Abstract
Growth factor stimulation and oncogenic transformation lead to increased glucose metabolism that may provide resistance to cell death. We have previously demonstrated that elevated glucose metabolism characteristic of stimulated or cancerous cells can stabilize the anti-apoptotic Bcl-2 family protein Mcl-1 through inhibition of GSK-3. Here we show that the pro-apoptotic Bcl-2 family protein, Puma, is also metabolically regulated. Growth factor deprivation led to the loss of glucose uptake and induction of Puma. Maintenance of glucose uptake after growth factor withdrawal by expression of the glucose transporter, Glut1, however, suppressed Puma up-regulation and attenuated growth factor withdrawal-induced activation of Bax, DNA fragmentation, and cell death. Conversely, glucose deprivation led to Puma induction even in the presence of growth factor. This regulation of Puma expression was a central component in cell death as a consequence of growth factor or glucose deprivation because Puma deficiency suppressed both of these cell death pathways. Puma induction in growth factor or glucose withdrawal was dependent on p53 in cell lines and in activated primary T lymphocytes because p53 deficiency suppressed Puma induction and delayed Bax and caspase activation, DNA fragmentation, and loss of clonogenic survival. Importantly, although p53 levels did not change or were slightly reduced, p53 activity was suppressed by elevated glucose metabolism to inhibit Puma induction after growth factor withdrawal. These data show that p53 is metabolically regulated and that glucose metabolism initiates a signaling mechanism to inhibit p53 activation and suppress Puma induction, thus promoting an anti-apoptotic balance to Bcl-2 family protein expression that supports cell survival.
Hematopoietic cells depend on extrinsic growth factors to maintain viability and prevent death by neglect (1, 2). This tight regulation of cell fate by the availability of growth factors is critical for hematopoietic homeostasis. Disturbance of the balance between cell death and cell survival can lead to diseases such as autoimmunity or cancer if growth factors are in excess and immunodeficiency if growth factors or their signaling mechanisms are limiting. In addition to survival, it has become clear that growth factors play prominent roles to regulate glucose uptake and metabolism (3–7). In the absence of necessary growth factors, a program of cellular atrophy is initiated that is characterized by decreased cell size, glucose uptake and metabolism, and mitochondrial potential (6). These changes in glucose metabolism occur prior to commitment to cell death and may play an important role in the initiation of apoptosis. Detailed mechanisms by which glucose metabolism affects cell death pathways, however, have not been completely resolved.
One key mechanism by which growth factors regulate glucose metabolism is through control of glucose uptake. In particular, glucose transporters (Gluts)2 and hexokinases (HKs) determine the first rate-limiting step of glucose metabolism. In hematopoietic cells, glucose is transported into cells through Glut1 and phosphorylated by mitochondrially bound hexokinase to become glucose-6-phosphate, which can then enter downstream pathways of glucose metabolism to generate energy as well as substrates for biosynthesis. Normally, when cells are withdrawn from growth factors, Glut1 is internalized and degraded in lysosomes, leading to decreased glucose uptake and metabolism prior to cell death (8, 9). Activated lymphocytes dramatically induce glucose uptake (10, 11), and cancer cells often overexpress Glut1 and maintain glucose metabolism in the absence of growth factors (12). Recently, the maintenance of glucose metabolism has been implicated in the regulation of cell survival because the loss of glucose uptake can promote activation of the pro-apoptotic protein Bax to cause cell death (5, 7, 13–15). Conversely, increased glucose metabolism or maintenance of glucose metabolism after growth factor withdrawal by expression of Glut1 alone or with HK1 was found to initiate a nutrient-dependent signaling pathway that phosphorylated and inactivated GSK-3 to protect cells from apoptosis (16). The full mechanisms by which glucose metabolism may regulate cell death, however, are not certain.
