Abstract
TEA domain (TEAD) transcription factors serve important functional roles during embryonic development and in striated muscle gene expression. Our previous work has implicated a role for TEAD-1 in the fast-to-slow fiber-type transition in response to mechanical overload. To investigate whether TEAD-1 is a modulator of slow muscle gene expression in vivo, we developed transgenic mice expressing hemagglutinin (HA)-tagged TEAD-1 under the control of the muscle creatine kinase promoter. We show that striated muscle-restricted HA-TEAD-1 expression induced a transition toward a slow muscle contractile protein phenotype, slower shortening velocity (Vmax), and longer contraction and relaxation times in adult fast twitch extensor digitalis longus muscle. Notably, HA-TEAD-1 overexpression resulted in an unexpected activation of GSK-3α/β and decreased nuclear β-catenin and NFATc1/c3 protein. These effects could be reversed in vivo by mechanical overload, which decreased muscle creatine kinase-driven TEAD-1 transgene expression, and in cultured satellite cells by TEAD-1-specific small interfering RNA. These novel in vivo data support a role for TEAD-1 in modulating slow muscle gene expression.
A functionally important characteristic of mammalian skeletal muscle is its diversity in fiber type composition, making each skeletal muscle uniquely suited for a particular function. This property is brought about by the differential transcription of contractile protein gene families during embryonic myogenesis and by the influence of nerve activity (1, 2). The structure of skeletal muscle is highly specialized for the generation of force that is required for various movement patterns and postural maintenance. The force-generating capacity of skeletal muscles occurs by means of an intricate organization of contractile proteins into myofibrils composed of repeating units called sarcomeres, the smallest force-producing unit of a muscle fiber (cell). Of the skeletal muscle contractile proteins, myosin heavy chain (MyHC)2 is a motor protein comprising the thick filament of sarcomeres, and it has been shown to represent a major determinant of the unloaded shortening velocity (Vmax) of a given muscle or muscle fiber (3). The MyHC protein is encoded by a multigene family that produces four distinct adult-stage MyHC isoforms termed fast type IIb, IIx/d, IIa, and slow type I (or β). Because each MyHC displays distinct biochemical (actin-activated ATPase activity) and physiological (force-velocity) properties, their expression pattern has been shown to contribute to the classification of four primary fiber types named fast IIb, IIx/d, IIa, and slow type I. Therefore, the amount and type of MyHC comprising the sarcomeres of a given skeletal muscle is of functional importance.
A noteworthy feature of adult skeletal muscle is its intrinsic ability to adapt its fiber-type composition to accommodate functional demands imposed by contractile usage patterns. In this regard, both human and animal studies have reported perturbation-specific skeletal muscle adaptations that involve mass, endurance, strength, power, metabolic properties, and MyHC expression pattern/fiber type (4). Recent inquiries into the molecular basis of skeletal muscle remodeling have provided evidence that stimulation of the calcium-activated calcineurin, calmodulin-dependent protein kinase, and protein kinase D1 (PKD1) signaling pathways cooperate to modulate the transcriptional activation of slow fiber genes through class II HDAC degradation and the activation of members of the MEF2 and nuclear factor of activated T-cells (NFAT) transcription factor families (5–7). In addition to MEF2 and NFAT, other transcriptional regulators, such as MusTRD/GTF3, Six1, Eya1, PGC-1α, PGC-1β, and PPARδ, have been implicated in fiber type-specific gene expression (8–13).
In our previous work aimed at identifying cis-acting element(s) that direct βMyHC slow fiber gene expression and mechanical overload (MOV) responsiveness, we delineated an A/T-rich element (βA/T-rich; –269/–258, 5′-GGAGATATTTTT-3′) that appears necessary for βMyHC transgene slow fiber expression and MOV responsiveness (14). By using the βA/T-rich element as bait in a one-hybrid screen of both adult skeletal muscle and MOV-plantaris cDNA libraries, we identified TEAD proteins as the functionally relevant βA/T-rich binding factor. Interestingly, additional experiments revealed specific TEAD protein binding to a subset of A/T-rich/MEF2 elements located within the control region of several other muscle-specific genes and revealed that this binding was enriched during MOV-induced fast to slow fiber type switching (15).
The vertebrate TEAD genes encode a family of transcription factors that include TEAD-1 (NTEF-1/TEF-1), TEAD-2 (ETEF-1/TEF-4), TEAD-3 (DTEF-1/TEF-5), and TEAD-4 (RTEF-1/TEF-3). All TEAD family members contain an evolutionarily conserved 72-amino acid DNA binding domain (TEAD) that is found in plant (ABAA), fly (Scalloped), and yeast (TEC1) transcription factors. TEAD proteins have been shown to serve a regulatory role by binding to canonical MCAT elements (5′-CATTCC(T/A)-3′) located in the promoter/enhancer region of several cardiac, smooth, and skeletal muscle genes and by combinatorial interactions with adjacently bound transcriptional regulators and ubiquitous and/or tissue-specific coactivators (16, 17). Although most mammalian tissues express more than one TEAD protein during embryonic and adult life, several recent studies involving single or double TEAD gene inactivation (TEAD-1, TEAD-2, and TEAD-4) have shown that these transcription factors serve both unique and overlapping functional roles during embryonic development (18–22). Thus, the regulatory specificity of each TEAD protein during development and in response to various stimuli is probably due to their capacity to recognize a broad spectrum of DNA elements, accessibility to target elements, interactions with various tissue-specific co-activator/co-binding proteins, post-translational modifications, or subtle gradients in their expression levels.
To investigate whether TEAD-1 functions as a modulator of basal and/or inducible slow muscle gene expression in vivo, we generated transgenic mice that expressed HA-tagged TEAD-1 under the control of the muscle creatine kinase (MCK) promoter. The mouse MCK promoter has been extensively studied in both cultured striated muscle cells and transgenic mice (23–25). In transgenic mice, a 3300-nucleotide region of the MCK 5′-flanking sequence was shown to be sufficient to drive expression of a reporter gene at high levels in skeletal muscle, at lower levels in the heart, and at barely detectable levels in nonmuscle tissue, which is similar in pattern and magnitude to the expression pattern of the endogenous MCK gene (23). In addition, we have previously shown that 48 h of MOV virtually repress the expression of the 3300-bp MCK promoter/reporter gene (26). On the basis of the latter data, we hypothesize that any phenotype resulting from MCK-driven TEAD-1 overexpression could in part be reversed by 48 h of MOV. We show that a persistent increase in TEAD-1 protein induced a change in MyHC and troponin complex protein expression pattern and contractile properties that more closely resemble slow oxidative muscle fibers. We further demonstrate that increased HA-TEAD-1 expression activated glycogen synthase kinase (GSK)-3α/3β, resulting in decreased nuclear β-catenin and NFATc1/c3. The latter effects could be reversed in vivo by 48 h of MOV, which decreased MCK-driven TEAD-1 transgene expression, and in cultured satellite cells by TEAD-1 siRNA. These data support a novel role for TEAD-1 in modulating a slow skeletal muscle gene program.
EXPERIMENTAL PROCEDURES
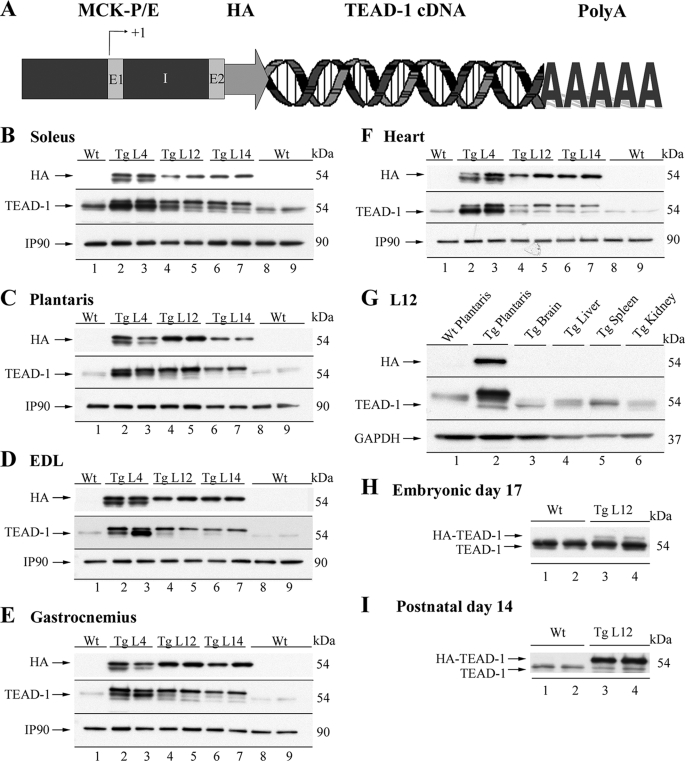
Generation and Screening of Transgenic Mice—Transgenic mice were generated by microinjection of purified transgene DNA into pronuclei of single cell fertilized embryos, as described previously (27). Transgene-positive mice (founders) were identified using PCR. Subsequent offspring derived from mating the founders were also screened for transgene incorporation by using PCR. In the present study, multiple independent transgenic lines representing the HA-TEAD-1 transgene were studied, and all lines were maintained in a heterozygous state by continual outbreeding to nontransgenic BDF1 mice. The transgene was designed as follows. The mouse TEAD-1 cDNA containing an in-frame HA tag at its 5′-end was subcloned at the 3′-end of the MCK regulatory sequences. The HA-tagged TEAD-1 cDNA was released from pBluescriptSK by XhoI digest and subcloned into the blunt-ended PstI site of plasmid pBSMCK (a generous gift from Dr. R. Kahn), which consisted of a 6.5 genomic region composed of 3.3 kb of the MCK promoter and enhancer 1, untranslated exon 1, 3 kb of intron 1, which included the enhancer 2 region, and the first 16 bp of exon 2 (28). The TEAD-1 cDNA was followed by the SV40 polyadenylation signal. The transgene was released from the plasmid by KpnI/SacII digest, gel-purified, and injected into fertilized eggs of BDF1 mice. The incorporation of an HA tag allowed us to easily make a distinction between endogenous TEAD-1 protein and the transgene-encoded HA-TEAD-1 protein by Western blot analysis (Fig. 1A).
