway NAM regenerates NAD through the action of two salvage pathway enzymes, nicotinamide phosphoribosyltransferase (NAMPT) and nicotinamide mononucleotide adenyltransferase (14, 15). Because NAMPT is known to have a rate-limiting affect on NAD production (16), evaluation of its expression is important to our understanding the cells metabolic state. These salvage pathway enzymes have been known to extend cellular life span, likely through their indirect effect on SirT1 activity (15, 17). Because nothing is known of the role SirT1, NAMPT, and NAD play in chondrocyte biology, we have explored their functions in chondrocytes derived from knee joints of osteoarthritic patients. The results obtained here show that SirT1 participates in the activation of cartilage-specific gene expression and is dependent on the NAD/NAMPT pathway.
EXPERIMENTAL PROCEDURES
Reagents, Cell Culture, and Transfections—Nicotinamide and resveratrol were purchased from Sigma. FK866 was obtained from the NIMH Chemical Synthesis and Drug Supply Program (NIH, Bethesda, MD). siRNAs for SirT1 were generated by Ambion (Austin, TX) and used according to the manufacturers recommendations. Custom stealth NAMPT siRNA was purchased from Invitrogen and used according to the manufacturers recommendations. The Dual Reporter Luciferase assay (Promega) was carried out to test transcription initiation of the collagen 2(α1) promoter (pGL2col2A.cg). This plasmid, a gift from Dr. M. Goldring, possesses the collagen 2(α1) promoter and enhancer linked to a luciferase reporter (21). Human chondrocytes were transfected with FuGENE 6 (Roche Applied Science) in the presence of Opti-MEM (Invitrogen), reduced serum media. 293 cells were transfected using ProFection calcium phosphate kit (Promega) and in accordance with the manufacturer's instructions.
Human chondrocytes were isolated from the knees of osteoarthritic patients undergoing total knee arthroplasty, supplied by the National Disease Research Interchange, Philadelphia, PA. Isolation was carried out according to Derfoul et al. (39). Cells were plated in 10-cm2 tissue culture dishes at a concentration of 3 × 106 cells/dish and were grown to confluence (passage 0 or P0). In our hands cartilage marker gene expression was maintained until about passage 4 at which time it declined. Thus, our experiments herein utilized chondrocytes up to passage 4 (P4). Monolayer cultures of human chondrocytes were maintained as previously described by Derfoul et al. (39).
All transfection experiments were initiated on 50% confluent monolayer cultures. If not otherwise indicated, chondrocyte transfections were carried out using the Amaxa Nucleofector technology (Gaithersburg, MD) in accordance with manufacturer's instructions.
Plasmids, Retroviral Infection, and Selection of Cell Lines—The retroviral expression plasmids, pHanPuro, pHanPuro-SirT1, and pHanPuro-SirT1-M(H355Y) were gifts of Dr. Vittorio Sartorelli (24). The SirT1 expression plasmid, pUSE-SirT1 was purchased from Upstate (Charlottesville, VA). The Sox9 expression plasmid pcDNA-Sox9 was a kind gift of Dr. Benoit de Crombrugghe. Retrovirus was generated, and infections were carried out as previously described (24, 40). Puromycin-resistant (1 μg/ml) colonies were pooled from each infection condition and used in the subsequent experiments.
NAD Assay—The NAD levels were measured using a BioVision NAD assay kit according to manufacturers instructions (Biovision, Mountain View, CA). NAD levels were calculated according to the equivalent protein quantity (μg protein) per data point.
SirT Activity Assay—SirT activity was determined using a BIOMOL assay (Plymouth Meeting, PA) was used according to manufacturers instructions, based on Borra et al. (41) with slight modifications. Briefly, 1 μm trichostatin A was added to the cell cultures 1 h before harvesting to block the class I, II, and IV HDACs. Whole cell extracts were then generated as described below with the addition of 1 μm trichostatin A. The extracts were added (15 μl) to an opaque multiwell plate with 1 μm trichostatin A and 1 mm acetylated Fluor-de-Lys substrate. As a negative control, 10 mm NAM was also added to some extracts. The extracts were then swirled for 20 min at room temperature, and the reactions were stopped by adding 50 μl of a developer solution supplemented with 1 μm trichostatin A and 10 mm NAM (final concentrations). The plate was read after a 10-min incubation at room temperature using a multiwell fluorometer (excitation 360 nm, emission 460 nm). A standard curve was generated using a deacetylated substrate Fluor-de-Lys (ranging from 1 to 40 μm). Experimental values are presented as pmol of converted substrate/μg of protein/min. The negative controls (10 mm NAM) were subtracted from each treatment to give the final values.
Chromatin Immunoprecipitation (ChIP) Analyses—Chromatin immunoprecipitation assays were carried out utilizing the EZ-ChIP kit (Upstate Millipore, Boston, MA), according to the manufacturers guidelines. After ChIP, DNA was extracted using a Qiaquick spin kit (Qiagen, Valencia, CA). PCR reactions were carried out using ProofStart PCR kit (Qiagen).
RT-PCR Analysis—RNeasy RNA purification columns (Qiagen) were used to isolate RNA; 300 ng of RNA per reaction was subjected to a one-step semiquantitative RT-PCR procedure (Invitrogen). Gels supplemented with 1 μg/ml ethidium bromide were subjected to densitometry using ImageJ Software. Values were normalized to GAPDH. Primer sequences and the predicted size of the PCR products are presented in supplementary Data (Table 1). RT-PCR was performed in triplicates on two experimental repetitions (n = 6).
Real-time PCR reactions were carried out using 10 ng of cDNA and Syber Green mix (Bio-Rad Laboratories, Hercules, CA), as previously described by Derfoul et al. (39). Quantitative analyses were performed using Bio-Rad iCycler software. Primers are specified in supplemental Table 1. For an additional control we tested each set of experiments for expression of nonlineage specific genes (collagen 1 (α1), fibronectin, NFκB (p65); supplemental Figs. A–H).
Protein Analysis and Immunoblotting—Whole cell protein extracts and immunoblotting procedures were carried out according to Perkins et al. (42). Primary and secondary antibodies used for immunoblotting, immunoprecipitation, and immunocytology are specified in supplemental Table 2.
Immunohistochemical Analyses—Monolayer cells were cultured in 6-well plates, rinsed with phosphate-buffered saline, and fixed in 4% paraformaldehyde for 10 min, and nonspecific antibody binding was blocked for 30 min in goat serum. They were then incubated with a 1:10 dilution of mouse anti-SirT1 primary antibody visualized using a broad-spectrum immunohistochemistry kit (Zymed Laboratories Inc., Carlsbad, CA).
Statistical Analyses—Statistical analysis was obtained using one-way analysis of variance, assuming confidence levels of 95% (p < 0.05) to be statistically significant. The least significant difference (LSD) test was carried out to determine the differences between two equivalent treatments within a group, assuming confidence levels of 95% (p < 0.05). Error bars indicate the S.D. around the mean value of data point. The symbol a was used to express statistical significance between SirT1 and pHan, b was used for SirT1 and SirT1-M, and c was use for pHan and SirT1-M. All transfections in the Col2-luciferase assays were compared with DNA control, and statistical significance is indicated as *, LSD, p < 0.05. The asterisk (*) was also used to express statistical significance in equivalent SirT1 transient transfection, reservatrol-treated cells, NAM-treated cells, and siRNA transfection comparisons (LSD, p < 0.05).
