Abstract
Poly(ADP-ribosyl)ation of transcription factors and coregulators, mediated by the poly(ADP-ribose) polymerase PARP-1, has been emerging as an important epigenetic mechanism that controls transcriptional dynamics in response to diverse intra- and extracellular signals. PARP-1 activity is also implicated in the regulation of mammalian lifespan. Herein we show that transcriptional down-regulation of androgen receptor (AR) in the aging rat liver and in oxidatively stressed hepatoma cells involves exchange of a PARP-1-associated, p/CAF-containing coactivator assembly for a p53-interacting, Groucho/TLE1-, and mSin3A-included corepressor complex at an age- and oxidant-responsive DNA element (age-dependent factor (ADF) element) in the AR promoter. The coregulator switch is mediated by B-Myb and c-Myb, which bind to the ADF element and physically associate with PARP-1 and the tumor suppressor p53. Heterogeneous nuclear ribonucleoprotein K, residing at the ADF element in association with PARP-1, may serve a platform role in stabilizing the activating complex. PARP-1 coactivated B-Myb- and c-Myb-mediated transactivation of the AR promoter, and p53 antagonized the B-Myb/c-Myb-induced AR promoter activation. PARP-1, heterogeneous nuclear ribonucleoprotein K, B-Myb, and c-Myb each serves as a positive regulator of cellular AR content, whereas p53 negatively regulates AR expression. Our results identify a shared, PARP-1-regulated sensing mechanism that coordinates transcriptional repression of AR during aging and in response to oxidative stress. This study may provide insights as to how advancing age and intracellular redox balance might influence androgen-regulated physiology.
Diverse physiology, involving both reproductive and non-reproductive processes, is regulated by the androgen receptor (AR),4 which is an inducible transcription factor and the transmitter of androgen signals to the nucleus. In the liver, AR influences a wide range of metabolic activities, especially those linked to glucose and lipid homeostasis, as evident from the deregulated liver metabolism in mice that have hepatocyte-specific AR deficiency (1), and those linked to steroid, drug, and nutrient metabolism, as evident from the AR/androgen-dependent regulation of hepatic phase I and phase II enzymes (2–4). A role for AR in liver carcinogenesis was initially recognized from the finding that testicular feminized (Tfm) mice, which lack functional AR, are resistant to liver cancer from carcinogen exposure (5). The male prevalence of liver cancer in humans (6) is attributed in part to the hepatic AR, which has been detected in clinical hepatocellular carcinoma at both initial and advanced stages of the disease (7). Increased AR expression from its transcriptional up-regulation occurs frequently in human prostate carcinoma (8). Therefore, it is important to delineate the regulatory factors that contribute to altered AR levels in response to a changing milieu of various AR-expressing tissues including the liver.
In the rat liver, reduced AR expression during aging, reaching a non-detectable level at late life, is transcriptionally coordinated (9, 10). Dietary calorie restriction, which retards age-related diseases and extends the invertebrate and vertebrate lifespan, also reverses loss of AR expression and restores androgen sensitivity of the aging liver (9, 11). In earlier studies, we had identified positive and negative changes in specific transcription regulatory activities that are linked to the loss of hepatic AR in old rats (10, 12). For example, NF-κB activity in the liver and in other tissues is known to rise with advancing age as a result of increased oxidative stress (12, 13), and AR gene transcription is negatively regulated by NF-κB (12, 14). Conversely the activity of a nuclear factor, which stimulates the promoter function of AR, declines gradually in the liver of aging rodents. This age-dependent factor or ADF (as per our designation) avidly binds to a 20-bp DNA element at around the -330 promoter/enhancer position in the rat AR gene. Inactivating point mutations within the 20-bp element abolished ADF binding to the cognate site (ADF element) and reduced AR promoter activity in transfected cells (10, 15, 16). ADF activity was also detected in non-hepatic cells, such as those from the rat and human prostate (PAIII and LNCaP, respectively), monkey kidney (COS-1), and human uterine cervix (HeLa). We have sought to characterize the molecular identity of ADF and delineate the coregulatory components that link reduced ADF activity with loss of AR in vivo.
Coregulators, which do not possess direct DNA binding activity, associate with DNA-bound transcription factors and serve as conduits in the relay of signals from target gene response elements to RNA polymerase II via the general transcription-promoting machinery. The nuclear enzyme PARP-1 (poly(ADP-ribose) polymerase-1) interacts with and coactivates many DNA-binding transcription factors such as the redox-sensitive and inflammation-responsive AP-1 and NF-κB and the protooncoprotein B-Myb, which regulates genes involved in cell cycle progression (17, 18). PARP-1 interacts with the mediator complex and thus may serve as a platform protein (19). Furthermore induced PARP-1 activity acts as a sensor in a promoter-specific corepressor to coactivator switch at a regulatory element (20). PARP-1 is also involved in other vital nuclear processes related to DNA repair, DNA replication/recombination, chromatin remodeling, genome stability, and cell death. Oxidative stress and other DNA damage signals superactivate PARP-1, which catalyzes the sequential transfer of ADP-ribose units from β-nicotinamide adenine dinucleotide (β-NAD+) to various nuclear proteins including itself (auto-modification) to cause attachment of polymeric ADP-ribosyl chains onto the acceptors. Proteins modified by ADP-ribosyl polymers are functionally altered. For example, poly(ADP-ribosyl)ation of the tumor suppressor protein p53 prevents its interaction with the nuclear export system, causing its nuclear accumulation, which facilitates target gene activation by p53 (21). PARP-1 activity correlates positively with the lifespan of various mammalian species (22), and lymphoblastoid cell lines derived from centenarians are enriched for PARP-1 activity (23). These results have advanced the notion that PARP-1 is important in mammalian aging.
The present study reveals the identity of the protein factors associated with the ADF element, demonstrates that oxidative stress can reduce AR mRNAs similar to that observed in aging, and identifies a shared regulatory strategy involving the Myb transcription factor-mediated exchange of PARP-1 for p53 at the ADF element, which coordinates transcriptional repression of AR in vivo at old age and in cells at a prooxidant state. PARP-1 is an essential component of the B-Myb/c-Myb-associated activating complex that governs AR gene stimulation. The multifunctional heterogeneous nuclear ribonucleoprotein K (hnRNPK) interacts with PARP-1 in vivo and may serve as a stabilizing docking platform for the activating complex. Exchange of PARP-1 and hnRNPK for p53 and p53-associated Groucho/TLE1 and mSin3A corepressors at regulated chromatin correlated with AR down-regulation in vivo and in cells challenged by oxidative stress. p53 interfered with B-Myb/c-Myb-mediated transactivation, and Myb/PARP-1 interaction was inhibited in the aging liver and in cells shifted to a prooxidant state. These results suggest that PARP-1 may provide for a sensing mechanism that triggers a coregulator exchange. AR levels were reduced in cells silenced for B-Myb, c-Myb, PARP-1, or hnRNPK and elevated after p53 knockdown. We conclude that reciprocal chromatin dynamics of p53 and PARP-1 at the ADF element, mediated by the Myb transcription factors, is integral to the regulatory network that orchestrates the loss of AR in aging and in response to oxidative stress.
EXPERIMENTAL PROCEDURES
Animal Care—Fisher 344 rats, young and old, were purchased from the National Institute of Aging and given rest for 1 week before sacrificing for tissue harvesting. Animal care and treatment protocols were approved by the institutional animal care committee.
Enrichment of ADF Activity and Mass Spectrometry—HeLa cells (from ATCC) were processed to prepare nuclear extracts as described previously (24). The extracts were dialyzed against 150 mm NaCl in buffer-1 (20 mm HEPES, pH 7.9, 0.2 mm EDTA, 2 mm EGTA, 2 mm dithiothreitol, 25% glycerol, 0.5 mm phenylmethylsulfonyl fluoride), and taken through a four-step enrichment protocol involving 1) DNA affinity chromatography on ADF element-coupled Sepharose 4B gel (Amersham Biosciences), 2) anion-exchange chromatography on Q-Sepharose CL-4B gel (Amersham Biosciences), 3) cation-exchange chromatography on a Mono S column (Amersham Biosciences), and 4) reverse DNA affinity chromatography on mutant ADF element-coupled Sepharose 4B gel. The mutant ADF element has three point mutations that cause complete loss of ADF activity. The sequences for the wild-type ADF element and its three-point mutant are provided under “Co-IP, Western Blotting, EMSA, and DNase I Footprinting.” Three tandem copies of the duplex ADF element or its three-point mutant were coupled to CNBr-activated Sepharose 4B (Amersham Biosciences) using standard, vendor-recommended conditions.
