Abstract
Yeast cells synthesize ∼3–6 million molecules of tRNA every cell cycle at a rate of ∼2–4 transcripts/gene/s. This high rate of transcription is achieved through many rounds of reinitiation by RNA polymerase (pol) III on stable DNA-bound complexes of the initiation factor TFIIIB. Studies in yeast have shown that the rate of reinitiation is increased by facilitated recycling, a process that involves the repeated reloading of the polymerase on the same transcription unit. However, when nutrients become limiting or stress conditions are encountered, RNA pol III transcription is rapidly repressed through the action of the conserved Maf1 protein. Here we examine the relationship between Maf1-mediated repression and facilitated recycling in a human RNA pol III in vitro system. Using an immobilized template transcription assay, we demonstrate that facilitated recycling is conserved from yeast to humans. We assessed the ability of recombinant human Maf1 to inhibit different steps in transcription before and after preinitiation complex assembly. We show that recombinant Maf1 can inhibit the recruitment of TFIIIB and RNA pol III to immobilized templates. However, RNA pol III bound to preinitiation complexes or in elongation complexes is protected from repression by Maf1 and can undergo several rounds of initiation. This indicates that recombinant Maf1 is unable to inhibit facilitated recycling. The data suggest that additional biochemical steps may be necessary for rapid Maf1-dependent repression of RNA pol III transcription.
Efficient in vitro transcription of tRNA genes and other genes with internal (type 2) promoters requires two multi-subunit transcription factors, TFIIIC and TFIIIB, in addition to RNA polymerase (pol)3 III (1, 2). These factors assemble onto the promoters of type 2 genes in an ordered and concerted process that involves conformational changes in both the DNA and the proteins (1, 3). The basic stepwise assembly pathway entails the binding of split promoter elements by each of the two globular domains of TFIIIC followed by the recruitment of TFIIIB, which envelops the DNA upstream of the transcription start site. The resulting complex is exceptionally stable and is able to recruit the polymerase for multiple rounds of transcription (4). Template competition experiments have shown that after the first cycle of transcription, the polymerase is committed to reinitiate on the same gene and does so more rapidly because it does not dissociate into the bulk solution (5, 6). This phenomenon is referred to as facilitated recycling and contributes to the high rate of RNA pol III transcription in yeast. Indeed, after the first round of transcription, the rate of subsequent rounds increases at least 5-fold (5). Facilitated recycling couples the termination of transcription with reinitiation in a manner that is not fully understood. However, the coupling of these two steps requires proper termination and is transcription factor-dependent. Transcription assays that employ run-off termination on truncated RNA pol III genes or factor-independent transcription of tailed linear templates do not allow efficient recycling (5, 7). Promoter-bound TFIIIB is sufficient for facilitated recycling of RNA pol III on short genes (∼100 bp). However, longer transcription units (>300 bp, e.g. the 7SL RNA gene) also require TFIIIC to achieve the high rates of reinitiation that characterize facilitated recycling (6). In humans, several activities have been shown to contribute to RNA pol III reinitiation. The transcript release factor La was found to facilitate RNA pol III transcription and template reutilization (8), whereas the NF1 protein acts through interactions with TFIIIC at the terminator region of the VA1 template to promote multiple rounds of transcription (9). Similarly, topoisomerase I and PC4 both enhance TFIIIC interactions with downstream promoter regions and stimulate multiple but not single round transcription from preformed preinitiation complexes (10). However, it is not yet known whether the fundamental mechanism of reinitiation in higher eukaryotes is analogous to the facilitated recycling pathway described in yeast.
Eukaryotic cells coordinately regulate transcription by all three nuclear RNA polymerases to maintain the appropriate protein synthetic capacity for cell growth and proliferation. Among the different mechanisms used to achieve coordinate control in higher eukaryotes, the Myc oncoprotein directly activates RNA pol II transcription of ribosomal protein genes and a wide variety of growth and cell cycle regulators along with transcription of rRNA and tRNA genes by RNA pols I and III (11, 12). In nondividing cells, positive signals for cell growth and proliferation are opposed by the action of numerous tumor suppressor proteins that repress transcription by RNA pols I and III (e.g. Rb, p53, ARF, PTEN) and regulate different subsets of protein coding genes. Notably, inactivation of these tumor suppressors in different types of cancer leads to the up-regulation of RNA pol I and RNA pol III transcription (11, 13, 14). Rather than being a simple consequence of cell transformation, a growing number of studies indicate that deregulation of RNA pol I and RNA pol III transcription plays an important role in the development of cancer and in the growth of tumors (11, 15, 16). Recent work has shown that several properties of tumor suppressors are shared by the Maf1 protein. In mammalian cells, Maf1 functions to directly repress transcription by RNA pol III and a subset of RNA pol II transcribed genes including the TATA-binding protein, TBP (17). Through its affect on TBP levels, Maf1 indirectly regulates transcription by RNA pol I (17). The transcription of many protein coding genes whose expression is limited by TBP is also likely to be affected indirectly by Maf1. Importantly, overexpression of Maf1 inhibits the growth of glioblastoma cells in soft agar, indicating that Maf1 is able to function as a tumor suppressor (17). Maf1 was identified genetically in yeast (18, 19) and was subsequently shown to function as an essential mediator of diverse nutritional and stress signaling pathways that repress transcription by RNA pol III (20). Although the protein is conserved from yeast to humans and functions as a repressor of RNA pol III transcription in these organisms, its ability to repress TBP gene transcription and thereby influence rDNA synthesis is not observed in yeast (20). This function appears to have evolved in higher eukaryotes. Much of what we currently know about the mechanism of repression by Maf1 derives from studies in the RNA pol III system (21).
The primary targets for Maf1 repression of RNA pol III transcription in yeast are TFIIIB and the polymerase. Recombinant Maf1 from budding and fission yeast binds to the Brf1 subunit of TFIIIB and blocks TFIIIB assembly directed by TFIIIC on tRNA gene promoters (22). The recombinant Maf1 proteins from yeast also inhibit RNA pol III transcription from preassembled TFIIIB-promoter complexes, presumably as a result of the direct interaction between Maf1 and the polymerase (22). Similarly, recombinant human Maf1 interacts with both Brf1 and RNA pol III in in vitro binding assays (23). The biological importance of these conserved interactions is demonstrated by the coimmunoprecipitation of endogenous yeast and human Maf1 proteins with Brf1 and RNA pol III from cell free extracts (19, 22, 24). Yeast and mammalian Maf1 proteins are phosphorylated under normal growing conditions and become dephosphorylated under repressing conditions (23–27). This process is important for the interaction of Maf1 with the polymerase (23, 26, 27). However, additional, as yet unknown steps are required to affect repression of RNA pol III transcription in vivo. This was first indicated by the properties of a nuclear-localized multi-phosphosite mutant of Maf1 (6SA) (25). Rather than being constitutively active, this mutant allele still requires the activation of cellular signaling pathways to effect repression.