FIGURE 1.
Schematic of TEAD-1 transgene and expression pattern of transgenic and endogenous TEAD-1. A hemagglutinin-tagged mouse TEAD-1 cDNA was subcloned at the 3′-end of a mouse muscle-specific creatine kinase promoter/enhancer cassette followed by SV40 polyadenylation signal (A). Western blot analysis was performed using 100 μg of total protein extract isolated from the soleus (B), plantaris (C), EDL (D), gastrocnemius (E), and heart (F) of WT and from three independent TEAD-1 transgenic mouse lines (lines 4, 12, and 14). Analysis using either anti-HA or anti-TEAD-1 antibody revealed the striated muscle-specific expression pattern of the TEAD-1 transgene (HA-tagged TEAD-1 protein). G, HA-tagged TEAD-1 protein was not detected in nonmuscle tissue (brain, liver, spleen, and kidney) of transgenic line 12. TEAD-1 transgenic line 12 embryonic day 17 (H) and neonatal (day 14 postbirth) (I) leg muscle tissue express HA-tagged TEAD-1 protein.
Mechanical Measurements—Force-velocity and power-load relationships on adult (5-month-old) sex-matched wild-type (WT) and HA-TEAD-1 transgenic mice (n = 4–6 mice/group) were determined using Dynamic Muscle Control software (Aurora Scientific) to elicit tetanic afterloaded contractions as previously described (29). Contractile measurements were completed with the assessment of maximal isometric contraction (Po). Following the completion of mechanical measurements, the extensor digitorum longus (EDL) muscle was removed from the experimental apparatus, blotted on filter paper, and immediately weighed. The total cross-sectional area (mm–2) for each muscle was determined by dividing muscle mass (mg) by the product of optimal fiber length and 1.06 mg mm–3, the density of mammalian skeletal muscle (30). Force-velocity and power-load curves were analyzed as described previously (31). The values for twitch and tetanic tension were normalized to cross-sectional area to obtain specific tension (millinewtons mm–2). In a similar fashion, power output (watts) was normalized with respect to muscle mass (watts kg–1) (30).
Protein Isolation for Western Blots—For total protein extracts, 50 mg of tissue was homogenized for 30 s on ice in 0.5 ml of buffer (50 mm Tris, pH 7.5, 10 mm EGTA, 5 mm EDTA, phenylmethylsulfonyl fluoride, phosphatase inhibitor mixture 1, 2, and proteinase inhibitor mixture (Sigma)), using an Omni International TH homogenizer. For isolation of nuclear protein, the homogenate was centrifuged at 14,000 × g for 10 min at 4 °C, the cytoplasmic fraction (supernatant) was removed, and the pellet was resuspended in 0.2 ml of buffer. Sonication for 10 s with a dismembranator (Fisher) of homogenates was followed by the addition of 1% Triton X-100 and incubation on ice for 30 min. Membrane fractions were isolated by homogenizing tissues in 1.5 ml of buffer (50 mm Tris-HCl, pH 7.4, 50 mm mannitol, 2 mm EDTA), centrifugation at 500 × g for 10 min at 4 °C, mixing the supernatant with 3.2 ml of buffer (50 mm Tris-HCl, pH 7.4, 300 mm mannitol, 2 mm EDTA), and centrifugation at 40,000 rpm for 45 min. Protein concentrations were determined using a protein assay kit (Bio-Rad), and extracts were stored at –80 °C. Protein separation and analysis was performed as previously described (15). Experimental and standard bands were scanned and quantified using Multi Gauge software, and the experimental data were normalized by dividing by the signal of the standard. The antibodies used in this study are as follows: TEAD-1 (BD Transduction Laboratories), HA (Cell Signaling), Akt, p-Akt, Akt1, and Akt2 (Cell Signaling), GSK-3α/β (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), p-Ser-GSK-3α and p-Ser-GSK-3β (Cell Signaling), glyceraldehyde-3-phosphate dehydrogenase (Cell Signaling), β-catenin (Cell Signaling), NFATc1 and NFATc3 (Santa Cruz Biotechnology), Serca2a (Badrilla), Serca1 (Santa Cruz Biotechnology), Myoglobin (Santa Cruz Biotechnology), Troponin I slow (Santa Cruz Biotechnology), histone H1 (Santa Cruz Biotechnology), IP90 anti-peptide antibody (Abcam), and anti-rabbit IgG (horseradish peroxidase-linked) and anti-mouse IgG (horseradish peroxidase-linked) (Cell Signaling).
Protein Extraction for High Resolution Gel Electrophoresis—Protein extraction and high resolution gel electrophoresis for MyHC isoform separation were performed as described previously (32). Briefly, 50 mg of muscle tissue of adult (5-month-old) and aged (11-month-old) wild-type and TEAD-1 Tg mice was processed, and 0.75 μg of total protein was separated on an 8% acrylamide gel by 104 V for 24 h at 4 °C. The gel was then stained with SilverSNAP stain (Thermo Scientific) according to the manufacturer's recommendations.
Animal Care and MOV Surgical Procedure—The imposition of a mechanical overload (bilateral) on the fast twitch plantaris muscle was accomplished as described by us previously (26) and approved by the Animal Care Committee for the University of Missouri-Columbia. The animals were housed in an Association for the Assessment and Accreditation of Laboratory Animal Care International-accredited animal facility.
Satellite Cell Isolation and siRNA Transfection—Freshly isolated satellite cells from adult mice were isolated and cultured as previously described (33). Briefly, muscle was dissected from the hind limbs, minced, digested in 400 units/ml collagenase type I (Worthington), diluted in Ham's F-12 medium (Invitrogen), filtered, and collected by centrifugation. Cells were cultured on treated plates (Nunc) coated with 0.66% gelatin unless otherwise noted. After 2 days in culture, 50 nm siRNA mouse TEFD1 (Dharmacon RNA Technologies, ON-TARGETplus SMARTpool) and siCONTROL Non-Targeting #1 were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol, and the protein was extracted after 36 h of incubation and analyzed by Western blot as described above.
RNA Samples and Isolation—EDL muscles of adult wild-type and transgenic (RTN12) mice were surgically removed and transferred to RNAlater (Ambion) according to the manufacturer's instructions. Total RNA was isolated from ∼20–50 mg of tissue from each animal using RNA STAT-60 according to the manufacturer's instructions (Tel Test Inc). Equal amounts of total RNA from each of four replicate tissue samples were pooled for cDNA synthesis after RNA quality control tests.
RNA Quality Control—Immediately prior to cDNA synthesis, the purity and concentration of RNA samples were determined from A260/280 readings using a dual beam UV spectrophotometer, and RNA integrity was determined by capillary electrophoresis using the RNA 6000 Nano Lab-on-a-Chip kit and the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) as per the manufacturer's instructions.
Reverse Transcription, PCR, and Real Time Quantitative PCR Analysis—Single strand cDNA was synthesized from 0.02 to 2.0 μg of each total RNA sample using the High Capacity cDNA synthesis kit (Applied Biosystems), according to the manufacturer's instructions. Conventional PCR for TEAD isoforms was performed using 20 nmol of the following primers for TEAD-1 (5′-AGAGCCCTGCCGAAAACATGG and 5′-TGGCTGTCCTGTCTGTATCAT), TEAD-2 (5′-GGAAGGCAGCGAAGAGGGC and 5′-CTTCACGTCTGGAACATTCCAT), TEAD-3 (5′-TGGACAAGGGTCTGGACAACG and 5′-AACCTTGAGGAGGAGGAGAAG), TEAD-4 (5′-ATTACCTCCAACGAGTGGAGC and 5′-CTGGCAAAGCTCCTTGCCAAA), and tubulin (5′-TCACTGTGCCTGAACTTACC and 5′-GGAACATAGCCGTAAACTGC), 2.5 mm MgCl2, 200 nm dNTPs, and 1 unit of Taq polymerase per reaction at 95 °C for 5 min at 95 °C, 65 °C, and 72 °C for 30 s each for 26–30 cycles, and at 72 °C for 10 min. The PCR product was analyzed on a 1.2% standard agarose gel and stained with ethidium bromide. Approximately 20 ng of each cDNA (9 μl) was mixed with 10 μl of TaqMan® Gene Expression 2× PCR Master Mix (Applied Biosystems) and 1 μl of each indicated TaqMan® gene expression assay (Applied Biosystems) in 384-well plates and analyzed on the 7900HT Fast Real Time PCR system according to the manufacturer's instructions (Applied Biosystems). Primary analysis of the acquired signal data were performed in SDS 2.3 and RQ Manager 1.2 (Applied Biosystems). Outlier reactions were removed after Grubb's test identification, and differential expression was calculated using the ΔΔCT method.