The immunoblots, ChIP, and co-immunoprecipitation assays represent two repetitions from two distinct cell lines or cell sources (n = 4). The SirT activity and luciferase assays were carried out in triplicate using 2 distinct cell lines generated from different transfections or cell sources (n = 6). The scanned RT-PCR presented in Figs. 1 and 2 were generated from 3 runs of 2 distinct cell lines (n = 6). Supporting quantitative PCR (qPCR) evidence for Figs. 1 and 2 are illustrated in the supplemental data for the equivalent experiments (n = 6, triplicates of two cell lines/treatment).
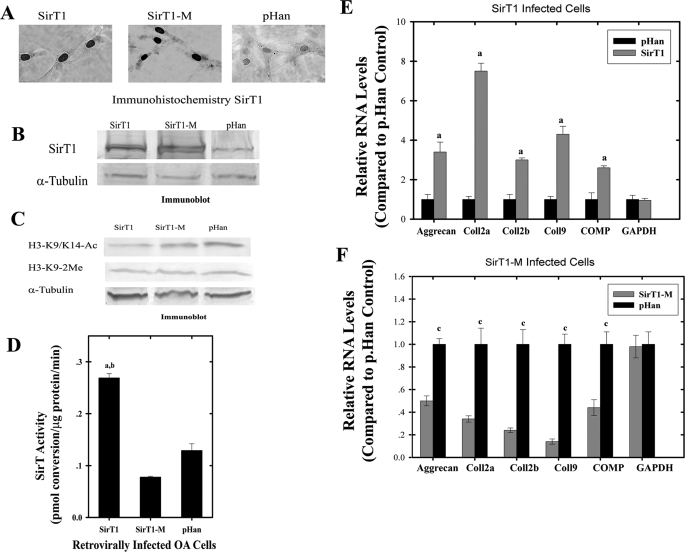
FIGURE 1.
Increased stable expression of SirT1 in OA chondrocytes leads to elevated cartilage-specific gene expression. Human OA chondrocytes were infected with a vector control (pHan), SirT1, or SirT1-M expressing retrovirus, selected in puromycin and resistent colonies pooled. A, the pooled cells were subjected to immunohistochemistry using a SirT1 antibody. B, extracts generated from the cell lines in A were used for immunoblotting with a SirT1 or tubulin antibody. C, cell extracts as in B were used in immunoblots with antibodies directed against AcH3K9 and 2MeH3K9. D, cell extracts as in B were tested for SirT activity. E and F, RNA isolated from the control, SirT1 (E). and SirT1-M (F) cells was used in RT-PCR reactions with the indicated human primers. The graphs show the -fold induction or repression of the indicated genes by SirT1 and SirT1M in relation to the levels of expression in the control cells (pHan). The error bars in the graphs indicate S.D., and statistical significance is indicated by a and b (LSD, p < 0.05). The displayed immunoblots are representatives of four experiments. RT-PCR was performed in triplicates for two experimental repetitions (n = 6). COMP, cartilage oligomeric matrix protein.
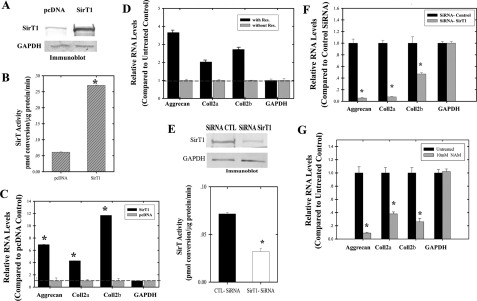
FIGURE 2.
Transient changes in SirT1 levels or activity alters cartilage gene expression. OA human chondrocytes (P0) were transiently transfected with a SirT1 expression plasmid. Extracts generated at 3 days post-transfection were used in immunoblot analysis (A) or SirT activity assays (B). C, RNA isolated from the transfected cells in A was used in RT-PCR assays with the indicated human primers. The graphs show relative induction of gene expression (compared with control). D, P0 and P1 OA chondrocytes were treated with resveratrol (1 μm) for 3 days at which time RNA was isolated and used in RT-PCR reactions with the indicated human primers. E, OA chondrocytes (P0) were transiently transfected with a SirT1 siRNA, and extracts were isolated and used in immunoblots for SirT1 and GAPDH (upper panel) or for assessment of SirT activity (lower panel). CTL, control. F, RNA was isolated from the cells expressing the SirT1siRNA in E which was used in RT-PCR reactions with the indicated human primers. G, OA chondrocytes were treated with NAM (10 mm) for 3 days at which time total RNA was isolated and used RT-PCR reactions with the indicated human primers. The graphs in F and G show relative repression of gene expression. The error bars in the graphs indicate the S.D., and the statistical significance is indicated by an asterisk (*) (LSD, p < 0.05). The displayed immunoblots are representative of four experiments. RT-PCR was performed in triplicate for two experimental repetitions (n = 6).
RESULTS
Elevation of SirT1 Positively Affects Cartilage Gene Expression—To test the hypothesis that cartilage-specific gene expression in chondrocytes is affected by SirT1, osteoarthritic (OA) human chondrocytes (P0-P2) were stably infected with a SirT1 expressing retrovirus. The vectors expressed either wild-type SirT1 or an enzymatically inactive mutant SirT1-M (SirT1H355Y). The vector alone (pHan) was used as a control. Immunohistochemistry assays (Fig. 1A) as well as immunoblot assays (Fig. 1B) confirmed SirT1 and SirT1-M overexpression and nuclear localization. As a test for SirT1 activity, the total levels of AcH3K9, a known substrate for SirT1 (18), were measured. From Fig. 1C it appears that the SirT1-expressing cells possess reduced AcH3K9 (∼3-fold) when compared with the SirT1-Mut and control cell lines. H3K9 dimethylation and tubulin, which served as a control, were unaffected in all cell lines. The graph in Fig. 1D indicates a significant increase of SirT activity in cells expressing wild type SirT1 compared with SirT1-M and pHan (LSD, p < 0.05, 2.5-fold). These data indicate that the ectopic SirT1 is active within these chondrocytes. Interestingly, cells expressing SirT1-M displayed lower SirT activity compared with control, suggesting that it acts in a dominant/negative way as previously described (19, 20).
We next examined the effects of SirT1 on cartilage-specific gene expression. RNA was harvested and used in RT-PCR reactions for markers of cartilage genes (aggrecan, collagen 2a(α1) and 2b(α1), collagen 9(α1), and cartilage oligomeric matrix protein (COMP)). Surprisingly, SirT1-expressing cells exhibited significantly higher expression values than the control cells (LSD, p < 0.05; Fig. 1E). When the same analyses were performed for SirT1-M-expressing cells, it was clear that cartilage gene expression was significantly repressed as compared with the control cells (LSD, p < 0.05; Fig. 1F). In all cell lines GAPDH remained unaffected. As additional controls, we assessed expression of the nonlineage-specific genes collagen 1(α1), fibronectin, and NFκB (p65) and found no changes in RNA levels between pHan, SirT1, and SirT1M cell lines (supplemental Figs. A and B). Additionally, qPCR was carried out on collagen 2a(α1), collagen 2b(α1), and aggrecan in the stably transfected cell lines (supplementary Figs. I and J). These data show collagen 2a(α1), collagen 2b(α1), and aggrecan to be induced by SirT1 and repressed by SirT1-M.