In this study, we have investigated the molecular mechanism by which human Maf1 represses RNA pol III transcription in vitro. Using fractionated HeLa cell components and an immobilized template transcription assay, we demonstrate facilitated recycling by human RNA pol III. Recombinant human Maf1 is a potent inhibitor of RNA pol III transcription in HeLa cell extracts. However, we find that human Maf1 is not capable of repressing single or multiple rounds of transcription in our reconstituted system once the polymerase has engaged the transcription factor-template complex. We show that Maf1 represses transcription primarily by preventing the recruitment of the polymerase to the TFIIIB-TFIIIC-DNA complex. To a lesser extent, human Maf1 also inhibits the binding of TFIIIB to promoter-bound TFIIIC.
EXPERIMENTAL PROCEDURES
Purification of Full-length Recombinant Human Maf1—Full-length human Maf1 was PCR-amplified from a pcDNA3.1 clone (17) and ligated into NdeI- and HindIII-digested pET-30a-(+) (Novagen) to produce a C-terminal hexahistidine-tagged hMaf1 protein. The plasmid was transformed into Escherichia coli BL21 (DE3) (Novagen), grown to an A600 of ∼0.7 at 37 °C, and induced with 0.1 mm isopropyl β-d-thiogalactopyranoside for 16 h at 15 °C. The hMaf1 protein was purified under native conditions on nickel-nitrilotriacetic acid-agarose (Qiagen) following the manufacturer's recommendations. The protein was dialyzed into Q100 buffer (20 mm Tris-actetate, pH 7.0, 5 mm magnesium acetate, 100 mm (NH4)2SO4, 0.1 mm EDTA, 1 mm dithiothreitol, 1 μg/ml leupeptin, 1 μg/ml pepstatin). The dialyzed protein was mixed with an equal volume of Q0 (no salt) buffer just before injection onto a Resource Q column (Amersham Biosciences). hMaf1 was eluted with a linear gradient from 50 mm (NH4)2SO4 to 500 mm (NH4)2SO4. The peak fractions were dialyzed into Q200 buffer and stored at –70 °C. Maf1 concentration was determined by absorbance at 280 nm using the calculated molar extinction coefficient.
Extract Preparation and Factor Purification—HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum at 37 °C in a 5% CO2 atmosphere. Nuclear extract and soluble S100 extract were prepared as described (28). The nuclear extract was fractionated by phosphocellulose chromatography (Whatman P11) as described previously (29) into fractions containing TFIIIB along with RNA pol III (P11:0.35) and TFIIIC (P11:0.6). S100 extract was applied to the phosphocellulose P11 column and eluted in one step at 0.35 m KCl as described (29). This eluate was subsequently loaded onto a DEAE Sephadex A25 column, and fractions containing TFIIIB (DEAE 0.15) and RNA pol III (DEAE 1.0) were separated as described (30). All of the fractions were dialyzed against KBC100 buffer (20 mm HEPES-KOH, pH 7.9, 100 mm KCl, 0.2 mm EDTA, 20% glycerol, 2 mm dithiothreitol, 0.2 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mm benzamidine) containing 5 mm MgCl2.
In Vitro Transcription—Standard 50-μl transcription reactions contained HeLa nuclear extract or protein fractions, 20 mm HEPES-KOH, pH 7.9, 60 mm KCl, 7 mm MgCl2, 10% glycerol, 600 μm of each ATP, GTP, and UTP, 25 μm CTP, 10 μCi of [α-32P]CTP (GE Healthcare), 10 units of RNasin ribonuclease inhibitor (Promega), and 0.5 μg of plasmid tRNALeu3 (YEp13) or 60 ng of immobilized VA1 DNA fragment (see below) as templates. Where indicated, the nuclear extract or protein fractions were preincubated with various concentrations of Maf1, BSA, or poly(dA-dT) for 10 min at room temperature. In single round reactions, Sarkosyl was added to 0.05% (final concentration). After transcription for various times at 30 °C, the RNA was subjected to electrophoresis in denaturing 6% polyacrylamide gels. The gels were dried and exposed to phosphorimaging screens (Molecular Dynamics) for capturing digital images and quantitation (ImageQuant).
Assembly of Complexes and Transcription on Immobilized Templates—The pUC18-VA1 construct contains a 0.28-kb SalI-SmaI fragment of the Adenovirus 2 VA1 gene from the parental transcription construct pVA1 (31) between the HindIII and EcoRI sites of pUC18. A 650-bp fragment containing the VA1 gene was generated from pUC18-VA1 by PCR using a 5′ end-biotinylated upstream primer (gctggcttaactatgcggcatatgagcagattgtac) and a downstream primer (cgcaacgcaattaatgtgagttagct). The fragment was immobilized on Streptavidin-coupled Dynabeads M-280 (Invitrogen) as described by the manufacturer. The Dynabead-immobilized VA1 template will be referred to subsequently as IT-VA1. For experiments in which stalled ternary complexes (TC) were used, IT-VA1 was preincubated in KBCM60 (KBC buffer containing 60 mm KCl and 7.5 mm MgCl2) with P11:0.35 + 0.6 fractions and ATP, CTP, GTP, but not UTP. TCs were washed three times with 200 μl of KBCM500/0.015 (KBCM with 500 mm KCl, 0.015% Nonidet P-40, and 0.1 mg/ml BSA) and subjected to one equilibrium wash with KBCM60/0.015. Preinitiation complexes (PICs) were formed by incubating IT-VA1 with P11:0.35 + 0.6 fractions in KBCM60 without NTPs. The complexes were washed three times with KBCM100/0.015 before a final wash with KBCM60/0.015. TFIIIC-DNA and TFIIIC-TFIIIB-DNA complexes were formed by incubating IT-VA1 with the P11:0.6 fraction alone or in the presence of the DEAE:0.15 fraction, respectively, in KBCM60. Washing conditions for these complexes were the same as for immobilized PICs. All of the binding reactions were carried out for 29 min at 30 °C, and immobilized complexes were transferred to fresh tubes after the first wash. BSA (0.1 mg/ml, RNase and DNase-free; GE Healthcare) carrier protein was included in all reactions in which preformed complexes were used. After binding reactions were complete, immobilized complexes were resuspended in KBCM60. As indicated in individual experiments, the reactions were supplemented with various concentrations of Maf1, BSA, or poly(dA-dT), and preincubations were allowed to proceed as described in the figure legends and text. When appropriate, Sarkosyl was added to a final concentration of 0.05%. For the experiment shown in Fig. 5, the DEAE:0.15 fraction was preincubated with Maf1 or Y for 10 min at room temperature before addition to the IT-VA1-bound TFIIIC. In the corresponding control reaction, IT-VA1 was preincubated with Maf1 or BSA and washed as described for the immobilized PICs. Where indicated, the reactions were supplemented with the RNA pol III DEAE:1.0 fraction. Transcription was initiated with complete set of nucleotides (NTPs) along with [α-32P]CTP. The reactions were carried out at 30 °C for the times indicated and then processed as described above.
