Abstract
Human T-cell leukemia virus type-1 is the causative agent for adult T-cell leukemia. Previous research has established that the viral oncoprotein Tax mediates the transformation process by impairing cell cycle control and cellular response to DNA damage. We showed previously that Tax sequesters huChk2 within chromatin and impairs the response to ionizing radiation. Here we demonstrate that DNA-dependent protein kinase (DNA-PK) is a member of the Tax·Chk2 nuclear complex. The catalytic subunit, DNA-PKcs, and the regulatory subunit, Ku70, were present. Tax-containing nuclear extracts showed increased DNA-PK activity, and specific inhibition of DNA-PK prevented Tax-induced activation of Chk2 kinase activity. Expression of Tax induced foci formation and phosphorylation of H2AX. However, Tax-induced constitutive signaling of the DNA-PK pathway impaired cellular response to new damage, as reflected in suppression of ionizing radiation-induced DNA-PK phosphorylation and γH2AX stabilization. Tax co-localized with phospho-DNA-PK into nuclear speckles and a nuclear excluded Tax mutant sequestered endogenous phospho-DNA-PK into the cytoplasm, suggesting that Tax interaction with DNA-PK is an initiating event. We also describe a novel interaction between DNA-PK and Chk2 that requires Tax. We propose that Tax binds to and stabilizes a protein complex with DNA-PK and Chk2, resulting in a saturation of DNA-PK-mediated damage repair response.
The human transforming retrovirus, human T-cell leukemia virus type 1 (HTLV-1),2 is the causative agent of adult T-cell leukemia (ATL) and HTLV-1-associated myelopathy/tropical spastic paraparesis as well as other subneoplastic conditions (1–5). Cellular transformation is attributed to expression of the viral oncoprotein Tax. Although the specific mechanism is not fully known, it is clear that Tax affects diverse cellular processes through direct interaction with various cellular proteins involved in cell cycle control and DNA damage repair response (6, 7). Recently, in an elegant ex vivo model, Sibon et al. (8) demonstrated that HTLV-1-infected (but preneoplastic) CD4+ T-lymphocytes displayed genomic instability that correlated with Tax expression. Thus, strong in vivo data exist implicating expression of Tax and the loss of genomic integrity as a pathway to development of ATL.
Studies showing increased mutation frequency in both Tax-expressing mammalian cells and yeast provide evidence of genomic instability induced by Tax (9–11). These mutations are of a random nature, suggesting impairment of the ability of the cell to repair accumulated DNA damage introduced during its normal life cycle (10). Furthermore, an increase in persistent DNA breaks is observed in Tax-expressing cells as micronuclei, which seems to occur because of Tax-induced loss of cellular DNA repair function (12–14). Several observations have prompted diverse models as to how Tax impairs cellular repair response. Tax represses transcription of DNA polymerase β, an enzyme involved specifically in base excision repair, and base excision repair is suppressed in HTLV-1-transformed cells (15, 16). Tax also suppresses nucleotide excision repair, which correlates with its ability to trans-activate proliferating cell nuclear antigen (17, 18). Transcriptional repression of human telomerase (hTert) by Tax may inhibit the addition of telomeric repeats to stabilize the ends of double-stranded DNA breaks (19). In addition, a reduction or loss of expression of two or more mismatch repair genes was observed in primary ATL leukemic cells (20).
There is also evidence that the ability of Tax to induce micronuclei is dependent on Ku86, a component of the non-homologous end-joining (NHEJ) pathway employed for DNA double strand break (DSB) repair (13). Ku86 and Ku70 form the regulatory subunit of DNA-PK, an enzyme with a critical role in NHEJ repair (21). We showed previously that Tax interacts with Chk2 (22), a downstream target of DNA-PK, and subsequently demonstrated Tax-induced impairment of the Chk2-mediated response to exogenous DNA damage (23). Collectively, these studies demonstrate the wide-ranging effects that Tax has on the capacity of the cell to respond appropriately to DNA damage.
In this study, we have identified a novel physical interaction between Tax and DNA-PK. We show that Tax co-localizes in nuclear speckles with forms of the catalytic subunit of DNA-PK (DNA-PKcs) that are phosphorylated at Ser-2056 and Thr-2609 and that these phosphorylated forms are increased in Tax-expressing cells. Tax is required for formation of a DNA-PK/Chk2 interaction and inhibition of DNA-PK ablated Tax activation of Chk2, implying that DNA-PK mediates this Tax activity. Tax expression alone resulted in increased steady-state levels of phospho-DNA-PKcs and phosphorylated histone 2AX (γ-H2AX). However, the ionizing radiation-induced foci (IRIF) formation and -fold induction of phospho-DNA-PK in response to ionizing radiation (IR) is repressed in the presence of Tax. We provide evidence that Tax elicits its action via physical binding and constitutive stabilization of phosphorylated DNA-PK. Collectively, these data demonstrate that through direct interaction with DNA-PK, Tax subverts a normal cellular DNA damage response by saturation of naive DNA-PK.
EXPERIMENTAL PROCEDURES
Plasmids—The S-tagged expression vectors STaxGFP and SGFP were constructed by inserting the tax-EGFP fusion or EGFP open reading frame, respectively, into the SmaI site of pTriEx4-Neo (Novagen, Madison, WI) in-frame with the amino-terminal S-tag and His tag (24). The Tax deletion mutant, STaxΔNLS, was constructed by removing the nuclear localization signal sequence encoding amino acids 29–52. Specifically, the deletion mutation was accomplished using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) and employing a forward primer (CAA GGC GAC TGG TGC CAG ATC ACC TGG GAC CCC) and a reverse primer (GGG GTC CCA GGT GAT CTG GCA CCA GTC GCC TTG). HpX plasmid expressing full-length Tax under the control of the HTLV-1 long-term repeat (25). S-tagged Chk2 and HA-tagged Chk2 were used to express the full length of Chk2 (23).
Cell Culture and Transfection—293T cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 in air in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (Invitrogen). Transfections were performed by standard calcium phosphate precipitation as described elsewhere (22).
Isolation of Tax-containing Protein Complexes—S-Tax-GFP expression plasmids were transfected into 293T cells. Cell lysates were incubated with 75-μl bed volume of S-protein-agarose (Novagen, Madison, WI) for 30 min at room temperature and then washed three times with 1 ml of bind/wash buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Triton X-100). The washed beads were eluted by resuspension in 150 μl of Laemmli sample buffer (Bio-Rad) with β-mercaptoethanol followed by boiling for 5 min. Eluates were separated in a 10% SDS one-dimensional polyacrylamide gel and visualized by SilverQuest silver staining (Invitrogen). Bands of interest were excised manually from the gel for further analysis.
Immunoprecipitation—Whole cell lysate containing ∼2 mg of protein in 500 μl was incubated with 5 μl of anti-Tax polyclonal antibody (25) at 4 °C overnight with constant rotation. The lysate was then incubated with 100 μl of protein A-Sepharose beads (Zymed Laboratories Inc., San Francisco, CA) while rotating for 1 h at 4 °C. The beads were washed three times with 1 ml each of 1× SNNTE buffer (5% sucrose, 500 mm NaCl, 1% Nonidet P-40, 50 mm Tris-HCl, pH 7.4, 5 mm EDTA). Proteins were eluted from beads by resuspension in 100 μl of Laemmli sample buffer (Bio-Rad) with β-mercaptoethanol and boiled for 5 min.
Immunoblot Analysis—Cell extracts were derived as described above. Total protein concentrations were determined by protein assay (Bio-Rad). An equal volume of sample loading buffer (Bio-Rad) with β-mercaptoethanol was added to the lysate and boiled for 5 min. Samples were normalized to total protein and separated through a 10% SDS-polyacrylamide gel. The proteins were transferred onto Immobilon-P (Millipore, Billerica, MA) membrane using a Trans-blot SD semi-dry transfer cell (Bio-Rad) at 400 mA for 50 min. After blocking in 5% nonfat milk in PBS/0.1% Tween-20, blots were incubated in primary antibody overnight followed by 1 h of incubation in secondary horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibody (Bio-Rad). Immunoreactivity was detected via Immun-Star enhanced chemiluminescence protein detection (Bio-Rad).