Bcl-2 family members are key regulators of apoptosis upon growth factor withdrawal, and anti-apoptotic glucose signaling may act through these proteins to affect cell death. In particular, growth factors can regulate the anti-apoptotic Bcl-2 family protein Mcl-1, a short-lived protein that is essential for hematopoietic cell survival (17, 18). When cells are deprived of necessary growth factors, GSK-3 becomes activated and phosphorylates Mcl-1 to target it for proteasomal degradation (19, 20). Decreased glucose (21) or inhibition of mitochondrial respiration (22) also leads to the loss of Mcl-1 protein. Glucose metabolism inhibits GSK-3 in highly glycolytic cells, however, preventing Mcl-1 phosphorylation and ubiquitination (16). In addition to regulation of Mcl-1, growth factor withdrawal leads to induction or activation of pro-apoptotic BH3-only proteins of the Bcl-2 family. In hematopoietic cells, the BH3-only protein Bim plays a critical role in the initiation of apoptosis in response to multiple death stimuli, including growth factor deprivation (23). The BH3-only protein, Puma, is also induced in growth factor-deprived hematopoietic cells, and cells deficient in Puma expression are resistant to death (24–26). Importantly, the combined loss of Bim and Puma leads to greater cell survival than loss of the individual proteins, indicating that they play an additive and only partially redundant role in growth factor withdrawal-induced cell death (25). The effect of glucose metabolism on these pro-apoptotic proteins, however, remains unknown.
The elevated glucose metabolism of activated lymphocytes and cancer cells may inhibit cell death through regulation of multiple Bcl-2 family members. Here we show that in addition to metabolic regulation of Mcl-1 by inhibition of GSK-3 (16), the pro-apoptotic BH3-only protein Puma is also responsive to changes in glucose metabolism. Increased glucose uptake attenuated Puma up-regulation and cell death in growth factor withdrawn cells. In contrast, glucose deprivation led to Puma induction even in the presence of growth factor. Although p53 protein levels did not increase when cells were deprived of growth factor, p53 was required for Puma induction and growth factor withdrawal-induced activation of Bax, DNA fragmentation, and cell death of IL-3-dependent cell lines as well as activated primary T lymphocytes. Elevated glucose uptake, however, suppressed growth factor-regulated p53 activity, indicating that p53 is responsive to the cellular metabolic state. Together these data show that elevated glucose metabolism characteristic of aerobic glycolysis in cancer cells or activated lymphocytes is sufficient to initiate anti-apoptotic signaling pathways that both maintain Mcl-1 and suppress p53-dependent induction of Puma.
EXPERIMENTAL PROCEDURES
Cells—Control, Glut1/HK1, and Bcl-xL expressing FL5.12 and 32D cells were generated and cultured in RPMI medium supplemented with 0.5 ng/ml recombinant mouse IL-3 (PeproTech Inc.; Rocky Hill, NJ) as previously described (5). Transient transfections were performed by nucleofection (Kit V; Amaxa Biosystems, Gaithersburg, MD). For growth factor withdrawal, the cells were washed three times in phosphate-buffered saline prior to resuspension in RPMI medium lacking IL-3. For glucose deprivations, the cells were washed three times and resuspended in glucose-free RPMI with addition of IL-3 and 10% dialyzed serum. Methyl-pyruvate (10 mm; Sigma) or etoposide (4 μm; Sigma) was added to some cultures.
Primary T Cell Purification and Culture—T cells from wild type and p53 nullizygous mice (p53tm1Tyj; The Jackson Laboratory; Bar Harbor, ME) were purified via negative selection from spleen (StemSep, Vancouver, Canada) and cultured in RPMI 1640 (Mediatech, Inc., Herndon, VA) supplemented with 10% fetal bovine serum (Gemini Bio-Products, Woodland, CA). T cell stimulation was achieved by culture of T cells on plates coated with 5 μg/ml anit-CD3ε (clone 145-2C11) and anti-CD28 (clone 37.51) (both from BD Pharmingen, San Diego, CA) with the addition of 5 ng/ml recombinate murine IL-2 (PeproTech Inc.) for 2 days. The cells were then washed off the plate and cultured on uncoated plates with IL-2 for two additional days. For IL-2 deprivation, the cells were washed three times in phosphate-buffered saline and cultured in medium in the absence of IL-2. Glucose deprivations were performed as described above except with IL-2 rather than IL-3.