FIGURE 5.
Maf1 prevents human TFIIIB assembly on DNA with low efficiency. A, upper scheme (TFIIIB/Maf1), IT-VA1 was incubated with the TFIIIC fraction (P11:0.6) for 29 min at 30 °C. TFIIIC-DNA complexes were washed with KBCM100/0.015 and mixed with a TFIIIB fraction (DEAE: 0.15) that had been separately preincubated with BSA or Maf1 for 10 min at room temperature. After a further 29-min incubation at 30 °C, the protein-IT-VA1 complexes were washed with KBCM100/0.015, and transcription was initiated with a RNA pol III fraction (DEAE 1.0) and NTPs. Lower scheme (Con.), IT-VA1 was preincubated with BSA or Maf1 for 29 min at 30 °C. After washing with KBCM100/0.015, TFIIIC- and TFIIIB-containing fractions were added, and the reactions were allowed to proceed as depicted in the upper scheme. B, reconstituted transcription assays. Upper panel, TFIIIB/Maf1; lower panel, control (Con.). Control lanes 1 and 2 show reactions with no additional protein and BSA (5 μg), respectively. Lanes 3–7 show reactions supplemented with 0.02, 0.1, 0.5, 2.5, and 5 μg of Maf1.
RESULTS
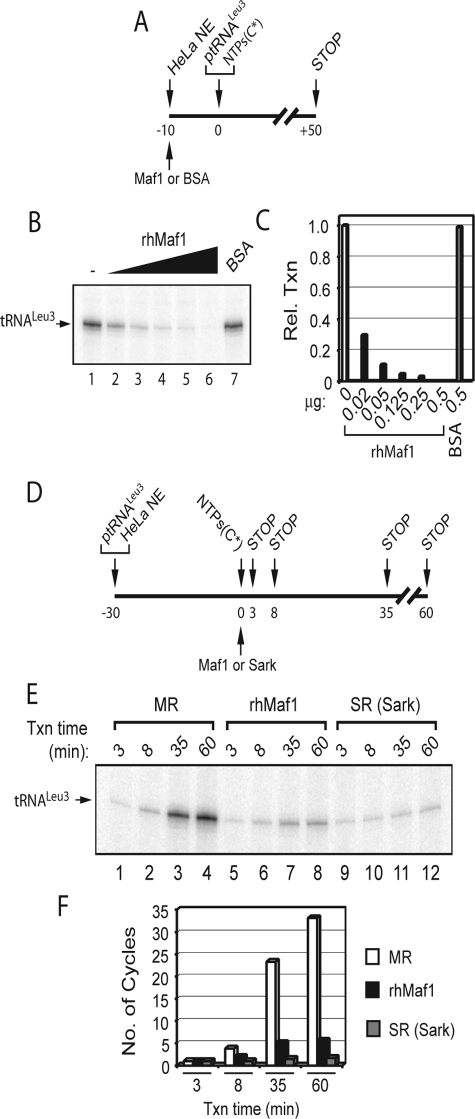
Human Maf1 Differentially Inhibits Transcription from Unassembled and Preassembled PICs—In vitro studies have shown that the addition of recombinant Maf1 to yeast and human cell-free systems represses RNA pol III transcription (22–24). To investigate the biochemical mechanism of repression by Maf1 in a human in vitro system, we initially used a yeast tRNALeu3 gene template (type 2 promoter) that is efficiently transcribed in a HeLa cell extract (32). Full-length human Maf1 was expressed in bacteria and purified to near homogeneity (see “Experimental Procedures”). Increasing amounts of this preparation were first assayed to determine its ability to inhibit RNA pol III transcription prior to the formation of a PIC (defined here as a promoter-bound complex of TFIIIC, TFIIIB, and pol III). When added to a HeLa nuclear extract before the template (Fig. 1A), the Maf1 protein efficiently inhibited transcription in a concentration-dependent manner (Fig. 1, B and C). Repression activity was specific to Maf1 because at the upper end of the titration, an equivalent amount of nuclease-free BSA had no effect on transcription (Fig. 1B, lane 7). We then used an amount of Maf1 that maximally inhibited transcription under these conditions (Fig. 1C; 0.5 μg) to test whether the formation of a PIC prior to Maf1 addition would affect the efficiency of repression. PICs were assembled under equilibrium conditions during a 30-min incubation of the nuclear extract with the template DNA (Fig. 1D). Maf1 was then added (or not) followed by nucleotides to initiate transcription and the reactions were stopped at various times over a 60-min time course. In contrast to the absolute effect of Maf1 on transcription before PIC assembly (Fig. 1B), Maf1 addition after PIC assembly reduced but did not abolish transcription (Fig. 1E, compare lanes 5–8 with lanes 1–4). To determine the number of transcription cycles occurring under these conditions, we performed the same experiment in the presence of the nonionic detergent Sarkosyl. Sarkosyl at a final concentration of 0.05% has been found to selectively inhibit the reassembly of PICs and reinitiation of transcription when it is added prior to the initiating nucleotides. Thus, these conditions allow only a single round (SR) of transcription (33). By titrating the amount of Sarkosyl in our reactions, we confirmed that a concentration of 0.05% was sufficient to limit transcription consistent with a single inititation event for each functional PIC (supplemental Fig. S1). In prolonged incubations, the absolute level of transcription remained essentially constant in the presence of 0.05% Sarkosyl (Fig. 1E, lanes 9–12). This defines the SR transcription value. Accordingly, multiple round (MR) reactions generated 24 and 33 cycles of transcription after 35 and 60 min, respectively, at a rate of ∼1 transcript every 90–110 s (Fig. 1F). In contrast, the reduced absolute level of transcription in reactions containing Maf1 (to ∼20% of the untreated control; Fig. 1E) resulted in only six cycles of transcription after 60 min (Fig. 1F). This may reflect an ability of Maf1 to decrease the overall rate of the transcription cycle on functional PICs. Alternatively, it could reflect some heterogeneity in the reactions with respect to the inhibition of transcription by Maf1. For example, the reassembly of PICs involving RNA pol III molecules in solution might be differentially sensitive to Maf1 relative to reinitiation by polymerase molecules that remain bound to the template following termination (i.e. facilitated recycling). We note however that although facilitated recycling is well established in yeast in vitro systems (5, 6), it has not been reported in a mammalian system.