The DNA affinity column was eluted with dialysis buffer containing step gradients of NaCl solution. The fractions from the 0.3 m NaCl eluate showing ADF activity were loaded onto a Q-Sepharose column, and pooled eluates from a linear salt gradient (0.3–0.4 m NaCl) showing peak ADF binding activity (assayed by EMSA) were passed through a Mono S cation-exchange column. After elution with a linear salt gradient (0.15–0.25 m NaCl), the pooled eluates possessing peak ADF activity were passed through mutant ADF element-coupled Sepharose 4B gel. Flow-through proteins containing unretained ADF activity were resolved by SDS-PAGE, and silver-stained protein bands were cut out of the gel for analysis by MALDI-TOF mass spectrometry to determine the identity of the ADF activity-copurified proteins. Mass spectrometry was performed at the institutional core facility.
Chromatin Immunoprecipitation (ChIP)—ChIP on liver tissues was performed as described by us previously (25). Briefly nuclei were prepared from formaldehyde-fixed tissue pieces, and the supernatant from lysed nuclei was sonicated (25) and analyzed on an agarose gel to ensure a <1-kbp size range of the fragmented chromatin DNAs. Soluble chromatin fragments in the supernatant were diluted and used for immunoprecipitation as described previously (14). H4IIE cells (seeded at 107cells/10-cm dish) were grown to 80% confluency and incubated in medium (vehicle) or tert-butyl hydroperoxide (TBH) for 30 or 60 min before fixing with formaldehyde. The purchased antibodies (Santa Cruz Biotechnology Inc.) used for ChIP analyses were: AR (sc-816), B-Myb (sc-724), c-Myb (sc-7874), hnRNPK (sc-16554), PARP-1 (sc-7150), p53 (sc-1311), TLE1 (sc-9121), mSin3A (sc-994), and p/CAF (sc-8999). Immunoprecipitated chromatin was washed with low salt, high salt, and LiCl solutions and finally with Tris-EDTA buffer. Complexes pulled down by protein A-Sepharose were eluted by the elution buffer and incubated with NaCl solution (at 200 mm final concentration) at 65 °C for at least 4 h to reverse protein-DNA cross-linking. 10 μl of the eluate (of 100 μl total) was used for PCR after DNA extraction. The remainder of the eluate was used for sequential ChIP as described previously (14).
PCR was performed on various DNA samples at equal inputs using Taq polymerase (Invitrogen) using the following primers to probe the ADF site in the rat AR genome: sense at -500, 5′-GACTCTCCCTTCTGCTTGTC; antisense at -300, 5′-GGAACCGGGCTTTAGACTC. For unrelated, rat AR intragenic region, the following primers were used: sense at +2241, 5′-TTCACTAATGTCAACTCCAGGA; antisense at +2688, 5′-CTTGCACAGAGA-TGATCTCTG.
Activation of PARP-1 Enzymatic Activity in Vitro—Rat liver nuclear extract (RLNE) was incubated with or without 500 μm β-NAD+ for 1 h at 37 °C using a buffer (10 mm Tris-HCl, pH 8, 1 mm MgCl2, 1 mm dithiothreitol) as described previously (26). Incubated nuclear extracts were used for co-immunoprecipitation (co-IP).
Cells, siRNAs, and mRNA Quantification—Rat hepatoma (H4IIE), human hepatoma (HepG2), and human embryonic kidney (293A) cells were cultured in Dulbecco's modified Eagle's medium. Human prostate carcinoma (LNCaP) cells were cultured in RPMI 1640 medium. Media contained 10% fetal bovine serum, 100 μg/5 ml penicillin, 100 units/5 ml streptomycin. Cell transfections were performed in 24-well flasks.
RNA interference oligos (Dharmacon) used were: human B-Myb (SMARTpool; catalog number M-010444-00-0005), c-Myb (SMARTpool; catalog number M-003910-00-0005), PARP-1 (SMARTpool; catalog number L-006656-00-0010), hnRNPK (SMARTpool; catalog number L-011692-00-0010), p53 (SMARTpool; catalog number L-003329-00-0010), and non-targeting, scrambled siRNAs (SMARTpool; catalog number D-001810-01-20). siRNAs (at 100 nm) were transfected into cells (6-well flasks) using Oligofectamine (Invitrogen). At 72 h post-RNA interference transfection, cell lysates were analyzed by Western blotting. Sequences for the RNA interference oligos (purchased from Dharmacon) used in this study are included in the supplemental material. Total cellular RNAs (isolated by TRIzol™) were assayed for AR mRNAs by reverse transcription qPCR using the SYBR Green PCR reagent kit (Bio-Rad). Primers to probe AR mRNAs (14) are described in the supplemental material.
Co-IP, Western Blotting, EMSA, and DNase I Footprinting—Co-IP was performed under standard conditions. Western blotting used the same antibodies as those used in ChIP.
Oligo DNA sequences used in EMSA were: ADF-AR, 5′-AGGCTTGTCTGTTAAAAAA (sense); and ADF-mut (three-point mutant), 5′-AGGCTCGTCTCTTAGAAAA (sense) (point mutations are at underlined bases). For duplex probe preparation, the sense strand was labeled at the 5′-end by T4 kinase and [γ-32P]dATP and then annealed to a 5-fold excess of cold antisense strand. The duplex CT-rich hnRNPK-binding element in the rat osteocalcin promoter used in the competition EMSA corresponds to 5′-CAGCATCTCCTGCCCCTCCTGCT (sense). The EMSA complex was competed out by the cold, homologous ADF sequence; by Myb/Stat6, which is the Myb-binding element in the Stat6 promoter; and by ADF/retinoic acid receptor, which is the ADF/Myb-like sequence that was identified by Basic Local Alignment Search Tool (BLAST) search on chromosome 3 contig within the human β-retinoic acid receptor gene (GenBank™ accession number NT_022517.17; location base, 25409343–25409317). DNase I footprinting was done as before (14).
Plasmids and GST-fused Proteins—Full-lengh B-Myb cDNA was prepared from HeLa cell total RNAs by real time PCR and was sequence-confirmed. Truncated B-Myb-(1–561) lacking the C-terminal inhibitory domain and B-Myb-(1–350) lacking the activation domain and C-terminal inhibitory domain were constructed from the full-length B-Myb cDNA by PCR. cDNAs were cloned into pCDNA 3.1 or into pGEX4T2. Three copies of the ADF element in tandem as an oligoduplex and three tandem copies of ADF three-point mutant oligoduplex were cloned in the basal TK-Luc vector. Sequences for the ADF element (-311 to -330) and its three-point mutant are shown above. GST fusions of B-Myb-(1–561), c-Myb (full length), and hnRNPK (full length) expressed in the Escherichia coli strain BL21RP were purified on a glutathione-Sepharose 4B column. Recombinant PARP-1 was purchased (Trevigen).
RESULTS
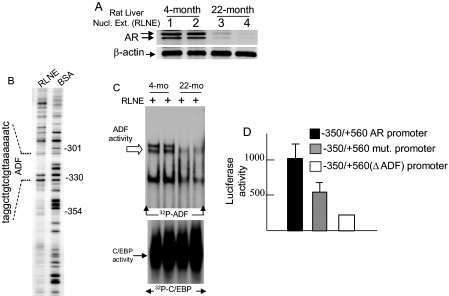
Loss of AR and Reduced ADF Activity in the Aging Rat Liver—The age-regulated loss of AR mRNAs in the rat liver reported earlier (9, 10) reflects a loss of AR protein levels as shown here (Fig. 1A). The closely migrating bands at ∼110 kDa correspond to AR in phosphorylated and unphosphorylated forms (27). Among multiple DNase I-protected footprints in the rat AR promoter produced by liver nuclear extracts (Fig. 1B), the footprint from positions -330 to -311 is relevant to the current study. The activity of the liver nuclear proteins that bind to the -330 to -311 element (ADF element) was markedly reduced in 22-month-old rats compared with the 4-month-old rats (Fig. 1C) similar to earlier findings (10). This ADF stimulates AR promoter function (Fig. 1D). The current study was undertaken to determine the identity of ADF and ADF-associated proteins and their roles in AR gene expression.
FIGURE 1.