Plasmid Constructs—shRNAi plasmids were constructed using previously described approaches (27). shRNAi sequences were: Puma, GAGGGTCATGTACAATCTCTTCCTCGAGCAAGAGATTGTACATGACCCTC; p53 shRNAi-a, GAGTATCTGGAAGACAGGCAGACTTCCTCGAGCAAGTCTGCCTGTCTTCCAGATACTC. p53 shRNAi-b, GAGACACAATCCTCCCGGTCCCTTCCTCGAGCAAGGGACCGGGAGGATTGTGTCTC. The sequences of Bim and green fluorescent protein shRNAi have been previously described (16). The sequences were cloned into pCR2.1 vector driven by the human U6 promoter as previously described (27). The p53 luciferase reporter construct was kindly provided by Dr. Ratna Ray (St. Louis University, St. Louis, MO). The pHRGTK Renilla construct was kindly provided by Dr. Michael Datto (Duke University, Durham, NC).
Glucose Uptake Assay—Glucose uptake was measured as previously described, with minor modifications (9). In brief, the cells were washed and resuspended in Krebs-Ringer-HEPES (at pH 7.4, 136 mm NaCl, 4.7 ml KCl, 1.25 mm CaCl2, 1.25 mm MgSO4, and 10 mm HEPES). 2-Deoxy-d-H3 glucose (2 μCi/reaction) was added, and the cells were incubated for 5 min at 37 °C. The reactions were quenched by the addition of ice-cold 200 μm phloretin (Calbiochem, Gibbstown, NJ) followed by centrifugation through an oil layer (1:1 Dow Corning 550 Silicon fluid from Motion Industries, Birmingham, AL; and dinonyl phthalate from Sigma-Aldrich). The cell pellets were washed and solubilized in 1 m NaOH, and radioactivity was measured using a scintillation counter.
Western Blots—The cells were lysed in radioimmune precipitation assay buffer with protease inhibitors (BD Pharmingen) on ice for 10 min and precleared by centrifugation. The protein concentrations were determined by bicinchoninic acid protein assay (Bio-Rad), and 35 μg of protein was run on a 10–20% SDS-PAGE gel (Bio-Rad). Antibodies used were rabbit anti-Bim (BD Pharmingen), rabbit anti-Puma (Cell Signaling Technology; Danvers, MA), mouse anti-actin (Sigma), mouse anti-p53 (Cell Signaling Technology), rabbit anti-Bid (Cell Signaling), mouse anti-Bad (BD Pharmingen), rabbit anti-Mcl-1 (BioLegend, San Diego, CA), rabbit anti-BiP (Cell Signaling Technology), mouse anti-p21 (BD Pharmingen), and rabbit anti-Bax (Cell Signaling). Secondary antibodies were anti-rabbit horseradish peroxidase-labeled antibody (Cell Signaling) and anti-rabbit and anti-mouse infrared-labeled antibody and were detected with ECL Plus (Pierce) or the Odyssey infrared imaging system (Licor). All of the images were uniformly contrasted, and some were digitally rearranged for ease of viewing (rearranged lanes indicated by spaces). For all immunoblots, the levels of proteins under analysis were quantified by normalization to actin and to the control sample. The numbers indicating these normalized quantifications of protein levels are presented under each lane throughout.
Cell Death Assays—A FACScan (BD Biosciences) and FLowJo (TreeStar, Ashland, OR) were used to analyze uptake of propidium iodide (1 μg/ml; Invitrogen). Bax activation was determined as previously described (5). Briefly, the cells were fixed in 0.25% paraformaldehyde for 5 min and stained with anti-active conformation Bax antibody (clone 6A7; BD Pharmingen) in 100 μg/ml digitonin (Sigma) in phosphate-buffered saline followed by staining with anti-mouse IgG1-PE (BD Pharmingen). DNA fragmentation analysis by quantification of subdiploid cells was performed simultaneously with Bax activation by the addition of propidium iodide (10 μg/ml) and RNase (Sigma) to fixed and permeabilized cells prior to flow cytometry. Caspase activation was determined by analysis of cell lysates for DEVDase activity as described (15). The samples were analyzed in triplicate for absorbance at 405 nm. The values given are Vmax values of absorbance over 30 min after start of reaction. Clonogenic survival was determined by culturing cells in the presence or absence of IL-3 for 12 h and then performing limiting dilution analysis of cell growth following readdition of IL-3 to determine the percentage of cells capable of responding to IL-3 readdition with clonogenic growth (16).