FIGURE 1.
Maf1 represses multiple round but not single round transcription by human RNA pol III. A, scheme to test the effect of Maf1 on HeLa nuclear extract (NE)-supported tRNALeu3 gene transcription. NE was preincubated with Maf1 or BSA for 10 min at room temperature. Cold nucleotides, [α-32P]CTP (C*), and plasmid tRNALeu3 template (0.5 μg) were added, and transcription was allowed to proceed for 50 min at 30 °C. B, lanes 1–6 show transcription supplemented with increasing amounts of recombinant human Maf1, (0–0.5 μg/reaction). Lane 7 contained NE with BSA (0.5 μg) as a negative control. C, the data in B were quantified and plotted relative to the unsupplemented NE. Rel Txn, relative transcription. D, as outlined, after preincubation of the plasmid template and NE, Maf1, or Sarkosyl and nucleotides were added, and the reactions were stopped after the times indicated. E, lanes 1–4 represent conditions that allow multiple round transcription (MR); lanes 5–8 were supplemented with Maf1 (0.5 μg); and lanes 9–12 contained Sarkosyl at a final concentration of 0.05% (SR Sark). F, transcript levels from E were quantified relative to the Sarkosyl-treated reaction (E, lane 9). Open bars, MR transcription; filled bars, Maf1-repressed transcription; shaded bars, SR transcription. One microgram of recombinant hMaf1/50-μl reaction corresponds to a concentration of 0.7 μm.
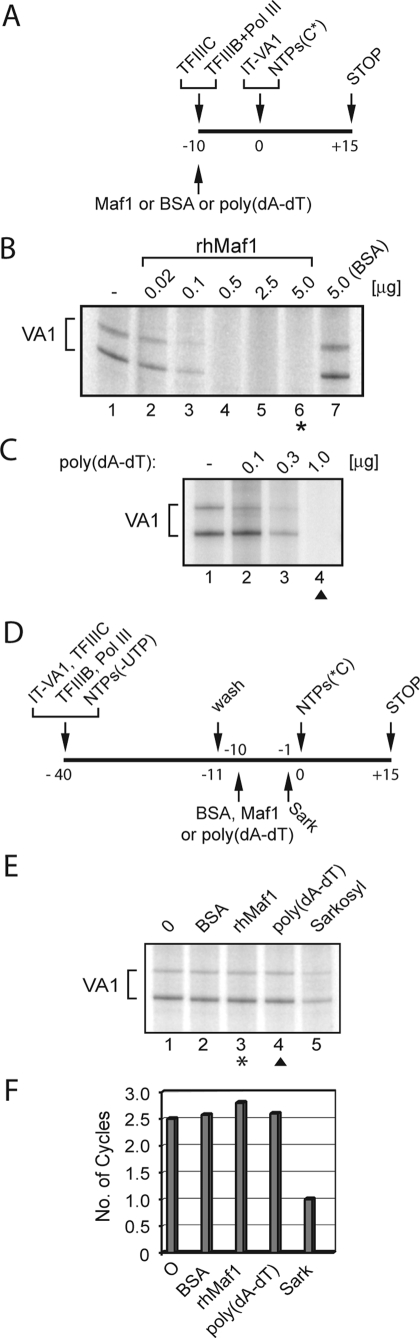
Facilitated Reinitiation of Transcription by Human RNA pol III on an Immobilized VA1 RNA Gene—To determine whether human RNA pol III is capable of facilitated recycling and to examine the effect of Maf1 on transcription of PICs, we developed an immobilized template transcription system. A linear VA1 promoter DNA fragment was generated by PCR using a biotinylated upstream oligonucleotide primer and immobilized on streptavidin-coated magnetic beads (IT-VA1). HeLa nuclear extract was fractionated on Phosphocellulose P11 to obtain fractions containing TFIIIB + pol III (P11:0.35) and TFIIIC (P11:0.6) that are able to actively transcribe the VA1 gene when combined (29). Importantly, these fractions are devoid of contaminating nucleotides, which is a prerequisite for stalling RNA pol III downstream of the transcription start site. Using the P11:0.35 + 0.6 fractions and ATP, GTP, and CTP but not UTP, we verified that transcription arrests after synthesis of the first six nucleotides on the VA1 template (data not shown). Two preliminary experiments were performed prior to analyzing the effect of Maf1 on transcription from purified stalled TCs. In the first, Maf1 was preincubated with the P11:0.35 + 0.6 fractions to determine its ability to repress reconstituted transcription from the immobilized VA1 template (Fig. 2A). Consistent with the experiment shown in Fig. 1B, IT-VA1 transcription was specifically and completely inhibited by Maf1 in a concentration-dependent manner (Fig. 2B). In the second experiment, we determined the ability of a nonspecific competitor DNA, poly(dA-dT), to sequester nonassembled transcription factors and polymerase, thereby preventing them from binding to the promoter. Inclusion of poly(dA-dT) into the P11:0.35 + 0.6 fractions before IT-VA1 addition (Fig. 2A) completely abolished transcription at the upper end of titration (Fig. 2C).
FIGURE 2.