To confirm that SirT1 had a positive effect on cartilage gene expression, transient expression experiments and drug treatments were also performed in low passage chondrocytes. A SirT1 expression vector was transiently expressed in chondrocytes. As shown in Fig. 2, A and B, SirT1 protein levels and SirT activity were markedly elevated in the transfected chondrocytes. Furthermore, the ectopically expressed SirT1 is completely nuclear as assessed by immunofluorescence (data not shown). Fig. 2C shows significantly higher gene expression values for the indicated genes in SirT1-expressing cells compared with the pcDNA control (LSD, p < 0.05). qPCR analysis for transiently transfected cells showed that collagen 2a(α1), collagen 2b(α1), and aggrecan were up-regulated by SirT1 (supplemental Fig. K). In a separate line of experiments, the chemical resveratrol (1 μm) was added to chondrocytes to directly activate SirT1 (19). As shown in Fig. 2D, this also resulted in the up-regulation of cartilage gene expression. qPCR analysis showed that aggrecan, collagen 2a(α1), collagen 2b(α1), and aggrecan were up-regulated by resveratrol (supplemental Fig. L). Taken together, these results show that by either transient overexpression or activation of SirT1, cartilage gene expression is enhanced in human chondrocytes. For an additional control we tested expression of the non-lineage specific genes collagen 1(α1), fibronectin, and NFκB (p65) in response to SirT1 transient transfection and resveratrol treatment. No effect was found on gene expression (supplemental Figs. C and F, respectively).
Inhibition of SirT1 Negatively Affects Cartilage Gene Expression—To perform the opposite of the above experiments, a SirT1 siRNA was transfected into chondrocytes to lower SirT1 protein levels. As shown in Fig. 2E, SirT1 levels and activity dropped by 3-fold after siRNA transfection. Fig. 2F shows that reducing SirT1 protein levels significantly lowered cartilage-specific gene expression (LSD, p < 0.05). qPCR analysis showed that SirT1 siRNA treatment repressed collagen 2(α1) and aggrecan (supplemental Fig. M). As a corollary to the above experiments, chondrocytes were also treated with NAM (10 mm), which is a direct feedback inhibitor of SirT1 (11, 12). Fig. 2G shows a clear reduction in cartilage gene expression in NAM-treated cells (LSD, p < 0.05). qPCR confirmed that NAM repressed expression of collagen 2(α1) and aggrecan in chondrocytes (supplemental Fig. M). In total these data indicate that reducing SirT1 levels or activity had a profound negative effect on cartilage gene expression. Furthermore, inspection of nonlineage-specific genes collagen 1(α1), fibronectin, and NFκB (p65) in SirT1 siRNA- and NAM-treated cells found no effect on gene expression (supplemental Figs. D and E, respectively) with the exception that SirT1 SiRNA-treated cells suppressed p65 expression.
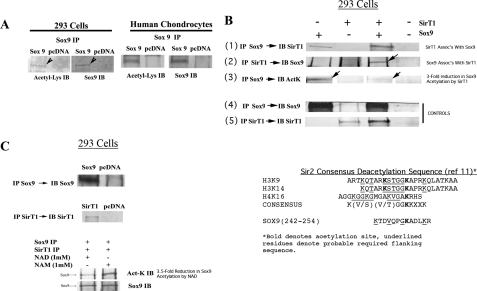
SirT1 Interacts with Sox9 and Enhances Transcription from the Collagen 2(α1) Promoter—The data above show that SirT1 positively regulates cartilage gene expression. Because it is well established that expression of many cartilage genes is regulated by the transcription factor Sox9 (1), it was hypothesized that SirT1 may interact with Sox9, thereby enhancing its transcriptional activity, possibly through deacetylation. To first determine whether Sox9 was acetylated, it was expressed in 293 cells (Fig. 3A, left panel) and in human chondrocytes derived from OA patients (Fig. 3A, right panel). It was then immunoprecipitated (IP) with a Sox9-specific antibody and then immunoblotted in duplicate lanes. Half the blot was incubated with a Sox9 antibody, and the other half was incubated with an acetyl-lysine-specific antibody. As shown in Fig. 3A, Sox9 is recognized by the acetyl-lysine antibody, indicating it is an acetylated protein.
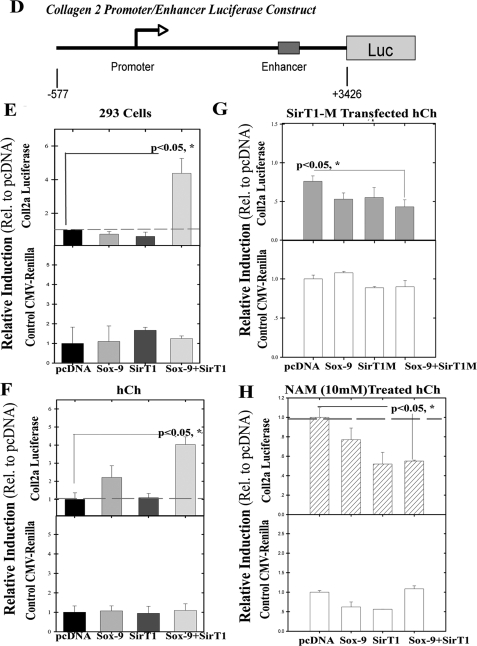
FIGURE 3.
SirT1 associates with Sox9, modulates its acetylation status, and enhances transcription from the collagen 2(α1) promoter. A, 293 cells and human P0 OA chondrocytes were transfected with the pcDNA or Sox9 expression plasmids. Extracts were generated and used in immunoprecipitations with a Sox9 antibody. The IPs were immunoblotted (IB) in duplicate lanes, and the blots were separated and probed with either a Sox9 antibody or an acetyl-lysine-specific antibody. B, 293 cells were transfected with Sox9, SirT1, or both the Sox9 and SirT1 expression plasmids. Extracts were generated and used in IP reactions with either a Sox9 or SirT1 antibody. Upper panels, the IPs were immunoblotted with either a Sox9, SirT1, or acetyl-lysine-specific antibody as indicated. Lower panels, control immunoblots showing the levels of SirT1 and Sox9 along with GAPDH in the transfected extracts. C, 293 cells were transfected with Sox9, SirT1, or pcDNA. The upper panel shows an IP for Sox9 followed by an immunoblot for Sox9 as well as an IP for SirT1 followed by an immunoblot for SirT1. Middle panel, the SirT1 and Sox9 IPs were incubated at room temperature for 3 h with either NAD or with NAM (1 mm) and then electrophoresed and blotted with either acetyl-lysine- or Sox9-specific antibodies. The displayed immunoblots are representatives of four experiments. Right panel, SirT1 consensus deacetylation site (11) is compared with a putative site for deacetylation within Sox9 (amino acids 242–254). This site is similar to that within histones H3(K19 and K14) and H4(K16). D, an outline of the collagen 2(α1)-promoter/enhancer luciferase construct utilized in the reporter assay. E and F, upper panels, SirT1 and Sox9 expression plasmids were transfected as indicated with the collagen 2(α1)-luciferase construct in 293 cells and human OA chondrocytes (hCh), respectively. The graphs show -fold induction of luciferase relative to the pcDNA control. G, upper panel, the SirT1M and Sox9 expression plasmids were transfected, as indicated with the collagen 2(α1)-luciferase construct in human OA chondrocytes. H, upper panel, the SirT1 and Sox9 expression plasmids were transfected into human OA chondrocytes with the collagen 2(α1)-luciferase construct in the presence of 10 mm NAM. The lower panels of E–H serve as control, where a cytomegalovirus-renilla promoter construct was cotransfected into the cells as indicated. All luciferase analyses were performed in triplicate for two experimental repetitions (n = 6).