Real Time PCR—Total RNA was isolated from cells using the TRIzol (Invitrogen) method. 2 μg of RNA was reverse transcribed with SuperScript II (Invitrogen). Quantitative reverse transcription-PCR was done by using SYBR Green Supermix (Bio-Rad) and was analyzed on an iCycler. β2-Microglobulin was used as the internal control. Primers are: Puma, forward primer, AGACAAGAAGAGCAGCATCGACAC, and reverse primer, TAGGCACCTAGTTGGGCTCCATTT; β2-microglobulin, forward primer, ACCGGCCTGTATGCTATCCAGAAA, and reverse primer, GGTGAATTCAGTGTGAGCCAGGAT.
Luciferase Assay—2.5 × 106 cells were transfected with 15 μg of luciferase reporter plasmid and 10 ng of Renilla plasmid. 18 h after transfection, the cells were washed and cultured in the presence or absence of IL-3 for 9 h. The cells were then lysed, and activities of firefly and Renilla luciferase were analyzed by a luminometer. Firefly luciferase activity was normalized to Renilla luciferase activity to adjust for transfection efficiency.
RESULTS
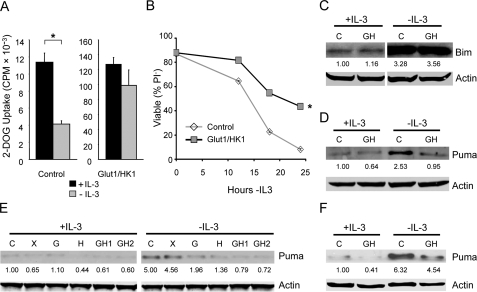
Increased Glucose Metabolism Attenuates Puma Up-regulation after Growth Factor Withdrawal—The early hematopoietic precursor cell line, FL5.12, is dependent on the cytokine IL-3 to maintain glucose metabolism and survival (6). Control of glucose uptake, in particular, plays an important role in the regulation of glucose metabolism and cell death. Prior to commitment to cell death, IL-3 withdrawal led to decreased glucose uptake in control cells (Fig. 1A, left panel). Expression of Glut1 and HK1 both elevated and sustained glucose uptake (Fig. 1A, right panel), rendering cells growth factor-independent for control of glucose uptake (16). The increased and growth factor-independent glucose uptake capacity of Glut1- and HK1-expressing cells provided a model of highly glycolytic cancer cells and activated lymphocytes. By expressing only these metabolic genes, the effects of elevated glucose uptake could be directly observed independent of oncogene- or growth factor-stimulated changes in cell signaling pathways that may otherwise complicate analyses of the role of cell metabolism in cell fate determination. Thus whereas Glut1/HK1-expressing cells have an apparent exaggerated glucose uptake capacity relative to control cells and do not simply model growth factor-independent regulation of glucose uptake, they represent highly glycolytic cells. Differences between control and Glut1/HK1 cells, therefore, demonstrate how high rates of glucose metabolism may affect cell signaling and survival pathways. Consistent with the hypothesis that elevated glucose uptake may affect cell signaling and survival, maintenance of elevated glucose uptake was sufficient to inhibit the death of Glut1/HK1 cells withdrawn from IL-3 as determined by propidium iodide exclusion (Fig. 1B) as well by Bax activation, cytochrome c release, and a stringent clonogenicity assay (16).
FIGURE 1.