RESULTS
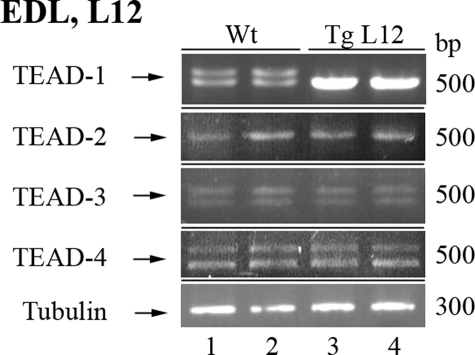
Persistent Expression of Transgenic HA-TEAD-1 Is Restricted to Striated Muscle and Does Not Induce Skeletal Muscle Hypertrophy—Our previous work has implicated a role for TEAD-1 in the fast-to-slow fiber type transition in the fast twitch plantaris muscle as a result of mechanical overload (14, 15). To determine the phenotypic consequences of a persistent increase in striated muscle TEAD-1 protein expression in vivo, we have generated multiple transgenic mouse lines that express hemagglutinin-tagged TEAD-1 (HA-TEAD-1) cDNA under the control of the mouse muscle-specific creatine kinase regulator sequences (Fig. 1A). A comparison of body weight, normalized skeletal muscle weight (muscle weight/tibia length or muscle weight/body weight), or cross-sectional area (data not shown) did not reveal a difference between age and gender-matched WT and TEAD-1 Tg mice. These data demonstrate that an increase in total TEAD-1 protein (endogenous and exogenous (transgenic)) was not sufficient to directly and/or indirectly induce skeletal muscle hypertrophy or to alter the growth rate of the TEAD-1 Tg mice. To determine the tissue-specific and developmental expression pattern of the MCK-HA-TEAD-1 transgene, we performed Western blot analysis using anti-HA antibody and total protein extracts isolated from skeletal muscles composed of varying proportions of fast and slow fiber types, nonmuscle tissues, and the heart of adult (5-month-old) WT and TEAD-1 Tg mice. Endogenous MCK gene expression is restricted to striated muscle (skeletal muscle and heart) and is first detected in skeletal muscle at approximately embryonic day 17. In addition, the magnitude of MCK gene expression in adult skeletal muscle follows a fiber type-specific pattern wherein the highest level of expression is detected in fast type IIb fibers, with successively lower levels occurring in the order IIx/d > IIa > I (β) (34, 35). Likewise, extensive study of the mouse MCK promoter has revealed that transgenic mice harboring a 3300-nucleotide region of MCK 5′-flanking sequence displayed an expression pattern that mimicked that of the endogenous MCK gene (23–25). By Western blot analysis, HA-TEAD-1 protein was detected only in the total protein extracts isolated from adult hearts, as well as adult, embryonic day 17 (fetal), and day 14 postnatal skeletal muscle of all three TEAD-1 Tg lines (4, 12, 14) examined (Fig. 1, B–H) (data not shown). The additional HA band that appears in lanes 2 and 3 for transgenic line 4 probably represents a truncated HA-TEAD-1 product due to degradation or a smaller product, initiation of translation at an alternative site, or truncation of the transgene during chromosomal integration at a second site. Moreover, HA-TEAD-1 protein was not detected in the cellular extracts obtained from the brain, liver, spleen, or kidney of TEAD-1 Tg mice (Fig. 1G). Consistent with the broad expression pattern of endogenous TEAD-1 protein, anti-TEAD-1-specific antibody detected endogenous TEAD-1 protein in the heart, skeletal muscle, brain, liver, spleen, and kidney of adult WT and TEAD-1 Tg mice (Fig. 1, B–H). Multiple bands were detected when using anti-TEAD-1 antibody, because this antibody recognizes a common epitope (amino acids 86–199) present in both HA-TEAD-1 and endogenous TEAD-1 protein (Fig. 1, B–H). Consistent with the pattern of MCK gene expression, HA-TEAD-1 protein was detected at the highest levels in the EDL and plantaris muscles, which contain a predominance of fast type II fibers (>90%), whereas comparatively lower levels were detected in the soleus muscle (∼60–70% type I fibers) and in the adult heart (Table 1). Importantly, although HA-TEAD-1 protein was detected in the heart of adult HA-TEAD-1 Tg (line 12) mice, a significant deficit in cardiac function was not detected, as assessed by echocardiography and MRI analysis.3 Since all lines displayed a similar striated muscle-restricted HA-TEAD-1 expression pattern, we have chosen to present only our analysis of HA-TEAD-1 Tg line 12. Since adult striated muscle has been shown to express multiple TEAD protein isoforms, we employed RT-PCR and qRT-PCR to assess whether the mRNA levels encoding other TEAD isoproteins in TEAD-1 Tg line 12 mice would be altered due to a persistent increase in TEAD-1 protein. With the exception of TEAD-1 mRNA levels, mRNAs encoding TEAD-2, TEAD-3, and TEAD-4 did not differ from the levels detected in WT mouse EDL muscle (Fig. 2A and Table 2). Collectively, these data demonstrate that the MCK-driven HA-TEAD-1 transgene mimicked the striated muscle-restricted expression pattern of the endogenous MCK gene. Additionally, the persistent increase in total TEAD-1 protein did not result in cardiac dysfunction; nor did it directly or indirectly alter the basal gene expression level of other members of the TEAD gene family.
TABLE 1.
Densitometry quantification of TEAD-1 overexpression in striated muscle tissue of transgenic line 12 (n = 6)
| TEAD-1 expression versus WT | |
|---|---|
| Plantaris | 23-fold ↑ |
| EDL | 37-fold ↑ |
| Soleus | 2.4-fold ↑ |
| Gastrocnemius | 17-fold ↑ |
| Heart | 3.3-fold ↑ |
FIGURE 2.
Expression analysis of the TEAD gene family in adult striated muscle. RT-PCR analysis of adult EDL muscle RNA (1 μg) isolated from WT and TEAD-1 transgenic line 12 (Tg) EDL muscle showed increased TEAD-1 mRNA levels without compensatory alteration of TEAD-2, TEAD-3, or TEAD-4 mRNA.
TABLE 2.
qRT-PCR analysis of TEAD1–4 mRNA expression in adult TEAD-1 Tg EDL muscle (line 12; n = 3) verified RT-PCR results showing an increase in HA-tagged TEAD-1 mRNA without a compensatory alteration in TEAD-2, TEAD-3, or TEAD-4 mRNA abundance
| Gene | qRT-PCR | Regulation |
|---|---|---|
| TEAD-1+HA-TEAD-1 | 18-fold | Increased |
| TEAD-1 | 1.2-fold | No change |
| TEAD-2 | 1.0-fold | No change |
| TEAD-3 | 1.2-fold | No change |
| TEAD-4 | 1.1-fold | No change |
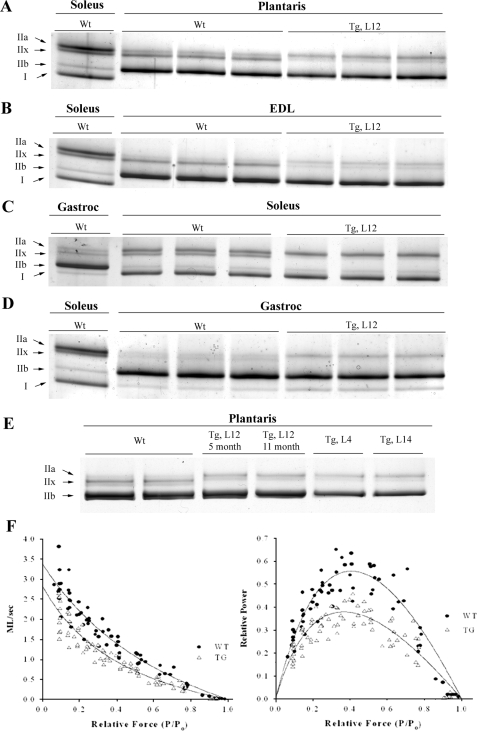
Mice Overexpressing TEAD-1 Display a Transition toward a Slower Muscle Contractile Phenotype—Previous work by us (15, 36) and others (16, 37–40) has provided in vitro evidence that the TEAD proteins participate in the regulation of several contractile proteins. To determine whether a persistent increase in TEAD-1 expression regulated MyHC gene expression in vivo, we performed a comparative high resolution gel electrophoresis analysis of myofibrillar protein isolated from either WT or TEAD-1 Tg line 12 mice. A striking observation was that the intensity of silver-stained bands representing the fast type IIx/d MyHC was basically absent in both fast and slow twitch muscles of the TEAD-1 Tg mice as compared with those of WT mice. Moreover, the plantaris, EDL, and gastrocnemius muscles of the TEAD-1 Tg mice displayed a small but reproducible decrease in the band intensity of fast type IIb MyHC and a small and reproducible increase in the band intensity of the fast type IIa MyHC. Interestingly, only the soleus and gastrocnemius muscles of the TEAD-1 Tg line 12 mice showed an increase in the band intensity of the slow type I (β) MyHC despite a significant increase in type I MyHC transcripts in the EDL muscle (Fig. 3, A–D, and Table 3). These data demonstrate that TEAD-1 overexpression resulted in an MyHC profile reflective of a slower phenotype in both fast and slow twitch skeletal muscles. Likewise, lines 4 and 14 displayed MyHC expression patterns that resembled that of line 12, indicating that the MyHC expression pattern observed due to TEAD-1 overexpression was not influenced by the transgene integration site. Furthermore, a comparative analysis of muscle extracts obtained from line 12 mice at 5 and 11 months of age did not reveal any differences in MyHC expression patterns (Fig. 3E).
FIGURE 3.
Representative MyHC protein expression pattern in various muscles and TEAD-1-induced contractile properties of the fast twitch EDL muscle. High resolution glycerol gel electrophoresis of protein extracts (0.75 μg) isolated from plantaris (A), EDL (B), soleus (C), and gastrocnemius (D) muscles of WT and TEAD-1 Tg (line 12) mice revealed that a persistent increase in TEAD-1 expression results in a fast-to-slow transition in skeletal muscle MyHC composition in both fast and slow twitch skeletal muscles. E, plantaris MyHC separation of WT and Tg lines 12, 4, and 14 as well as 5- and 11-month Tg line 12 show no difference between Tg lines and age. F, cumulative force-velocity and power-load relationships of EDL muscles isolated from WT and TEAD-1 Tg mice (line 12). Shortening velocities and power output were reduced over most relative loads in the TEAD-1 Tg EDL muscle, as evidenced by a ∼40% decrease in peak normalized power.
TABLE 3.
Densitometry quantification of MyHC isoform expression in striated muscle tissue of wild-type versus transgenic line 12 (n = 6)
| MyHC isoform | Plantaris | EDL | Soleus | Gastrocnemius |
|---|---|---|---|---|
| IIa | 1.5-fold ↑a | 1.3-fold ↑a | 1.15-fold ↑a | 1.25-fold ↑b |
| IIx | 67% ↓a | 81% ↓c | 95% ↓a | 89% fold ↓b |
| IIb | 1.0 no change | 1.0 no change | 1.0 no change | 10% ↓b |
| I | NDd | ND | 1.2 fold ↑a | 1.2 fold ↑ |
p < 0.005.
p < 0.05.
p < 0.0005.