Loss of AR and ADF activity in the liver nuclei of old rats. A, Western blot of AR. B, DNase I footprinting of the rat AR promoter by RLNE. C, ADF activity in RLNE revealed by EMSA. The control EMSA used an end-radiolabeled 25-bp consensus sequence for CCAAT/enhancer-binding protein (C/EBP) binding. D, activity of wild-type and mutant rat AR promoters in transfected HepG2 cells. The mutant AR promoter has a deleted ADF element (ΔADF) or three point mutations within the ADF element. Luciferase activities were averages (±S.D.) of three independent duplicate experiments. Numbers in A refer to individual animals of a given age: 1 and 2, each 4 months old; 3 and 4, each 22 months old. Nucl. Ext., nuclear extract; BSA, bovine serum albumin; mo, month; mut., mutant.
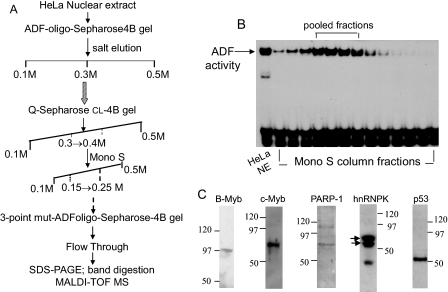
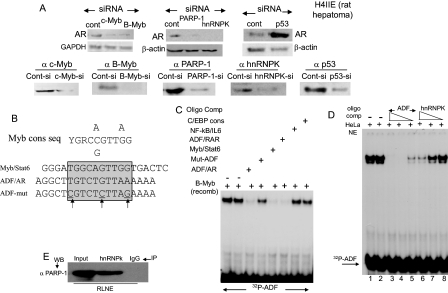
Proteomics Identification of ADF-interacting Nuclear Proteins—Nuclear proteins, which associated directly or indirectly with the ADF element, were enriched within HeLa cell nuclear extracts using a four-step enrichment scheme (Fig. 2A). The first step was affinity chromatography with a Sepharose 4B gel slurry that was coupled to a concatamer of the ADF element (-330 to -311). The pooled eluates at 0.3 m NaCl possessing ADF activity (assessed by EMSA) were processed by anion-exchange chromatography on a Q-Sepharose CL-4B column and then by cation-exchange chromatography on a Mono S column. The salt gradient-eluted protein fractions were examined for ADF activity (shown here for the Mono S column eluates (Fig. 2B)). The final step involved reverse affinity chromatography with Sepharose 4B gel coupled to a mutant ADF oligo (three point mutations), which does not bind to the ADF factor. Unbound proteins collected in the flow-through were separated by SDS-PAGE and visualized by silver staining. EMSA of the pooled fractions before and after reverse affinity chromatography revealed that ADF activity was not retained by the ADF mutant column. Several protein bands (∼116–35 kDa) were associated with ADF activity in the flow-through (data not shown), indicating that these proteins may be closely linked to ADF activity.
FIGURE 2.
Enrichment of ADF activity from HeLa nuclear extracts and proteomics identification of ADF-copurified proteins. A, four-step enrichment scheme. B, ADF activity of Mono S column fractions assayed by EMSA. C, Western blot confirmation of the identity of the proteins that copurified with the ADF activity and were characterized by MALDI-TOF mass spectrometry (MS). The closely migrating hnRNPK bands (noted by arrows) represent post-translationally modified isoforms of hnRNPK. Apparent molecular masses of these isoforms (∼65 kDa) are in agreement with the reported molecular weight of intracellular hnRNPK (44). The third band may be due to a proteolytic fragment of hnRNPK or an unrelated protein. NE, nuclear extract.
MALDI mass spectrometry revealed that PARP-1 (∼113 kDa), B-Myb (∼97 kDa), c-Myb (∼75 kDa), hnRNPK (∼65 kDa), and p53 (∼53 kDa) are the major proteins that copurified with ADF activity. Additional ADF-associated proteins, which stained as exceedingly faint bands, were of uncertain identity because of low matching scores; they await further exploration.
Western blotting further showed the presence of PARP-1, hnRNPK, the Myb transcription factors B-Myb and c-Myb, and p53 in the flow-through (Fig. 2C). hnRNPK was detected as two closely migrating post-translationally modified bands at ∼65 kDa (noted by arrows) representing two hnRNPK isoforms. Predicted molecular masses of these isoforms are 50,976 kDa (isoform-1, 463 amino acids) and 51,028 kDa (isoform-2, 464 amino acids). The additional, faster migrating band arose either from a proteolytic fragment of hnRNPK or from an unrelated protein that cross-reacted with the hnRNPK antibody. The identity of these ADF-associated proteins, validated from Western blot results, enabled us to explore how PARP-1, hnRNPK, p53, and the Myb proteins might be linked to ADF activity and to regulation of the AR gene in vivo.
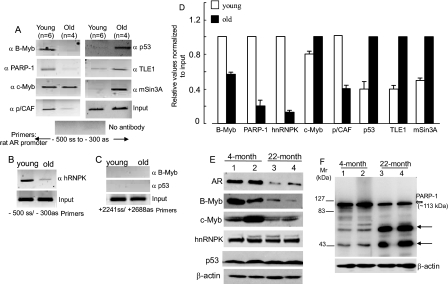
Chromatin Dynamics of ADF-copurified Proteins in Young and Old Rats—We examined whether PARP-1, hnRNPK, B-Myb, c-Myb, and p53 are associated with the ADF element in vivo and whether they are differentially engaged at regulated liver chromatin in young versus old rats. ChIP revealed that B-Myb, PARP-1, and hnRNPK all associated with the ADF element with higher abundance in the samples from the young rats compared with those from the old; the reverse was true for p53 (Fig. 3, A, B, and D). c-Myb association with the ADF site was similar for the young and old rats. Lack of signal in the absence of a primary antibody (Fig. 3A) and lack of chromatin IP from a non-related intragenic region of the rat AR by antibodies to B-Myb and p53 (Fig. 3C) ensured specificity of the ChIP assay. Pooled livers were used (Fig. 3, A and D) to control for animal-to-animal variations. Nevertheless individual liver samples also showed similar age-regulated differential recruitment of the above factors (shown here for hnRNPK (Fig. 3B)).
FIGURE 3.
Factor recruitment to the ADF site in liver chromatin (analyzed by ChIP) and nuclear levels of these factors (analyzed by Western blotting) in young and old rats. A, pooled livers from each age group. n = number of animals per pool. B, livers from individual young and old rats. C, negative control using primers amplifying the intragenic +2241 to +2688 bp region in the rat genomic AR. D, real time qPCR of chromatin-immunoprecipitated DNAs. Averages (±S.D.) of three experiments are shown. Input DNAs were at equal amounts. E, Western blot on RLNE (50 μg) of the young and old rats. F, Western blotting of RLNE for PARP-1. Numbers in E and F refer to individual animals: 1 and 2, two rats, each 4 months old; 3 and 4, two rats, each 22 months old. ss, sense; as, antisense.
The coactivator p/CAF (a histone acetyltransferase) was enriched at the ADF site in the young but not in old rats (Fig. 3, A and D), indicating that p/CAF is part of the coactivator complex mediating AR gene transcription. The p300 coactivator was absent in this region. The presence of p53 at the ADF site correlated with the loss of AR expression in old animals. Because p53-mediated transcriptional repression can involve histone deacetylase and the mSin3A corepressor (28), we checked for mSin3A occupancy. Indeed mSin3A occupied the ADF site in old animals but not in the young (Fig. 3, A and D). HDAC-1/HDAC-2-containing mSin3A corepressor complexes are present in mammalian cells, and p53 has been detected in association with mSin3A and HDAC-1 (29). Neither HDAC-1 nor HDAC-2, however, was present at the ADF site (data not shown). Whether another HDAC family member(s), as part of the mSin3A corepressor complex, would occupy the ADF site in the liver chromatin of old rats awaits future experiments.
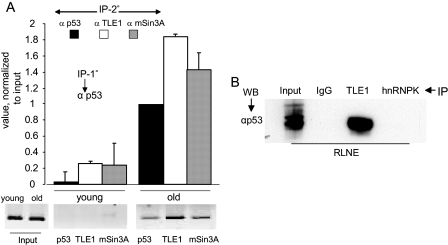
We examined whether the Groucho/TLE corepressor would play a role in p53-mediated negative AR regulation. TLE proteins mediate gene repression by interacting with DNA-binding transcription factors through a WD domain (30) without interacting with DNA (31). We reasoned that p53 may heterodimerize with TLE because bioinformatics suggests that the conserved WD domain of TLE is a potential interactor with a 13-amino acid amphipathic α-helical transactivation domain of p53 (32). Indeed TLE1 was detected at the ADF site in the liver chromatin of old rats (Fig. 3, A and D). Association of p53 with TLE1 in liver nuclear extract as a co-immunoprecipitable complex was also observed (see Fig. 9B). TLE1 and mSin3A, the interacting partners of p53, are thus part of the p53-associated corepressor complex at the ADF element.