Expression of Glut1 and HK1 attenuates growth factor withdrawal-induced Puma up-regulation. A, glucose uptake was measured in control and Glut1/HK1-expressing FL5.12 cells in the presence of IL-3 or after 10-h IL-3 withdrawal. B–D, control and Glut1/HK1 cells were cultured in the presence or absence of IL-3 and cell viability by propidium iodide uptake (B), and the levels of Bim (C) and Puma (D) proteins were determined. E and F, levels of Puma protein in independent FL5.12 clones (E) and 32D cells (F) were determined in the presence or absence of IL-3. lanes C, control; lanes G, Glut1; lanes H, HK1; lanes GH, Glut1/HK1; lanes X, Bcl-xL. The values are the means from triplicate samples, and the error bars indicate standard deviations. Statistical significance is shown by an asterisk to indicate Students t test p < 0.01 for cells with (+) and without (-) IL-3 (A) and for death curves (B) in which p < 0.01 for multiple time points for test samples relative to control samples.
It was unclear whether in addition to regulation of the anti-apoptotic protein, Mcl-1 (16), increased glucose metabolism affected pro-apoptotic BH3-only proteins that are critical to initiate cell death upon growth factor withdrawal. To address this, control and Glut1/HK1 cells were cultured in the presence of IL-3 or withdrawn from IL-3 for 10 h, and the expression of the pro-apoptotic BH3-only proteins Bim and Puma was analyzed by immunoblot (Fig. 1, C and D). The BH3-only protein Bad was also analyzed, but the inactive phosphorylated form of Bad was undetectable, and endogenous unphosphorylated Bad was unchanged by IL-3 withdrawal or glucose uptake (data not shown). Levels of both Bim and Puma increased in control cells after IL-3 deprivation. Although control and Glut1/HK1 cells had similar induction of Bim, Puma up-regulation was attenuated in Glut1/HK1 cells. This suppression of Puma induction upon IL-3 withdrawal occurred in multiple independently derived cell clones that expressed Glut1 and HK1, either individually or together, when compared with control or Bcl-xL-expressing cells (Fig. 1E). Similar inhibition of Puma expression by elevated glucose uptake was also seen in other IL-3-dependent cell lines, such as 32D cells (Fig. 1F). These data suggest, therefore, that Puma induction after growth factor withdrawal is inhibited by elevated glucose metabolism.
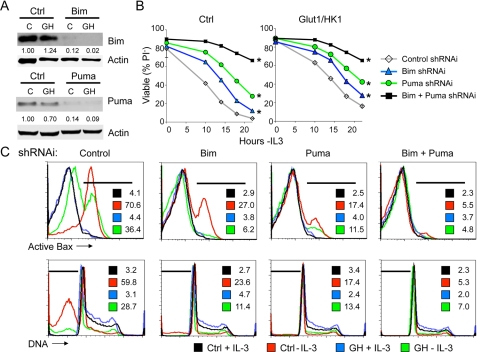
Increased Glucose Metabolism Depends on Inhibition of Puma to Attenuate Cell Death—Expression of Glut1 and HK1 attenuated the up-regulation of Puma, but not Bim, upon growth factor withdrawal, suggesting that Puma may play a role in glucose-mediated anti-apoptotic signaling. We have shown previously that expression of Bim was sufficient to induce cell death even in the presence of IL-3 (16). Puma has also been implicated in promoting cell death upon growth factor withdrawal (24–26). To determine whether Bim and Puma expression was required for cell death following growth factor withdrawal, the cells were transiently transfected with vector control, Bim, or Puma shRNAi. Protein levels were determined 1 day after transfection (Fig. 2A). The requirement for each protein in IL-3 withdrawal-induced cell death and glucose-mediated protection from this death pathway was next tested. A time course analysis of cell death by measurement of loss of cellular membrane integrity and uptake of the vital dye, propidium iodide (PI), showed that Bim deficiency delayed cell death in both control and Glut1/HK1 cells (Fig. 2B). Bim-deficient Glut1/HK1 cells remained more resistant to cell death, however, than Bim-deficient control cells, indicating that although Bim played a role in growth factor withdrawal-induced cell death, glucose metabolism provided additional protection from death. Inhibition of Puma expression also attenuated cell death as measured by PI uptake. Combined absence of Bim and Puma led to even greater ability to maintain membrane integrity and resist PI uptake. Consistent with these results suggesting roles for both Bim and Puma in regulation of apoptosis, analysis of control and Glut1/HK1 cells transfected with control, Bim, Puma, or Bim together with Puma shRNAi showed that both Bim and Puma are critical for Bax activation and DNA fragmentation following growth factor withdrawal (Fig. 2C). Therefore, inhibition of Puma induction appears to be a critical mechanism for glucose metabolism to protect cells against growth factor withdrawal-induced apoptosis.