Liquid Chromatography-MS/MS Analysis—Protein bands were excised from one-dimensional polyacrylamide gels. Gel slices were cut into 1–2-mm cubes, washed three times with 500 μl of ultra-pure water, and incubated in 100% acetonitrile for 45 min. The material was dried in a SpeedVac, rehydrated in a 12.5 ng/μl modified sequencing grade trypsin solution (Promega, Madison, WI), and incubated in an ice bath for 40–45 min. The excess trypsin solution was then removed and replaced with 40–50 μl of 50 mm ammonium bicarbonate, 10% acetonitrile, pH 8.0, and the mixture was incubated overnight at 37 °C. Peptides were extracted twice with 25 μl of 50% acetonitrile, 5% formic acid and dried in a SpeedVac. Digests were resuspended in 20 μl of Buffer A (5% acetonitrile, 0.1% formic acid, 0.005% heptafluorobutyric acid), and 3–6 μl was loaded onto a 12-cm × 0.075-mm fused silica capillary column packed with 5 μm diameter C-18 beads (The Nest Group, Southboro, MA). Peptides were eluted for 55 min by applying a 0–80% linear gradient of Buffer B (95% acetonitrile, 0.1% formic acid, 0.005% heptafluorobutyric acid) at a flow rate of 130 μl/min with a pre-column flow splitter, resulting in a final flow rate of ∼200 nL/min directly into the source. A ThermoFinnigan LCQ™ Deca XP (Thermo Scientific, San Jose, CA) was run in an automated collection mode with an instrument method composed of a single segment and four data-dependent scan events with a full MS scan followed by three MS/MS scans of the highest intensity ions. Normalized collision energy was set at 30, and activation Q was 0.250, with minimum full scan signal intensity at 5 × 105 and a minimum MS2 intensity at 1 × 104. Dynamic exclusion was turned on utilizing a 3-min repeat count of 2, with the mass width set at 1.50 Da. Sequence analysis was performed with TurboSEQUEST™ (Thermo Scientific) using an indexed human subset data base of the nonredundant protein data base from the National Center for Biotechnology Information (NCBI) Web site.
RT-PCR—Total RNA was extracted from cells transfected with Hpx Tax expression vector or mock-transfected by harvesting in TRIzol reagent (Invitrogen) followed by chloroform extraction. The aqueous layer was transferred to a fresh tube with isopropanol, and the mixture was applied to an RNeasy column (Qiagen, Valencia, CA). RNase-free DNase was added to the wash buffer, and RNA was eluted with RNase-free water. Gene expression was measured using the Access RT-PCR system (Promega) for coupled reverse transcription and PCR amplification according to the manufacturer's protocol. Briefly, 10 ng of RNA template was reverse-transcribed using AMV reverse transcriptase for first strand cDNA synthesis and Tfl DNA polymerase for second strand cDNA synthesis and DNA amplification. 18S rRNA was amplified as an internal control for equal total RNA using primers 5′-TGACTCTAGATAACCTCGGG-3′ (forward) and 5′-CCCAAGATCCAACTACGAGC-3′ (reverse). A 348 bp fragment of DNA-PKcs cDNA (3325–3672 bp) was amplified using primers 5′-AGGGAAGAAGAGTCTCTGGTGG-3′ (forward) and 5′-ATTAGGGGATCTGTTGCCTGGC-3′ (reverse). Semiquantitation was achieved by limiting dilution of products.
Immunofluorescence—Cells were seeded onto 22-mm-diameter coverslips in 6-well plates at 1 × 105 cells/well. Transfections were performed as described above, and 48 h later cells were washed twice with PBS, fixed in 4% paraformaldehyde, and permeabilized with methanol. Coverslips were incubated with primary antibody in 3% bovine serum albumin/PBS overnight at 4 °C followed by two washes in PBS/0.1% Tween 20 and two washes in PBS. Coverslips were then incubated in secondary antibody with the addition of 1 μm TOPRO-3-iodide (Molecular Probes, Eugene, OR) for 1 h at room temperature followed by two washes in 3% bovine serum albumin/PBS and two washes in PBS. Coverslips were then mounted in Vectashield containing DAPI (Vector Laboratories, Burlingame, CA). Fluorescent images were acquired using a Zeiss LSM 510 confocal microscope at ×40 magnification with a 2.8× zoom and imaged with Image Browser software (Carl Zeiss, Jena, Germany).
Nuclear Extracts—Confluent 150-mm plates of transfected 293T cells were washed twice in cold PBS and harvested by scraping in 3 ml of Buffer C (10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 0.5% Nonidet P-40, 0.5 mm dithiothreitol, protease inhibitor mixture (Roche Applied Science)). Cells were allowed to lyse by incubation for 10 min on ice. Nuclei were centrifuged at 2000 rpm for 10 min at 4 °C. The supernatant was discarded, and 1 ml of Buffer D (20 mm HEPES, pH 7.9, 25% glycerol, 420 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 0.5 mm dithiothreitol, protease inhibitor mixture) was added to the pellet. The resuspended pellet was incubated on ice for 30 min and mixed gently every 5 min by pipetting up and down. The sample was centrifuged at 14,000 rpm for 30 min at 4 °C. The supernatant was dialyzed against Buffer E (20 mm HEPES, pH 7.9, 20% glycerol, 100 mm KCl, 0.2 mm EDTA, 0.5 mm phenylmethylsulfonyl fluoride, 0.5 mm dithiothreitol) for 5 h to overnight at 4 °C. Nuclear extracts were stored in aliquots at -80 °C.
Chromatin-bound Fraction—Cells were washed twice with cold PBS and fractionated into soluble cytoplasmic, soluble nuclear, and chromatin-bound fractions as described in our previous study (23). The final chromatin pellet was resuspended in M-Per protein lysis buffer by pulling the sample through a 21-gauge needle for 10 times to break the protein-DNA binding. The chromatin-bound proteins (P3) were collected by further centrifugation (10 min, 12,000 × g). Protein concentration was measured by the Bradford assay as described above.
DNA-dependent Protein Kinase Assay—Protein kinase activity was assayed using the SignaTECT® DNA-dependent protein kinase assay system (Promega) following the manufacturer's protocol. In some reactions, extracts were preincubated with 10 μm DNA-PK inhibitor NU7026 (Calbiochem) for 1 h on ice before being added to the reaction. Samples were incubated at 30 °C for 10 min. Termination buffer containing guanidine hydrochloride was then added, and 10 μl of each reaction mixture was spotted onto SAM2® biotin capture membrane, containing biotin-binding streptavidin matrix. After washing with 2 m NaCl, membranes were dried, and incorporated 32P-phosphorylated substrate was measured by scintillation counting.
Chk2 Kinase Assay—pCDNA4-Chk2 in combination with pCDNA3-Tax (provided by Ralph Grassmann, Institute for Clinical Virology, Erlangen, Germany) or control plasmid expressing nonspecific protein was subjected to in vitro transcription/translation using the rabbit reticulocyte lysate system (Promega). Standard 50-μl reactions were performed following the manufacturer's protocol. 8 μl of the in vitro translation product was mixed with 300 μl of NETN buffer (20 mm Tris-HCl, pH 8.0, 0.1 m NaCl, 1 mm EDTA, 0.5% Nonidet P-40, protease inhibitor mixture (Roche Applied Science)) for immunoprecipitation using 2 μg of anti-Xpress tag antibody (Invitrogen) for 3 h. Precipitates were washed twice with NETN buffer lacking protease inhibitors followed by a final wash with 1× kinase assay buffer (20 mm Tris, pH 7.5, 10 mm MgCl2, 10 mm MnCl2, 1 mm dithiothreitol). In some reactions, precipitated Chk2 immune complexes were preincubated with 10 μm DNA-PK inhibitor II (Calbiochem) for 1 h on ice before being added to the kinase reaction. Reactions were incubated at 30 °C for 10 min in 1× kinase assay buffer supplemented with 2 μm unlabeled ATP and 10 μCi of [γ-32P]ATP (Pierce). The reaction mixture was resolved on a 10% SDS-polyacrylamide gel, dried, and subjected to phosphorimaging using a Typhoon scanner (GE Healthcare). Relative intensity of the bands was calculated by densitometry.