To determine whether SirT1 and Sox9 associate, 293 cells were transfected with the SirT1 and/or Sox9 expression plasmids, and co-IP experiments were performed. Sox9 was first immunoprecipitated from the extracts, and the IPs were immunoblotted for SirT1 (Fig. 3B, panel 1). The data show that SirT1 associates with Sox9. The data in panel 1 also show that when only Sox9 was expressed, endogenous SirT1 was able to associate with it (first lane in panel 1). Next, SirT1 was immunoprecipitated, and the IPs were blotted for Sox9. The data in Fig. 3B, panel 2 (note the arrow) show that Sox9 associates with SirT1. Together these data indicate that Sox9 and SirT1 are able to interact in vivo.
When Sox9 was immunoprecipitated from the extracts and the IPs were blotted for acetylated lysine, a band was detected (Fig. 3B, panel 3, first lane, note the arrow). Interestingly, when SirT1 was coexpressed with Sox9, the intensity of the acetylated Sox9 band was diminished (Fig. 3B, panel 3, third lane, note the arrow). Densitometry scans of these bands indicate that the amount of acetylated Sox9 was reduced by 3-fold when SirT1 was coexpressed.
The IP controls show efficient expression and immunoprecipitation of both Sox9 and SirT1 (Fig. 3B, panels 4 and 5). Additional controls demonstrated that Sox9 and SirT1 protein levels were unaffected by coexpression in 293 cells (data not shown).
To further demonstrate that SirT1 may directly deacetylate Sox9, 293 cells were transfected with Sox9 or SirT1, and the individual proteins were immunoprecipitated (Fig. 3C, top two panels). As in Firestein et al. (43), the IPs were then mixed equally and then incubated with either NAD or NAM (SirT1 is only active with NAD). The IPs were then electrophoresed and immunoblotted for either acetyl-lycine or Sox9. As shown in the bottom two panels of Fig. 3C, in the presence of NAD there was a 3.5-fold reduction in the amount of Sox9 recognized by the acetyl-lysine antibody, whereas the total level of Sox9 was invariant. These data suggest that SirT1 is active in deacetylating Sox9 and is dependent on NAD.
As shown in Fig. 3C (right side), the sequence spanning residues 242–254 of Sox9 corresponds approximately to those predicted by Blander and Guarente (11) to be a consensus SirT1 deacetylation site. This site does not represent the nuclear localization sequence of Sox9 and is outside the high mobility group DNA binding domain. Current efforts are under way to determine whether lysine residue 249 is acetylated, and, further, if it is a target of SirT1.
To determine the functional consequences of the Sox9/SirT1 association, studies were performed with the collagen 2(α1) promoter/enhancer linked to luciferase (21), as this is a well known Sox9-responsive gene (1). 293 cells were transfected with the SirT1 and/or Sox9 expression plasmids in the presence of the collagen 2(α1)-luciferase construct. An outline of the collagen 2(α1)-luciferase construct is presented in Fig. 3D. As shown in Fig. 3, E and F, for 293 cells and human OA chondrocytes, respectively, SirT1 alone did not affect transcription from collagen 2(α1) promoter, whereas Sox9 alone was able to activate transcription in human chondrocytes. When both SirT1 and Sox9 were coexpressed, they were able to transactivate the promoter ∼4-fold, which was found to be statistically significant in both cell types (LSD, p < 0.05). As a control, the cytomegalovirus promoter expressing the renilla reporter gene was not affected by Sox9, SirT1, or Sox9+SirT1 (Fig. 3, E and F, lower graph panel). As an additional control, human chondrocytes were transfected with SirT1 mutant (H355Y) expression plasmid in the presence or absence of the Sox9 plasmid (Fig. 3G). The mutant was unable to transactivate the collagen 2(α1) promoter. In a separate experiment human chondrocytes were treated with 10 mm NAM (a repressor of SirT1, Fig. 3H).
The results show a reduction in the transcription of collagen 2(α1) in the presence of 10 mm NAM. In fact, as shown in Fig. 3H, in either the presence or absence of exogenous SirT1, NAM is able to block the transactivating ability of Sox9. Overall, these data suggest that active SirT1 is required for transactivation of the collagen 2(α1) promoter/enhancer.
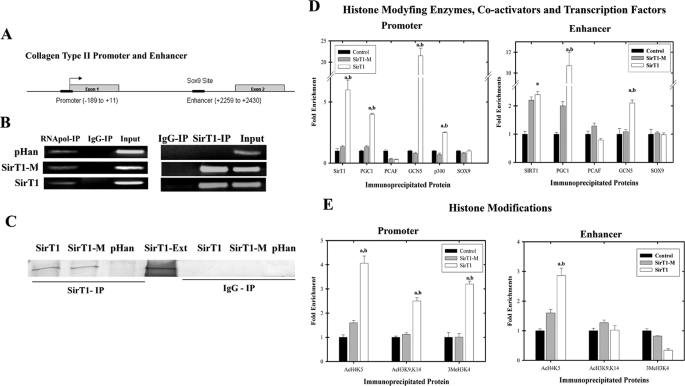
Wild-type SirT1, but Not the Inactive SirT1 Mutant, Targets GCN5, p300, and PGC1α to the Collagen 2(α1) Promoter—Because SirT1 and Sox9 interact in vivo and affect transcription from the collagen 2(α1) promoter, ChIP analyses were used to determine whether SirT1 is targeted to regulatory regions in the collagen 2(α1) gene. Fig. 4A outlines the promoter sequence as well as the Sox9 binding site (enhancer) within the first intron of the collagen 2(α1) used in the ChIP analysis. The Sox9 binding site spans the 48-base pair sequence +2364 to +2412 in the human collagen type 2(α1) first intron, which is required for cartilage-specific transcription (1). Nucleotide positions are as per Bell et al. (22).
FIGURE 4.