Maf1 does not repress facilitated reinitiation by human RNA pol III on an immobilized VA1 DNA template. A, preincubation mixtures consisted of P11 fractions containing TFIIIC (P11:0.6), TFIIIB and RNA pol III (P11:0.35) together with Maf1, or nonspecific competitor DNA, poly(dA-dT). The immobilized VA1 DNA fragment (IT-VA1, 0.14 pmol of DNA) was added, and transcription was initiated with nucleotides. B, Maf1-mediated repression of IT-VA1 transcription. Recombinant Maf1 (0.0, 0.02, 0.1, 0.5, 2.5, and 5 μg) or BSA (5 μg) was tested as depicted in A. The transcripts initiated from the VA1 promoter are indicated at the left. One microgram of recombinant hMaf1/50-μl reaction corresponds to a concentration of 0.7 μm. C, competitor poly(dA-dT) (0.0 0.1, 0.3, and 1 μg) was tested as depicted in A. The asterisk in B (lane 6) and arrowhead in C (lane 4) indicate the amounts of Maf1 and poly(dA-dT), respectively, used in E. D, effect of recombinant Maf1 on transcription of TCs. IT-VA1 was preincubated with TFIIIC, TFIIIB, and RNA pol III fractions (P11: 0.35 + 0.6) in the presence of ATP, CTP, and GTP for 29 min at 30 °C. The supernatant was removed, and TCs were washed with KBCM500/0.015. The purified TCs were divided into aliquots and supplemented with transcription buffer only, BSA (5 μg), Maf1 (5 μg), or poly(dA-dT) (1 μg). The reactions were incubated for 10 min at room temperature. After 9 min, Sarkosyl was added (final concentration, 0.05%) to a buffer-supplemented reaction, and the incubation continued. Transcription in all of the reactions was resumed following addition of a complete set of NTPs and [α-32P]CTP (C*). E, IT-VA1 transcription in the presence of BSA, Maf1, poly(dA-dT), or Sarkosyl is compared with an unsupplemented reaction. F, data in E were expressed relative to the value obtained for the Sarkosyl-treated reaction.
To establish a transcription system that supports MR transcription only from recruited factors and polymerase, immobilized stalled ternary complexes were preassembled from the P11:0.35 + 0.6 fractions, washed in 0.5 m KCl, equilibrated in transcription buffer, and supplemented with all four NTPs, including [α-32P]CTP (Fig. 2D). RNA pol III-transcribed RNAs from this template were 156 and 200 nucleotides in length (the latter results from read-through of the first termination site) and were identical in size to the transcripts generated by HeLa extracts using VA1 plasmid templates (data not shown). A comparison of untreated and Sarkosyl-treated reactions revealed a small but reproducible difference in IT-VA1 transcription (Fig. 2E, lanes 1 and 5). Quantitation confirmed that the untreated reaction had completed two or three transcription cycles during the 15-min incubation (Fig. 2F). We then examined the effect of adding excess amounts of Maf1 or poly(dA-dT) (Fig. 2, B and C) on transcription of the immobilized TCs (Fig. 2D). Importantly, transcription remained at similar levels as in the unsupplemented or BSA-supplemented reactions (Fig. 2, E, lanes 1–4, and F), indicating that neither the factors nor the polymerase were released from the template in the course of transcription. Two to three rounds of transcription were also observed in the presence of Maf1 after a 50-min incubation (supplemental Fig. S2). This indicates that the 15-min reactions (Fig. 2, D–F) have reached a stable end point. Taken together, these data indicate that human RNA pol III can undergo facilitated recycling. Moreover, our data suggest that recombinant human Maf1 is unable to inhibit this type of processive reinitiation.
Maf1 Does Not Repress Facilitated Recycling of Human RNA pol III from Preinitiation Complexes—We next asked whether Maf1 is able to inhibit transcription from purified PICs. To assemble PICs, IT-VA1 was incubated with the P11:0.35 + 0.6 fractions containing the transcription factors and polymerase (Fig. 3A). We then determined the stability of the resulting complexes by subjecting them to extensive washes with various concentrations of KCl (0.06–0.5 m). Finally, nucleotides were provided to initiate transcription. Consistent with the anticipated lower stability of PICs compared with ternary complexes, these assays revealed that a maximum of 0.1 m KCl could be used for washing the PICs without diminishing transcription activity (data not shown). Ternary complexes, on the other hand, could withstand a 5-fold higher washing stringency. In subsequent reactions, washed PICs were preincubated with BSA or varying amounts of Maf1 in the presence of excess of poly(dA-dT) to ensure that any DNA-binding factor that dissociated from the template would not rebind (Fig. 3A, PIC scheme). A direct comparison of transcription from these PICs with transcription from stalled ternary complexes (assembled as in Fig. 3A, TC scheme) showed a very similar pattern (Fig. 3B, compare lanes 1–7 with lane 8 in both panels). Regardless of the amount of Maf1 tested (up to 5 μg), two or three transcription cycles were reproducibly completed (Fig. 3C). Collectively, the data demonstrate that polymerase stalling is not required to make transcription complexes resistant to Maf1. Rather, once polymerase-containing complexes are formed on the promoter, Maf1 is unable to inhibit RNA pol III transcription.
FIGURE 3.
Facilitated recycling of human RNA pol III from preinitiation complexes is resistant to Maf1. A, upper scheme, immobilized PICs were formed on the VA1 gene (IT-VA1) by incubation of TFIIIC, TFIIIB, and RNA pol III fractions (P11: 0.35 + 0.6) for 29 min at 30 °C. Unbound material was removed by extensive washing with KBCM100/0.015. The purified PICs were challenged with poly(dA-dT) (1 μg) and BSA (5 μg) or Maf1 (0.02–5 μg). In a separate reaction, Sarkosyl treatment was carried out as described in Fig. 2D. Transcription was initiated with the addition of nucleotides and continued for 15 min at 30 °C. Lower scheme, immobilized TCs were prepared and assayed as described in Fig. 2D. The reactions contained BSA (5 μg) or Maf1 (0.0–5 μg) or Sarkosyl as indicated. B, MR transcription from PICs (upper panel) and TCs (lower panel) supplemented with 0.02, 0.1, 0.5, 2.5, and 5 μg of Maf1 (lanes 3–7). Control lanes 1 and 2 contained no additional protein and BSA (5 μg), respectively. SR transcription in the presence of Sarkosyl is shown in lane 8. C, the data in B are expressed relative to the Sarkosyl-treated reactions (lane 8).
