Abstract
IMP dehydrogenase (IMPDH) catalyzes the pivotal step in guanine nucleotide biosynthesis. Here we show that both IMPDH type 1 (IMPDH1) and IMPDH type 2 are associated with polyribosomes, suggesting that these housekeeping proteins have an unanticipated role in translation regulation. This interaction is mediated by the subdomain, a region of disputed function that is the site of mutations that cause retinal degeneration. The retinal isoforms of IMPDH1 also associate with polyribosomes. The most common disease-causing mutation, D226N, disrupts the polyribosome association of at least one retinal IMPDH1 isoform. Finally, we find that IMPDH1 is associated with polyribosomes containing rhodopsin mRNA. Because any perturbation of rhodopsin expression can trigger apoptosis in photoreceptor cells, these observations suggest a likely pathological mechanism for IMPDH1-mediated hereditary blindness. We propose that IMPDH coordinates the translation of a set of mRNAs, perhaps by modulating localization or degradation.
IMP dehydrogenase (IMPDH)2 catalyzes the reaction that controls the entry of purines into the guanine nucleotide pool, and thus controls proliferation (1). The enzyme is a homotetramer; each monomer is composed of a catalytic (β/α)8 barrel and a subdomain containing two CBS domains (named for the related domain in cystathionine β-synthase) (Fig. 1). Deletion of the subdomain has no effect on enzymatic activity (2, 3), and the function of the subdomain in IMPDH is currently under debate. CBS domains act as adenosine nucleotide-binding modules in several proteins (4–9), and a similar role has been proposed for the CBS domains of IMPDH (5), but we and others have been unable to confirm this function in IMPDH (10–13). Notably, the CBS domains of IMPDH share little sequence identity with the other proteins, so it would not be surprising if their function has diverged. The subdomain does appear to coordinately regulate the adenine and guanine nucleotide pool in Escherichia coli, although the molecular mechanism of this process has not yet been elucidated (11). We have discovered that IMPDH binds single-stranded nucleic acids and that the subdomain mediates this interaction (10, 15). IMPDH associates with RNA in tissue culture cells, which suggests that this housekeeping enzyme is involved in translation, splicing, or some other feature of RNA metabolism (10, 15). Others report that IMPDH binds DNA and may be involved in gene expression (16, 17). These observations suggest that IMPDH has a “moonlighting” function involving nucleic acid that is mediated by the subdomain.
FIGURE 1.
The adRP/LCA-causing mutations of IMPDH1. A, positions of the disease-associated mutations are depicted on a monomer of IMPDH from Streptococcus pyogenes, which corresponds to the canonical IMPDH1(514) (Protein Data Bank accession number 1ZFJ (30); note that the CBS domains are disordered in the structure of human IMPDH1 (Protein Data Bank accession number 1JCN), so that several of the positions of mutation are not observed). Magenta denotes mutations that are clearly pathogenic; red, likely pathogenic; green, possibly pathogenic (27). Molecular graphics images were produced using the University of California, San Francisco, Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH Grant P41 RR-01081) (14). B, scheme showing the differences between IMPDH1(514), IMPDH1(546), and IMPDH1(595).
Mammals have two IMPDH genes, encoding IMPDH1 and IMPDH2, and most tissues express both isozymes (18, 19). In contrast, only IMPDH1 appears to be expressed in the retina; in addition, retina contains distinct IMPDH1 isoforms generated by alternative mRNA splicing as follows: IMPDH1(546) (major) and IMPDH1(595) (minor) (Fig. 1; these proteins are also known as IMPDH1α/IMPDH1(13b) and IMPDHγ/IMPDH1(A+13b), respectively; the canonical enzyme is hereafter designated IMPDH1(514) (20, 21)). Both retinal isoforms IMPDH1(546) and IMPDH1(595) contain a 32-residue C-terminal extension. IMPDH1(595) has an additional 49-residue extension on the N terminus (20, 21). These extensions have no effect on the activity of purified enzyme (22). Surprisingly, although the subdomain region is identical in all of the IMPDH1 isoforms, nucleic acid does not readily associate with the retinal isoforms (22). This observation suggests that the C-terminal extension blocks the nucleic acid-binding site, perhaps by interacting with the subdomain (22). Retinal cells may contain additional protein factors that interact with the C-terminal extension to regulate nucleic acid binding.
Most intriguingly, mutations in the subdomain and the neighboring region of the barrel domain of IMPDH1 cause autosomal dominant retinitis pigmentosa (adRP) and are also associated with a more severe hereditary blindness, Leber congenital amaurosis (LCA) (23–25) (for the sake of simplicity, we will refer to the mutations of IMPDH1 that cause retinal disease as “RP-linked”; Fig. 1; mutations are designated according to the sequence of IMPDH1(514) (23, 25–28)). The D226N mutation alone accounts for ∼1% of all adRP cases (27).