RESULTS
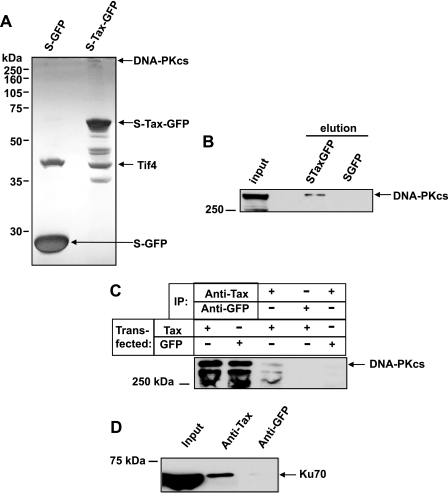
Identification of DNA-PK in Cellular Tax Complexes—We previously described a tandem affinity-tagged Tax expression system and demonstrated the expression of biologically active full-length Tax protein fused to amino-terminal S-tags to facilitate affinity purification, and carboxyl-terminal GFP to enable monitoring of protein expression (24). In the current study, we purified S-tagged Tax protein along with interacting cellular factors and resolved these protein complexes by SDS-PAGE and visualization by silver staining (Fig. 1A). Individual bands corresponding to proteins that interact with Tax were excised from the gel, and the proteins were identified by LC-MS/MS analysis. S-Tax-GFP was identified from a band of the expected size on the gel, as indicated (Fig. 1A). In addition, a high molecular weight protein that specifically bound Tax was identified as DNA-PKcs. The S peptide-binding protein EIF4 was also identified. The remaining bands were identified as degraded forms of S-Tax-GFP. The sequence identification was confirmed by resolving replicate samples on SDS-PAGE and immunoblotting with antibody specific to DNA-PKcs (Fig. 1B).
FIGURE 1.
DNA-PK binds to Tax. A, S-GFP and S-Tax-GFP (indicated) were affinity-purified using S-protein-agarose beads as described. Protein complexes were normalized to total protein (30 μg), subjected to SDS-PAGE separation, and visualized by silver staining. The indicated bands, representing the proteins of interest, were excised and sequence-identified by liquid chromatography-MS/MS. B, the same samples were subjected to immunoblotting with anti-DNA-PKcs antibody. The band representing DNA-PKcs is indicated. Crude lysate (30 μg of total protein) from STaxGFP-transfected cells was loaded as a control (Input). C, co-immunoprecipitation of Tax and DNA-PKcs. Cell lysates (normalized to total protein) from STaxGFP-or SGFP-transfected cells as indicated (Transfected) were immunoprecipitated with anti-Tax or anti-GFP polyclonal antibodies as indicated (IP). The first two lanes are total lysate (30 μg) and represent input controls. The resulting precipitates were subjected to SDS-PAGE and immunoblot analysis using anti-DNA-PKcs monoclonal antibody. D, co-immunoprecipitation of Tax and Ku70. Lysates from Tax-transfected cells (Input) were immunoprecipitated with anti-Tax or anti-GFP polyclonal antibodies as indicated. The resulting precipitates were subjected to SDS-PAGE and immunoblot analysis using anti-Ku70 monoclonal antibody.
As an orthogonal approach to confirmation of the Tax/DNA-PK interaction, we conducted co-immunoprecipitation with anti-Tax antibody. Tax complexes were immunoprecipitated from whole cell lysates with anti-Tax antibody, and subsequent immunoblotting with anti-DNA-PKcs antibody revealed that DNA-PKcs is specifically present in the Tax complex (Fig. 1C). DNA-PKcs could not be detected when an equal concentration of control anti-GFP antibody was substituted for Tax antibody or when S-GFP was substituted for Tax in the whole cell lysate, substantiating the specificity of the Tax/DNA-PKcs interaction.
The DNA-PK holoenzyme consists of a catalytic subunit as well as two regulatory subunits, Ku70 and Ku86 (26, 27). In our co-immunoprecipitation experiments we demonstrated that Ku70 is isolated from Tax-expressing cells with anti-Tax antibody, indicating that the Ku70 regulatory subunit of DNA-PK is also in the Tax complex (Fig. 1D). Similar experiments were conducted to ascertain the presence of Ku86, but we were unable to demonstrate the presence of Ku86 in the Tax complex (data not shown). Thus, the DNA-PK catalytic subunit and at least one regulatory subunit are in a complex with Tax protein.
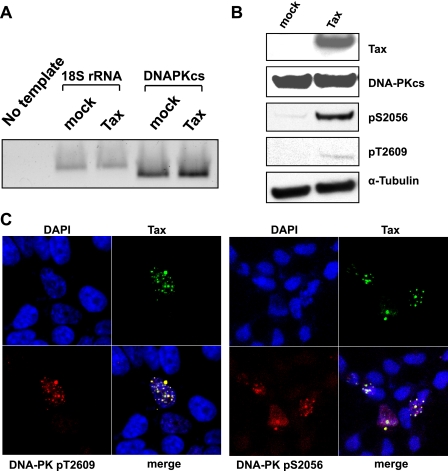
Induction of Phosphorylated DNA-PK by Tax—The activation of DNA-PK occurs at the initial stages of DNA damage recognition (28), and phosphorylation of DNA-PKcs at Ser-2056 and Thr-2609 is a critical autophosphorylation event required for kinase activity and repair activity in vivo (29–31). Tax did not alter steady-state levels of endogenous DNA-PK RNA as measured by semiquantitative RT-PCR (Fig. 2A). In addition, immunoblot analysis showed that the steady-state levels of native DNA-PKcs protein were unaffected by Tax (Fig. 2B). However, when we assessed phosphorylated forms of DNA-PKcs in the presence of Tax, we observed a dramatic increase in steady-state levels (Fig. 2B). Thus, phosphorylation of endogenous DNA-PK at Ser-2056 and Thr-2609 resulted from the expression of Tax alone without external DNA damage agents. Furthermore, these phosphorylated forms of DNA-PK co-localized with Tax in nuclear structures, which we have previously termed Tax speckled structures (TSS) (25). Phosphorylated DNA-PK is present in characteristic nuclear foci only in the Tax-expressing cells (Fig. 2C). Collectively, data from immunoprecipitation and immunofluorescence experiments demonstrate that Tax induces the phosphorylated form of DNA-PKcs and, specifically, interacts with it at discrete nuclear foci.
FIGURE 2.
Induction of phosphorylated DNA-PKcs by Tax. A, semiquantitative RT-PCR using primers specific for DNA-PKcs or control primers for 18S rRNA (indicated) for normalization to amplify cDNA from untransfected (mock) or Tax-transfected (Tax) cells. DNA-PKcs-specific primers were included in a negative control reaction with no template DNA. B, nuclear extracts from mock-transfected (mock) or Tax-transfected (Tax) cells were subjected to SDS-PAGE and immunoblot analysis with the indicated antibodies. α-Tubulin was used as control to demonstrate equal protein loading. C, cells were transiently transfected with STaxGFP (green) under conditions to achieve ∼20% transfection efficiency. The cells were then fixed, permeabilized, and immunostained with mouse anti-DNA-PKcs (pT2609, red) or rabbit anti-DNA-PKcs (pS2056, red) followed by the corresponding Alexa-594-conjugated anti-mouse or anti-rabbit secondary antibody. Cells were counterstained with DAPI and TOPRO-3-iodide to stain the nuclei (blue). The merged image shows the colocalized area represented in white (merge).
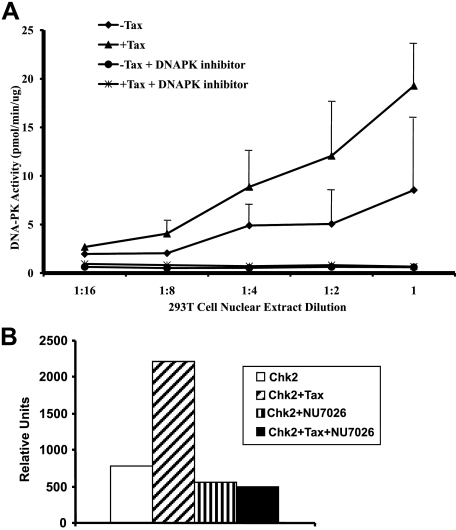
Tax Activation of DNA-PK Kinase Activity—A critical activity of DNA-PK is its ability to initiate signal cascades via its selective kinase function. Our observation that Tax expression resulted in an increase in the phosphorylated form of DNA-PK is consistent with activation of the protein as a kinase. However, we wanted to directly confirm that Tax expression resulted in enhanced DNA-PK activity. Using an in vitro kinase assay, we clearly showed that in the presence of Tax, DNA-PK displayed increased kinase activity (Fig. 3A). This assay measures phosphorylation of a p53 peptide substrate that is a known target of DNA-PK. The specificity of the assay for DNA-PK is demonstrated by the loss of kinase activity in the presence of the DNA-PK inhibitor NU7026 (Fig. 3A). This inhibitor has an IC50 for DNA-PK that is 500-fold lower than ATM/ATR and 50-fold lower than all other phosphatidylinositol 3-kinases, and it has no effect on Chk2 (32).