FIGURE 9.
p53 dependence of TLE1 and mSin3A occupancy at the ADF element in the liver chromatin and p53/TLE1 interaction in liver nuclear extract. A, sequential ChIP using anti-p53 antibody for the first step ChIP and antibodies to p53, TLE1, and mSin3A for the second step ChIP. Analysis was performed on individual liver samples from two young (4-month-old) rats and two old (22-month-old) rats. The bar graphs represent average values (±S.D.) from q-PCR results (conducted in duplicate) derived from liver samples of two individual animals from each age group. Representative semiquantitative PCR results for one young and one old animal are shown below. Signals from the liver chromatin of young animals were at the background level (or non-existent) when chromatin fragments were immunoprecipitated with anti-p53 antibody at the first step ChIP. B, co-IP of p53 and TLE1 from RLNE. WB, Western blot.
Reciprocal chromatin engagement of Myb/PARP-1/hnRNPK and p53/mSin3A/TLE1 at the ADF site prompted us to compare the steady state levels of these proteins in the liver nuclear extracts from young versus old rats. The age-dependent precipitous decline in AR expression was associated with markedly lower B-Myb levels in 22-month-old animals. In contrast, hnRNPK and p53 levels were similar in the young and old (Fig. 3E). The c-Myb level in a 4-month-old (animal-1) did not differ significantly from that in the 22-month-old rats (animal-3 and animal-4), whereas young animal-2 showed higher c-Myb levels (Fig. 3E) possibly because of interanimal variations in the liver expression of c-Myb.
PARP-1 at ∼113 kDa was detected at a somewhat reduced abundance in the old animals (Fig. 3F). Importantly the higher mobility protein bands (two arrows) are at much increased levels in the 22-month-old rats. PARP-1 undergoes caspase-dependent cleavage to yield 89- and 24-kDa fragments (33), and the 24-kDa fragment is known to exert a dominant negative effect on PARP-1 function, including its role in gene transcription. However, we rule out the possibility that the additional bands in the old rats originated from known caspase activity because their electrophoretic migration was faster than 83-kDa and slower than 43-kDa positions. The DNA-binding domain of PARP-1, which is a known dominant negative inhibitor of PARP-1 activity, has a predicted molecular mass of >100 kDa, and therefore, this DNA-binding domain fragment cannot represent either of the higher mobility protein bands. Significance of these bands in old liver nuclei awaits further exploration.
Role of ADF-copurified Proteins in Cellular AR Levels and Identity of ADF Element-binding Proteins—Individual depletion of PARP-1, B-Myb, c-Myb, or hnRNPK in H4IIE rat hepatoma cells using siRNAs reduced endogenous AR levels (Fig. 4A), suggesting positive roles for these proteins in AR expression. In contrast, p53 is a negative regulator of AR as evident from increased AR content in p53-silenced H4IIE cells. Efficacy of the siRNA-mediated specific silencing of these proteins is shown (Fig. 4A). Control, non-targeting siRNAs (a pool of four scrambled sequences, purchased from Dharmacon) did not cause any change in target protein levels (B-Myb, c-Myb, hnRNPK, PARP-1, and p53) and in the level of AR in H4IIE cells. AR levels in LNCaP cells (a human prostate carcinoma cell line) also decreased when B-Myb, c-Myb, and hnRNPK were silenced (data not shown). Elevated AR expression in p53-silenced LNCaP cells has been reported (34).
FIGURE 4.
AR levels regulated by B-Myb/c-Myb/PARP-1/hnRNPK/p53 and identity of the factor(s) directly binding to the ADF element. A, a pool of specifically targeting siRNAs (at 100 nm final concentration) was transfected into H4IIE cells using Oligo fectamine. Sequences foreach pool are provided in the supplemental material. Cell lysates (50 μg of total protein/lane) were analyzed by Western blotting. Control siRNAs are a non-targeting, scrambled siRNA pool (Dharmacon) used to ensure that experimental conditions did not trigger off-target knockdown. B, consensus binding sequence for the Myb family protooncoproteins. C, EMSA produced by GST-B-Myb and radiolabeled ADF element (-311 to -330). Competition EMSA used various cold oligoduplexes. D, competition for the EMSA complex between radiolabeled ADF element and HeLa nuclear extracts. Competition was with cold ADF sequence (rat AR promoter) at 200-, 50-, and 10-fold molar excess (lanes 3–5, respectively) or with cold duplex CT element (rat osteocalcin promoter (53)) at 200-, 50-, and 10-fold molar excess (lanes 6–8, respectively). E, co-IP of PARP-1 and hnRNPK from RLNE. cons, consensus; seq, sequence; C/EBP, CCAAT/enhancer-binding protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; si, siRNA; mut, mutant; WB, Western blot; NE, nuclear extract; comp, competition; recomb, recombinant; Cont, control; RAR, retinoic acid receptor.
To address which of the above proteins would directly bind to the ADF sequence, we searched the transcription factor data base (TRANSFAC) for sequence homology to the ADF element and found that the consensus binding sequence for the Myb family transcription factors is highly homologous to the ADF-binding sequence in the rat AR promoter (Fig. 4B). Recombinant B-Myb specifically bound to the ADF element in EMSA (Fig. 4C) because the EMSA complex was competed off by the cold homologous oligoduplex, by the Myb element in the Stat6 promoter (35), and by the ADF-like element in the human retinoic acid receptor-β gene, but no competition was observed with the mutant ADF oligo (three point mutations), with the NF-κB element in the IL6 promoter, and with the consensus sequence cognate to CCAAT/enhancer-binding protein. Recombinant c-Myb also bound to the ADF element (data not shown).
hnRNPK, which copurified with ADF activity, interacted with the ADF sequence as well albeit much more weakly than B-Myb/c-Myb (at least 20-fold weaker) as demonstrated by a competition EMSA (Fig. 4D, compare lanes 3–5 with lanes 6–8). hnRNPK/PARP-1 interaction within rat liver nuclear extract was also observed (Fig. 4E). Possible implication for weak hnRNPK binding to the ADF element will be considered under the “Discussion.”
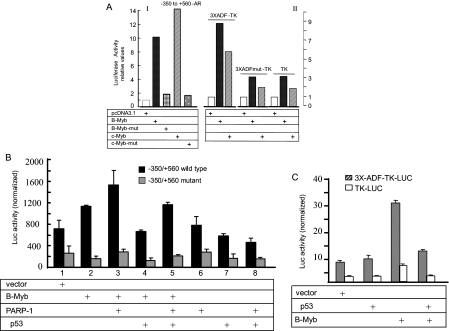
B-Myb and c-Myb activated the natural AR promoter (Fig. 5A, I) and induced the ADF element on the TK promoter (Fig. 5A, II) in transfected cells. A truncated B-Myb (amino acids 1–350, lacking an activation domain) or a frameshift mutant of the c-Myb plasmid (abrogating c-Myb protein production (36)) reduced promoter activity, indicating specific promoter activation (Fig. 5A, I).
FIGURE 5.
B-Myb/c-Myb-mediated transactivation of the AR promoter and functional interaction of Myb with PARP-1 and p53. A, stimulation of the AR promoter by B-Myb and c-Myb in transfected HepG2 cells. Luciferase expression was driven by the natural rat AR promoter (I) and by the ADF element (three copies in tandem) linked to the downstream TK promoter (II). The average of two independent transfections, each induplicate, is shown for each bar graph. B, PARP-1 coactivated and p53 repressed B-Myb-mediated transactivation of the natural rat AR promoter activity in 293A cells. Point mutations within the ADF sequence reduced the basal AR promoter activity and obliterated both promoter induction by B-Myb and coactivation of B-Myb by PARP-1. C, p53-mediated repression of the ADF element activity on the heterologous TK promoter in HepG2 cells. Averages (±S.D.) of three normalized luciferase activities are shown for B and C. Error bars are standard errors, calculated from standard deviation and number of replicate measures. c-Myb activity was also coactivated by PARP-1 and repressed by p53 (data not presented). Luciferase (Luc) activities were normalized to constant protein amounts. mut, mutant.