Maf1 Efficiently Inhibits Transcription from Preassembled TFIIIC-TFIIIB-DNA Complexes—To examine the effect of Maf1 on different steps in PIC assembly, we prepared separate RNA pol III and TFIIIB fractions. HeLa S100 extract was applied to Phosphocellulose P11 to obtain the P11:0.35 fraction that was further chromatographed on DEAE-Sephadex A25 to yield fractions DEAE:0.15 (TFIIIB) and DEAE:1.0 (pol III) (30). TFIIIC- and TFIIIB-containing fractions (P11:0.6 and DEAE: 0.15, respectively) were used to assemble complexes on the IT-VA1. After a washing step (0.1 m KCl) to remove unbound factors, the complexes were exposed (or not) to BSA or increasing amounts of Maf1 before the addition of RNA pol III (DEAE:1.0) and nucleotides to initiate transcription (Fig. 4A). Strong inhibition of transcription activity was observed (Fig. 4B); at the highest concentration of Maf1, transcription was reduced to 13% of the unsupplemented reaction (Fig. 4C). However, this level of repression is noticeably less potent than that observed when the factors and polymerase are incubated with Maf1 before the template (Fig. 2, A and B). Repression by Maf1 from preassembled TFIIIB-TFIIIC-DNA complexes (Fig. 4A) is likely to reflect, at least in part, its inhibitory binding to the polymerase in solution (19, 22–24). However, another possibility is that the retention of Maf1 by promoter-bound TFIIIB, via its interactions with Brf1 (22–24), could prevent the recruitment of RNA pol III into a productive complex. To examine this possibility, complexes containing TFIIIC and TFIIIB were assembled on IT-VA1 and treated with Maf1 as described above. Then unbound Maf1 was removed in a second washing step before the addition of RNA pol III and NTPs (Fig. 4D). Titration of Maf1 resulted in a modest concentration-dependent reduction in transcription (Fig. 4E). At the highest Maf1 concentrations, transcription reached a plateau at ∼50% of the untreated level, implying that half of the complexes were able to retain Maf1 under these conditions (Fig. 4F). By comparing transcription without and with the additional wash to remove unbound Maf1 (Fig. 4, C and F), it is apparent that the efficiency of repression is higher for Maf1 binding to the polymerase off the DNA. However, the ability to chromatin immunoprecipitate human Maf1 on target promoters in vivo (17, 24) suggests that transcriptional repression by Maf1 bound to TFIIIB-DNA complexes is likely to be biologically significant.
FIGURE 4.
Maf1 inhibits the recruitment of human RNA pol III to immobilized TFIIIB-TFIIIC-DNA complexes. A, stable binary complexes were formed on the immobilized VA1 template (IT-VA1) during a 29-min incubation at 30 °C with fractions containing TFIIIC (P11: 0.6) and TFIIIB (DEAE: 0.15). Following washes with KBCM100/0.015, BSA or Maf1 was added. The reactions were complemented with RNA pol III (DEAE 1.0) and NTPs to initiate transcription. B, transcripts from reactions supplemented with 0.02, 0.1, 0.5, 2.5, and 5 μg of Maf1 are shown in lanes 3–7. Control lanes 1 and 2 contained no additional protein and BSA (5 μg), respectively. C, the bar graph shows the transcription levels in B relative to the reaction without additional protein (open bar). D, reactions were assembled as in A, except that incubations with BSA and Maf1 were conducted for 14 min, and the mixtures were washed with KBCM100/0.015 before the addition of RNA pol III and NTPs. E, Maf1 retained by binary complexes reduces RNA pol III transcription. The additions of BSA and Maf1 are identical to B. F, transcripts were quantified as in C. Rel Txn, relative transcription.
Human Maf1 Inhibits TFIIIB Recruitment to DNA-bound TFIIIC, albeit with Low Efficiency—Previous studies in yeast have shown that recombinant Maf1 can interact with the Brf1 subunit of TFIIIB in solution and thereby inhibit the assembly of TFIIIB onto TFIIIC-DNA complexes (22). Because the interaction between Maf1 and Brf1 is conserved in humans (23, 24), it is similarly expected to be inhibitory for transcription. Taking advantage of the sequential assembly of TFIIIC and then TFIIIB onto DNA, we followed the binding of each factor to IT-VA1 with a washing step and assayed for an effect of preincubating the TFIIIB fraction (DEAE:0.15) with Maf1 on complex assembly using transcription activity as functional readout (Fig. 5A, TFIIIB/Maf1). As expected, we observed a concentration-dependent inhibition of transcription by Maf1 at the upper end of titration (Fig. 5B, upper panel, lanes 6 and 7). As a control experiment to exclude the possibility of an inhibitory effect caused by nonspecific binding of Maf1 to DNA or beads, we incubated Maf1 or BSA with IT-VA1 and employed the same sequential washing steps before and after the addition of TFIIIC and TFIIIB (Fig. 5A, Con). Consistent with expectations, this regimen had no effect on transcription (Fig. 5B, lower panel). Comparing the efficiency of repression resulting from preincubation of Maf1 with the TFIIIB versus polymerase fractions (Fig. 5, TFIIIB/Maf1 versus Fig. 4, A–C), it appears that the polymerase is more sensitive to inhibition by Maf1 in this system.
Human Maf1 Targets RNA pol III in Solution but Not in Stalled Ternary Complexes—Evidence gained from our previous experiments (Figs. 2, D–F, 3, and 4, A–C) supports a concept that RNA pol III engaged in DNA-bound complexes (PICs or TCs) is resistant to the action of Maf1, whereas the free pool of RNA pol III can be efficiently targeted by Maf1 to repress transcription. To demonstrate these properties in a single experiment, purified stalled TCs formed with a limiting amount of RNA pol III were examined for their response to Maf1 in the presence of an additional quantity of RNA pol III that was added to the washed complexes (Fig. 6A). Relative to reactions containing the washed TCs but no extra polymerase, RNA pol III supplementation increased transcription ∼2-fold (Fig. 6, B and C, compare + Pol III lanes 1, 2, and 7 with [minus] Pol III lane 1′). Importantly, the increased level of transcription resulting from RNA pol III supplementation was sensitive to inhibition by Maf1 (Fig. 6, B and C) unlike transcription from washed TCs (Figs. 2F and 3C). At the upper end of the Maf1 titration, transcription was reduced to the same level as in the non-supplemented reaction (Fig. 6, B and C). Similar to other experiments (Figs. 2F and 3C), the addition of excess Maf1 (5 μg) to washed TCs in the absence or the presence of RNA pol III supplementation resulted in three cycles of transcription. This is consistent with our previous observations that the initial rounds of transcription from PICs and TCs are resistant to repression by Maf1.