The presence of the retinal isoforms may account for the tissue specificity of disease. Nonetheless, the underlying pathological mechanism of IMPDH1-mediated retinal disease remains perplexing. Guanine nucleotides are critical components of photoreceptor signaling, so it is tempting to attribute disease to loss of enzymatic activity and the consequential decrease in guanine nucleotides. However, several observations suggest that this mechanism does not apply. First, the RP-linked mutations have no effect on the enzymatic activity of the canonical IMPDH1(514) or the retinal isoforms in vitro (10, 22, 27, 29), nor do they alter the localization of these proteins in cells grown in tissue culture (10, 22, 29). Furthermore, mice with a heterozygous null allele in IMPDH1 have no phenotype, and homozygous null mice display only a mild retinopathy (29). Finally, IMPDH inhibitors are widely used in immunosuppressive chemotherapy (e.g. CellCept), yet side effects involving retinal degeneration have not been reported. Others have proposed that these mutations increase the propensity of IMPDH1 to form aggregates (29), but we have not found this to be true in our constructs (10). Instead, we find that RP-causing mutations decrease the affinity and specificity of nucleic acid binding in the canonical IMPDH1(514), suggesting that perturbation of this function underlies retinal disease (10, 27).
Here we show that IMPDH1 and the retinal isoforms associate with polyribosomes and that this interaction is mediated by the subdomain. The most commonly occurring adRP-causing mutation, D226N, blocks the polyribosome association of IMPDH1(595). Furthermore, we find that IMPDH is associated with polyribosomes translating rhodopsin mRNA in bovine retina. Because essentially any perturbation of rhodopsin expression triggers apoptosis in photoreceptor cells (31), the misregulation of polyribosomes translating rhodopsin provides an attractive mechanism for retinal disease. These observations indicate that IMPDH has a previously unsuspected role in the regulation of translation.
EXPERIMENTAL PROCEDURES
Materials—Superfect was obtained from Qiagen (Valencia, CA). Anti-GFP affinity-purified antibody was obtained from Invitrogen. Anti-CBP80 antibody was a gift of the Dr. Melissa J. Moore (Brandeis University), and anti-rhodopsin antibody 1D4 was the generous gift of Dr. Daniel Oprian (Brandeis University). Puromycin was purchased from Sigma. Anti-IMPDH BRD5 antibodies were obtained as described previously (20). The full-length rhodopsin cDNA was synthesized by Epoch Biolabs (Sugar Land, TX) following the sequence of Entrez data base NM_000539 and cloned into a plasmid under control of a cytomegalovirus promoter.
Polyribosome Analysis of IMPDH—Polyribosome profiles were performed with minor modifications of standard protocols (32, 33). C-terminal EGFP-tagged IMPDH1 was expressed in mammalian cells under control of the mammalian cytomegalovirus promoter as described previously (10). Cells (1 × 107) were treated with 100 μg/ml cycloheximide and lysed on ice with polyribosome extraction buffer (PEB, 0.5% Triton X-100, 15 mm Tris-HCl, pH 8, 15 mm MgCl2, 60 mm NaCl, 100 μg/ml cycloheximide, and 1 mg/ml heparin). The clarified lysate was fractionated on a 10–50% sucrose gradient, precipitated with trichloroacetic acid, and resolved by SDS-PAGE. Immunoblots were probed with anti-GFP antibody or anti-IMPDH serum and alkaline phosphatase-conjugated anti-rabbit secondary antibody. When indicated, 100 μg/ml puromycin was added to cultures 2 h before harvesting. For RNase disruption of polyribosomes, lysates were treated with 60 units/ml micrococcal nuclease and 1 mm CaCl2 for 25 min on ice prior to loading on sucrose gradients. Data are representative of at least two individual experiments.
Polyribosome Association of Retinal IMPDH1 Isoforms—Plasmids expressing IMPDH1(546)-GFP and IMPDH1(595)-GFP were expressed in HEK293 cells as described previously (22). Polyribosomes were harvested by centrifugation through a 30% sucrose cushion. The pellets were dissolved by PEB and analyzed by Western blot analysis with the primary mouse monoclonal anti-IMPDH antibody AS37-P (Antibody Solutions, Mountain View, CA). Immunodetection was performed using goat anti-mouse IgG secondary antibody conjugated with horseradish peroxidase (Millipore) and visualized with the ECL Plus system (Amersham Biosciences).
Identification of IMPDH-associated mRNAs—Five bovine retinas were lysed in PEB and 1 mm dithiothreitol. The mixture was incubated on ice for 10 min, and then cell debris was removed by centrifugation. The supernatant was applied to Sepharose CL-B column with bed volume of 110 ml. Fractions in the void volume were collected and loaded onto IMP-agarose. IMPDH was eluted with 2 mm IMP in 50 mm Tris-HCl, pH 7.5, 0.5 mm dithiothreitol, 10% glycerol. RNA was isolated using TRIzol LS reagent (Invitrogen), and cDNA was prepared using Super SMART PCR cDNA synthesis kit (Clontech) and cloned into Topo cloning kit (Invitrogen).