FIGURE 3.
Tax activation of DNA-PK activity. A, titration (indicated) of nuclear extracts from cells transfected with Tax expression vector (+Tax, red) or mock-transfected control cells (-Tax, blue) were prepared. DNA-PK enzyme activity was quantitated in the presence or absence of DNA-PK inhibitor NU7026 (indicated). Plotted are the average values of three experiments. B, DNA-PK activity mediates Tax activation of Chk2 kinase. Kinase assays were performed using in vitro transcribed/translated Chk2 and Tax (Chk2+Tax) or Chk2 alone (Chk2) in the presence or absence of DNA-PK inhibitor (indicated). The reaction mixtures were subjected to SDS-PAGE and quantitated using phosphorimaging analysis. Plotted are the average values from three experiments. The experimental error was <5%.
One of the known endogenous phosphorylation targets of DNA-PK is the DNA damage response protein Chk2, in which DNA-PK mediates phosphorylation of Chk2 at Thr-68 in response to DNA damage (33). Our laboratory and others have shown that Tax interacts with Chk2 (22, 34) and increases its kinase activity (23). To determine the role of DNA-PK in mediating Tax activation of Chk2, we measured the in vitro kinase activity of Chk2. The basal activity of Chk2 in rabbit reticulocyte lysates has been attributed previously to the presence of DNA-PK (33). Using this system we observed a decrease in Tax-induced Chk2 activation in the presence of the DNA-PK inhibitor NU7026 (Fig. 3B). These data establish that activation of DNA-PK in Tax-expressing cells is sufficient for the observed increase in Chk2 kinase activity.
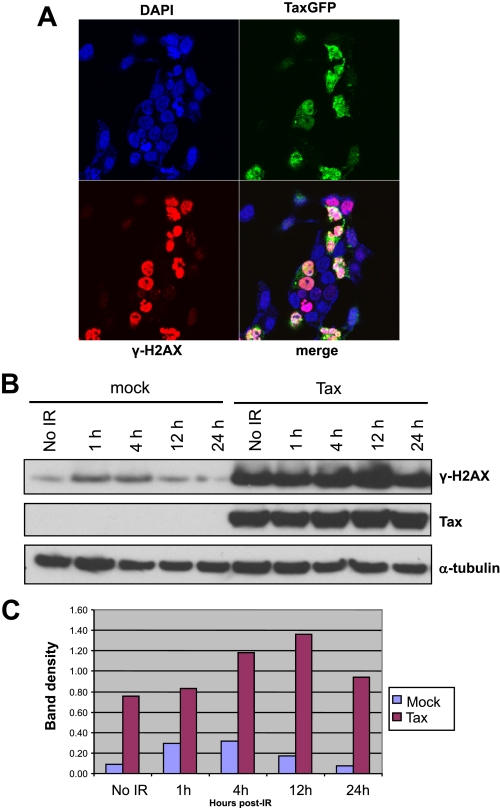
Tax Expression Is Coincident with DNA-PK Phosphorylation and γ-H2AX Expression—The formation of γ-H2AX nuclear foci at the site of DNA strand breaks is an early cellular response to DNA damage (35). Direct IR-induced phosphorylation of H2AX can be carried out efficiently by DNA-PK (36). Because we observed increased DNA-PK activity in Tax-expressing cells, we asked whether phosphorylation of the DNA-PK substrate H2AX was increased concomitantly. Strikingly, immunofluorescence experiments using confocal microscopy showed that Tax-expressing cells have a strong induction of γ-H2AX as compared with non-Tax-expressing cells in the absence of an external source of DNA damage (Fig. 4A). The images were digitally overexposed to reveal background staining of the non-Tax-expressing cells. Upon proper digital exposure, Tax and H2AX presented the same speckled pattern shown in Fig. 2.
FIGURE 4.
Tax-expressing cells display altered IRIF formation. A, Tax expression results in induction of γH2AX foci in the absence of exogenous ionizing radiation. Cells were fixed, permeabilized, and immunostained with rabbit anti-γ-H2AX, and the nuclei were counterstained with DAPI/TOPRO-3-iodide (indicated; blue). Shown are the expression of Tax-GFP (indicated; green), γH2AX (indicated; red), and the merged expression of each protein (white). B, Cells were either mock-transfected or transfected with Tax-expressing vector (indicated). The cultures were subjected to 5 Gy of IR and then harvested at 1, 4, 12, and 24 h (indicated). Cell extracts were subjected to SDS-PAGE and immunoblot analysis for Tax and γH2AX (indicated). Endogenous α-tubulin was used for normalization of extracts. C, phosphorimaging analysis of the blots shown in B. Shown is the semiquantitation of the γ-H2AX induction.
Immunoblotting of nuclear lysates from the same cells imaged in Fig. 4A with antibody to γ-H2AX showed a nearly 8-fold increase in phosphorylated H2AX in Tax-expressing cells compared with mock-transfected cells (Fig. 4, B and C, No IR). Thus, in the absence of any exogenous DNA damage, Tax expression results in increased steady-state levels of γ-H2AX. We next addressed the impact that Tax expression would have on the γ-H2AX response to exogenous DNA damage. Nuclear lysates of mock-transfected cells harvested at various time points post-irradiation show peak H2AX phosphorylation at 1 h post-IR, decreasing at 12 h, and returning to pre-IR levels before 24 h (Fig. 4, B and C). The increase in γ-H2AX is directly related to the availability of naive unphosphorylated H2AX and is a common feature of cells with a competent DNA damage response. In contrast, the temporal regulation of γ-H2AX induction is delayed in Tax-expressing cells such that peak levels of H2AX phosphorylation are not reached until 12 h post-IR (Fig. 4, B and C). Furthermore, although the peak γ-H2AX induction is 3.5-fold higher than pre-IR levels in mock-transfected cells, Tax-expressing cells achieve only a 1.4-fold increase in γ-H2AX in response to IR (Fig. 4C). These results are consistent with a model in which Tax-expressing cells have an impaired capacity to respond to new damage.
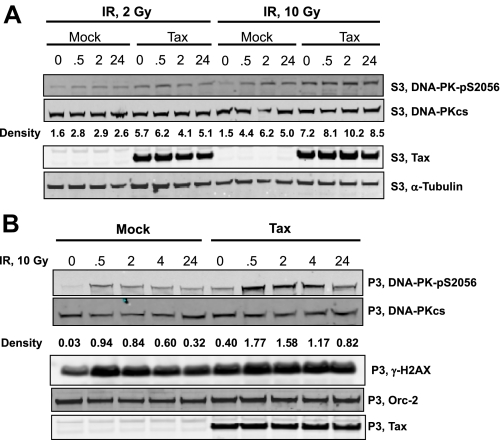
Quantitative Response to IR Damage is Reduced in Tax-expressing Cells—Autophosphorylation of DNA-PK at Ser-2609 is believed to be an initiating event for activation of kinase activity. We evaluated the change in phospho-DNA-PK(pS2609) as a quantitative measure of the response to exogenous damage in Tax-expressing cells. As expected, we show that nuclear levels of phospho-DNA-PK(pS2609) increase directly proportional to the extent of exposure to IR (Fig. 5A). The -fold increase in phospho-DNA-PK(pS2609) in response to 10 Gy (2.9-fold) is greater than to 2 Gy (1.8-fold) in mock-transfected cells. Also note that the -fold increase in phospho-DNA-PK(pS2609) between Tax-expressing cells and non-expressing cells (4.8-fold) in the absence of IR is comparable to the increase in response to 10 Gy IR. However, in Tax-expressing cells, the -fold increase in phospho-DNA-PK(pS2609) is suppressed at both low and high IR exposure (1.1-fold). Thus, Tax suppresses the quantitative change in phosphorylated DNA-PK in response to exogenous sources of DNA damage.