FIGURE 6.
Maf1 targets only nonrecruited human RNA pol III. A, IT-VA1 ternary complexes were prepared, washed and then BSA, Maf1 or Sarkosyl was added as described in the legend for Fig. 3C. Transcription was initiated with NTPs in the presence or absence of additional RNA pol III and allowed to proceed for 50 min at 30 °C. B, upper panel (+Pol III) shows RNA pol III-supplemented reactions where 0.02, 0.1, 0.5, 2.5, and 5 μg of Maf1 were tested in lanes 3–7. Lanes 1 (no added protein) and 2 (BSA; 5 μg) serve as negative controls. Sarkosyl in lane 8 limits transcription to a single round. Lanes 1′ and 2′ contain Maf1 (5 μg) and Sarkosyl, respectively, and are the same as lanes 7 and 8 except that no additional RNA pol III was added (lower panel, –Pol III). C, the data in B are expressed relative to the corresponding Sarkosyl-treated reactions (lanes 8 and 2′).
DISCUSSION
Although a considerable amount of information has been accumulated over the last few years regarding transcriptional regulation by Maf1 (21), a detailed mechanistic description of how this important regulator functions to repress transcription is lacking. Toward this goal, we have used transcription from a VA1 template attached to a solid support to assess Maf1 inhibition of biochemically defined steps preceding and following transcription initiation by human RNA pol III. Consistent with studies in yeast and the conservation of Maf1 interactions with Brf1 and RNA pol III from yeast to humans (22, 23), our experiments revealed differences in the efficiency of repression by Maf1 between unassembled and preassembled initiation complexes. Transcription is fully inhibited by relatively small amounts of human Maf1 when the recombinant protein is added to the factors and polymerase prior to the template (Figs. 1, A–C, and 2, A–C) (24). As suggested by different order of addition experiments, this reflects a relatively high affinity interaction of human Maf1 with RNA pol III in solution (estimated to be 10–50 nm; Figs. 1, A–C, and 4, A–C) and a lower affinity interaction of Maf1 with Brf1 in the TFIIIB fraction (Fig. 5). We conclude that human Maf1 can prevent TFIIIB and, even more efficiently, RNA pol III from forming active complexes, either by directly interfering with their associations or by causing them to assemble in an inactive conformation. In addition, we detected a functional interaction between Maf1 and preexisting TFIIIB-TFIIIC-DNA complexes, suggesting that Maf1 binding to Brf1 in the DNA-bound complex can inhibit RNA pol III recruitment or function.
The high stability of TFIIIB complexes on RNA pol III genes in vitro and in vivo (1, 34) indicates that most RNA synthesis on these genes occurs through reinitiation of transcription. This process is highly efficient and in yeast results in the production of ∼3–6 million molecules of tRNA (35) from 274 genes over a generation time of ∼100 min (i.e. 2–4 transcripts/gene/sec). Facilitated recycling of RNA pol III is thought to play an important role in achieving this high level of transcription (5, 6). Biochemical studies including template commitment assays have shown that recycling of the polymerase on the same template (i.e. without dissociation) increases the rate of reinitiation by yeast RNA pol III 5–10-fold compared with reactions where the polymerase must first be recruited from solution (5). We have employed an immobilized template assay to show that human RNA pol III is also retained in the original transcription complex and is able to direct several rounds of transcription without dissociating after each round of synthesis (Fig. 2). Thus, facilitated recycling is a conserved property of RNA pol III in yeast and humans.
The high transcription activity of RNA pol III genes is tightly regulated to ensure metabolic economy and to maintain the appropriate levels of 5 S rRNA, tRNA, and other transcripts to meet the protein synthetic and other needs of the cell. Accordingly, efficient repression of transcription would seem to require the ability to interrupt the process of facilitated recycling. We were therefore surprised to find that the TFIIIC-TFIIIB-pol III complexes assembled on the immobilized VA1 promoter were completely resistant to inhibition by Maf1 and allowed multiple rounds of transcription, equivalent to control reactions. Similarly, stalled ternary complexes, where the polymerase was arrested after the synthesis of a short transcript, were refractory to Maf1 concentrations that prevented transcription under conditions requiring polymerase recruitment. Release of the stalled polymerase by the addition of NTPs again produced multiple rounds of transcription despite the presence of Maf1. These experiments show that human RNA pol III is susceptible to Maf1 only when the polymerase is free in solution and that RNA pol III bound to preinitiation complexes or in elongation complexes is protected from repression by Maf1 for at least several rounds of transcription in our in vitro system (Fig. 7).
FIGURE 7.
A model for human Maf1 involvement in repression of RNA polymerase III. A, efficient repression of transcription by Maf1 is achieved by preventing RNA pol III recruitment to promoter-bound TFIIIB and TFIIIC. B, once RNA pol III is recruited into PICs or transcription is initiated and the polymerase is stalled at TCs, Maf1 is unable to inhibit multiple round transcription mediated by facilitated recycling.
How do we reconcile these in vitro results with the concept of robust Maf1-dependent repression of reinitiation in vivo? One possibility is that the recombinant Maf1 used for our experiments and/or other aspects of the in vitro system may not recapitulate the repression of facilitated recycling that is thought to operate in vivo. In support of this, previous studies in yeast have shown that a nuclear-localized, dephosphorylated (6SA mutant) form of Maf1 is not sufficient for repression and that additional (presumably) post-translational events are necessary (21, 25). An understanding of these events in yeast or in humans is currently lacking. This information will be crucial in evaluating whether facilitated recycling can be interrupted by Maf1 in vitro. Alternatively, Maf1 may in fact not inhibit facilitated recycling in vivo. In this scenario, Maf1-dependent repression could be achieved if mechanisms exist to limit the number of rounds of facilitated reinitiation (e.g. stochastic dissociation of the polymerase or regulatory events impacting this process). Support for this possibility is provided by the in vitro properties of an RNA pol III variant, pol IIIΔ, which is defective in termination and facilitated recycling because of the absence of the subunits C37, C53, and C11 (36). Through in vitro complementation experiments, the terminator recognition defect of pol IIIΔ was corrected by the addition of a recombinant C37-C53 complex. Facilitated recycling was restored by the further addition of recombinant C11. Notably, this effect did not require the stimulatory RNA cleavage activity of the C11 subunit. These and other data have lead to a model in which a C11-dependent conformational change in the polymerase is important for facilitated recycling (36). As an extension to this model, we suggest that cellular events may impact this conformational change to alter the relative rates of recycling and dissociation and provide an opportunity for Maf1 to repress the dissociated polymerase.