IMPDH Is Associated with Polyribosomes Containing Rhodopsin mRNA—Approximately 10 g of bovine retina was lysed as described above within 3 h of harvest. Polyribosomes were isolated by centrifugation through a 30% sucrose cushion in PEB. The polyribosomes were resuspended in 1 ml of PEB. Polyribosome mixture (250 ml) was treated with monoclonal antibody RET-P1 (Abcam), which recognizes the rhodopsin N-terminal peptide, or 1D4, which recognizes the opsin C-terminal peptide. Antibody complexes were isolated with protein G Dynabeads and polyribosomes, and the bound proteins were recovered by treatment with 10% SDS. Alternatively, polyribosomes were disrupted by treatment with 2 mm EDTA, 50 mm Tris-HCl, pH 7.5, and 100 mm KCl. The eluted protein was analyzed by 10% SDS-PAGE and immunoblotting visualized with anti-IMPDH antibody or anti-ribosomal protein L7a antibody (Cell Signaling Technologies, Danvers, MA).
Co-immunoprecipitation of IMPDH1(546) and Rhodopsin mRNA—HEK293T cells were co-transfected with plasmids directing expression of rhodopsin and IMPDH(546)-GFP (Transfectin, Bio-Rad). Cells were collected 48 h after transfection, washed with ice-cold phosphate-buffered saline, and lysed in PEB. Immunoprecipitation was performed as in Ref. 34. Briefly, magnetic beads (New England Biolabs) coated with anti-GFP antibody were mixed with clarified cell lysate in NT2 buffer (50 mm Tris, 150 mm NaCl, 1 mm MgCl2, 0.05% Nonidet P-40), supplemented with 200 units of RNase Inhibitor (New England Biolabs), 1 mm dithiothreitol, and 15 mm EDTA. Following precipitation, RNA was recovered with TRIzol LS and DNase-treated (RQ1, Promega) according to manufacturer protocols. Reverse transcription-PCR was performed using SuperScriptII One-step kit (Invitrogen) with primers for rhodopsin (5′-ccccatcaacttcctcacgctctac-3′ and 5′-cgatgaccatgatgatgaccatgc-3′).
RESULTS
IMPDH Associates with Polyribosomes—The co-precipitation of RNA with IMPDH suggested that IMPDH might be involved in translation or translation regulation (10). To determine whether IMPDH associates with polyribosomes, lysates were prepared from cycloheximide-treated HeLa cells and fractionated by sucrose gradient sedimentation; polyribosome-containing fractions were identified by monitoring absorbance at 254 nm (Fig. 2). When purified recombinant IMPDH1(514) is subjected to sucrose gradient sedimentation, the protein migrates in the lower density fractions, as expected for the homotetramer. Also as expected, the 80-kDa cap-binding protein (CBP80) was observed in the cytosolic, ribosomal subunits and monosome-containing fractions, but was excluded from higher density fractions (Fig. 2) (35). Unlike CBP80, endogenous IMPDH is found throughout the gradient. These observations indicate that endogenous IMPDH is part of a large complex. Puromycin terminates nascent peptide chains and disrupts polyribosomes (Fig. 2) (36), and puromycin also disrupts the IMPDH complex (Fig. 2). Similarly, EDTA and RNase treatment disrupt both polyribosomes and the endogenous IMPDH complex (data not shown). Although the anti-IMPDH antisera does not distinguish between IMPDH isozymes, IMPDH2 predominates in HeLa cells (37, 38). Thus, IMPDH2, and possibly IMPDH1, associates with polyribosomes in HeLa cells.
FIGURE 2.
Polyribosome profiles of HeLa cell extracts. Lysates were prepared and fractionated on a sucrose gradient as described under “Experimental Procedures.” A typical profile is shown, with the corresponding Western blot with anti-IMPDH antisera immediately below. For the remaining panels, fractions were obtained similarly, and Western blots were probed with antibody against the designated protein. Puromycin disrupts both polyribosomes and the high molecular weight IMPDH complex. A single blot was split and probed with anti-CBP80 antibody and anti-IMPDH antisera. CBP80 is found in the cytosolic ribosomal subunits and monosome-containing fractions, but excluded from higher density fractions, whereas IMPDH is found throughout the gradient. The bottom panel shows the sedimentation of purified recombinant (r) IMPDH1(514).
Subdomain Mediates the Association of IMPDH1(514) with Polyribosomes—To demonstrate that IMPDH1(514) associates with polyribosomes, we expressed a C-terminally EGFP-tagged version in HeLa cells. We have previously shown that the EGFP tag has no effect on the enzymatic or nucleic acid binding properties of IMPDH1(514) (10), and we have also established conditions where IMPDH1-GFP is expressed at approximately the same levels as the endogenous IMPDH (10). IMPDH1-GFP is also found in the polyribosome-containing fractions of the sucrose gradient (Fig. 3A), whereas GFP migrates in the cytosolic fractions. As above, treatment with puromycin and RNase disrupts the IMPDH1-GFP complex, indicating the IMPDH1-GFP associates with polyribosomes (Fig. 3, B and C). Deletion of the subdomain substantially decreases the fraction of IMPDH1-GFP found in the polyribosomes (Fig. 3D), indicating that the CBS domains mediate the polyribosome interaction. This observation suggests that the RP-linked mutations of IMPDH1 may perturb the polyribosome interaction. The most common RP-linked mutation, D226N, does not change the polyribosome distribution of IMPDH1-GFP. However, this assay would be unable to detect a subtle change in polyribosome distribution.
FIGURE 3.