FIGURE 5.
Constitutive expression of phospho-DNA-PK correlates with prolonged γH2AX expression. 293T cells were either mock-transfected (mock) or transfected with a Tax expression plasmid (Tax). At 24 h post-transfection cells were exposed to the indicated dose of ionizing radiation for the specified amount of time. A, soluble nuclear extracts were derived from the above cells and subjected to SDS-PAGE. The gels were then analyzed by Western blotting for expression of DNA-PK(pS2056), DNA-PKcs, Tax, and α-tubulin as indicated. Relative densitometry was performed for the intensity of DNA-PK(pS2056) normalized to α-tubulin. B, chromatin extracts were derived from the above cells and subjected to SDS-PAGE. The gels were then analyzed by Western blotting for expression of DNA-PK(pS2056), DNA-PKcs, γH2AX, Orc-2, and Tax as indicated. Relative densitometry was performed for the intensity of DNA-PK(pS2056) normalized to α-tubulin.
We extended this analysis to the chromatin-bound fraction of DNA-PK. Following chromatin isolation, extracts were examined by Western analysis for the presence of phospho-DNA-PK(pS2609) and γ-H2AX (Fig. 5B). As was the case for nuclear fraction, Tax expression alone induced an increase in phosphorylation of DNA-PK as well as γ-H2AX. This increase was comparable with that seen following 10 Gy exposure in non-Tax-expressing cells. However, the quantitative response to IR in Tax-expressing cells was severely muted. The relative change in DNA-PK phosphorylation, calculated based upon densitometry, is shown in Fig. 5B. In response to IR, non-Tax-expressing cells displayed an ∼30-fold increase in chromatin-bound phospho-DNA-PK(pS2609), whereas Tax-expressing cells responded to the same IR exposure with an ∼4-fold increase. These data support the conclusion that Tax expression saturates the DNA-PK-mediated damage response.
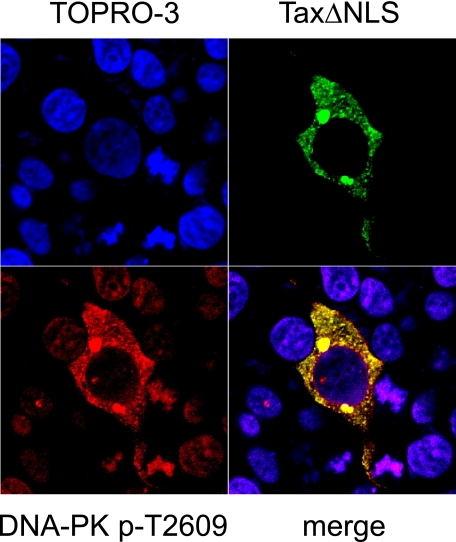
Tax Expression Dictates Compartmentalization of Phosphorylated DNA-PK—One potential model for describing the ability of Tax to misappropriate the DNA-PK-initiated damage response is via molecular sequestration of DNA-PK by Tax. Using immunofluorescence confocal microscopy, we examined the localization of the phospho-DNA-PK(pT2609) in the presence of wild type Tax and a Tax mutant that is deleted of the native nuclear localization signal and expressed exclusively in the cytoplasm (TaxΔNLS). Interestingly, phospho-DNA-PK relocates to the cytoplasm in the presence of the cytoplasm-restricted Tax deletion mutant (Fig. 6). The interaction between the cytoplasm-restricted Tax mutant and phospho-DNA-PK is stable enough to redistribute DNA-PK to the cytoplasm. Thus, it appears that the binding between Tax and DNA-PK is robust enough to account for and facilitate cellular redistribution via molecular sequestration.
FIGURE 6.
Redistribution of DNA-PK by Tax deletion mutants. Cells were transfected with a Tax deletion mutant (TaxΔNLS; green), fixed, permeabilized, and immunostained with mouse anti-DNA-PKcs (DNA-PK pThr-2609; red) followed by Alexa-594-conjugated anti-mouse secondary antibody. Nuclei were counterstained with DAPI/TOPRO-3-iodide (blue). The merged image shows the colocalized area represented in yellow or white (merge).
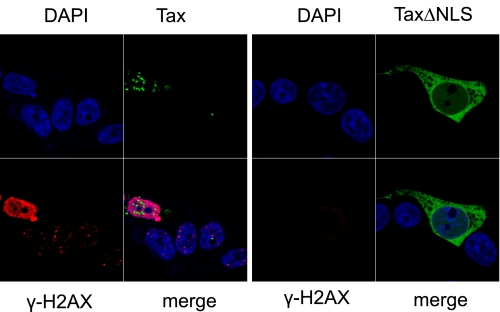
We have suggested that Tax binds to DNA-PK and co-opts the protein into a phosphorylated constitutively active state. The result is induction of γ-H2AX foci in the absence of exogenous DNA damage. If this model is correct then the Tax mutant that is unable to target to nuclear speckles should not activate the foci even though this mutant can still bind to phospho-DNA-PK. To test this hypothesis we expressed TaxΔNLS in cells and observed the resulting activation of H2AX. In Fig. 7 we show that the dramatic increase in steady-state levels of γ-H2AX due to Tax expression is not reproduced by the nuclear excluded Tax mutant. This failure is observable at early time points, indicating that the initial events of H2AX activation are ablated. Although the cytoplasmic DNA-PK remains phosphorylated at extended times post-IR (8 h), there is no indication of induction in γH2AX (see supplemental Fig. 1). This result supports our contention that Tax binding to DNA-PK initiates stable recruitment of γ-H2AX foci.
FIGURE 7.
Nuclear excluded Tax mutant protein fails to induce γH2AX foci. Cells were transfected with expression vectors for Tax or TaxΔNLS (as indicated). The cells were fixed, permeabilized, and immunostained with rabbit anti-γ-H2AX, and the nuclei were counterstained with DAPI/TOPRO-3-iodide (indicated; blue). Tax-expressing cells (green) and γH2AX-expressing cells (red) are indicated.
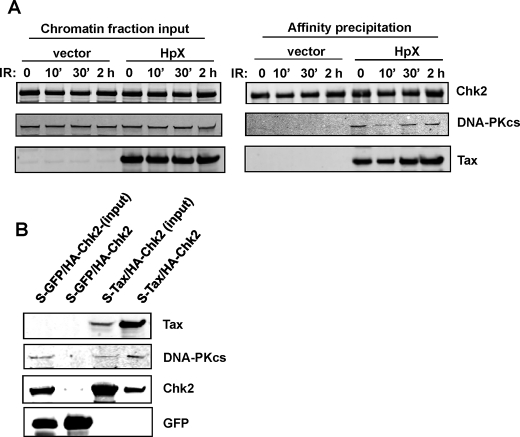
Tax Induces Binding of DNA-PK and Chk2 within the Chromatin Fraction—Upon recognition of damaged DNA, repair response factors accumulate at the site of damage and populate the cellular chromatin fraction (37–40). If Tax targets the damage repair response machinery, we would expect that the Tax·DNA-PK·Chk2 complex would be “stabilized” in the chromatin fraction. To test this hypothesis we expressed S-Chk2 into 293T cells and assessed the interaction with endogenous DNA-PKcs in the chromatin fraction. As shown in Fig. 8A, Chk2, DNA-PK, and Tax are all present in the cellular chromatin fraction. Following affinity isolation of S-Chk2 from the chromatin, we did not observe co-precipitation with DNA-PK in the absence of Tax. However, in the presence of Tax, DNA-PK and Tax co-precipitated with S-Chk2. Interestingly, IR alone did not result in Chk2/DNA-PK stable interaction. We also confirmed the triple complex by co-expression of S-Tax and HA-Chk2 followed by isolation of S-Tax (Fig. 8B). Again, Tax specifically recruited Chk2 and DNA-PK within a chromatin complex. These results demonstrate the existence of a Tax-dependent protein complex between Tax, Chk2, and DNA-PKcs.