AR promoter transactivation by B-Myb was further enhanced by PARP-1 (Fig. 5B), confirming earlier reports that PARP-1 is a coactivator of B-Myb (37). p53 antagonized B-Myb-mediated transactivation of the AR promoter (Fig. 5B) as well as transactivation of the ADF element in a heterologous TK promoter context (Fig. 5C). c-Myb-induced transactivation of the AR promoter was also antagonized by p53 (data not shown). Antagonism occurred in the absence of p53 binding to DNA because no EMSA complex of p53 with the ADF element was produced.
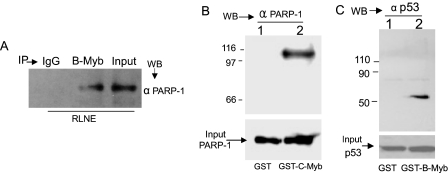
PARP-1-coactivated B-Myb and c-Myb functions involved physical association of these proteins (Fig. 6, A and B). Thus, B-Myb and PARP-1 were present in the same complex within rat liver nuclear extract as shown by co-IP (Fig. 6A and see Fig. 8A). PARP-1 also co-immunoprecipitated with c-Myb from cell lysates (supplemental Fig. S-A), indicating that, similar to B-Myb, c-Myb physically associated with PARP-1 in vivo. Direct association of c-Myb with PARP-1 is evident from GST pulldown (Fig. 6B) because recombinant PARP-1 was retained by GST-c-Myb but not GST alone (Fig. 6B). Direct p53 association with B-Myb is also shown (Fig. 6C). Additionally physical interaction of p53 with B-Myb and c-Myb is evidenced by the result that both Myb proteins co-immunoprecipitated p53 from rat liver nuclear extract (see below, Fig. 8B). p53 co-immunoprecipitated with c-Myb from cell lysates as well (supplemental Fig. S-B), further indicating interaction between p53 and the Myb proteins in vivo.
FIGURE 6.
Association of B-Myb/c-Myb with PARP-1 and p53. A, PARP-1/B-Myb interaction in RLNE demonstrated by co-IP. B, pulldown of recombinant PARP-1 by GST-c-Myb, which was immobilized onto glutathione-Sepharose 4B gel. PARP-1 was not retained by immobilized GST that was not fused to c-Myb. C, GST pulldown of recombinant p53 (produced using a rabbit reticulocyte based in vitro transcription/translation system) by glutathione-Sepharose 4B gel-immobilized GST-B-Myb. WB, Western blot.
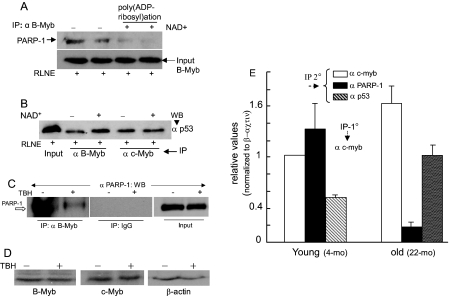
FIGURE 8.
Effects of β-NAD+, oxidative stress, and aging on Myb interaction with PARP-1 and p53. RLNE was incubated at 37 °C with β-NAD+ (500 μm)-containing buffer to promote PARP-1-mediated poly(ADP-ribosyl)ation and then immunoprecipitated with B-Myb antibody (A) and B-Myb and c-Myb antibodies (B) followed by Western blotting as shown. Duplicate lanes in A correspond to duplicate experiments. C, B-Myb/PARP-1 interaction in H4IIE cells in the presence or absence of TBH (4 μm). D, Western blot assay of B-Myb, c-Myb, andβ-actin levels in H4IIE cells that were treated with vehicle or TBH. 50 μg of protein was used per lane. E, sequential ChIP (first ChIP with anti-c-Myb) on liver chromatin of young (4 months) and old rats (22 months). Two rats from each group were analyzed separately, and q-PCR was run in triplicate. Combined data from each age is presented as the average ± S.D.
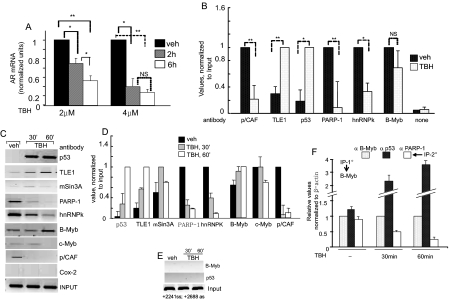
Loss of AR mRNAs in Cells Subjected to Oxidative Stress and Associated Changes in Regulatory Dynamics—Oxidative stress is a hallmark of aging and various age-dependent maladies (38). How changes in the intracellular redox state might impact AR expression is thus an important question. TBH, which is metabolized within cells to produce reactive oxygen species (39), was used to stress H4IIE cells. TBH caused a dose- and time-dependent reduction of AR mRNA levels (Fig. 7A). LNCaP prostate cancer cells showed a similar decline in AR mRNAs after TBH treatment (data not shown). Concomitant with the down-regulation of AR mRNAs, PARP-1, hnRNPK, and the global coactivator p/CAF were depleted from the regulatory site. Repression of AR correlated with recruitment of p53 and the corepressor Groucho/TLE1 to the ADF site (Fig. 7B). This is similar to the reciprocal chromatin dynamics between p53 and PARP-1/hnRNPK observed in old animals (Fig. 3). Unlike aging, however, B-Myb occupancy did not show any significant change in the stressed cells (Fig. 7B). Overall therefore, the ADF element responds to both aging and oxidative stress to regulate AR gene activity.
FIGURE 7.
Oxidative stress regulation of AR mRNA levels and chromatin dynamics of ADF-associated proteins. A, real time qPCR of AR mRNAs in H4IIE cells treated with TBH. Averages (±S.D.) are for three experiments. B, ChIP using H4IIE cells treated with vehicle or TBH (4 μm, 60 min). Primers covering the -500 to -300 bp region (rat AR promoter) were used for PCR. For statistical assessment in A and B,* = p < 0.05 and ** = p < 0.005. C and D, kinetics of factor occupancy at the ADF site in response to TBH treatment of H4IIE cells for 0, 30, and 60 min (′). DNAs from chromatin IP were analyzed by semi-qPCR (C) and real time qPCR (D). E, negative controls. PCR amplified a non-regulated chromatin region in the intragenic AR from +2241 to +2688. F, sequential ChIP; first ChIP with anti-B-Myb and second ChIP with indicated antibodies. C, D, and F used PCR primers at -500 (forward) and -300 (reverse). veh, vehicle; NS, not significant.
The kinetics of loading and unloading of the above factors onto regulated chromatin was compared in cells that had received TBH versus vehicle (Fig. 7, C and D). The enriched presence of p53, TLE1, and mSin3A was observed at the regulated site within 30 min of TBH treatment. Decreased mSin3A levels at 60 min most likely reflect a cyclic pattern of chromatin occupancy (40). Results also show that PARP-1 occupied the ADF site as part of the B-Myb-associated coactivator complex (Fig. 7, C, D, and F) in cells at a normal redox state when AR mRNA levels are high. In response to oxidative stress, PARP-1 dissociated from the ADF site, and hnRNPK and p/CAF were concomitantly lost from this site (Fig. 7, C and D). Oxidative stress did not significantly change chromatin occupancy of B-Myb (Fig. 7, B, C, and D). At 60 min, the c-Myb level at the regulated region decreased slightly. As negative controls, non-related anti-COX-2 antibody did not immunoprecipitate chromatin fragments harboring the ADF element (Fig. 7C), and anti-p53/anti-B-Myb antibodies did not pull down chromatin fragments lacking the ADF-binding site (Fig. 7E).
p53 and B-Myb co-occupied the chromatin fragments that were depleted of PARP-1 in oxidant-stressed cells as revealed from sequential ChIP assay (Fig. 7F). This co-occupancy along with the results that p53 repressed stimulation of AR promoter activity by B-Myb (Fig. 5B) and also by c-Myb (data not shown) suggests that oxidative stress-induced loss of PARP-1 from the ADF site promotes negative functional interaction between p53 and B-Myb.