The association of IMPDH with polyribosomes is mediated by the subdomain. HeLa cells were transfected to express wild-type (WT) IMPDH1(514) tagged with EGFP at the C terminus (IMPDH1-GFP, WT) or IMPDH lacking subdomain (ΔSD-IMPDH1-GFP, ΔSD). Lysates were prepared, and fractions were aliquoted as described in Fig. 2 and probed with antibodies against GFP. A, IMPDH1-GFP; B, IMPDH1-GFP + puromycin; C, IMPDH1-GFP + RNase; D, GFP alone, ΔSD-IMPDH1-GFP, IMPDH1-GFP/D226N, and wild type. The IMPDH1-GFP/D226N and WT are from parallel transfections.
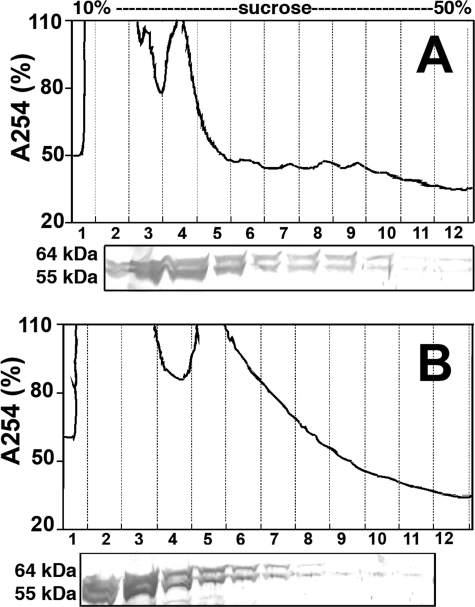
IMPDH Interacts with Polyribosomes in Retinal Cells—The C-terminal extensions of IMPDH1(546) and IMPDH1(595) appear to block nucleic acid binding in vitro (22), suggesting that the retinal isoforms might not associate with polyribosomes. To determine whether IMPDH1(546) and/or IMPDH1(595) interacts with polyribosomes in photoreceptor cells, we prepared lysates from freshly dissected bovine retina. These lysates contain 55- and 64-kDa proteins that cross-react with anti-IMPDH antibodies (Fig. 4); and as reported previously (21), the 64-kDa protein, but not the 55-kDa protein, also reacts with antibody against the N-terminal extension of IMPDH1(595) (data not shown), indicating that these proteins are the bovine orthologs of IMPDH1(546) and IMPDH1(595). Both bovine IMPDH1 isoforms are found in the polyribosome containing fractions, and RNase disrupts this association (Fig. 4). Thus, both IMPDH1(546) and IMPDH1(595) associate with polyribosomes despite the fact that these proteins are poor nucleic acid-binding proteins in vitro. These observations suggest that photoreceptors may contain additional proteins that interact with C-terminal extension to regulate polyribosome association.
FIGURE 4.
The retinal isoforms IMPDH1(546) and IMPDH1(595) associate with polyribosomes in bovine retinal cells. Lysates were prepared from freshly dissected bovine retina and fractionated as described above. Immunoblots were probed with anti-IMPDH serum BRD5. A, no pretreatment; B, RNase-treated retinal lysate.
RP-causing Mutation D226N Decreases the Association of IMPDH1(595) with Polyribosomes—To assess the effect of the D226N mutation on the polyribosome association of IMPDH1(546) and IMPDH1(595), the retinal isoforms were tagged with EGFP at the C terminus and expressed in HEK293 cells as described previously (22). Total polyribosomes were isolated by centrifugation through a sucrose cushion. As expected, endogenous IMPDH, IMPDH1(546)-GFP, and IMPDH1(595)-GFP are found in the pellet with the polyribosomes (Fig. 5). When polyribosomes are disrupted by treatment with puromycin, endogenous IMPDH and the retinal isoforms no longer sediment. Therefore, IMPDH1(546)-GFP and IMPDH1(595)-GFP associate with polyribosomes in tissue culture as observed in retina. The D226N variant of IMPDH1(546)-GFP also associates with polyribosomes (Fig. 5A), although the mutation may cause a subtle decrease in the polyribosome association of this retinal isoform (Fig. 5B; note that this effect is not statistically significant). In contrast, the D226N mutation blocks the association of IMPDH1(595)-GFP with polyribosomes (Fig. 5, A and B). Therefore, the D226N mutation decreases the polyribosome association of at least one retinal isoform.
FIGURE 5.
The D226N mutation decreases the association of IMPDH1(595) with polyribosomes. A, lysates were prepared from HEK cells expressing GFP-tagged retinal IMPDH1 isoforms, and total polyribosomes were collected by sedimentation through a sucrose cushion. Samples were analyzed by immunoblotting with monoclonal antibody recognizing IMPDH. B, composite data from three experiments as in A. The endogenous IMPDH band was used to normalize the amount of GFP-tagged IMPDH1 in the polyribosomes. The units are arbitrary.
IMPDH1 Associates with Polyribosomes Translating Rhodopsin mRNA in the Retina—To identify the mRNAs in IMPDH-associated polyribosomes, total polyribosomes were isolated from five bovine retinas. The IMPDH-containing polyribosomes were recovered by affinity chromatography on IMP-agarose and the RNA converted into cDNA and cloned. Seven clones were recovered, of which two contained known bovine mRNAs (rhodopsin and DEAH box polypeptide 29), four contained 12 S rRNA, and one contained an unidentified mRNA. Mutations in rhodopsin are the most common cause of hereditary visual disease (39), so the association of IMPDH1 with polyribosomes translating rhodopsin provides an obvious link to adRP and LCA. Therefore, we sought to validate this interaction with a nascent chain immunoprecipitation experiment.