ND, not detectable.
It is well established that the contractile velocity of a given muscle or muscle fiber is strongly correlated to the MyHC composition of its sarcomeres (3, 41). To directly ascertain whether the putative TEAD-1-induced shifts in MyHC isoform profile resulted in alterations in the overall power-generating capabilities of the fast twitch extensor digitorum longus muscle of adult TEAD-1 Tg line 12 mice, we performed a comparative characterization of the force-velocity and power-load relationship between WT (n = 6) and TEAD-1 Tg (n = 6) EDL muscle. Shortening velocities were slower in the TEAD-1 Tg mice as compared with the WT mice at all relative loads less than isometric (Po; Fig. 3E). This effect resulted in a marked depression in the relative power output in the EDL muscle of TEAD-1 Tg mice (0.32 ± 0.02 versus 0.51 ± 0.03 P/Po·ml/s; Fig. 3F). Concomitant with the reduction in relative power output, overexpression of TEAD-1 elicited a ∼40% reduction in both peak absolute power (1.2 ± 0.2 versus 2.3 ± 0.4 watts; p < 0.05) and normalized power output (55.9 ± 6.4 versus 93.6 ± 10.7 watts kg–1; p < 0.05).
To further investigate the effect of TEAD-1 overexpression on the contractile properties of the fast twitch EDL muscle, we assessed twitch kinetics. The contractile properties of the TEAD-1 Tg line 12 and WT EDL muscle are summarized in Table 4. The EDL muscle of TEAD-1 Tg mice (n = 6) displayed significantly prolonged twitch duration (92.0 ± 3.0 versus 79.0 ± 4.0 ms) and half-relaxation time (18.0 ± 1.0 versus 14.0 ± 1.0 ms) when compared with WT EDL (n = 6) muscle. No significant difference in either twitch tension or tetanic tension between WT and TEAD-1 Tg mice were observed. The slowing of twitch kinetics is consistent with changes in the MyHC isoform profile as well as altered kinetics of Ca2+ handling by the sarcoplasmic reticulum and Ca2+ binding to the thin filament or a combination of all three (42).
TABLE 4.
Summary of contractile properties shows significantly longer contraction and relaxation times in TEAD-1 Tg EDL muscle (line 12)
| Measurement | WT | Tg, L12 |
|---|---|---|
| Body weight (BW) (mg) | 37.6 ± 4.9 | 31.2 ± 2.0 |
| Muscle weight (MW) (mg) | 11.1 ± 1.1 | 10.1 ± 0.4 |
| MW/BW (mg g-1) | 0.30 ± 0.01 | 0.33 ± 0.02 |
| Lo (mm) | 12.5 ± 0.1 | 12.2 ± 0.1 |
| Cross-sectional area (mm2) | 1.9 ± 0.2 | 1.8 ± 0.1 |
| Twitch tension (micronewtons mm-2) | 28.8 ± 1.6 | 26.5 ± 2.6 |
| Tetanic tension (micronewtons mm-2) | 184.2 ± 9.1 | 203.2 ± 13.9 |
| TR50 (ms)a | 14 ± 1 | 18 ± 1b |
| CT (ms)c | 79 ± 4 | 92 ± 3b |
TR50, half-relaxation time.
p < 0.05.
CT, twitch duration.
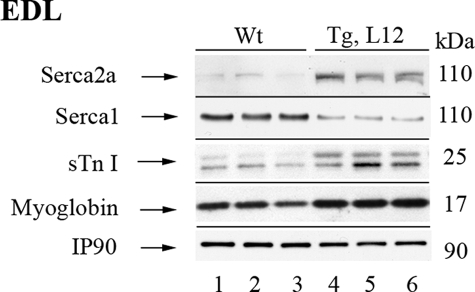
To determine whether TEAD-1 overexpression resulted in the transcription of genes encoding proteins involved in Ca2+ handling by the sarcoplasmic reticulum and Ca2+ binding to the sarcomeric thin filament, we used qRT-PCR to obtain an assessment of transcripts encoding the slow sarcoplasmic reticulum Ca2+-ATPase (SERCA2a), the fast sarcoplasmic reticulum Ca2+-ATPase (SERCA1), and the slow isoforms of troponin T, troponin I, and troponin C. When compared with the WT EDL, transcripts encoding the slow sarcoplasmic reticulum Ca2+ ATPase and the slow isoforms of troponin T, troponin I, and troponin C were increased in the TEAD-1 Tg line 12 EDL muscle 4.7-, 3.6-, 7.1-, and 6.1-fold, respectively, whereas transcripts encoding fast SERCA1 decreased 1.7-fold (Table 5). Consistent with an increase in mRNA transcripts encoding sTnI and SERCA2a, Western blot analysis using protein extract obtained from the EDL muscle of TEAD-1 Tg mice and anti-sTnI- or anti-SERCA2a-specific antibodies detected higher levels of sTnI and SERCA2a protein. Likewise, Western blot analysis using anti-SERCA1 antibody confirmed decreased fast SERCA1 expression (Fig. 4). In addition, Western blot analysis detected an increase in myoglobin protein, an oxygen binding protein that is highly expressed only in oxidative skeletal muscle fibers (Type I > IIa > IIx/d), providing further evidence for a fast-to-slow fiber-type transition in the EDL muscle of TEAD-1 Tg mice (Fig. 4). Together, these results provide strong in vivo evidence suggestive of a regulatory role for TEAD-1 in the transcription of genes encoding proteins highly represented in slow oxidative myofibers.
TABLE 5.
Summary of qRT-PCR (1 μg of RNA) and Western blot (100 μg of protein) analysis of adult EDL muscle from wild-type and TEAD-1 Tg (line 12) mice indicates a regulatory role for TEAD-1 in the transcription of these genes
| Gene | qRT-PCR | Regulation |
|---|---|---|
| MyHC IIx | 5.0-fold | Decreased |
| MyHC IIb | 1.4-fold | Decreased |
| MyHC IIa | 2.0-fold | Increased |
| MyHC I (β) | 13.7-fold | Increased |
| Troponin C, slow | 6.1-fold | Increased |
| Troponin I, slow | 7.1-fold | Increased |
| Troponin T1, slow | 3.6-fold | Increased |
| ATPase, Ca2+-transporting, slow | 4.7-fold | Increased |
| ATPase, Ca2+-transporting, fast | 1.7-fold | Decreased |
FIGURE 4.
Contractile properties and expression profile of genes encoding proteins highly represented in slow oxidative myofibers. Western blot analysis using 100 μg of protein extract obtained from adult wild-type (WT) and TEAD-1 Tg (line 12) EDL muscle confirmed qRT-PCR analysis, revealing a fast-to-slow shift via increase in slow troponin I and SERCA2a while fast SERCA1 decreased.
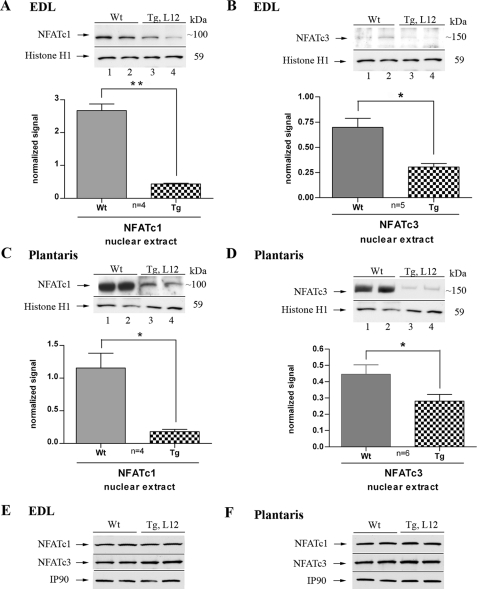
TEAD-1 Overexpression Decreases Nuclear NFAT Levels—Previous work has shown that activation of the calcium-dependent phosphatase calcineurin results in the nuclear translocation of members of the NFAT transcription factor family and their subsequent interaction with other transcriptional activators to mediate slow muscle gene expression (6, 43). To determine if TEAD-1 overexpression altered the levels of nuclear NFAT protein, we performed a Western blot analysis using both anti-NFATc1- and anti-NFATc3-specific antibodies and nuclear extracts obtained from the EDL and plantaris muscle of WT and TEAD-1 line 12 Tg mice. There was a significant decrease in NFATc1 and NFATc3 protein in the nuclear extracts isolated from the EDL and plantaris muscles of TEAD-1 Tg mice when compared with those isolated from WT mice. In contrast, we did not detect a difference in NFATc1 and NFATc3 protein by Western blot when using total cellular extracts (Fig. 5, A–F).
FIGURE 5.
Western blot analysis of NFATc1 and NFATc3 expression in fast twitch skeletal muscle. Densitometry quantification of nuclear protein extracts (100 μg) obtained from adult WT and TEAD-1 Tg (line 12) EDL and plantaris muscle show a significant decrease of nuclear NFAT protein in transgenic versus wild type EDL (A and B) and plantaris (C and D) muscle. Comparative Western blot analysis using 100 μg of total protein extract obtained from either adult WT control or TEAD-1 Tg (line 12) control revealed no difference in total level of NFAT protein in EDL (E) and plantaris (F).