Impact of Poly(ADP-ribosyl)ation on PARP-1 and p53 Interaction with B-Myb/c-Myb—Because peroxidative DNA damage associated with aging and a prooxidant state of cells is known to robustly induce basal PARP-1 activity, we asked how might Myb interaction with PARP-1 be affected by advanced age or oxidative stress. Protein poly-(ADP-ribosyl)ation by PARP-1 is known to be increased in the presence of excess β-NAD+ in an in vitro assay (26, 41). Incubation of rat liver nuclear extract with 500 μm β-NAD+ and subsequent co-IP assay showed that B-Myb/PARP-1 interaction was reduced (Fig. 8A), whereas p53 interacted more avidly with B-Myb (Fig. 8B). p53/c-Myb interaction did not change under this condition. Most importantly, TBH-induced oxidative stress markedly reduced PARP-1 interaction with B-Myb in H4IIE cells as revealed by co-IP (Fig. 8C). Furthermore TBH-induced oxidative stress did not alter B-Myb and c-Myb levels in the hepatoma cells (Fig. 8D). This contrasts to the age-associated reduction of the B-Myb level in liver nuclei, whereas no significant change in the c-Myb level was observed. Thus, it appears that c-Myb plays a key role in recruiting p53 to the ADF element in the liver chromatin of the old animals, whereas in the young animals, both c-Myb and B-Myb are involved in this regard. Increased PARP-1 activity in the presence of excess β-NAD+ or oxidative stress is expected to convert PARP-1 to an automodified form, leading to the likelihood that the bulky anionic branched-chain ADP-ribosyl polymers attached to PARP-1 would negatively influence PARP-1/B-Myb interaction. A greater proportion of PARP-1-free B-Myb may then associate with p53, which will be manifested in increased B-Myb/p53 co-IP (Fig. 8B). Whether pharmacologic inhibition of PARP-1 activity would prevent the oxidative stress-induced reduction of Myb/PARP-1 interaction will be important to determine.
Aging is associated with reduced Myb/PARP-1 interaction as well as revealed by sequential ChIP that compared co-immunoprecipitation of PARP-1 and c-Myb from the ADF site of liver chromatin in the young versus old rats (Fig. 8E). Following first step chromatin immunoprecipitation with the anti-c-Myb antibody, re-ChIP on liver chromatin was performed with the anti-PARP-1 or anti-p53 antibody. The control experiment in which the first step ChIP was performed without any antibody (no antibody control) produced only background signal (not shown here). In the liver of the old rats, concurrent occupancy of c-Myb and PARP-1 at the ADF element of the same chromatin fragments was markedly reduced, whereas p53 was present at elevated levels at the c-Myb-loaded chromatin fragments (Fig. 8D). Taken together, these results strongly indicate that Myb/PARP-1 interaction is inhibited by aging and oxidative stress.
The results of two-step ChIP in Figs. 7F and 8E reveal that p53 occupancy at the ADF element, which occurs in response to oxidative stress and aging, is dependent upon the presence of the Myb oncoproteins (B-Myb and c-Myb) at the regulated chromatin. Myb dependence of PARP-1 occupancy at this region was similarly observed. Recruitment of TLE1 and mSin3A to the ADF element is in turn p53-dependent because both associated with the liver chromatin fragments that immunoprecipitated with a p53-specific antibody (Fig. 9A). Similarly co-occupancy of p53, TLE1, and mSin3A at the regulated chromatin was observed in TBH-treated H4IIE hepatoma cells (data not shown). Furthermore p53 interacted with TLE-1 but not hnRNPK within rat liver nuclear extract (Fig. 9B), and p53/mSin3A interaction is known to occur in mammalian cells (28).
DISCUSSION
Certain coregulators are closely linked to invertebrate and vertebrate aging as highlighted by the finding that increased deacetylase activity of sirtuin-1 (SIRT1), an NAD+-dependent class-III HDAC, is associated with improved metabolic functions and extended longevity (42), and higher poly(ADP-ribosyl)ating activity of the transcriptional coactivator PARP-1 was detected in centenarians and in various long lived mammalian species. Coregulator function is also linked to oxidative stress, which is a prominent trait in aging, as evident from the coactivator role of the β-catenin ortholog Bar-1, which amplifies the activity of the Forkhead box-containing protein, O-sub-family (FOXO) transcription factor Daf-16 in Caenorhabditis elegans (43). Daf-16 is a positive contributor to the C. elegans lifespan because it stimulates expression of the antioxidant enzyme superoxide dismutase and thus plays a role in bolstering oxidative stress resistance in the worm. The Bar-1-coactivated Daf-16 also induces cell cycle arrest, leading to cell survival under oxidative stress (43). In the current study, we show that, similar to aging in vivo, AR gene expression is down-regulated by oxidative stress in a cell culture model. Furthermore our study sheds light on specific DNA-binding transcription factors and interacting ancillary partners and coregulators, which are involved in the loss of AR expression during physiological aging and the oxidative stress response.
Included among the ADF-copurified proteins were PARP-1, the protooncoproteins B-Myb and c-Myb, and hnRNPK, which is an RNA- and DNA-binding protein involved in diverse cellular processes (44). Knockdown of these proteins reduced AR expression, indicating their roles as positive regulators of cellular AR content. Increased AR levels in p53-silenced cells indicated a negative role for p53 in AR expression. The 20-bp ADF element was identified by EMSA as the cognate binding site for B-Myb/c-Myb. B-Myb and c-Myb are transcription factors and members of the Myb protooncoprotein family (45). B-Myb is expressed ubiquitously and plays roles in cell growth control, differentiation, and cancer. c-Myb is involved in epithelial cell function; in addition, c-Myb plays a key role in fetal hematopoiesis (46). A-Myb, the third member of the Myb family, is expressed in germinal B lymphocytes, in male testes, and in the female reproductive system (45). A-Myb did not copurify with ADF activity.
We conclude that the ADF-copurified positive and negative regulators of AR are also involved in the induction and repression of the ADF element in vivo in a chromatin environment because residency of these proteins at or near the ADF site in liver chromatin correlated with up- or down-regulation of AR gene transcription. We demonstrate that 1) B-Myb, c-Myb, PARP-1, hnRNPK, and the global coactivator p/CAF occupy the ADF site in young adult rats and in rat hepatoma cells that are at redox equilibrium, 2) p53 and the p53-associated corepressors mSin3A and Groucho/TLE1 occupy regulated chromatin in old rats and in hepatoma cells that show a prooxidant shift because of TBH exposure, and 3) PARP-1 and hnRNPK are absent from the ADF site in the aging liver and in oxidatively stressed cells.
Disengagement of B-Myb from the regulated chromatin of the aging liver correlated with reduced B-Myb levels in the liver nuclear extracts from old rats. c-Myb occupancy, however, continued at the ADF element in the aging rodent. Repression of the ADF element by oxidative stress in hepatoma cells paralleled the persistent presence of both c-Myb and B-Myb at regulated chromatin. Data presented herein also show that 1) hnRNPK weakly binds to the ADF element (Fig. 4D), and PARP-1 co-immunoprecipitates with hnRNPK (Fig. 4E); 2) Myb/PARP-1 interaction is inhibited in the presence of excess β-NAD+ (Fig. 8A) or oxidative stress (Fig. 8C); and 3) p53 interferes with Myb-mediated AR promoter transactivation (Fig. 5, B and C), and p53 can interact with B-Myb and c-Myb (Fig. 6C and supplemental Fig. S-B).
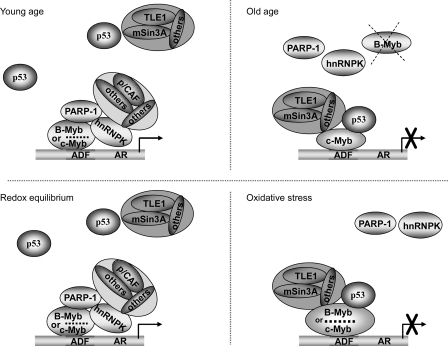
Collectively our results favor a model predicting how the transcription factors B-Myb/c-Myb, bound to the ADF element, might facilitate reciprocal chromatin dynamics between PARP-1 and p53 to coordinate loss of AR expression during aging and oxidative stress (Fig. 10). The model suggests that the DNA-bound B-Myb and c-Myb are part of an activating complex that includes the Myb coactivator PARP-1 and the multifunctional hnRNPK, which facilitates PARP-1 recruitment through physical association and stabilizes coactivator assembly by serving as a docking platform. The model also predicts that dissociation of PARP-1 from B-Myb and c-Myb because of aging and/or oxidative stress would destabilize the activating complex. In this setting, active repression of the ADF element would be set in motion after the recruitment of p53, which associates with the DNA-bound B-Myb and c-Myb. p53 affinity for TLE1 (Fig. 9B) and for mSin3A (28) in turn would facilitate congregation of corepressors at the ADF element.
FIGURE 10.