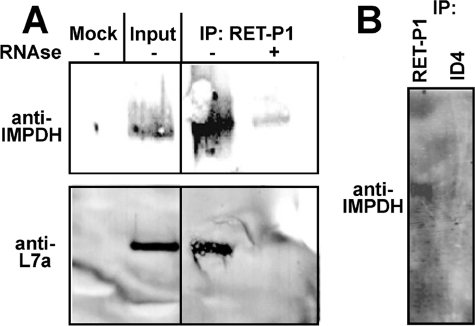
Polyribosomes translating rhodopsin mRNA were isolated from total polyribosomes using monoclonal antibody RET-P1, which recognizes the N-terminal peptide of rhodopsin. Ribosomal protein L7a co-precipitated with RET-P1, confirming that polyribosomes were obtained. IMPDH1 co-precipitated with these polyribosomes (Fig. 6). No IMPDH was recovered when the polyribosomes were disrupted by RNase prior to immunoprecipitation. Similarly, EDTA disrupts the interaction between IMPDH1 and polyribosomes (data not shown). No IMPDH1 co-precipitated when monoclonal antibody 1D4, which recognizes the C-terminal peptide of rhodopsin, was utilized (Fig. 6). These experiments demonstrate that IMPDH1 associates with polyribosomes translating rhodopsin mRNA.
FIGURE 6.
IMPDH1 is associated with polyribosomes translating rhodopsin mRNA. A, polyribosomes were isolated from bovine retinal lysate by sedimentation through a sucrose cushion as described under “Experimental Procedures.” Polyribosomes expressing rhodopsin were isolated by immunoprecipitation (IP) with monoclonal antibody RET-P1, which recognizes the N-terminal peptide of rhodopsin. The resulting precipitates were analyzed by SDS-PAGE and immunoblotting. The input sample contains both IMPDH1 and ribosomal protein L7a. Both IMPDH and L7a co-precipitate with RET-P1. Neither IMPDH1 nor L7a are observed in mock immunoprecipitations where RET-P1 is omitted or when the sample is pretreated with RNase to disrupt the polyribosomes. B, samples are prepared as in A. IMPDH1 precipitates with RET-P1 but not with monoclonal antibody 1D4, which recognizes the C-terminal segment of rhodopsin.
IMPDH1 Associates with Polyribosomes Translating Rhodopsin mRNA in Cell Culture—To further confirm this interaction, IMPDH1(546)-GFP was expressed in HEK293 cells together with rhodopsin mRNA. IMPDH1(546)-GFP was immunoprecipitated with anti-GFP antibody (Fig. 7A). Rhodopsin mRNA co-precipitated (Fig. 7B), confirming that IMPDH1(546)-GFP associates with polyribosomes containing rhodopsin mRNA.
FIGURE 7.
Co-immunoprecipitation of IMPDH1(546)-GFP and rhodopsin mRNA. HEK293 cells were co-transfected with constructs directing the expression of IMPDH1(546)-GFP and rhodopsin mRNA. IMPDH1(546)-GFP was immunoprecipitated with anti-GFP antibody. A, Western blot of the immunoprecipitate showing efficient pulldown of IMPDH1(546)-GFP. HEK cells transfected with constructs expressing only rhodopsin (-) and rhodopsin and IMPDH1(546)-GFP (+); ft, flow-through. B, reverse transcription (RT) followed by PCR demonstrates the presence of rhodopsin mRNA in the immunoprecipitate. Rho, rhodopsin; ft, flow-through.
DISCUSSION
The IMPDH reaction controls the guanine nucleotide pool, which in turn controls cell proliferation, and the success of IMPDH-targeted drugs validates the link between enzymatic activity and proliferation (40). Yet the presence of the subdomain provokes the tantalizing suggestion that IMPDH has cellular functions that extend beyond enzymatic activity. Several disparate observations reinforce this view; IMPDH associates with nucleic acids (10, 15, 17) and with lipid vesicles (41), and forms large intracellular aggregates when guanine nucleotides are depleted (42, 43). A recent report suggests that IMPDH controls the balance between adenine and guanine nucleotides via the subdomain (11). We now demonstrate that both IMPDH1 and IMPDH2 associate with polyribosomes, which suggests that these housekeeping enzymes also have an unanticipated role in the regulation of translation, either directly or by modulating mRNA localization or degradation.
Although the association of IMPDH with polyribosomes may be mediated by rRNA and/or other protein factors, because IMPDH binds single-stranded nucleic acids (10, 15), we favor a model whereby the canonical IMPDH interacts directly with mRNA. In addition, given that the retinal IMPDH1 isoforms are poor nucleic acid-binding proteins, we further hypothesize that additional factors interact with the C-terminal extension, freeing the canonical IMPDH1 to form the polyribosome interaction.