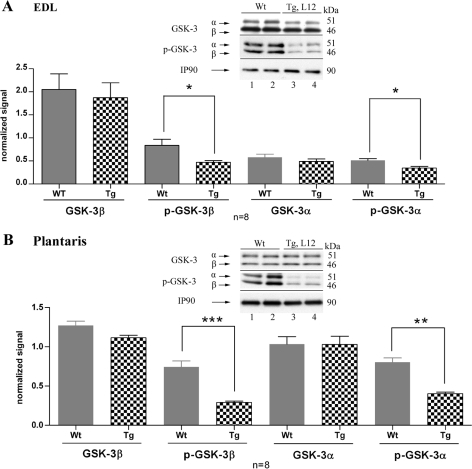
TEAD-1 Overexpression Increases Glycogen Synthase Kinase-3 Activity—In contrast to the phosphatase calcineurin, phosphorylation of NFAT by GSK-3β has been shown to exclude NFAT from the nucleus and by this means decreases activation of NFAT target genes (44, 45). To determine whether TEAD-1 overexpression leads to altered GSK-3 activity we performed a Western blot analysis using anti-GSK3α- or anti-GSK-3β-specific antibody to assess GSK-3 content and activity. No significant difference in total GSK-3α or GSK-3β was detected in total protein extract obtained from TEAD-1 Tg line 12 EDL or plantaris muscle when compared with those obtained from WT mice (Fig. 6, A and B). The activity of GSK-3 is tightly regulated, whereby phosphorylation of Ser-21 of GSK-3α and Ser-9 of GSK-3β decreases kinase activity, whereas dephosphorylation of these serine residues activates kinase activity (46, 47). Western blot analysis using either phospho-GSK-3α (Ser-21)-specific or phospho-GSK-3β (Ser-9)-specific antibodies revealed a significant decrease in GSK-3α and GSK-3β phosphorylation (activation) in the protein extracts obtained from the EDL and plantaris muscles of TEAD-1 Tg mice when compared with those obtained from WT mice (Fig. 6, A and B).
FIGURE 6.
Western blot analysis of total and phosphorylated GSK-3α and -β expression in fast twitch skeletal muscle. Densitometry quantification of total protein extracts (100 μg) obtained from adult WT and TEAD-1 Tg (line 12) EDL and plantaris muscle reveals no difference in total GSK-3α and GSK-3β protein and a significant decrease in the phosphorylation status of GSK-3α and GSK-3β in transgenic versus wild type EDL (A) and plantaris (B) muscle.
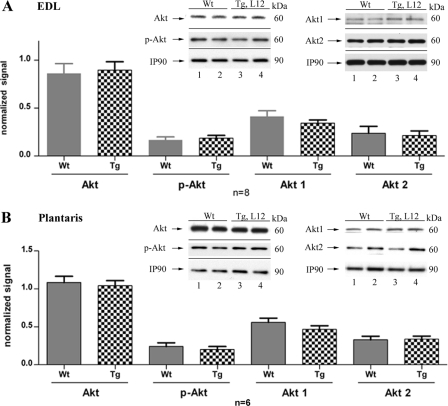
Signaling by Akt family members (Akt1, -2, and -3) has been shown to inhibit GSK-3α/β activity via phosphorylation of Ser (Ser-21 and Ser-9) residues (46, 48). To evaluate whether TEAD-1 overexpression altered Akt signaling activity, we performed a Western blot analysis using an anti-Akt antibody that recognizes all three Akt isoforms (Akt1, Akt2, and Akt3) as well as antibodies that specifically recognize either Akt1 or Akt2 protein. No difference in total Akt (Akt1 to -3), Akt1, or Akt2 protein was detected between the cellular extracts isolated from the EDL and plantaris muscles of WT and TEAD-1 Tg line 12 mice (Fig. 7, A and B). Similarly, phospho-Ser473 Akt antibody did not detect significant differences in Akt phosphorylation between WT and TEAD-1 Tg EDL and plantaris muscle extracts (Fig. 7, A and B). These data show that activation of GSK-3α and GSK-3β in the EDL and plantaris muscles of TEAD-1 transgenic mice was not due to decreases in total Akt or Akt activity (phospho-Akt).
FIGURE 7.
Western blot analysis of total and phosphorylated Akt protein in fast-twitch skeletal muscle. Densitometry quantification of total protein extracts (100 μg) obtained from WT and TEAD-1 Tg (line 12) EDL and plantaris muscle revealed no difference in expression and phosphorylation levels of Akt in transgenic mice in EDL (A) and plantaris (B) muscle.
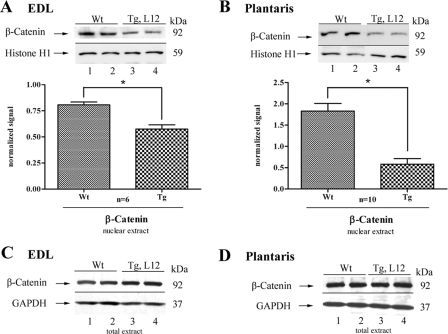
Increased GSK-3 Activity Alters Nuclear Levels of Its Downstream Target β-Catenin—It is well established that activated GSK-3β is a negative regulator of the transcriptional regulatory protein, β-catenin. GSK-3-mediated phosphorylation of β-catenin leads to its proteosome-mediated degradation, resulting in altered transcriptional regulation of β-catenin target genes (46, 49). Immunoblot analysis using anti-β-catenin-specific antibody detected reduced levels of β-catenin in the nuclear extracts obtained from the EDL and plantaris muscles of TEAD-1 Tg line 12 mice. Although we detected a decrease in nuclear β-catenin, no differences where obtained between WT and HA-TEAD-1 line 12 mice when using total cellular extracts, which is probably due to the large amount of total cellular β-catenin (Fig. 8, A–D).
FIGURE 8.
Western blot analysis of nuclearβ-catenin expression in fast-twitch skeletal muscle. Densitometry quantification of nuclear protein extracts (100 μg) obtained from WT and TEAD-1 Tg (line 12) EDL and plantaris muscle revealed a significant decrease in nuclear β-catenin protein abundance in transgenic EDL (A) and plantaris (B) muscle. Comparative Western blot analysis using 100μg of total protein extract obtained from either adult WT control or TEAD-1 Tg (line 12) control revealed no difference in total level of β-catenin protein in EDL (C) and plantaris (D).
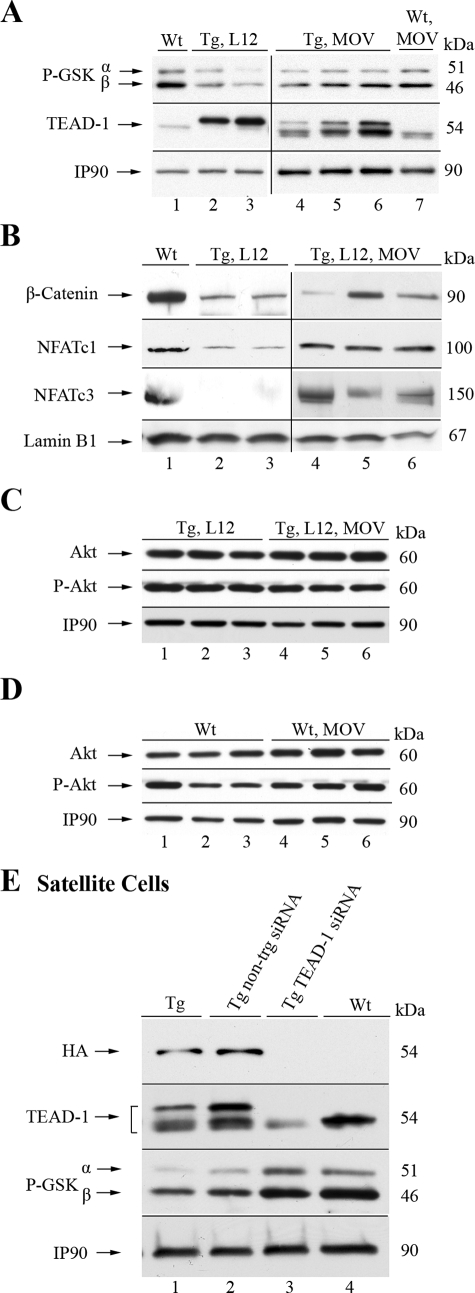
The Effects of Activated GSK-3 Are Reversed by MOV and TEAD-1 siRNA—Previously, we have demonstrated that 48 h of MOV was sufficient to nearly silence the expression of a 3300-bp mouse MCK promoter-reporter transgene (–3300 to +7-CAT) in the plantaris muscle of transgenic mice (26). Since expression of the HA-TEAD-1 transgene is controlled in part by a 3300-bp MCK promoter (see Fig. 1A), we tested the hypothesis that 48 h of MOV would return the levels of phospho-GSK-3α/β back to WT levels, thereby restoring nuclear NFAT and β-catenin levels. Immunoblot analysis using protein extracts isolated from 48-h MOV-plantaris muscle of TEAD-1 Tg line 12 mice and an anti-TEAD-1-specific antibody showed a remarkable decrease in HA-TEAD-1 protein when compared with plantaris muscle protein extract obtained from TEAD-1 Tg mice that had not been subjected to MOV (Fig. 9A). Most strikingly, 48 h of MOV resulted in a rescue of GSK-3α and GSK-3β phosphorylation level that was not different from that detected in total protein extract isolated from the plantaris muscle of WT and WT MOV mice (Fig. 9A, lane 1 versus lanes 4–7). The latter indicates that 48 h of MOV did not alter the phosphorylation status of GSK-3α/β. Furthermore, Western blot analysis using nuclear extract isolated from 48-h TEAD-1 Tg MOV-plantaris muscle showed that the nuclear levels of NFATc1, NFATc3, and β-catenin were similar to those detected in nuclear extracts isolated from the plantaris muscle of WT mice (Fig. 9B, lane 1 versus lanes 4–6). Since Akt is a primary upstream regulator of GSK-3 activity, we performed a Western blot analysis to determine if 48 h of MOV activated Akt signaling. The levels of total and phosphorylated Akt in TEAD-1 Tg MOV-plantaris total protein extract did not differ significantly from those detected in plantaris extract isolated from TEAD-1 Tg mice not subjected to MOV (Fig. 9C, lanes 1–3 versus lanes 4–6). Similar results were obtained when assessing total and phosphorylated Akt in total protein extract obtained from WT versus WT-MOV plantaris muscle (Fig. 9D, lanes 1–3 versus lanes 4–6).
FIGURE 9.