Model illustrating a likely scenario for AR gene regulation during aging and oxidative stress. AR gene transcription in young rat liver cells and in cells at redox equilibrium is positively regulated at the ADF site through the involvement of ADF-bound B-Myb/c-Myb and a B-Myb/c-Myb-associated coactivator complex, which includes PARP-1, hnRNPK, and p/CAF. AR gene repression at old age and in cells at a prooxidant state is coordinated at the ADF site by B-Myb and/or c-Myb bound to a p53-associated corepressor complex that includes Groucho/TLE1 and mSin3A corepressors.
The members of the human TLE corepressor family (TLE 1 through 4) are orthologs of the Drosophila Groucho. The TLE proteins are recognized as important players in diverse developmental processes (31). TLE usually converts transcriptional activators to repressors. Sin3A is a large multidomain protein, which acts as a scaffold upon which other components of the corepressor complex are known to gather. TLE associates with the core mSin3A corepressor complex via specific bridge proteins (47). Thus TLE1 and mSin3A in association with p53, which is anchored at the ADF element through its protein/protein interaction with the DNA-bound B-Myb/c-Myb, provide for a mechanism to promote corepressor assembly that drives repression of the ADF element in vivo. Whether the p53-associated B-Myb and c-Myb would interact directly with corepressors or indirectly via p53 remains to be determined. In this regard, it is noteworthy that the corepressors NCoR (nuclear receptor corepressor) and SMRT (silencing mediator of retinoid and thyroid receptors) were found to directly interact with B-Myb but not c-Myb, and this corepressor interaction was shown to maintain B-Myb in a transcriptionally inactivated state (48).
Silencing of hnRNPK decreased AR expression (Fig. 4A). Nevertheless hnRNPK function is linked to both gene induction and gene repression (44). It has been reported that the 5′-untranslated region of the AR promoter harbors a suppressor element, which is regulated in the androgen-dependent LNCaP cells by the single-stranded DNA- and RNA-binding protein Pur-α. Pur-α was shown to act in combination with hnRNPK at the suppressor element, and Pur-α knockdown elevated AR protein levels in LNCaP cells (49). However, the outcome of hnRNPK silencing with regard to AR expression was not described in this study. At any rate, it is likely that hnRNPK plays context-dependent positive or negative roles at any particular regulatory element in the AR gene, although the overall consequence of hnRNPK depletion in cells is to confer loss of AR expression as revealed by our results in hepatoma cells (Fig. 4A) and in LNCaP cells (data not shown). hnRNPK is thought to function as a docking platform to integrate cross-talk among various signal transduction cascades (44), thereby regulating diverse processes such as chromatin remodeling, transcription, RNA splicing, mRNA transport, and translation. Consistent with the known PARP-1-mediated poly(ADP-ribosyl)ation of hnRNPK (50), hnRNPK/PARP-1 interaction can be demonstrated (Fig. 4E). Because hnRNPK binds to the ADF element, albeit much more weakly than the Myb proteins (Fig. 4D), we hypothesize that hnRNPK transiently binds to the ADF element to facilitate recruitment of PARP-1, p/CAF, and other coactivators at the activating complex, which is further stabilized by the interaction of PARP-1 with B-Myb and c-Myb, both of which bind to the ADF element with high affinity.
Poly(ADP-ribosyl)ation of histones, transcription factors, and other nuclear proteins by PARP-1 is gaining increasing recognition as an important mode of epigenetic regulation similar to other post-translational modifications such as phosphorylation, acetylation, and ubiquitination (20, 51). A sensing function of PARP-1 is thought to be involved in the transcriptional activation of the proneural protein MASH1 in neural stem cells following growth factor stimulation (52). In this case, PARP-1 was identified as part of a TLE1-associated corepressor complex, which interacts with the DNA-bound HES, a basic helix-loop-helix transcription factor, to keep MASH1 transcriptionally silenced, preventing neural differentiation. Upon growth factor stimulation, activation of PARP-1 by a calcium-dependent protein kinase leads to the loss of PARP-1 and other corepressors from the regulated site and the subsequent corepressor to coactivator exchange that results in MASH1 derepression. Our data show that increased PARP-1 activity caused by the presence of excess β-NAD+ in vitro, by intracellular oxidative stress, or during aging led to a reduced Myb/PARP-1 interaction (Fig. 8, A and C). We propose that the peroxidative DNA damage induced by aging and oxidative stress and the resulting poly(ADP-ribosyl) modification of PARP-1 itself serve as a trigger for the dissociation of PARP-1 from the ADF element. hnRNPK is expected to leave the regulated region in the absence of PARP-1 because these two proteins interact in vivo. PARP-1-free B-Myb and c-Myb at this point are available to interact with p53 and p53-associated corepressors, and as a result, they are incorporated as part of a corepressor assembly. Thus, we suggest that PARP-1 is the sensor molecule, which causes role reversal for B-Myb and c-Myb from gene activators to gene repressors at the ADF element during aging and oxidative stress.
In future work, characterization of the signaling cascade(s) that impinges upon PARP-1 activity to cause repression of the ADF element in vivo in a chromatin environment, leading to the loss of AR expression, should be insightful in knowing how tissue androgen sensitivity is impacted by aging and oxidative stress. The information may also help illuminate the pathways to androgen-regulated physiologic deficits in the elderly and in diseases that manifest a prooxidant shift of the intracellular redox balance.
Supplementary Material
Acknowledgments
c-Myb cDNAs were generous gifts of Dr. Tim Bender (University of Virginia, Charlottesville, VA). We thank Dr. Mandakini Patel for contributions and Dr. Z. Dave Sharp (The University of Texas Health Science Center at San Antonio) for critical review of the manuscript. Mass spectrometry was performed by The University of Texas Health Science Center at San Antonio Core Facility (National Institutes of Health Grant 7P30CA54174-14).
This work was supported, in whole or in part, by National Institutes of Health Grants AG-10486 and AG-19660. This work was also supported by a merit review grant from Veterans Affairs. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S-A and S-B and sequences of siRNA oligos and primers.
Footnotes
The abbreviations used are: AR, androgen receptor; ADF, age-dependent factor; p/CAF, p300/CBP-associating factor (CBP, cAMP-responsive element-binding protein (CREB)-binding protein); hnRNPK, heterogeneous nuclear ribonucleoprotein K; PARP-1, poly(ADP-ribose) polymerase-1; β-NAD+, β-nicotinamide adenine dinucleotide; EMSA, electrophoretic mobility shift assay; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; ChIP, chromatin immunoprecipitation; TBH, tert-butyl hydroperoxide; RLNE, rat liver nuclear extract; siRNA, small interfering RNA; GST, glutathione S-transferase; IP, immunoprecipitation; HDAC, histone deacetylase; TK, thymidine kinase; qPCR, quantitative PCR; TLE, transducin-like enhancer of split.