The expression of functionally related proteins may be coordinated by specific mRNA-binding proteins that direct the localization, translation, and/or degradation of mRNA (44, 45). Evidence for the existence of such “RNA regulons” is accumulating (46–51), and we suggest that IMPDH1 and IMPDH2 act in this manner. Importantly, polyribosome association is mediated by the subdomain, which is also the site of the greatest structural divergence between IMPDH1 and IMPDH2, so perhaps IMPDH1 and IMPDH2 recognize different mRNAs and organize distinct RNA regulons. Furthermore, the subdomain is found in IMPDHs from almost every organism, so similar post-transcriptional processes may be present in prokaryotes and archaea.
The subdomain is also the site of the RP-causing mutations of IMPDH1. Our experiments show that polyribosome association is blocked by the most common RP-causing mutation, D226N, in IMPDH1(595). The association of IMPDH1(546) may also be perturbed, although the errors in the data are too large to make this claim with confidence. Nonetheless, given that the symptoms of RP often develop over decades, even a subtle perturbation may be sufficient to cause disease. These observations proffer the idea that IMPDH1 regulates the expression of a protein(s) critical for photoreceptor function by modulating translation, localization, or degradation of mRNA. Perturbation of each of these processes is known to induce apoptosis (52, 53). Furthermore, our experiments demonstrate that retinal IMPDH1 is associated with polyribosomes translating rhodopsin mRNA. Mutations in rhodopsin are the most common cause of hereditary blindness; disease can result from the mislocalization and/or misfolding of rhodopsin (31, 54), a decrease in rhodopsin expression (55), or even a mere 20% increase in rhodopsin (56). Mislocalization, misfolding, and protein levels are all intimately linked to translation and regulated by RNA-binding proteins. Therefore, the association of IMPDH1 with polyribosomes translating rhodopsin mRNA provides a ready explanation for IMPDH1-mediated visual disease.
Acknowledgments
We thank our former Brandeis colleagues Dr. Melissa J. Moore (University of Massachusetts Medical School) for invaluable discussions and resources and Dr. Michael A. Welte (University of Rochester) for help with confocal fluorescence microscopy. We also thank Dr. Daniel Oprian (Brandeis University) for the gift of monoclonal antibody 1D4.
This work was supported, in whole or in part, by National Institutes of Health Grants GM54403 and EY17325 (to L. H.) and EY014170 (to S. P. D.), and GM07956 (Macromolecular Structure and Mechanism Training Grant to S. E. M.). This work was also supported by grant from the Foundation Fighting Blindness (to S. P. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IMPDH, inosine 5′-monophosphate dehydrogenase; CBS, cystathionine β-synthase; IMPDH1, human IMPDH type 1; IMPDH2, human IMPDH type 2; RP, retinitis pigmentosa; adRPA, autosomal dominant RPA; LCA, Leber congenital amaurosis; GFP, green fluorescent protein; EGFP, enhanced GFP.
References
- 1.Weber, G., Nakamura, H., Natsumeda, Y., Szekeres, T., and Nagai, M. (1992) Adv. Enzyme Regul. 32 57-69 [DOI] [PubMed] [Google Scholar]
- 2.Nimmesgern, E., Black, J., Futer, O., Fulghum, J. R., Chambers, S. P., Brummel, C. L., Raybuck, S. A., and Sintchak, M. D. (1999) Protein Expression Purif. 17 282-289 [DOI] [PubMed] [Google Scholar]
- 3.Gan, L., Petsko, G. A., and Hedstrom, L. (2002) Biochemistry 41 13309-13317 [DOI] [PubMed] [Google Scholar]
- 4.Janosik, M., Kery, V., Gaustadnes, M., Maclean, K. N., and Kraus, J. P. (2001) Biochemistry 40 10625-10633 [DOI] [PubMed] [Google Scholar]
- 5.Scott, J. W., Hawley, S. A., Green, K. A., Anis, M., Stewart, G., Scullion, G. A., Norman, D. G., and Hardie, D. G. (2004) J. Clin. Investig. 113 274-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer, S., Savaresi, S., Forster, I. C., and Dutzler, R. (2007) Nat. Struct. Mol. Biol. 14 60-67 [DOI] [PubMed] [Google Scholar]
- 7.Day, P., Sharff, A., Parra, L., Cleasby, A., Williams, M., Horer, S., Nar, H., Redemann, N., Tickle, I., and Yon, J. (2007) Acta Crystallogr. Sect. D Biol. Crystallogr. 63 587-596 [DOI] [PubMed] [Google Scholar]