Enzymatically active SirT1 recruits GCN5, PGC1α, and p300 to the collagen 2(α1) promoter and elevates histone modifications at this site. A, the collagen 2(α1) promoter and enhancer positions used in ChIP assays are indicated (black bars). B, chondrocytes stably expressing SirT1, SirT1-M, and the pHan control were processed for ChIP analyses using antibodies for SirT1, GAPDH, and an IgG control. PCR was then performed with primers flanking the enhancer within the collagen 2(α1) first intron (SirT1 IP). In the right panel, as an additional control, RNA polymerase II was immunoprecipitated, and the GAPDH promoter was amplified. The RT-PCR products shown are representative of three separate experiments (n = 3). C, protein extracts generated from the ChIP analyses were immunoblotted with SirT1. SirT1-Ext indicates protein extracts isolated from SirT1 stably expressing cells. D and E, ChIP analyses as in B using primers for either the enhancer or promoter sequence of collagen 2(α1) were carried out with the indicated cell lines. The chromatin was immunoprecipitated with antibodies specific for SirT1, PGC1α, GCN5, PCAF, p300, and Sox9 (D) or antibodies specific for AcH3K9/K14, AcH4K5, and 3MeH3K4 (E). The DNA products were amplified by quantitative PCR. An IgG control for D and E showed no PCR product and was subtracted from the equivalent antibody ChIP value. The error bars in the graphs indicate the S.D., and the statistical significance is indicated by an a (SirT1 compared with pHan) or b (SirT1-M compared with SirT1-M) (LSD, p < 0.05). The displayed immunoblots are representative of four experiments. qPCR was performed in triplicates using IPs from 2 distinct cell lines.
The ChIP assays were performed on the pHan-, SirT1-, and SirT1-M-expressing chondrocytes. Using a SirT1-specific antibody in the ChIP analyses, a PCR product encompassing the Sox9 enhancer site was significantly elevated in the SirT1 and SirT1-M cells relative to the pHan control (Fig. 4B), indicating that the SirT1 and SirT1-M proteins are targeted to this site in vivo. Also, an antibody to RNA polymerase produced an equal signal in the PCR reactions for the GAPDH promoter in the three cell lines, whereas the IgG control produced no signal (Fig. 4B). To confirm that SirT1 was immunoprecipitated in the ChIP assay, the precipitates were immunoblotted and probed with a SirT1 antibody. Fig. 4C confirms SirT1 and SirT1M were precipitated in the corresponding cells lines.
To better elucidate the mechanism by which SirT1 transactivates collagen 2(α1), we investigated additional potential effectors such as histone modifying enzymes in the ChIP assays. As shown in Fig. 4D (right panel), SirT1 and SirT1-M are enriched on the enhancer sequence when compared with the levels in the control cells, correlating with the interaction of SirT1 with Sox9. Furthermore, active SirT1 appears to recruit the transcriptional coactivators PGC1α and GCN5 to the enhancer site as cells overexpressing SirT1 have markedly elevated PGC1α and GCN4 on the enhancer relative to the control and SirT1-M cells. SirT1-M moderately increases the level of PGC1α on the enhancer, which may be due to the fact that SirT1 and PGC1α are known to interact (37). However, that wild-type SirT1 leads to a 5-fold increase in PGC1α on the enhancer compared with SirT1-M argues that active SirT1 is necessary for optimal recruitment of PGC1α to this site. In total these data indicate that active SirT1 can direct the targeting of PGC1α and GCN5 to the enhancer. ChIP analysis of the promoter sequence (Fig. 4D, left panel) shows that wild-type SirT1, but not the mutant SirT1-M, is targeted to the promoter sequence. Furthermore, PGC1α, GCN5, and p300 are enhanced on the promoter sequence but only in the cells expressing active SirT1. These data indicate that PGC1α, GCN5, and p300 are recruited to the promoter by active SirT1.
When histone modifications were assessed by ChIP analysis on the promoter region, as shown in Fig. 4E (left panel), it was clear that AcH3K9/K14, AcH4K5, and 3MeH3K4 levels were increased within the promoter in the SirT1-expressing cells compared with the control and SirT1-Mutant cells. These data would indicate that the recruitment of PGC1α, GCN5, and p300 enzymes to the promoter has a functional outcome of enhanced histone acetylation. We have yet to identify the histone methyltransferase that leads to an increase in 3MeH3K4. ChIP analysis on the enhancer region (Fig. 4E, right panel) showed an increase in AcH4K5 but no change in AcH3K9/K14 or 3MeH3K4 in the SirT1 cells. These data in total indicate that through recruitment of activator/coactivator proteins, SirT1 enhances acetylation and methylation of critical histone residues primarily in the region of the promoter. These enhanced histone modifications appear to require active SirT1 as the SirT1 mutant does not affect histone modification.
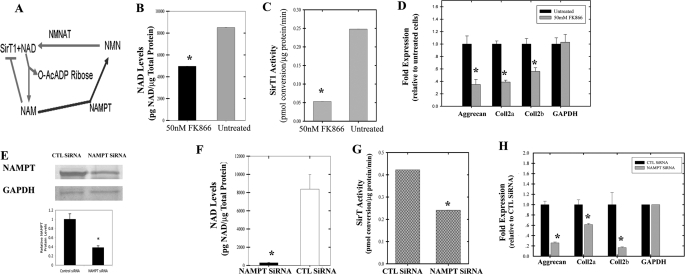
Inhibition of NAMPT Reduces Cartilage Gene Expression—An important aspect of SirT1 function is the requirement of NAD as a cofactor enzymatic activity. NAD is typically thought to arise from oxidative phosphorylation, which occurs at low levels in human chondrocytes (13). A separate mechanism to generate NAD is via the NAD salvage pathway (14, 15). Fig. 5A outlines NAD generation from NAM by the salvage pathway enzymes NAMPT and nicotinamide mononucleotide adenyltransferase. Because NAMPT is thought to be rate-limiting in the reaction (16), we tested whether NAMPT plays a role in the regulation of cartilage gene expression. The NAMPT-specific inhibitor FK866 was, therefore, used on low passage chondrocytes. FK866 blocks the activity of NAMPT leading to a reduction in NAD levels and an elevation in NAM in vivo (23). When NAD was assessed in FK866-treated chondrocytes, it was apparent that the inhibitor lowered NAD levels by 43% (LSD, p < 0.05; Fig. 5B), and SirT activity levels were reduced by 5-fold (Fig. 5C). When cartilage-specific gene expression was examined, as shown in Fig. 5D, the levels of aggrecan, collagen 2a(α1), and collagen 2b(α1) dropped significantly (LSD, p < 0.05) in the FK866-treated cells.
FIGURE 5.
Inhibition of NAMPT leads to a reduction in NAD levels, SirT activity, and cartilage-specific gene expression. A, generation of NAD from NAM by the salvage pathway enzymes (nicotinamide mononucleotide adenyltransferase (NMNAT) and NAMPT). OA chondrocytes (P0) were treated with NAMPT inhibitor FK866 (50 nm) for 2 days. Cell extracts were generated and used in an NAD/NADH assay (B) or a SirT activity assay (C). NAD levels and SirT activity was carried out in triplicate from 2 different cell lines (n = 6). D, total RNA was isolated from the cells treated as in (B and C) and was used in RT-PCR reactions with the indicated primers. The graph shows relative levels of gene expression. E, OA chondrocytes (P0 and P1) were transiently transfected with a NAMPT siRNA. Extracts were isolated at 2 days and used in immunoblots for NAMPT or GAPDH (upper panel). The lower panel shows NAMPT protein levels from the immunoblots. Extracts generated from the cells in E were used to determine NAD levels (F) and SirT activity (G). H, total RNA was isolated from the cells in E, which was used in qPCR reactions with the human primers. qPCR was performed twice using 2 different cell lines (n = 4). The graph shows relative levels of gene expression. Statistical significance is indicated by an asterisk (*) (LSD, p < 0.05). The displayed immunoblots are representatives of four experiments. CTL, control.