FIGURE 8.
Chromatin-bound DNA-PK co-precipitates with Chk2 in the presence of Tax. A, cells were co-transfected with S-Chk2 expression plasmids and either Tax (HpX) or control (vector) plasmid. The co-transfected cells were then subjected to IR (10 Gy) for the times indicated. In the left panel (input fraction), the chromatin fraction (20 μg of total protein) was separated by SDS-PAGE and analyzed by Western blotting using anti-Chk2, anti-DNA-PK, and anti-Tax antibodies (as indicated). In the right panel (affinity precipitation), the fractions (350 μg of total protein) were subjected to affinity isolation of S-Chk2 followed by separation on SDS-PAGE and Western analysis using anti-Chk2, anti-DNA-PK, and anti-Tax antibodies (as indicated). B, cells were co-transfected with HA-Chk2 and either S-GFP or S-Tax plasmids. Whole cell lysates were extracted 48 h post-transfection. The whole cell protein lysates (700 μg) were subjected to affinity isolation of either S-GFP or S-Tax followed by separation on SDS-PAGE and Western analysis using anti-Tax, anti-Chk2, and anti-DNA-PK antibodies (as indicated).
DISCUSSION
DNA-PK is a key initiating enzyme in the NHEJ pathway of DNA DSB repair (21, 26, 41, 42). Impairment of the DSB repair pathway can lead to gross genomic abnormalities exacerbated by premature entry of cells into mitosis before DSBs are repaired, by the rejoining of DSBs on different chromosomes, fusion of chromosome ends with eroded telomeres, or defective repair (43). Along with playing a key role in DNA repair via NHEJ, DNA-PK is also involved in other important responses to DSBs, including cell cycle arrest and apoptosis (21). Because of the central role it plays in genomic integrity, DNA-PK has been suggested as a potential target for Tax-induced genomic instability (13). Our observation that Tax binds specifically to DNA-PK provides a critical component in the development of this model for impaired cellular responses to DNA damage and presentation of genomic instability.
DNA-PK consists of an ∼470-kDa catalytic subunit (DNA-PKcs) and a regulatory subunit composed of the Ku70 and Ku86 heterodimer. The activation of DNA-PK is tightly regulated so that recognition of DNA damage activates the response cascade. Specifically, initial DNA end synapsis occurs prior to (and in fact it activates) DNA-PKcs autophosphorylation and kinase activation (29, 44, 45). Autophosphorylation at Ser-2056 and Thr-2609, which is facilitated by Ku, activates the kinase and is a key event for the regulation of DNA-PK function (29–31, 46). However, the relationship among autophosphorylation, kinase activation, and a functional damage response is not a simple on-off mechanism. For instance, it has been observed that activated autophosphorylated DNA-PKcs must separate from the DNA ends to allow for ligation (47). This insures that DNA-PK is activated only in the presence of damaged DNA, facilitated by the presence of the Ku heterodimer at the broken DNA ends (42). Thus, in addition to initiating the kinase signal cascade, autophosphorylation of DNA-PKcs promotes disassembly of the protein-DNA complex at the end of the repair process and presumably initiates recycling of the component proteins for future repair events (41, 48, 49). Clearly, appropriate dephosphorylation and timely factor recycling are critical in providing DNA-PK in a naive ready state for response to new damage. Disruption in the delicate coordinated timing of DNA repair and DNA-PKcs autophosphorylation/activation would result in significant impairment of the DNA repair response (50). In fact, sustained hyperphosphorylation of DNA-PKcs at Ser-2056 and Thr-2609 results in increased sustained damage, indicating that prolonged phosphorylation is detrimental to DNA-PK repair function (51). Interestingly, elevated DNA-PK activity has been correlated previously with a number of human cancers with both metastatic and multidrug-resistant phenotypes (52–55). In fact, constitutive activation of γH2AX, ATM, and Chk2 is a hallmark of Bloom syndrome, a classic cancer-predisposed state typified by genomic instability (56). Thus our observation that Tax induced sustained activation of DNA-PK is consistent with a model for impaired damage response. If Tax binding to DNA-PK interferes with recycling of repair factors for additional rounds of repair, the result could be manifest as genomic instability and supportive of cellular transformation and development of ATL.
To assess the perturbation in DNA-PK function and evaluate potential quantitative failure in the repair response, we used the IR-induced H2AX foci model. The spatial organization of the cellular DNA damage repair response is reflected in the formation and resolution of these IRIF (57, 58). Treatment of cells with IR induces DNA-PK autophosphorylation and γ-H2AX foci formation, which normally begin to resolve by 8 h post-IR (29–31, 35). As expected, our model demonstrated potent IRIF formation in response to damage that was maximal at 2 h and resolved by 12 h following ionizing radiation. Conversely, Tax-expressing cells displayed a prolonged duration of IRIF as evidenced by phosphorylated DNA-PK and H2AX. Most significantly, however, may be the finding that the -fold response to IR was dramatically reduced in Tax-expressing cells, implying a reduced capacity to respond to DNA damage. The likelihood of such a scenario was bolstered by our observation of decreased survival of Tax-expressing cells following exposure to IR.
In addition to the direct effects upon DNA damage recognition and repair, DNA-PK is an early mediator of cell cycle arrest via activation of Chk2 (33). Our laboratory has shown that constitutive expression of hyperphosphorylated Chk2 by Tax leads to delayed cell cycle progression through the G2/M checkpoint (22) and an impaired response to ionizing radiation (23). Persistent activation of cell cycle checkpoints, such as we observed in Tax-expressing cells, is a common early step in tumorigenesis that is generally associated with genomic instability (43, 59). Our discovery that DNA-PK, a known upstream activator of Chk2, is a major component of Tax·Chk2 complexes provides for a clearer mechanistic understanding of how Tax impacts cellular damage response and checkpoint activation. Furthermore, we clearly show that Tax induces a stable interaction between DNA-PK and Chk2. Thus, we propose that Tax physically stabilizes phosphorylated DNA-PK and subsequently phosphorylated Chk2, which results in a saturation of DNA-PK signaling and, by inference, supports a Bloom syndrome-like state.
Another target of DNA-PK is the tumor suppressor p53, which has been shown to be phosphorylated by DNA-PK on serine residues 15 and 37 (60). Incidentally, there is significant evidence that Tax can inactivate p53 function through a proposed mechanism involving hyperphosphorylation of p53 at serines 15 and 392, which interferes with the function of p53 as a transcriptional activator (61, 62). Although it has been proposed that DNA-PK could mediate the phosphorylation mechanism through which Tax inactivates p53 (61, 62), a separate study found that Tax-mediated p53 inactivation still occurs in DNA-PKcs-deficient cells (63). However, because of overlapping roles of DNA-PK with other DNA damage response proteins, ATM and ATR (28, 64), we would suggest that other redundant damage response mediators compensated for the lack of DNA-PK in these knock-out systems. Thus, it is entirely possible that DNA-PK mediates the observed effects of Tax on p53.
DNA-PK is also critical for telomere capping, and DNA-PK-deficiency promotes increased chromosomal instability with telomeric fusions (65–68). Telomerase inactivation, via Tax-mediated repression of human telomerase reverse transcriptase (hTert) (19), would promote unstabilized telomeres, which could result in end-to-end chromosomal fusions in the context of increased DNA-PK activity, leading to the cytogenetic abnormalities observed in HTLV-1-infected cells (69). Consequently, Tax-expressing cells display frequent unstabilized DNA breaks detected as unprotected free 3′-OH DNA ends (12, 13).