- 8.Jamsen, J., Tuomunen, H., Salminen, A., Belogurov, G. A., Magretova, N. N., Baykov, A. A., and Lahti, R. (2007) Biochem. J. 408 327-333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennetts, B., Rychkov, G. Y., Ng, H. L., Morton, C. J., Stapleton, D., Parker, M. W., and Cromer, B. A. (2005) J. Biol. Chem. 280 32452-32458 [DOI] [PubMed] [Google Scholar]
- 10.Mortimer, S. E., and Hedstrom, L. (2005) Biochem. J. 390 41-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pimkin, M., and Markham, G. D. (2008) Mol. Microbiol. 69 342-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carr, S. F., Papp, E., Wu, J. C., and Natsumeda, Y. (1993) J. Biol. Chem. 268 27286-27290 [PubMed] [Google Scholar]
- 13.Holmes, E., Pehlke, D., and Kelley, W. (1974) Biochim. Biophys. Acta 364 209-217 [DOI] [PubMed] [Google Scholar]
- 14.Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., and Ferrin, T. E. (2004) J. Comp. Chem. 25 1605-1612 [DOI] [PubMed] [Google Scholar]
- 15.McLean, J. E., Hamaguchi, N., Belenky, P., Mortimer, S. E., Stanton, M., and Hedstrom, L. (2004) Biochem. J. 379 243-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuno, K., Miyamoto, T., Yamaguchi, K., Abu Sayed, M., Kajiwara, T., and Hatano, S. (1995) Biosci. Biotechnol. Biochem. 59 231-235 [DOI] [PubMed] [Google Scholar]
- 17.Cornuel, J. F., Moraillon, A., and Gueron, M. (2002) Biochimie (Paris) 84 279-289 [DOI] [PubMed] [Google Scholar]
- 18.Jain, J., Almquist, S. J., Ford, P. J., Shlyakhter, D., Wang, Y., Nimmesgern, E., and Germann, U. A. (2004) Biochem. Pharmacol. 67 767-776 [DOI] [PubMed] [Google Scholar]
- 19.Senda, M., and Natsumeda, Y. (1994) Life Sci. 54 1917-1926 [DOI] [PubMed] [Google Scholar]
- 20.Bowne, S. J., Liu, Q., Sullivan, L. S., Zhu, J., Spellicy, C. J., Rickman, C. B., Pierce, E. A., and Daiger, S. P. (2006) Investig. Ophthalmol. Vis. Sci. 47 3754-3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spellicy, C. J., Daiger, S. P., Sullivan, L. S., Zhu, J., Liu, Q., Pierce, E. A., and Bowne, S. J. (2007) Mol. Vis. 13 1866-1872 [PubMed] [Google Scholar]
- 22.Xu, D., Cobb, G. C., Spellicy, C. J., Bowne, S. J., Daiger, S. P., and Hedstrom, L. (2008) Arch. Biochem. Biophys. 472 100-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowne, S. J., Sullivan, L. S., Blanton, S. H., Cepko, C. L., Blackshaw, S., Birch, D. G., Hughbanks-Wheaton, D., Heckenlively, J. R., and Daiger, S. P. (2002) Hum. Mol. Genet. 11 559-568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennan, A., Aherne, A., Palfi, A., Humphries, M., McKee, A., Stitt, A., Simpson, D. A., Demtroder, K., Orntoft, T., Ayuso, C., Kenna, P. F., Farrar, G. J., and Humphries, P. (2002) Hum. Mol. Genet. 11 547-557 [DOI] [PubMed] [Google Scholar]
- 25.Wada, Y., Sandberg, M. A., McGee, T. L., Stillberger, M. A., Berson, E. L., and Dryja, T. P. (2005) Investig. Ophthalmol. Vis. Sci. 46 1735-1741 [DOI] [PubMed] [Google Scholar]
- 26.Kennan, A., Aherne, A., Bowne, S. J., Daiger, S. P., Farrar, G. J., Kenna, P. F., and Humphries, P. (2003) Adv. Exp. Med. Biol. 533 13-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowne, S. J., Sullivan, L. S., Mortimer, S. E., Hedstrom, L., Zhu, J., Spellicy, C. J., Gire, A. I., Hughbanks-Wheaton, D., Birch, D. G., Lewis, R. A., Heckenlively, J. R., and Daiger, S. P. (2006) Investig. Ophthalmol. Vis. Sci. 47 34-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grover, S., Fishman, G. A., and Stone, E. M. (2004) Ophthalmology 111 1910-1916 [DOI] [PubMed] [Google Scholar]
- 29.Aherne, A., Kennan, A., Kenna, P. F., McNally, N., Lloyd, D. G., Alberts, I. L., Kiang, A. S., Humphries, M. M., Ayuso, C., Engel, P. C., Gu, J. J., Mitchell, B. S., Farrar, G. J., and Humphries, P. (2004) Hum. Mol. Genet. 13 641-650 [DOI] [PubMed] [Google Scholar]
- 30.Zhang, R.-G., Evans, G., Rotella, F. J., Westbrook, E. M., Beno, D., Huberman, E., Joachimiak, A., and Collart, F. R. (1999) Biochemistry 38 4691-4700 [DOI] [PubMed] [Google Scholar]
- 31.Mendes, H. F., van der Spuy, J., Chapple, J. P., and Cheetham, M. E. (2005) Trends Mol. Med. 11 177-185 [DOI] [PubMed] [Google Scholar]