To further support these findings, a siRNA approach was utilized to lower NAMPT levels. As evident in Fig. 5E, transfection of the cells with an NAMPT siRNA led to a reduction in protein levels by about 3-fold (LSD, p < 0.05), whereas the GAPDH protein control was unaffected. SirT activity and NAD levels in NAMPT siRNA-treated cells significantly dropped as compared with the control siRNA-treated cells (LSD, p < 0.05; Fig. 5, F and G, respectively). Finally, when cartilage-specific gene expression was examined, as shown in Fig. 5H, the levels of aggrecan, collagen 2a(α1), and collagen 2b(α1) dropped significantly in NAMPT siRNA-treated cells (LSD, p < 0.05). As additional controls, we assessed expression of the non-lineage specific gene collagen 1(α1), fibronectin, and NFκB p65 in the FK866 and NAMPT siRNA-treated cells and found no changes in RNA levels (supplemental Figs. G and H, respectively). In total these data indicate that NAMPT plays a positive role in regulating NAD levels, SirT activity, and cartilage-specific gene expression in human chondrocytes.
DISCUSSION
SirT1 was originally defined as a silencer of gene expression (11) and has been demonstrated to negatively affect transcription via deacetylation of both histones (18) and transcription factors, e.g. MyoD, p53, and NFκB (19, 20, 23, 25). It was, therefore, initially expected that SirT1 might act in a negative capacity in OA human chondrocytes to repress cartilage-specific gene expression. To test this hypothesis, we analyzed the effect of SirT1 expression on chondrocytes isolated from adult human OA articular cartilage. In both transient and stable expression assays we find that elevation of wild-type SirT1 levels enhances cartilage-specific gene expression. Collagen 2(α1), collagen 9(α1), and aggrecan mRNAs were significantly elevated by SirT1, whereas other non-lineage-specific genes remained unchanged. Furthermore, when SirT1 activity was enhanced by the addition of the chemical activator resveratrol, cartilage gene expression was also increased. In contrast, when SirT1 activity was blocked by the chemical inhibitor NAM or when SirT1 levels were decreased by a SirT1 siRNA, cartilage gene expression was decreased. Taken together, these results indicate that SirT1 is a positive regulator of cartilage-specific gene expression in chondrocytes. It should be noted that in these studies we saw no evidence of a toxic effect of SirT1 expression on chondrocytes. Cells expressing SirT1 grew normally and showed no sign of elevated apoptosis.
In contrast to the effects of wild-type SirT1 on cartilage gene expression, the inactive mutant SirT1-M repressed expression of these same cartilage genes. Thus, this mutant appears to act in a dominant/negative fashion with respect to gene expression, similar to previous reports regarding cell survival and caloric restriction (19, 20). It is possible that SirT1-M interferes with the activity of the endogenous enzyme. If the mammalian SirT1 forms homotrimeric complexes similar to the yeast enzyme (26, 27), the SirT1-M could complex with the endogenous wild-type SirT1, resulting in trimers possessing reduced activity, thereby negatively effecting cartilage gene expression.
Because SirT1 has no inherent DNA binding ability, it was thought that its effect is mediated through Sox9 transcription factor, which regulates expression of many cartilage-specific genes (1). Indeed, coimmunoprecipitation experiments demonstrated that the two proteins associate in vivo and that only active SirT1 can transactivate the collagen 2(α1) promoter/enhancer and only in conjunction with Sox9. Consistent with this observation, SirT1 appears to deacetylate Sox9. Therefore, Sox9 appears similar to a growing number of other transcription factors, such as p53, NFkB, MyoD, and FOXO, in that it can be modified by acetylation/deacetylation. By ChIP analyses it was clear that SirT1 is targeted to both the Sox9 enhancer site and to the promoter region of the collagen 2(α1) gene. Its positioning at the enhancer is likely due to its association with Sox9.
SirT1 targeting the promoter may depend on an association with a promoter-specific transcription factor such as Sp1, which is known to bind to collagen 2(α1) promoter sites (29, 30). It is also possible that a looping mechanism, as outlined in Fig. 6, places enhancer-bound SirT1 next to the promoter (31). Importantly, expression of wild-type SirT1 in chondrocytes leads to the recruitment of the HATs (i.e. GCN5 and p300) to the collagen 2(α1) promoter. GCN5 and p300 are two histone acetyltransferases that are known activators of transcription (2). Also, p300 has been demonstrated to interact with Sox9 and enhance collagen 2(α1) transcription (32). Thus, the association of GCN5 and p300 on the collagen 2(α1) promoter is consistent with enhanced acetylation of H3K9/K14 and H4K5 at this site in the SirT1-expressing cells. In addition to histone acetylation on the collagen 2(α1) promoter, we also observe enhanced levels of 3MeH3K4, which is an essential histone modification for transcription initiation and indicates that histone methyltransferases are also targeted to the promoter. Importantly, these chromatin marks (AcH3K9/K14, AcH4K5, 3MeH3K4) are present near the transcription initiation sites of most actively transcribed genes (2, 33–35) and are in line with the elevated transcription of the collagen 2(α1) gene by SirT1.
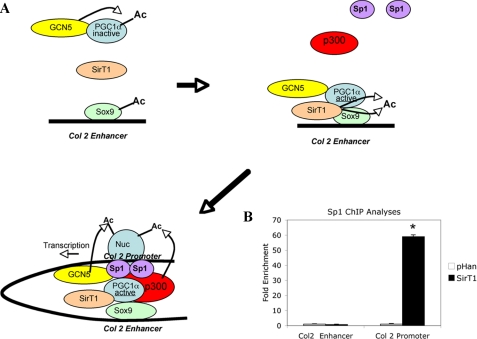
FIGURE 6.
Suggested model for the transcriptional activation of collagen 2(α1) by SirT1. A, PC1α and p300 are required for Col2 transcription and are known to interact with Sox9 (36, 44). GCN5 is known to acetylate and inactivate PGC1α (45), whereas SirT1 has been shown to bind, deacetylate, and thereby activate PGC1α (38, 46). The association of SirT1 with PGC1α and Sox9 and the subsequent deacetylation of both proteins may aid in the activation of PGC1α and the assembly or recruitment of p300 and Sp1 to the promoter via a looping mechanism. It remains to be determined whether deacetylation of PGC1α is the critical step in recruitment of proteins to the promoter as previously suggested by Rodgers et al. (47). Both p300 and GCN5 have the capacity to acetylate the critical histones on the promoter (Nuc, nucleosome). B, chondrocytes stably expressing SirT1 and the pHan control were processed for ChIP analyses using antibodies for the transcription factor Sp1. PCR was then performed with primers flanking either the promoter or enhancer within the collagen 2(α1) gene. The DNA products were amplified by quantitative PCR. An IgG control showed no PCR product and was subtracted from the equivalent antibody ChIP value. The error bars in the graphs indicate the S.D., and the statistical significance is indicated by an asterisk (*; LSD, p < 0.05).