MOV- and TEAD-1-specific siRNA repress TEAD-1 transgene expression and reverse the effects of activated GSK-3α/β on nuclear abundance of NFATc1/c3 and β-catenin. A–D, comparative Western blot analysis using 100 μg of total protein extract obtained from either adult WT and MOV-treated (WT MOV) or TEAD-1 Tg line 12 (Tg, L12) control and MOV-treated plantaris muscles (Tg, L12, MOV) revealed that the phosphorylation of GSK-3α/β returned to WT levels, thereby restoring nuclear NFAT and β-catenin levels, whereas Akt was not activated. E, Western blot analysis of protein extract obtained from satellite cells isolated from either WT or TEAD-1 Tg (line 12) muscle treated with nontargeting or TEAD-1 siRNA showed silencing of HA-TEAD-1 expression concurrent with a return of GSK-3α/β phosphorylation status to WT levels.
Mouse skeletal muscle satellite cells reportedly appear shortly before the end of gestation (2, 50, 51). Since we have detected HA-TEAD-1 protein in the hind limb skeletal muscle of embryonic day 17 (fetal) TEAD-1 Tg line 12 mice (Fig. 1H), it was important to examine if the HA-TEAD-1 transgene was expressed in adult skeletal muscle myoblasts (satellite cells) of these mice and, if so, whether TEAD-1 overexpression in these cells resulted in an altered phosphorylation status of GSK-3α/β. Western blot analysis using TEAD-1-specific antibody and total protein extract obtained from differentiated satellite cells isolated from either WT or TEAD-1 Tg mice (3 months old) revealed the presence of endogenous TEAD-1 (Fig. 9E, lanes 1–4), whereas the HA-tagged TEAD-1 protein only appeared in the protein extracts of differentiated satellite cells isolated from TEAD-1 Tg line 12 mice (Fig. 9E, lanes 1 and 2 versus lane 4). Transfection of TEAD-1 Tg satellite cells with TEAD-1-specific siRNA markedly decreased the levels of total cellular TEAD-1 (endogenous and HA-TEAD-1) protein as compared with transfection with nontargeting siRNA (Fig. 9E, lane 2 versus lane 3) or in comparison with WT TEAD-1 levels (Fig. 9E, lane 3 versus lane 4). Importantly, Western blot analysis revealed that the phosphorylation status of GSK-3α/β was strikingly decreased in protein extracts isolated from TEAD-1 Tg satellite cells when compared with protein extracts isolated from WT satellite cells (Fig. 9E, lanes 1 and 2 versus lane 4). GSK-3α/β phosphorylation levels returned to WT levels in TEAD-1 Tg satellite cells treated with TEAD-1 siRNA (Fig. 9E, lane 3 versus 4), whereas those treated with nontargeting siRNA maintained reduced levels of phospho-GSK-3α/β (Fig. 9E, lane 2 versus lane 3). When considered collectively, these data provide convincing evidence that TEAD-1 participates in slow muscle contractile protein gene expression in vivo and reveal a regulatory role for TEAD-1 in GSK-3α/β signaling.
DISCUSSION
This study provides the first in vivo evidence indicating that TEAD-1 participates in slow skeletal muscle gene expression, and it reveals that TEAD-1 unexpectedly activated GSK-3α/β, resulting in decreased nuclear β-catenin and NFATc1/c3 protein. We have verified the link between increased TEAD-1 expression and activation of GSK-3α/β by using TEAD-1 siRNA in culture satellite cells and in vivo by using MOV, both of which decreased TEAD-1 transgene expression, thereby restoring WT levels of phospho-GSK-3α/β and nuclear β-catenin and NFATc1/c3. TEAD-1 activation of GSK-3α/β signaling in satellite cells may have future applications to novel therapeutic strategies in regenerative medicine.
Although numerous in vitro studies have provided evidence that TEAD proteins regulate basal and adaptive skeletal muscle contractile protein gene expression, their in vivo role in this capacity has not been examined in adult skeletal muscle. We show herein that a persistent increase in TEAD-1 expression in adult skeletal muscle induced a transition toward a slow muscle contractile protein phenotype. This was evidenced by a shift toward a slower MyHC profile that was distinguished by the virtual loss of fast type IIx/d MyHC protein in both fast and slow twitch muscles, a variable decrease in type IIb MyHC protein, and an increase in type IIa and/or type I MyHC protein for each of three independent Tg lines (Fig. 3E). Most importantly, the physiological significance of the shift toward a slower MyHC expression pattern was underscored by a corresponding decrease in the shortening velocity of the fast twitch EDL muscle, which is consistent with a large body of evidence demonstrating that each MyHC isoprotein confers a different ATPase activity and contractile velocity (3, 41, 52). In addition to the transition toward a slower MyHC expression pattern in the fast twitch EDL muscle, we have shown a striking increase in the abundance of mRNAs encoding the slow isoforms of the thin filament troponin complex proteins (TnC, TnT, and TnI (troponin C, troponin T, and troponin I, respectively)) and the slow sarcoplasmic calcium ATPase (SERCA2a) that was paralleled by increases in myoglobin, slow TnI, and SERCA2a protein with a decrease in fast SERCA1 mRNA and protein. This fast-to-slow transition in EDL gene expression was paralleled by significantly longer contraction and relaxation times, which corresponds well with previous studies demonstrating that the troponin complex proteins and sarcoplasmic calcium-ATPase regulate the kinetics of calcium-activated muscle contraction (29). It is notable that the skeletal muscle phenotype displayed by the TEAD-1 Tg mice shows a remarkable correlation to our previous work wherein we showed that MOV not only induced TEAD-1 protein expression but also increased the expression of myoglobin, slow TnI, and βMyHC protein concurrent with a significant increase in type I fibers (15, 53, 54). In addition, trends in MyHC gene expression similar to those reported herein have been reported to occur in both human and rodent fast twitch muscle in response to resistance and endurance exercise (55). It is also noteworthy that the transcriptional activation of myoglobin and slow TnI in fast twitch muscle of transgenic mice expressing constitutively active forms of calcineurin, CaMK, or PDK1 has been used as a standard to confirm the induction of a slow gene program (5, 56, 57). Collectively, these data provide in vivo evidence that TEAD-1 serves a role in slow and oxidative skeletal muscle contractile protein gene expression and point toward a role for this transcription factor in human and rodent skeletal muscle adaptations in response to various modes of exercise.
An interesting and rather unexpected finding from our study was that although the fast-twitch EDL muscle of TEAD-1 Tg mice displayed increased levels of βMyHC mRNA (∼14-fold; this is a muscle that normally does not express βMyHC to any appreciable degree), increased βMyHC protein expression was only detected in the gastrocnemius and soleus muscles, indicating that βMyHC protein production was being regulated by a muscle-specific post-transcriptional mechanism. There are two reasonable explanations that can account for this finding. Micro-RNAs (small noncoding RNAs) have been shown to regulate gene expression post-transcriptionally by binding target mRNAs and in this manner prevent and/or decrease protein synthesis of the encoded protein (58). In this regard, several striated muscle-specific micro-RNAs have been shown to serve crucial regulatory roles during skeletal muscle development and in cardiac gene expression and stress responsiveness (59, 60). For example, the hearts from miR-208 nullizygous mice displayed up-regulation of numerous fast twitch skeletal muscle genes and were unable to induce βMyHC gene expression in response to pressure overload, indicating a specific role for miR-208 in maintaining the cardiac phenotype and in βMyHC gene expression as a result of stress. Although speculative, it is conceivable that the fast twitch EDL muscle expresses a set of micro-RNAs that target mRNAs encoding various slow muscle proteins (e.g. βMyHC), thereby maintaining its fast fiber phenotype. Equally plausible, the βMyHC protein may have been degraded as an inappropriately expressed protein. The ubiquitin-proteasome system participates in the regulation of numerous cellular processes by proteolytic degradation of regulatory proteins of signaling and gene expression pathways and serves as the primary site of skeletal muscle contractile protein maintenance and turnover under basal conditions and in response to physiological and pathological stimuli (61). In this regard, mice deficient for two E3 ligases, muscle RING finger 1 and 3 (MuRF1 and MuRF3), develop skeletal muscle pathologies characterized by subsarcolemma accumulation of MyHC (βMyHC and fast type IIa MyHC), indicating a role for MuRF1 and MuRF3 in the maintenance of basal contractile protein turnover (62). Thus, degradation of the βMyHC protein may have served as a compensatory response to retain the fast fiber phenotype of the EDL muscle. Clearly, skeletal muscle fiber type conversion, be it fast-to-slow or slow-to-fast, requires coordinate regulation at multiple levels, given that it involves a multitude of changes in muscle-specific gene expression, and regulation can be imposed at any point along the gene expression pathway. Consistent with this concept, recent inquiries into the molecular mechanisms involved in establishing, maintaining, and modulating skeletal muscle fiber types have led to a proposed model wherein the cooperative interactions between three signaling pathways (calcineurin, CaMKs, and PKD1) modulate the transcriptional activation of slow muscle fiber genes through class II HDAC degradation and the activation of members of the MEF2 and NFAT transcription factor families (5–7, 43).
Another intriguing and unexpected finding was that the TEAD-1-induced transition toward a slower muscle contractile phenotype was accomplished in a seemingly NFAT-independent manner. Our Western blot analyses showed a reduction in the phosphorylation status of both isoforms of glycogen synthase kinase-3 (GSK-3α and GSK-3β; dephosphorylation activates GSK kinase activity) concurrent with decreased nuclear levels of NFATc1 and NFATc3. The observed decrease in the phosphorylation status of GSK-3α/β could not be attributed to an alteration in signaling from Akt, a primary upstream regulator of GSK phosphorylation, since we did not detect changes in total or phosphorylated levels of Akt. Importantly, previous work has shown that GSK-3β works in opposition to the protein phosphatase calcineurin. Once activated by a sustained increase in intracellular calcium, calcineurin dephosphorylates cytoplasmic NFAT, resulting in its nuclear translocation and subsequent activation of NFAT-dependent gene expression, whereas GSK-3β phosphorylates NFAT, affecting its nuclear abundance and, by this means, decreases NFAT target gene expression (63). Consistent with our result showing that GSK-3α/β can regulate nuclear NFAT levels, it has been shown that overexpression of a constitutively active form of GSK-3β increased NFAT export from the nucleus of cardiomyocytes (64). Furthermore, the inhibition of GSK-3β has been shown to slow the export of NFATc1 from the nucleus of single fibers isolated from the mouse flexor digitorum brevis muscle following the cessation of electrically stimulated import of NFATc1 (65). It is also worth noting that although several studies have provided evidence that calcineurin signaling is important for fiber type switching, activation of the slow gene program can occur by an NFAT-independent mechanism (13, 56, 57, 66–69).