- 32.Nott, A., Le Hir, H., and Moore, M. J. (2004) Genes Dev. 18 210-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johannes, G., and Sarnow, P. (1998) RNA (N. Y.) 4 1500-1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tenenbaum, S. A., Lager, P. J., Carson, C. C., and Keene, J. D. (2002) Methods (San Diego) 26 191-198 [DOI] [PubMed] [Google Scholar]
- 35.Chiu, S. Y., Lejeune, F., Ranganathan, A. C., and Maquat, L. E. (2004) Genes Dev. 18 745-754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blobel, G., and Sabatini, D. (1971) Proc. Natl. Acad. Sci. U. S. A. 68 390-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amit, I., Citri, A., Shay, T., Lu, Y., Katz, M., Zhang, F., Tarcic, G., Siwak, D., Lahad, J., Jacob-Hirsch, J., Amariglio, N., Vaisman, N., Segal, E., Rechavi, G., Alon, U., Mills, G. B., Domany, E., and Yarden, Y. (2007) Nat. Genet. 39 503-512 [DOI] [PubMed] [Google Scholar]
- 38.Carson, J. P., Zhang, N., Frampton, G. M., Gerry, N. P., Lenburg, M. E., and Christman, M. F. (2004) Cancer Res. 64 2096-2104 [DOI] [PubMed] [Google Scholar]
- 39.Hartong, D. T., Berson, E. L., and Dryja, T. P. (2006) Lancet 368 1795-1809 [DOI] [PubMed] [Google Scholar]
- 40.Hedstrom, L. (1999) Curr. Med. Chem. 6 545-560 [PubMed] [Google Scholar]
- 41.Whitehead, J. P., Simpson, F., Hill, M. M., Thomas, E. C., Connolly, L. M., Collart, F., Simpson, R. J., and James, D. E. (2004) Traffic 5 739-749 [DOI] [PubMed] [Google Scholar]
- 42.Ji, Y., Gu, J., Makhov, A. M., Griffith, J. D., and Mitchell, B. S. (2006) J. Biol. Chem. 281 206-212 [DOI] [PubMed] [Google Scholar]
- 43.Gunter, J. H., Thomas, E. C., Lengefeld, N., Kruger, S. J., Worton, L., Gardiner, E. M., Jones, A., Barnett, N. L., and Whitehead, J. P. (2008) Int. J. Biochem. Cell Biol. 40 1716-1728 [DOI] [PubMed] [Google Scholar]
- 44.Keene, J. D., and Tenenbaum, S. A. (2002) Mol. Cell 9 1161-1167 [DOI] [PubMed] [Google Scholar]
- 45.Keene, J. D. (2007) Nat. Rev. Genet 8 533-543 [DOI] [PubMed] [Google Scholar]
- 46.Tenenbaum, S. A., Carson, C. C., Lager, P. J., and Keene, J. D. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 14085-14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, Y., Liu, C. L., Storey, J. D., Tibshirani, R. J., Herschlag, D., and Brown, P. O. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 5860-5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eystathioy, T., Chan, E. K., Tenenbaum, S. A., Keene, J. D., Griffith, K., and Fritzler, M. J. (2002) Mol. Biol. Cell 13 1338-1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho, Y., Gruhler, A., Hellbut, A., Bader, G. D., Moore, L., Adams, S.-L., Millar, A., Taylor, P., Bennett, K., Boutliller, K., Yang, L., Wolting, C., Donaldson, I., Scchandorff, S., Shewnarane, J., Vo, M., Taggart, J., Goudreault, M., Muskat, B., Alfarano, C., Dewar, D., Lin, Z., Michalickova, K., Willems, A. R., Sasl, H., Nielsen, P. A., Rasmussen, K. J., Andersen, J. R., Johansen, L. E., Hansens, L. K., Jespersen, H., Podtelejnikov, A., Neilsen, E., Crawford, J., Poulsens, V., Sorensen, B. D., Matthiesen, J., Hendrickson, R. C., Gleeson, F., Pawson, T., Moran, M. F., Durocher, D., Mann, M., Hogue, C. W. V., Figeys, D., and Tyers, M. (2002) Nature 415 180-18311805837 [Google Scholar]
- 50.Shepard, K. A., Gerber, A. P., Jambhekar, A., Takizawa, P. A., Brown, P. O., Herschlag, D., DeRisi, J. L., and Vale, R. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 11429-11434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerber, A. P., Herschlag, D., and Brown, P. O. (2004) PLoS Biol. 2 E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holcik, M., and Sonenberg, N. (2005) Nat. Rev. Mol. Cell Biol. 6 318-327 [DOI] [PubMed] [Google Scholar]
- 53.Gunawardena, S., and Goldstein, L. S. (2004) J. Neurobiol. 58 258-271 [DOI] [PubMed] [Google Scholar]
- 54.Sung, C. H., and Tai, A. W. (2000) Int. Rev. Cytol. 195 215-267 [DOI] [PubMed] [Google Scholar]
- 55.Humphries, M. M., Rancourt, D., Farrar, G. J., Kenna, P., Hazel, M., Bush, R. A., Sieving, P. A., Sheils, D. M., McNally, N., Creighton, P., Erven, A., Boros, A., Gulya, K., Capecchi, M. R., and Humphries, P. (1997) Nat. Genet. 15 216-219 [DOI] [PubMed] [Google Scholar]
- 56.Tan, E., Wang, Q., Quiambao, A. B., Xu, X., Qtaishat, N. M., Peachey, N. S., Lem, J., Fliesler, S. J., Pepperberg, D. R., Naash, M. I., and Al-Ubaidi, M. R. (2001) Investig. Ophthalmol. Vis. Sci. 42 589-600 [PubMed] [Google Scholar]