In addition to GCN5 and p300, PGC1α was also detected in both the collagen 2(α1) promoter and enhancer regions. PGC1α is a transcriptional coactivator known to associate with both Sox9 (36) and SirT1 (37, 38) and has been shown to enhance collagen 2(α1) transcription (36). Interestingly, PGC1α has been shown to be affected by acetylation (47). GCN5 can associate with PGC1α and acetylate it, thereby inactivating it (45). SirT1 can reverse this modification and by deacetylation lead to PGC1α activation (38, 46). Thus, SirT1 and GCN5 act in opposing ways (47) to regulate PGC1α activity, as outlined in Fig. 6A. SirT1-mediated deacetylation of Sox9 and PGC1α may, therefore, be one of the first critical steps in transcription of collagen 2(α1).
Our ChIP findings demonstrate that the inactive mutant SirT1H355Y was not able to recruit these activators/coactivators to the promoter, indicating that enzymatic activity of SirT1 is essential for this assembly function. We suggest that the association of SirT1 with PGC1α and Sox9 on the enhancer in collagen 2(α1) functions via a looping mechanism (Fig. 6A) to recruit the activator p300 and the transcription factor Sp1 to the promoter. Both p300 and Sp1 are needed for optimal collagen 2(α1) transcription (30, 44). That Sp1 is targeted to the collagen 2(α1) promoter is revealed by the ChIP assay. We find that Sp1 shows significantly enhanced binding to the promoter in the presence of SirT1 (Fig. 6B). Thus, SirT1 is able to recruit activators, coactivators, and a transcription factor to the collagen 2(α1) promoter. p300 and GCN5 would then be likely candidates to acetylate the promoter-associated histones H3 and H4, necessary for transcription initiation. One likely prediction from these studies is that p300 and/or Sp1 may have a higher affinity for deacetylated Sox9 and PGC1α, which remains to be determined.
Because NAD is required for activity of SirT1, we also investigated the effect of the rate-limiting salvage pathway enzyme NAMPT (17) on cartilage-specific gene expression. Blocking NAMPT by either the specific inhibitor FK866 or NAMPT siRNA resulted in a reduced NAD levels, SirT activity, and chondrocyte-specific gene expression. This would suggest that NAMPT is critically involved in the maintenance of NAD pools and that it indirectly affects cartilage-specific gene expression through SirT1 activity. Furthermore, in our preliminary studies comparing knee articular cartilage from normal and OA patients, we observed an elevation in SirT1 levels and a decrease NAMPT levels in normal versus OA samples (supplemental Fig. O). This may suggest that certain aspects of OA development are due to an imbalance in SirT1 and NAMPT.
In conclusion, the data presented here show for the first time that SirT1 plays a positive role in the regulation of cartilage-specific gene expression in human articular chondrocytes and that at least for collagen 2(α1) promoter SirT1 activates transcription by recruitment of a number of activator/coactivator proteins. Furthermore, our data show a strong supporting role for the NAD salvage pathway enzyme in SirT1-mediated transcriptional activation.
Supplementary Material
Acknowledgments
We thank National Disease Research Interchange for providing the human cartilage tissue samples, Dr. Vittorio Sartorelli for the kind gift of the SirT1 and SirT1Mut retroviral plasmids, and Dr. M. Goldring for the gift of the PGL2Col2A.cg luciferase plasmid. We also thank Dr. Vittorio Sartorelli for insight and Dr. Assia Derfoul for assistance with real-time PCR. We acknowledge Dr. Lucilia Pereira-Mouries for editorial input on the manuscript.
This work was supported, in whole or in part, by the National Institutes of Health (Intramural Research Program of the NIAMS). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. A–O.
Footnotes
The abbreviations used are: HAT, histone acetyltransferase; HDAC, histone deacetylase; NAM, nicotinamide; qPCR, quantitative PCR; NAMPT, nicotinamide phosphoribosyltransferase; siRNA, small interfering RNA; ChIP, chromatin immunoprecipitation; RT, reverse transcription; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; LSD, least significant difference; OA, osteoarthritic; IP, immunoprecipitate.
References
- 1.Lefebvre, V., and Smits, P. (2005) Birth Defects Res. C Embryo Today 75 200-212 [DOI] [PubMed] [Google Scholar]
- 2.Roth, S. Y., Denu, J. M., and Allis, C. D. (2001) Annu. Rev. Biochem. 70 81-120 [DOI] [PubMed] [Google Scholar]
- 3.Hasan, S., and Hottiger, M. O. (2002) J. Mol. Med. 80 463-474 [DOI] [PubMed] [Google Scholar]
- 4.Ruijter, A., Gennip, A., Caron, H. N., Kemp, S., and Kuilenburg, A. (2003) Biochem. J. 370 737-749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnstone, R. W., and Licht, J. D. (2003) Cancer Cell 4 13-18 [DOI] [PubMed] [Google Scholar]
- 6.McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2002) Curr. Opin. Cell Biol. 14 763-772 [DOI] [PubMed] [Google Scholar]
- 7.Young, D. A., Lakey, R. L., Pennington, C. J., Jones, D., Kevorkian, L., Edwards, D. R., Cawston, T. E., and Clark, I. M. (2005) Arthritis Res. Ther. 7 503-512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, Y. L., Lee, M. Y., Wang, A. J., and Yao, L. F. (2003) Mol. Ther. 8 707-717 [DOI] [PubMed] [Google Scholar]
- 9.Nishida, K., Komiyama, T., Miyazawa, S., Shen, Z. N., Furumatsu, T., Doi, H., Yoshida, A., Yamana, J., Yamamura, M., Ninomiya, Y., Inoue, H., and Asahara, H. (2004) Arthritis Rheum. 50 3365-3376 [DOI] [PubMed] [Google Scholar]
- 10.Vega, R. B., Matsuda, K., Oh, J., Barbosa, A. C., Yang, X., Meadows, E., McAnally, J., Pomajzi, C., Shelton, J. M., Richardson, J. A., Karsenty, G., and Olson, E. N. (2004) Cell 119 555-566 [DOI] [PubMed] [Google Scholar]
- 11.Blander, G., and Guarente, L. (2004) Annu. Rev. Biochem. 73 417-435 [DOI] [PubMed] [Google Scholar]