As with NFAT, we also demonstrate a notable decrease in the nuclear levels of β-catenin, which is another downstream target of activated GSK-3β. This result is consistent with previous work, wherein a decrease in β-catenin was detected in embryonic stem cells isolated from double homozygous GSKα/β(21A/21A/9A/9A) knock-in mice expressing constitutively active forms of GSK-3α/β due to a serine-to-alanine substitution that prevents phosphorylation and thus inactivation of GSK-3α and GSK-3β (70). GSK-3β phosphorylation of β-catenin targets it for degradation by the ubiquitin proteosome, thereby controlling its nuclear level and transcriptional activity of β-catenin/TCF/Lef-responsive promoters (46, 47). Although β-catenin has been shown to be necessary for skeletal muscle development (71), we did not detect a difference in skeletal muscle growth rate or normalized weight between wild type and TEAD-1 Tg mice despite the early expression pattern of the TEAD-1 transgene (embryonic day 17), indicating that β-catenin is required at an earlier time during myogenesis or that the overall magnitude of decrease in nuclear β-catenin in our experiment was not sufficient to alter muscle growth. Although our data did not provide evidence as to whether decreased nuclear β-catenin contributed to the observed transition to a slower contractile protein phenotype, a role in this capacity is conceivable based on the numerous TCF/Lef elements contained within the promoter regions of the MyHC, troponin complex, and SERCA2a genes.
Our current study also provides substantial evidence that the persistent increase in TEAD-1 expression is responsible for the reduced level of phospho-GSK-3α/3β and nuclear NFATc1/3 and β-catenin. This was evidence by employing 48 h of MOV, which resulted in a significant decrease in MCK-driven HA-tagged TEAD-1 transgene expression and a return of phospho-GSK-3α/3β, nuclear NFATc1/c3, and β-catenin to WT levels in the absence of a significant change in total or phospho-Akt. Of direct relevance to the latter study is our previous work showing that 48 h of MOV is sufficient to repress endogenous and transgenic MCK reporter gene expression (26). However, since MOV is associated with an increase in motor nerve activity, a stimulus previously shown to regulate nucleocytoplasmic shuttling of NFAT via the calcium-dependent calcineurin-NFAT pathway (65, 72, 73), it could be argued that the observed return of nuclear NFATc1/c3 to wild type levels in our study was due to heightened neural activity imposed by MOV. Nevertheless, the connection between increased TEAD-1 expression and dephosphorylation of GSK-3α/β was further substantiated when we demonstrated that adult stage myoblast (satellite cells) isolated from TEAD-1 Tg mice displayed WT levels of phospho-GSK-3α/β concurrent with a striking decrease in MCK-driven HA-TEAD-1 expression as a result of transfection with TEAD-1-specific siRNA. Although beyond the scope of the current study, it will be intriguing to determine the role TEAD-1 serves in satellite cell (skeletal muscle adult stem cells) regulation, given that satellite cells contribute to postnatal skeletal muscle growth and play an essential role in skeletal muscle homeostasis and regeneration after injury and during aging (74).
In summary, the present study provides the first in vivo evidence that TEAD-1 participates in slow skeletal muscle gene expression and revealed a novel link between increased TEAD-1 expression and activation of GSK-3α/β, which, in turn, decreased nuclear NFATc1/c3 and β-catenin protein. It is expected that this putative pathway (TEAD-1 → GSK-3α/β → β-catenin/NFATc1/c3) would work in conjunction with the well established calcium-dependent signaling pathways to activate a complete array of gene programs necessary to accomplish a complete fast-to-slow phenotype switch in adult skeletal muscle. However, our data also support the notion that TEAD-1 can induce a transition toward a slower contractile protein phenotype independent of activation of calcium-dependent pathways, conceivably via direct TEAD element binding and combinatorial interactions with adjacently bound transcriptional regulators and coactivators. Additional work will be required to elucidate 1) the phosphatase that dephosphorylates GSK-3α/β, 2) the transcription factors and coactivators that interact with the TEAD proteins in vivo to mediate skeletal muscle-fiber-specific gene expression under basal conditions and during remodeling in response to MOV/exercise, and 3) the full complement of TEAD target genes, given the crucial transcriptional role recently reported for the TEAD proteins in neural cells and smooth and striated muscle (17–22, 75). Finally, our current results suggest a possible regulatory role of TEAD-1 in satellite cells. Because satellite cells are basically solely responsible for regeneration of adult skeletal muscle, increased TEAD-1 expression potentially may be exploited for the development of novel therapeutic strategies in regenerative medicine.
Acknowledgments
We thank Drs. Dawn Cornelison and Mark Hannink for critical review of the manuscript and Dr. Fadia Haddad for critical advice on MyHC gel separation. The transgenic mice were produced by the University of Missouri-Columbia Transgenic Facility.
This work was supported, in whole or in part, by National Institutes of Health, NIAMS, Grants AR41464 and AR47197 (to R. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MyHC, myosin heavy chain; MOV, mechanical overload; TEAD, TEA domain; HA, hemagglutinin; MCK, muscle creatine kinase; WT, wild type; siRNA, small interfering RNA; EDL, extensor digitorum longus; Tg, transgenic; RT, reverse transcription; qRT, quantitative reverse transcription; NFAT, nuclear factor of activated T-cells; GSK, glycogen synthase kinase.
R. W. Tsika, et al. manuscript in preparation.
References
- 1.Schiaffino, S., and Reggiani, C. (1996) Physiol. Rev. 76 371–423 [DOI] [PubMed] [Google Scholar]
- 2.Biressi, S., Molinaro, M., and Cossu, G. (2007) Dev. Biol. 308 281–293 [DOI] [PubMed] [Google Scholar]
- 3.Barany, M. (1967) J. Gen. Physiol. 50 (suppl.) 197–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booth, F. W., and Baldwin, K. M. (1996) in Handbook of Physiology, Section 12: Exercise: Regulation and Integration of Multiple Systems (Rowell, L. B., and Sheperd, J. T., eds) pp. 1075–1123, American Physiology Society, Bethesda, MD
- 5.Kim, M. S., Fielitz, J., McAnally, J., Shelton, J. M., Lemon, D. D., McKinsey, T. A., Richardson, J. A., Bassel-Duby, R., and Olson, E. N. (2008) Mol. Cell. Biol. 28 3600–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potthoff, M. J., Olson, E. N., and Bassel-Duby, R. (2007) Curr. Opin. Rheumatol. 19 542–549 [DOI] [PubMed] [Google Scholar]
- 7.Potthoff, M. J., Wu, H., Arnold, M. A., Shelton, J. M., Backs, J., McAnally, J., Richardson, J. A., Bassel-Duby, R., and Olson, E. N. (2007) J. Clin. Invest. 117 2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grifone, R., Laclef, C., Spitz, F., Lopez, S., Demignon, J., Guidotti, J. E., Kawakami, K., Xu, P. X., Kelly, R., Petrof, B. J., Daegelen, D., Concordet, J. P., and Maire, P. (2004) Mol. Cell. Biol. 24 6253–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Issa, L. L., Palmer, S. J., Guven, K. L., Santucci, N., Hodgson, V. R., Popovic, K., Joya, J. E., and Hardeman, E. C. (2006) Dev. Biol. 293 104–115 [DOI] [PubMed] [Google Scholar]
- 10.Arany, Z., Lebrasseur, N., Morris, C., Smith, E., Yang, W., Ma, Y., Chin, S., and Spiegelman, B. M. (2007) Cell Metab. 5 35–46 [DOI] [PubMed] [Google Scholar]
- 11.Schuler, M., Ali, F., Chambon, C., Duteil, D., Bornert, J. M., Tardivel, A., Desvergne, B., Wahli, W., Chambon, P., and Metzger, D. (2006) Cell Metab. 4 407–414 [DOI] [PubMed] [Google Scholar]
- 12.Barish, G. D., Narkar, V. A., and Evans, R. M. (2006) J. Clin. Invest. 116 590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calvo, S., Vullhorst, D., Venepally, P., Cheng, J., Karavanova, I., and Buonanno, A. (2001) Mol. Cell. Biol. 21 8490–8503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vyas, D. R., McCarthy, J. J., and Tsika, R. W. (1999) J. Biol. Chem. 274 30832–30842 [DOI] [PubMed] [Google Scholar]
- 15.Karasseva, N., Tsika, G., Ji, J., Zhang, A., Mao, X., and Tsika, R. (2003) Mol. Cell. Biol. 23 5143–5164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larkin, S. B., and Ordahl, C. P. (1999) in Heart Development (Harvey, R. P., and Rosenthal, N., eds) pp. 307–329, Academic Press, Inc., San Diego