Supplementary Material
Acknowledgments
We thank Karen Puglia for technical assistance and Robyn Moir for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM42728. This work was also supported by funds from the Albert Einstein College of Medicine. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: pol, polymerase; PIC, preinitiation complex; TC, ternary complex; BSA, bovine serum albumin; SR, single round; MR, multiple round; NE, nuclear extract.
References
- 1.Geiduschek, E. P., and Kassavetis, G. A. (2001) J. Mol. Biol. 310 1–26 [DOI] [PubMed] [Google Scholar]
- 2.Schramm, L., and Hernandez, N. (2002) Genes Dev. 16 2593–2620 [DOI] [PubMed] [Google Scholar]
- 3.Liao, Y., Moir, R. D., and Willis, I. M. (2006) Mol. Cell Biol. 26 5946–5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kassavetis, G. A., Braun, B. R., Nguyen, L. H., and Geiduschek, E. P. (1990) Cell 60 235–245 [DOI] [PubMed] [Google Scholar]
- 5.Dieci, G., and Sentenac, A. (1996) Cell 84 245–252 [DOI] [PubMed] [Google Scholar]
- 6.Ferrari, R., Rivetti, C., Acker, J., and Dieci, G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 13442–13447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrari, R., and Dieci, G. (2008) Cell Mol. Biol. Lett. 13 112–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maraia, R. J., Kenan, D. J., and Keene, J. D. (1994) Mol. Cell Biol. 14 2147–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, Z., Bai, L., Hsieh, Y. J., and Roeder, R. G. (2000) EMBO J. 19 6823–6832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, Z., and Roeder, R. G. (1998) Mol. Cell 1 749–757 [DOI] [PubMed] [Google Scholar]
- 11.White, R. J. (2005) Nat. Rev. Mol. Cell Biol. 6 69–78 [DOI] [PubMed] [Google Scholar]
- 12.Gomez-Roman, N., Felton-Edkins, Z. A., Kenneth, N. S., Goodfellow, S. J., Athineos, D., Zhang, J., Ramsbottom, B. A., Innes, F., Kantidakis, T., Kerr, E. R., Brodie, J., Grandori, C., and White, R. J. (2006) Biochem. Soc. Symp. 141–154 [DOI] [PubMed]
- 13.Zhang, C., Comai, L., and Johnson, D. L. (2005) Mol. Cell Biol. 25 6899–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woiwode, A., Johnson, S. A., Zhong, S., Zhang, C., Roeder, R. G., Teichmann, M., and Johnson, D. L. (2008) Mol. Cell Biol. 28 4204–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall, L., Kenneth, N. S., and White, R. J. (2008) Cell 133 78–89 [DOI] [PubMed] [Google Scholar]
- 16.Johnson, S. A., Dubeau, L., and Johnson, D. L. (2008) J. Biol. Chem. 283 19184–19191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, S. S., Zhang, C., Fromm, J., Willis, I. M., and Johnson, D. L. (2007) Mol. Cell 26 367–379 [DOI] [PubMed] [Google Scholar]
- 18.Boguta, M., Czerska, K., and Zoladek, T. (1997) Gene (Amst.) 185 291–296 [DOI] [PubMed] [Google Scholar]
- 19.Pluta, K., Lefebvre, O., Martin, N. C., Smagowicz, W. J., Stanford, D. R., Ellis, S. R., Hopper, A. K., Sentenac, A., and Boguta, M. (2001) Mol. Cell Biol. 21 5031–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Upadhya, R., Lee, J., and Willis, I. M. (2002) Mol. Cell 10 1489–1494 [DOI] [PubMed] [Google Scholar]
- 21.Willis, I. M., and Moir, R. D. (2007) Trends Biochem. Sci. 32 51–53 [DOI] [PubMed] [Google Scholar]
- 22.Desai, N., Lee, J., Upadhya, R., Chu, Y., Moir, R. D., and Willis, I. M. (2005) J. Biol. Chem. 280 6455–6462 [DOI] [PubMed] [Google Scholar]
- 23.Reina, J. H., Azzouz, T. N., and Hernandez, N. (2006) PLoS ONE 1 e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodfellow, S. J., Graham, E. L., Kantidakis, T., Marshall, L., Coppins, B. A., Oficjalska-Pham, D., Gerard, M., Lefebvre, O., and White, R. J. (2008) J. Mol. Biol. 378 481–491 [DOI] [PubMed] [Google Scholar]
- 25.Moir, R. D., Lee, J., Haeusler, R. A., Desai, N., Engelke, D. R., and Willis, I. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 15044–15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oficjalska-Pham, D., Harismendy, O., Smagowicz, W. J., Gonzalez, d. P., Boguta, M., Sentenac, A., and Lefebvre, O. (2006) Mol. Cell 22 623–632 [DOI] [PubMed] [Google Scholar]
- 27.Roberts, D. N., Wilson, B., Huff, J. T., Stewart, A. J., and Cairns, B. R. (2006) Mol. Cell 22 633–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dignam, J. D., Lebovitz, R. M., and Roeder, R. G. (1983) Nucleic Acids Res. 11 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segall, J., Matsui, T., and Roeder, R. G. (1980) J. Biol. Chem. 255 11986–11991 [PubMed] [Google Scholar]
- 30.White, R. J., Gottlieb, T. M., Downes, C. S., and Jackson, S. P. (1995) Mol. Cell Biol. 15 1983–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dean, N., and Berk, A. J. (1988) Mol. Cell Biol. 8 3017–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Standring, D. N., Venegas, A., and Rutter, W. J. (1981) Proc. Natl. Acad. Sci. U. S. A. 78 5963–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovelman, R., and Roeder, R. G. (1990) Genes Dev. 4 646–658 [DOI] [PubMed] [Google Scholar]
- 34.Huibregtse, J. M., and Engelke, D. R. (1989) Mol. Cell Biol. 9 3244–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phizicky, E. M. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 11127–11128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landrieux, E., Alic, N., Ducrot, C., Acker, J., Riva, M., and Carles, C. (2006) EMBO J. 25 118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.