- 12.Sauve, A. A., Wolberger, C., Schramm, V. L., and Boeke, J. D. (2006) Annu. Rev. Biochem. 75 435-465 [DOI] [PubMed] [Google Scholar]
- 13.Terkeltaub, R., Johnson, K., Murphy, A., and Ghosh, S. (2002) Mitochondrion 1 301-319 [DOI] [PubMed] [Google Scholar]
- 14.Rongvaux, A., Andris, F., Van Gool, F., and Leo, O. (2003) BioEssays 25 683-690 [DOI] [PubMed] [Google Scholar]
- 15.Yang, H., Lavu, S., and Sinclair, D. A. (2006) Exp. Gerontol. 41 718-726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Revollo, J. R., Körner, A., Mills, K. F., Satoh, A., Wang, T., Garten, A., Dasgupta, B., Sasaki, Y., Wolberger, C., Townsend, R. R., Milbrandt, J., Kiess, W., and Imai, S. (2007) Cell Metab. 6 341-34317983577 [Google Scholar]
- 17.van der Veer, E., Ho, C., O'Neil, C., Barbosa, N., Scott, R., Cregan, S. P., and Pickering, J. G. (2007) J. Biol. Chem. 282 10841-10845 [DOI] [PubMed] [Google Scholar]
- 18.Imai, S. I., Armstrong, C. M., Kaeberlein, M., and Guarente, L. (2000) Nature 403 795-800 [DOI] [PubMed] [Google Scholar]
- 19.Luo, J., Nikolaev, A. Y., Imai, S. I., Chen, D., Su, F., Shiloh, A., Guarente, L., and Gu, W. (2001) Cell 107 137-148 [DOI] [PubMed] [Google Scholar]
- 20.Vaziri, H., Dessain, S. K., Eaton, E. N., Imai, S. I., Frye, R. A., Pandita, T. K., Guarente, L., and Weinberg, R. A. (2001) Cell 107 149-159 [DOI] [PubMed] [Google Scholar]
- 21.Tan, L., Peng, H., Osaki, M., Choy, B. K., Auron, P. E., Sandell, L. J., and Goldring, M. B. (2003) J. Biol. Chem. 278 17688-17700 [DOI] [PubMed] [Google Scholar]
- 22.Bell, D. M., Leung, K. K., Wheatley, S. C., Ng, L. J., Zhou, S., Ling, K. W., Sham, M. H., Koopman, P., Tam, P. P., and Cheah, K. S. (1997) Nat. Genet. 16 174-178 [DOI] [PubMed] [Google Scholar]
- 23.Hasmann, M., and Schemainda, I. (2003) Cancer Res. 63 7436-7442 [PubMed] [Google Scholar]
- 24.Fulco, M., Schiltz, R. L., Iezzi, S., King, M. T., Zhao, P., Kashiwaya, Y., Hoffman, E., Veech, R. L., and Sartorelli, V. (2003) Mol. Cell 12 51-62 [DOI] [PubMed] [Google Scholar]
- 25.Yeung, F., Hoberg, J. E., Ramsey, C. S., Keller, M. D., Jones, D. R., Frye, R. A., and Mayo, M. W. (2004) EMBO J. 23 2369-2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao, K., Chai, X., Clements, A., and Marmorstein, R. (2003) Nat. Struct. Biol. 10 864-871 [DOI] [PubMed] [Google Scholar]
- 27.Liou, G. G., Tanny, J. C., Kruger, R. G., Walz, T., and Moazed, D. (2005) Cell 121 515-527 [DOI] [PubMed] [Google Scholar]
- 28.Lefebvre, V., Li, P., and de Crombrugghe, B. (1998) EMBO J. 17 5718-5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dharmavaram, R. M., Liu, G., Mowers, S. D., and Jimenez, S. A. (1997) J. Biol. Chem. 272 26918-26925 [DOI] [PubMed] [Google Scholar]
- 30.Ghayor, C., Charjichristos, C., Herrouin, F. J., Ala-Kokko, L., Suske, G., Pujol, J. P., and Galera, P. (2001) J. Biol. Chem. 276 36881-36895 [DOI] [PubMed] [Google Scholar]
- 31.Vakoc, C. R., Letting, D. L., Gheldof, N., Sawado, T., Bender, M. A., Groudine, M., Weiss, M. J., Dekker, J., and Blobel, G. A. (2005) Mol. Cell 17 453-462 [DOI] [PubMed] [Google Scholar]
- 32.Tsuda, M., Takahashi, S., Takahashi, Y., and Asahara, H. (2003) J. Biol. Chem. 278 27224-27229 [DOI] [PubMed] [Google Scholar]
- 33.Trojer, P., and Reinberg, D. (2006) Cell 125 213-217 [DOI] [PubMed] [Google Scholar]
- 34.Mikkelsen, T. S., Ku, M., Jaffe, D. B., Issac, B., Lieberman, E., Giannoukos, G., Alvarez, P., Brockman, W., Kim, T. K., Koche, R. P., Lee, W., Mendenhall, E., O'Donovan, A., Presser, A., Russ, C., Xie, X., Meissner, A., Wernig, M., Jaenisch, R., Nusbaum, C., Lander, E. S., and Bernstein, B. E. (2007) Nature 448 553-560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guenther, M. G., Levine, S. S., Boyer, L. A., Jaenisch, R., and Young, R. A. (2007) Cell 130 77-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawakami, Y., Tsuda, M., Takahashi, S., Taniguchi, N., Esteban, C. R., Zemmyo, M., Furumatsu, T., Lotz, M., Belmonte, J. C. I., and Asahara, H. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2414-2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nemoto, S., Fergusson, M. M., and Finkel, T. (2005) J. Biol. Chem. 280 16456-16460 [DOI] [PubMed] [Google Scholar]
- 38.Rodgers, J. T., Lerin, C., Haas, W., Gygi, S. P., Spiegelman, B. M., and Puigserver, P. (2005) Nature 434 113-117 [DOI] [PubMed] [Google Scholar]
- 39.Derfoul, A., Miyoshi, A. D., Freeman, D. E., and Tuan, R. S. (2007) Osteoarthritis Cartilage 15 646-655 [DOI] [PubMed] [Google Scholar]
- 40.Kipnes, J., Carlberg, A. L., Loredo, G. A, Lawler, J., Tuan, R. S., and Hall, D. J. (2003) Osteoarthritis Cartilage 11 442-454 [DOI] [PubMed] [Google Scholar]
- 41.Borra, M. T., Smith, B. C., and Denu, J. M. (2005) J. Biol. Chem. 280 17187-17195 [DOI] [PubMed] [Google Scholar]
- 42.Perkins, G. L., Derfoul, A., Ast, A., and Hall, D. J. (2005) Differentiation 73 199-211 [DOI] [PubMed] [Google Scholar]
- 43.Firestein, R., Blander, G., Michan, S., Oberdoerffer, P., Ogino, J. S., Campbell, J., Bhimavarapu, A., Luikenhuis, S., Cabo, R., Fuchs, C., Hahn, W. C., Guarente, L. P., and Sinclair, D. (2008) PLoS One 4 e2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furumatsu, T., Tsuda, M., Taniguchi, N., Tajima, Y., and Asahara, H. (2005) J. Biol. Chem. 280 8343-8350 [DOI] [PubMed] [Google Scholar]
- 45.Lerin, C., Rodgers, J. T., Kalume, D. E., Kim, S. H., Pandey, A., and Puigserver, P. (2006) Cell Metab. 3 429-438 [DOI] [PubMed] [Google Scholar]
- 46.Gerhart-Hines, Z., Rodgers, J. T., Bare, O., Lerin, C., Kim, S. H., Mostoslavsky, R., Alt, F. W., Wu, Z., and Puigserver, P. (2007) EMBO J. 26 1913-1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodgers, J. T., Lerin, C., Gerhart-Hines, Z., and Puigserver, P. (2008) FEBS Lett. 582 46-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.