An immediate question to address is why a retrovirus would target the NHEJ DNA repair pathway. To fully explore this issue it is important to recall that clonal expansion is a critical component of HTLV-1 biology. Specifically, cell-free de novo infection by HTLV-1 is a rare event, and numeric propagation of infected cells occurs via clonal expansion (70–72). Thus, a survival advantage to the virus would be realized via facilitation of cellular expansion. A DSB is especially harmful to cells, in large part because of the propensity to form genetic translocations (reviewed in Ref. 73). As a result of the severity of gene translocations, a failure to repair DSBs primarily leads to cell death or neoplastic transformation. Because the opportunity for DSBs increases with cell replication, clonal expansion would result in increased exposure to DSB (74). In this scenario, suppression of NHEJ may constitute a selective advantage by avoiding recognition-initiated apoptosis. Interestingly, NHEJ repair gene knockouts uniformly result exclusively in T-cell tumors (75–77), suggesting that the link among NHEJ, defective DSB repair, and clonal expansion may be particularly linear in the HTLV-1 host cell type. The isolation of V(D)J inversion events, a hallmark of DSB repair defects in lymphocytes, from patient-isolated ATL cells is strong evidence for the intimate role of NHEJ function and ATL development (78). Thus, resistance to damage repaired via the NHEJ pathway by HTLV-1 Tax may contribute to the endurance of HTLV-infected cells. So in this scenario HTLV-1 avoids DNA damage-mediated apoptotic host elimination at the risk of increased host genomic instability and cellular transformation. However, this is likely an evolutionarily beneficial trade-off because ATL events arise in less than 3% of infected individuals.
Supplementary Material
Acknowledgments
We thank Edward Johnson and Ann Campbell for helpful comments.
This work was supported, in whole or in part, by National Institutes of Health Public Health Service Grant CA-76595 from NCI. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: HTLV-1, human T-cell leukemia virus type 1; ATL, adult T-cell leukemia; DNA-PK, DNA-dependent protein kinase; DNA-PKcs, catalytic subunit of DNA-PK; NHEJ, non-homologous end joining; DSB, double strand break; IRIF, ionizing radiation-induced foci; HA, hemagglutinin; GFP, green fluorescent protein; PBS, phosphate-buffered saline; MS/MS, tandem mass spectrometry; RT, reverse transcription; DAPI, 4′,6-diamidino-2-phenylindole; Gy, gray.
References
- 1.Gessain, A., Barin, F., Vernant, J. C., Gout, O., Maurs, L., Calender, A., and de The, G. (1985) Lancet 2 407-410 [DOI] [PubMed] [Google Scholar]
- 2.Osame, M., Usuku, K., Izumo, S., Ijichi, N., Amitani, H., Igata, A., Matsumoto, M., and Tara, M. (1986) Lancet 1 1031-1032 [DOI] [PubMed] [Google Scholar]
- 3.Poiesz, B. J., Ruscetti, F. W., Gazdar, A. F., Bunn, P. A., Minna, J. D., and Gallo, R. C. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 7415-7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takatsuki, K. (2005) Retrovirology 2 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida, M., Miyoshi, I., and Hinuma, Y. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 2031-2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grassmann, R., Aboud, M., and Jeang, K. T. (2005) Oncogene 24 5976-5985 [DOI] [PubMed] [Google Scholar]
- 7.Marriott, S. J., and Semmes, O. J. (2005) Oncogene 24 5986-5995 [DOI] [PubMed] [Google Scholar]
- 8.Sibon, D., Gabet, A. S., Zandecki, M., Pinatel, C., Thete, J., Delfau-Larue, M. H., Rabaaoui, S., Gessain, A., Gout, O., Jacobson, S., Mortreux, F., and Wattel, E. (2006) J. Clin. Investig. 116 974-983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, B., Liang, M. H., Kuo, Y. L., Liao, W., Boros, I., Kleinberger, T., Blancato, J., and Giam, C. Z. (2003) Mol. Cell. Biol. 23 5269-5281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyake, H., Suzuki, T., Hirai, H., and Yoshida, M. (1999) Virology 253 155-161 [DOI] [PubMed] [Google Scholar]
- 11.Saggioro, D., D'Agostino, D. M., and Chieco-Bianchi, L. (1999) Exp. Cell Res. 247 525-533 [DOI] [PubMed] [Google Scholar]
- 12.Majone, F., and Jeang, K. T. (2000) J. Biol. Chem. 275 32906-32910 [DOI] [PubMed] [Google Scholar]
- 13.Majone, F., Luisetto, R., Zamboni, D., Iwanaga, Y., and Jeang, K. T. (2005) Retrovirology 2 45-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majone, F., Semmes, O. J., and Jeang, K. T. (1993) Virology 193 456-459 [DOI] [PubMed] [Google Scholar]
- 15.Jeang, K. T., Widen, S. G., Semmes, O. J., and Wilson, S. H. (1990) Science 247 1082-1084 [DOI] [PubMed] [Google Scholar]
- 16.Philpott, S. M., and Buehring, G. C. (1999) J. Natl. Cancer Inst. 91 933-942 [DOI] [PubMed] [Google Scholar]
- 17.Haoudi, A., and Semmes, O. J. (2003) Virology 305 229-239 [DOI] [PubMed] [Google Scholar]
- 18.Kao, S. Y., and Marriott, S. J. (1999) J. Virol. 73 4299-4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabet, A. S., Mortreux, F., Charneau, P., Riou, P., Duc-Dodon, M., Wu, Y., Jeang, K. T., and Wattel, E. (2003) Oncogene 22 3734-3741 [DOI] [PubMed] [Google Scholar]
- 20.Morimoto, H., Tsukada, J., Kominato, Y., and Tanaka, Y. (2005) Am. J. Hematol. 78 100-107 [DOI] [PubMed] [Google Scholar]
- 21.Burma, S., and Chen, D. J. (2004) DNA Repair (Amst.) 3 909-918 [DOI] [PubMed] [Google Scholar]
- 22.Haoudi, A., Daniels, R. C., Wong, E., Kupfer, G., and Semmes, O. J. (2003) J. Biol. Chem. 278 37736-37744 [DOI] [PubMed] [Google Scholar]
- 23.Gupta, S. K., Guo, X., Durkin, S. S., Fryrear, K. F., Ward, M. D., and Semmes, O. J. (2007) J. Biol. Chem. 282 29431-29440 [DOI] [PubMed] [Google Scholar]
- 24.Durkin, S. S., Ward, M. D., Fryrear, K. A., and Semmes, O. J. (2006) J. Biol. Chem. 281 31705-31712 [DOI] [PubMed] [Google Scholar]
- 25.Semmes, O. J., and Jeang, K. T. (1996) J. Virol. 70 6347-6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, G. C., and Jackson, S. P. (1999) Genes Dev. 13 916-934 [DOI] [PubMed] [Google Scholar]
- 27.Doherty, A. J., and Jackson, S. P. (2001) Curr. Biol. 11 R920-924 [DOI] [PubMed] [Google Scholar]
- 28.Yang, J., Yu, Y., Hamrick, H. E., and Duerksen-Hughes, P. J. (2003) Carcinogenesis 24 1571-1580 [DOI] [PubMed] [Google Scholar]
- 29.Chan, D. W., Chen, B. P., Prithivirajsingh, S., Kurimasa, A., Story, M. D., Qin, J., and Chen, D. J. (2002) Genes Dev. 16 2333-2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, B. P., Chan, D. W., Kobayashi, J., Burma, S., Asaithamby, A., Morotomi-Yano, K., Botvinick, E., Qin, J., and Chen, D. J. (2005) J. Biol. Chem. 280 14709-14715 [DOI] [PubMed] [Google Scholar]
- 31.Cui, X., Yu, Y., Gupta, S., Cho, Y. M., Lees-Miller, S. P., and Meek, K. (2005) Mol. Cell. Biol. 25 10842-10852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veuger, S. J., Curtin, N. J., Richardson, C. J., Smith, G. C., and Durkacz, B. W. (2003) Cancer Res. 63 6008-6015 [PubMed] [Google Scholar]