References
- 1.Lin, H.-Y., Yu, I.-C., Wang, R.-S., Chen, Y.-T., Liu, N.-C., Altuwaijri, S., Hsu, C.-L., Ma, W.-L., Jokinen, J., Sparks, J. D., Yeh, S., and Chang, C. (2008) Hepatology 47 1924-1935 [DOI] [PubMed] [Google Scholar]
- 2.Song, C. S., Jung, M. H., Kim, S. C., Hassan, T., Roy, A. K., and Chatterjee, B. (1998) J. Biol. Chem. 273 21856-21866 [DOI] [PubMed] [Google Scholar]
- 3.Cai, Y., Dai, T., Ao, Y., Konishi, T., Chuang, K. H., Lue, Y., Chang, C., and Wan, Y.-J. Y. (2003) Endocrinology 144 2311-2318 [DOI] [PubMed] [Google Scholar]
- 4.Nantermet, P., Harada, S., Liu, Y., Cheng, S., Johnson, C., Yu, Y., Kimme, D., Holder, D., Hodor, P., Phillips, R., and Ray, W. J. (2008) Endocrinology 149 1551-1561 [DOI] [PubMed] [Google Scholar]
- 5.Kemp, C. J., Leary, C. N., and Drinkwater, N. R. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 7505-7509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Serag, H. B., and Rudolph, K. L. (2007) Gastroenterology 132 2557-2576 [DOI] [PubMed] [Google Scholar]
- 7.Boix, L., Castells, A., Bruix, J., Sole, M., Bru, C., Fuster, J., Rivera, F., and Rodes, J. (1995) J. Hepatol. 22 616-622 [DOI] [PubMed] [Google Scholar]
- 8.Pienta, K. J., and Bradley, D. (2006) Clin. Cancer Res. 12 1665-1670 [DOI] [PubMed] [Google Scholar]
- 9.Song, C. S., Rao, T. R., Demyan, W. F., Mancini, M. A., Chatterjee, B., and Roy, A. K. (1991) Endocrinology 128 349-356 [DOI] [PubMed] [Google Scholar]
- 10.Supakar, P. C., Song, C. S., Jung, M. H., Slomczynska, M. A., Kim, J. M., Vellanoweth, R. L., Chatterjee, B., and Roy, A. K. (1993) J. Biol. Chem. 268 26400-26408 [PubMed] [Google Scholar]
- 11.Roy, A. K., Vellanoweth, R. L., Chen, S., Supakar, P. C., Jung, M. H., Song, C. S., and Chatterjee, B. (1996) Exp. Gerontol. 31 83-94 [DOI] [PubMed] [Google Scholar]
- 12.Supakar, P. C., Jung, M. H., Song, C. S., Chatterjee, B., and Roy, A. K. (1995) J. Biol. Chem. 270 837-842 [DOI] [PubMed] [Google Scholar]
- 13.Helenius, M., Hänninen, M., Lehtinen, S. K., and Salminen, A. (1996) Biochem. J. 318 603-608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko, S. Y., Shi, L. H., Kim, S. Y., Song, C. S., and Chatterjee, B. (2008) Mol. Endocrinol. 22 273-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Supakar, P. C., and Roy, A. K. (1996) Biol. Signals 5 170-179 [DOI] [PubMed] [Google Scholar]
- 16.Roy, A. K., Oh, T., Rivera, O., Mubiru, J., Song, C. S., and Chatterjee, B. (2002) Ageing Res. Rev. 1 367-380 [DOI] [PubMed] [Google Scholar]
- 17.D'Amours, D., Desnoyers, S., D'Silva, I., and Poirier, G. G. (1999) Biochem. J. 342 249-266 [PMC free article] [PubMed] [Google Scholar]
- 18.Kraus, W. L., and Lis, J. T. (2003) Cell 113 677-683 [DOI] [PubMed] [Google Scholar]
- 19.Pavri, R., Lewis, B., Kim, T. K., Dilworth, F. J., Erdjument-Bromage, H., Tempst, P., de Murcia, G., Evans, R., Chambon, P., and Reinberg, D. (2005) Mol. Cell 18 83-96 [DOI] [PubMed] [Google Scholar]
- 20.Rosenfeld, M. G., Lunyak, V. V., and Glass, C. K. (2006) Genes Dev. 20 1405-1428 [DOI] [PubMed] [Google Scholar]
- 21.Kanai, M., Hanashiro, K., Kim, S. H., Hanai, S., Boulares, A. H., Miwa, M., and Fukasawa, K. (2007) Nat. Cell Biol. 9 1175-1183 [DOI] [PubMed] [Google Scholar]
- 22.Grube, K., and Bürkle, A. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 11759-11763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murias, M. L., Muller, M., Schachter, F., and Burkle, A. (1998) J. Mol. Med. 76 346-354 [DOI] [PubMed] [Google Scholar]
- 24.Dignam, J. D., Lebowitz, R. M., and Roeder, R. G. (1983) Nucleic Acids Res. 11 1475-1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seo, Y. K., Chung, Y. T., Kim, S. Y., Echchgadda, I., Song, C. S., and Chatterjee, B. (2007) Gene (Amst.) 386 218-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendoza-Alvarez, H., and Alvarez-Gonzalez, R. (2001) J. Biol. Chem. 276 36425-36430 [DOI] [PubMed] [Google Scholar]
- 27.Wang, L. G., Liu, X. M., Kreis, W., and Budman, D. R. (1999) Biochem. Biophys. Res. Commun. 259 21-28 [DOI] [PubMed] [Google Scholar]
- 28.Murphy, M., Ahn, J., Walker, K. K., Hoffman, W. H., Evans, R. M., Levine, A. J., and George, D. L. (1999) Genes Dev. 13 2490-2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanikawa, J., Nomura, T., Macmillan, E. M., Shinagawa, T., Jin, W., Kokura, K., Baba, D., Shirakawa, M., Gonda, T. J., and Ishii, S. (2004) J. Biol. Chem. 279 55393-55400 [DOI] [PubMed] [Google Scholar]
- 30.Jennings, B. H., Pickles, L. M., Wainwright, S. M., Roe, S. M., Pearl, L. H., and Ish-Horowicz, D. (2006) Mol. Cell 22 645-655 [DOI] [PubMed] [Google Scholar]
- 31.Chen, G., and Courey, A. J. (2000) Gene (Amst.) 249 1-16 [DOI] [PubMed] [Google Scholar]
- 32.Yaklichkin, S., Vekker, A., Stayrook, S., Lewis, M., and Kessler, D. S. (2007) BMC Genomics 8 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Germain, M., Affar, E. B., D'Amours, D., Dixit, V. M., Salvesen, G. S., and Poirier, G. G. (1999) J. Biol. Chem. 274 28379-28384 [DOI] [PubMed] [Google Scholar]
- 34.Alimirah, F., Panchanathan, R., Chen, J, Zhang, X., Ho, S.-M., and Choubey, D. (2007) Neoplasia 9 1152-1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moticelli, S., Ghittoni, R., Kabesch, M., and Vercelli, D. (2001) Mol. Immunol. 38 1129-1138 [DOI] [PubMed] [Google Scholar]
- 36.Miglarese, M. R., Richardson, A. F., Aziz, N., and Bender, T. P. (1996) J. Biol. Chem. 271 22697-22705 [DOI] [PubMed] [Google Scholar]
- 37.Cervellera, M. N., and Sala, A. (2000) J. Biol. Chem. 275 10692-10696 [DOI] [PubMed] [Google Scholar]
- 38.Finkel, T., and Holbrook, N. J. (2000) Nature 408 239-247 [DOI] [PubMed] [Google Scholar]
- 39.Ochi, T., and Miyaura, S. (1989) Toxicology 55 69-82 [DOI] [PubMed] [Google Scholar]
- 40.Métivier, R., Penot, G., Hübner, M. R., Reid, G., Brand, H., Koš, M., and Gannon, F. (2003) Cell 115 751-763 [DOI] [PubMed] [Google Scholar]
- 41.Kim, M. Y., Mauro, S., Gevry, N., Lis, J. T., and Kraus, W. L. (2004) Cell 119 803-814 [DOI] [PubMed] [Google Scholar]
- 42.Bordone, L., and Guarente, L. (2005) Nat. Rev. Mol. Cell Biol. 6 298-305 [DOI] [PubMed] [Google Scholar]
- 43.Bowerman, B. (2005) Science 308 1119-1120 [DOI] [PubMed] [Google Scholar]
- 44.Bomsztyk, K., Denisenko, O., and Ostrowski, J. (2004) BioEssays 26 629-638 [DOI] [PubMed] [Google Scholar]
- 45.Sala, A. (2005) Eur. J. Cancer 41 2479-2484 [DOI] [PubMed] [Google Scholar]
- 46.Ness, S. A. (2003) Blood Cells Mol. Dis. 31 192-200 [DOI] [PubMed] [Google Scholar]
- 47.Yochum, G. S., and Ayer, D. E. (2001) Mol. Cell. Biol. 21 4110-4118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li, X., and McDonnell, D. P. (2002) Mol. Cell. Biol. 22 3663-3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, L. G., Johnson, E. M., Kinoshita, Y., Babb, H. S., Buckley, M. T., Liebes, L. F., Melamed, J., Liu, X.-M., Kurek, R., Ossowski, L., and Ferrari, A. C. (2008) Cancer Res. 68 2678-2688 [DOI] [PubMed] [Google Scholar]
- 50.Gagne, J.-P., Hunter, J. M., Labrecque, B., Chabot, B., and Poirier, G. G. (2003) Biochem. J. 371 331-340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lonard, D. M., and O'malley, B. W. (2007) Mol. Cell 27 691-700 [DOI] [PubMed] [Google Scholar]
- 52.Ju, B.-G., Solum, D., Song, E. J., Lee, K.-J., Rose, D. W., Glass, C. K., and Rosenfeld, M. G. (2004) Cell 119 815-829 [DOI] [PubMed] [Google Scholar]
- 53.Stains, J. P., Lecanda, F., Towler, D. A., and Civitelli, R. (2005) Biochem. J. 385 613-623 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.