- 17.Yoshida, T. (2008) Arterioscler. Thromb. Vasc. Biol. 28 8–17 [DOI] [PubMed] [Google Scholar]
- 18.Sawada, A., Kiyonari, H., Ukita, K., Nishioka, N., Imuta, Y., and Sasaki, H. (2008) Mol. Cell. Biol., in press [DOI] [PMC free article] [PubMed]
- 19.Kaneko, K. J., Kohn, M. J., Liu, C., and DePamphilis, M. L. (2007) Genesis 45 577–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishioka, N., Yamamoto, S., Kiyonari, H., Sato, H., Sawada, A., Ota, M., Nakao, K., and Sasaki, H. (2008) Mech. Dev. 125 270–283 [DOI] [PubMed] [Google Scholar]
- 21.Yagi, R., Kohn, M. J., Karavanova, I., Kaneko, K. J., Vullhorst, D., DePamphilis, M. L., and Buonanno, A. (2007) Development 134 3827–3836 [DOI] [PubMed] [Google Scholar]
- 22.Chen, Z., Friedrich, G. A., and Soriano, P. (1994) Genes Dev. 8 2293–2301 [DOI] [PubMed] [Google Scholar]
- 23.Johnson, J. E., Wold, B. J., and Hauschka, S. D. (1989) Mol. Cell. Biol. 9 3393–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen, Q. G., Buskin, J. N., Himeda, C. L., Fabre-Suver, C., and Hauschka, S. D. (2003) Transgenic Res. 12 337–349 [DOI] [PubMed] [Google Scholar]
- 25.Nguyen, Q. G., Buskin, J. N., Himeda, C. L., Shield, M. A., and Hauschka, S. D. (2003) J. Biol. Chem. 278 46494–46505 [DOI] [PubMed] [Google Scholar]
- 26.Tsika, R. W., Hauschka, S. D., and Gao, L. (1995) Am. J. Physiol. 269 C665–C674 [DOI] [PubMed] [Google Scholar]
- 27.Tsika, R. W. (1994) Exercise Sport Sci. Rev. 22 361–388 [PubMed] [Google Scholar]
- 28.Bruning, J. C., Michael, M. D., Winnay, J. N., Hayashi, T., Horsch, D., Accili, D., Goodyear, L. J., and Kahn, C. R. (1998) Mol. Cell 2 559–569 [DOI] [PubMed] [Google Scholar]
- 29.Sonnemann, K. J., Fitzsimons, D. P., Patel, J. R., Liu, Y., Schneider Martin, F., Moss, R. L., and Ervasti, James, M. (2006) Dev. Cell 11 387–397 [DOI] [PubMed] [Google Scholar]
- 30.Lynch, G. S., Hinkle, R. T., Chamberlain, J. S., Brooks, S. V., and Faulkner, J. A. (2001) J. Physiol. (Lond.) 535 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herron, T. J., Korte, F. S., and McDonald, K. S. (2001) Am. J. Physiol. 281 H1217–H1222 [DOI] [PubMed] [Google Scholar]
- 32.Talmadge, R. J., and Roy, R. R. (1993) J. Appl. Physiol. 75 2337–2340 [DOI] [PubMed] [Google Scholar]
- 33.Cornelison, D. D. W., Wilcox-Adelman, S. A., Goetinck, P. F., Rauvala, H., Rapraeger, A. C., and Olwin, B. B. (2004) Genes Dev. 18 2231–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trask, R. V., and Billadello, J. J. (1990) Biochim. Biophys. Acta 1049 182–188 [DOI] [PubMed] [Google Scholar]
- 35.Lyons, G. E., Muhlebach, S., Moser, A., Masood, R., Paterson, B. M., Buckingham, M. E., and Perriard, J. C. (1991) Development 113 1017–1029 [DOI] [PubMed] [Google Scholar]
- 36.Vyas, D. R., McCarthy, J. J., Tsika, G. L., and Tsika, R. W. (2001) J. Biol. Chem. 276 1173–1184 [DOI] [PubMed] [Google Scholar]
- 37.Larkin, S. B., Farrance, I. K., and Ordahl, C. P. (1996) Mol. Cell. Biol. 16 3742–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler, A. J., and Ordahl, C. P. (1999) Mol. Cell. Biol. 19 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart, A. F., Larkin, S. B., Farrance, I. K., Mar, J. H., Hall, D. E., and Ordahl, C. P. (1994) J. Biol. Chem. 269 3147–3150 [PubMed] [Google Scholar]
- 40.Farrance, I. K., and Ordahl, C. P. (1996) J. Biol. Chem. 271 8266–8274 [DOI] [PubMed] [Google Scholar]
- 41.Bottinelli, R., Betto, R., Schiaffino, S., and Reggiani, C. (1994) J. Physiol. 478 341–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moss, R. L., Diffee, G. M., and Greaser, M. L. (1995) Rev. Physiol. Biochem. Pharmacol. 126 1–63 [DOI] [PubMed] [Google Scholar]
- 43.Bassel-Duby, R., and Olson, E. N. (2006) Annu. Rev. Biochem. 75 19–37 [DOI] [PubMed] [Google Scholar]
- 44.Beals, C. R., Sheridan, C. M., Turck, C. W., Gardner, P., and Crabtree, G. R. (1997) Science 275 1930–1934 [DOI] [PubMed] [Google Scholar]
- 45.Neal, J. W., and Clipstone, N. A. (2001) J. Biol. Chem. 276 3666–3673 [DOI] [PubMed] [Google Scholar]
- 46.Doble, B. W., and Woodgett, J. R. (2003) J. Cell Sci. 116 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forde, J. E., and Dale, T. C. (2007) Cell Mol. Life Sci. 64 1930–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manning, B. D., and Cantley, L. C. (2007) Cell 129 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eastman, Q., and Grosschedl, R. (1999) Curr. Opin. Cell Biol. 11 233–240 [DOI] [PubMed] [Google Scholar]
- 50.Relaix, F., Rocancourt, D., Mansouri, A., and Buckingham, M. (2005) Nature 435 948–953 [DOI] [PubMed] [Google Scholar]
- 51.Hawke, T. J., and Garry, D. J. (2001) J. Appl. Physiol. 91 534–551 [DOI] [PubMed] [Google Scholar]
- 52.Bottinelli, R., Schiaffino, S., and Reggiani, C. (1991) J. Physiol. 437 655–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsika, G. L., Wiedenman, J. L., Gao, L., McCarthy, J. J., Sheriff-Carter, K., Rivera-Rivera, I. D., and Tsika, R. W. (1996) Am. J. Physiol. 271 C690–C699 [DOI] [PubMed] [Google Scholar]
- 54.Wiedenman, J. L., Rivera-Rivera, I., Vyas, D., Tsika, G., Gao, L., Sheriff-Carter, K., Wang, X., Kwan, L. Y., and Tsika, R. W. (1996) Am. J. Physiol. 270 C1111–C1121 [DOI] [PubMed] [Google Scholar]
- 55.Baldwin, K. M., and Haddad, F. (2001) J. Appl. Physiol. 90 345–357 [DOI] [PubMed] [Google Scholar]
- 56.Wu, H., Naya, F. J., McKinsey, T. A., Mercer, B., Shelton, J. M., Chin, E. R., Simard, A. R., Michel, R. N., Bassel-Duby, R., Olson, E. N., and Williams, R. S. (2000) EMBO J. 19 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, H., Rothermel, B., Kanatous, S., Rosenberg, P., Naya, F. J., Shelton, J. M., Hutcheson, K. A., DiMaio, J. M., Olson, E. N., Bassel-Duby, R., and Williams, R. S. (2001) EMBO J. 20 6414–6423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krutzfeldt, J., and Stoffel, M. (2006) Cell Metab. 4 9–12 [DOI] [PubMed] [Google Scholar]
- 59.Kim, H. K., Lee, Y. S., Sivaprasad, U., Malhotra, A., and Dutta, A. (2006) J. Cell Biol. 174 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Rooij, E., and Olson, E. N. (2007) J. Clin. Invest. 117 2369–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naujokat, C., and Saric, T. (2007) Stem Cells 25 2408–2418 [DOI] [PubMed] [Google Scholar]
- 62.Fielitz, J., Kim, M. S., Shelton, J. M., Latif, S., Spencer, J. A., Glass, D. J., Richardson, J. A., Bassel-Duby, R., and Olson, E. N. (2007) J. Clin. Invest. 117 2486–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crabtree, G. R., and Olson, E. N. (2002) Cell 109 (suppl.) 67–79 [DOI] [PubMed] [Google Scholar]
- 64.Haq, S., Choukroun, G., Kang, Z. B., Ranu, H., Matsui, T., Rosenzweig, A., Molkentin, J. D., Alessandrini, A., Woodgett, J., Hajjar, R., Michael, A., and Force, T. (2000) J. Cell Biol. 151 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen, T., Cseresnyes, Z., Liu, Y., Randall, W. R., and Schneider, M. F. (2007) J. Physiol. (Lond.) 579 535–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delling, U., Tureckova, J., Lim, H. W., De Windt, L. J., Rotwein, P., and Molkentin, J. D. (2000) Mol. Cell. Biol. 20 6600–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Naya, F. J., Mercer, B., Shelton, J., Richardson, J. A., Williams, R. S., and Olson, E. N. (2000) J. Biol. Chem. 275 4545–4548 [DOI] [PubMed] [Google Scholar]
- 68.Swoap, S. J. (1998) Am. J. Physiol. 274 C681–C687 [DOI] [PubMed] [Google Scholar]
- 69.Parsons, S. A., Wilkins, B. J., Bueno, O. F., and Molkentin, J. D. (2003) Mol. Cell. Biol. 23 4331–4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McManus, E. J., Sakamoto, K., Armit, L. J., Ronaldson, L., Shpiro, N., Marquez, R., and Alessi, D. R. (2005) EMBO J. 24 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cossu, G., and Borello, U. (1999) EMBO J. 18 6867–6872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tothova, J., Blaauw, B., Pallafacchina, G., Rudolf, R., Argentini, C., Reggiani, C., and Schiaffino, S. (2006) J. Cell Sci. 119 1604–1611 [DOI] [PubMed] [Google Scholar]
- 73.Schiaffino, S., Sandri, M., and Murgia, M. (2007) Physiol. (Bethesda) 22 269–278 [DOI] [PubMed] [Google Scholar]
- 74.Kuang, S., Gillespie, M. A., and Rudnicki, M. A. (2008) Cell Stem Cell 2 22–31 [DOI] [PubMed] [Google Scholar]
- 75.Gan, Q., Yoshida, T., Li, J., and Owens, G. K. (2007) Circ. Res. 101 883–892 [DOI] [PubMed] [Google Scholar]