- 33.Li, J., and Stern, D. F. (2005) J. Biol. Chem. 280 12041-12050 [DOI] [PubMed] [Google Scholar]
- 34.Park, H. U., Jeong, S. J., Jeong, J. H., Chung, J. H., and Brady, J. N. (2006) Oncogene 25 438-447 [DOI] [PubMed] [Google Scholar]
- 35.Takahashi, A., and Ohnishi, T. (2005) Cancer Lett. 229 171-179 [DOI] [PubMed] [Google Scholar]
- 36.Stiff, T., O'Driscoll, M., Rief, N., Iwabuchi, K., Lobrich, M., and Jeggo, P. A. (2004) Cancer Res. 64 2390-2396 [DOI] [PubMed] [Google Scholar]
- 37.Bekker-Jensen, S., Lukas, C., Kitagawa, R., Melander, F., Kastan, M. B., Bartek, J., and Lukas, J. (2006) J. Cell Biol. 173 195-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bewersdorf, J., Bennett, B. T., and Knight, K. L. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 18137-18142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park, E. J., Chan, D. W., Park, J. H., Oettinger, M. A., and Kwon, J. (2003) Nucleic Acids Res. 31 6819-6827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yaneva, M., Kowalewski, T., and Lieber, M. R. (1997) EMBO J. 16 5098-5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meek, K., Gupta, S., Ramsden, D. A., and Lees-Miller, S. P. (2004) Immunol. Rev. 200 132-141 [DOI] [PubMed] [Google Scholar]
- 42.Collis, S. J., DeWeese, T. L., Jeggo, P. A., and Parker, A. R. (2005) Oncogene 24 949-961 [DOI] [PubMed] [Google Scholar]
- 43.Burma, S., Chen, B. P., and Chen, D. J. (2006) DNA Repair (Amst.) 5 1042-1048 [DOI] [PubMed] [Google Scholar]
- 44.Budman, J., Kim, S. A., and Chu, G. (2007) J. Biol. Chem. 282 11950-11959 [DOI] [PubMed] [Google Scholar]
- 45.Meek, K., Douglas, P., Cui, X., Ding, Q., and Lees-Miller, S. P. (2007) Mol. Cell. Biol. 27 3881-3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding, Q., Reddy, Y. V., Wang, W., Woods, T., Douglas, P., Ramsden, D. A., Lees-Miller, S. P., and Meek, K. (2003) Mol. Cell. Biol. 23 5836-5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reddy, Y. V., Ding, Q., Lees-Miller, S. P., Meek, K., and Ramsden, D. A. (2004) J. Biol. Chem. 279 39408-39413 [DOI] [PubMed] [Google Scholar]
- 48.Chan, D. W., and Lees-Miller, S. P. (1996) J. Biol. Chem. 271 8936-8941 [DOI] [PubMed] [Google Scholar]
- 49.Merkle, D., Douglas, P., Moorhead, G. B., Leonenko, Z., Yu, Y., Cramb, D., Bazett-Jones, D. P., and Lees-Miller, S. P. (2002) Biochemistry 41 12706-12714 [DOI] [PubMed] [Google Scholar]
- 50.Uematsu, N., Weterings, E., Yano, K., Morotomi-Yano, K., Jakob, B., Taucher-Scholz, G., Mari, P. O., van Gent, D. C., Chen, B. P., and Chen, D. J. (2007) J. Cell Biol. 177 219-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wechsler, T., Chen, B. P., Harper, R., Morotomi-Yano, K., Huang, B. C., Meek, K., Cleaver, J. E., Chen, D. J., and Wabl, M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 1247-1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chu, G. (1997) J. Biol. Chem. 272 24097-24100 [DOI] [PubMed] [Google Scholar]
- 53.Hosoi, Y., Watanabe, T., Nakagawa, K., Matsumoto, Y., Enomoto, A., Morita, A., Nagawa, H., and Suzuki, N. (2004) Int. J. Oncol. 25 461-468 [PubMed] [Google Scholar]
- 54.Liu, L. X., Jiang, H. C., Liu, Z. H., Zhu, A. L., Zhou, J., Zhang, W. H., Wang, X. Q., and Wu, M. (2003) Hepatogastroenterology 50 1496-1501 [PubMed] [Google Scholar]
- 55.Um, J. H., Kwon, J. K., Kang, C. D., Kim, M. J., Ju, D. S., Bae, J. H., Kim, D. W., Chung, B. S., and Kim, S. H. (2004) J. Pharmacol. Exp. Ther. 311 1062-1070 [DOI] [PubMed] [Google Scholar]
- 56.Rao, V. A., Conti, C., Guirouilh-Barbat, J., Nakamura, A., Miao, Z. H., Davies, S. L., Sacca, B., Hickson, I. D., Bensimon, A., and Pommier, Y. (2007) Mol. Cancer Res. 5 713-724 [DOI] [PubMed] [Google Scholar]
- 57.Rouse, J., and Jackson, S. P. (2002) Science 297 547-551 [DOI] [PubMed] [Google Scholar]
- 58.Shiloh, Y. (2003) Nat. Rev. Cancer 3 155-168 [DOI] [PubMed] [Google Scholar]
- 59.Bartkova, J., Horejsi, Z., Koed, K., Kramer, A., Tort, F., Zieger, K., Guldberg, P., Sehested, M., Nesland, J. M., Lukas, C., Orntoft, T., Lukas, J., and Bartek, J. (2005) Nature 434 864-870 [DOI] [PubMed] [Google Scholar]
- 60.Lees-Miller, S. P., Sakaguchi, K., Ullrich, S. J., Appella, E., and Anderson, C. W. (1992) Mol. Cell. Biol. 12 5041-5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pise-Masison, C. A., Radonovich, M., Sakaguchi, K., Appella, E., and Brady, J. N. (1998) J. Virol. 72 6348-6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pise-Masison, C. A., Mahieux, R., Jiang, H., Ashcroft, M., Radonovich, M., Duvall, J., Guillerm, C., and Brady, J. N. (2000) Mol. Cell. Biol. 20 3377-3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van, P. L., Yim, K. W., Jin, D. Y., Dapolito, G., Kurimasa, A., and Jeang, K. T. (2001) J. Virol. 75 396-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Durocher, D., and Jackson, S. P. (2001) Curr. Opin. Cell Biol. 13 225-231 [DOI] [PubMed] [Google Scholar]
- 65.Bailey, S. M., and Murnane, J. P. (2006) Nucleic Acids Res. 34 2408-2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Espejel, S., Franco, S., Sgura, A., Gae, D., Bailey, S. M., Taccioli, G. E., and Blasco, M. A. (2002) EMBO J. 21 6275-6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gilley, D., Tanaka, H., Hande, M. P., Kurimasa, A., Li, G. C., Oshimura, M., and Chen, D. J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 15084-15088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goytisolo, F. A., Samper, E., Edmonson, S., Taccioli, G. E., and Blasco, M. A. (2001) Mol. Cell. Biol. 21 3642-3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Itoyama, T., Chaganti, R. S., Yamada, Y., Tsukasaki, K., Atogami, S., Nakamura, H., Tomonaga, M., Ohshima, K., Kikuchi, M., and Sadamori, N. (2001) Blood 97 3612-3620 [DOI] [PubMed] [Google Scholar]
- 70.Derse, D., Heidecker, G., Mitchell, M., Hill, S., Lloyd, P., and Princler, G. (2004) Front. Biosci. 9 2495-2499 [DOI] [PubMed] [Google Scholar]
- 71.Furukawa, Y., Fujisawa, J., Osame, M., Toita, M., Sonoda, S., Kubota, R., Ijichi, S., and Yoshida, M. (1992) Blood 80 1012-1016 [PubMed] [Google Scholar]
- 72.Mortreux, F., Gabet, A. S., and Wattel, E. (2003) Leukemia (Basingstoke) 17 26-38 [DOI] [PubMed] [Google Scholar]
- 73.Khanna, K. K., and Jackson, S. P. (2001) Nat. Genet. 27 247-254 [DOI] [PubMed] [Google Scholar]
- 74.Mills, K. D., Ferguson, D. O., and Alt, F. W. (2003) Immunol. Rev. 194 77-95 [DOI] [PubMed] [Google Scholar]
- 75.Custer, R. P., Bosma, G. C., and Bosma, M. J. (1985) Am. J. Pathol. 120 464-477 [PMC free article] [PubMed] [Google Scholar]
- 76.Jhappan, C., Morse, H. C., III, Fleischmann, R. D., Gottesman, M. M., and Merlino, G. (1997) Nat. Genet. 17 483-486 [DOI] [PubMed] [Google Scholar]
- 77.Li, G. C., Ouyang, H., Li, X., Nagasawa, H., Little, J. B., Chen, D. J., Ling, C. C., Fuks, Z., and Cordon-Cardo, C. (1998) Mol. Cell 2 1-8 [DOI] [PubMed] [Google Scholar]
- 78.Saitou, M., Sadamori, N., and Isobe, M. (2001) J. Hum. Genet. 46 706-711 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.