FIGURE 2.
Increased glucose metabolism depends on inhibition of Puma induction to attenuate cell death. A, control (lanes C) and Glut1/HK1 (lanes GH) cells were transfected with control (Ctrl), Bim (top panel), or Puma (bottom panel) shRNAi plasmid, and expression was determined 10 h after IL-3 withdrawal. B and C, cell viability was analyzed over time after IL-3 withdrawal by uptake of the vital dye PI (B) and after 15 h by flow cytometric analysis (C) for active conformation Bax (top row) and DNA fragmentation (bottom row). The percentage of cells in areas indicated by the bars are provided within each plot. The values are the means for cell survival from triplicate samples; error bars indicate standard deviations. Statistical significance is shown by an asterisk to indicate Students t test p < 0.01 for death curves with multiple time points for test samples relative to control samples.
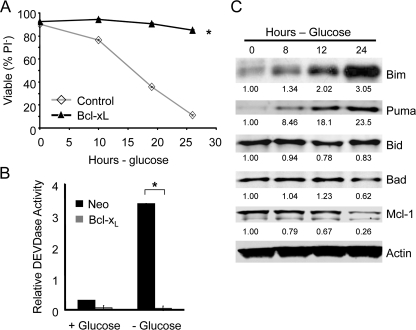
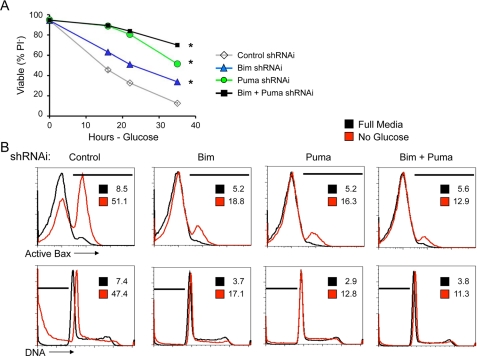
Glucose Deprivation Causes Metabolic Stress to Induce Puma and Initiate Apoptosis—Maintenance of glucose uptake was sufficient to suppress Puma induction even after growth factor signaling ceased, suggesting a metabolic mechanism for Puma regulation. It was possible, therefore, that glucose deprivation may promote Puma induction even in the presence of growth factors. Glucose deprivation led to apoptosis and caspase activation that was comparable with that observed upon IL-3 withdrawal and was strongly suppressed by expression of the anti-apoptotic Bcl-2 family protein, Bcl-xL (Fig. 3, A and B). To determine how glucose deprivation may affect expression of BH3-only proteins after 1 day in the absence of glucose, Bcl-xL-expressing cells were deprived glucose and analyzed. In this approach, cell death was prevented by Bcl-xL, allowing observation of Bcl-2 family members and pro-apoptotic BH3-only proteins. Analysis of Bim, Puma, Bid, and Bad showed that glucose deprivation led to induction of both Bim and Puma (Fig. 3C). Bid and Bad levels were unchanged. In addition, Mcl-1 levels, which are maintained by Glut1 expression in IL-3 withdrawal, decreased after 1 day of glucose withdrawal. These changes in Bcl-2 family expression and induction of Puma and Bim, in particular, were critical to cause apoptosis after glucose deprivation because shRNAi of Bim and Puma each protected glucose-deprived cells from apoptosis compared with cells transfected with control shRNAi as measured by loss of membrane integrity and uptake of PI (Fig. 4A) as well as activation of Bax and DNA fragmentation (Fig. 4B). Thus cell death upon loss of glucose metabolism is regulated by Bcl-2 family members, with up-regulation of Bim and Puma being essential to allow efficient apoptosis.
FIGURE 3.
Glucose deprivation leads to apoptosis and induction of Puma and Bim. A and B, control and Bcl-xL expressing cells were withdrawn from glucose and viability was measured by PI exclusion over time (A) and caspase 3 activity after 12 h (B). C, Bcl-xL expressing cells were analyzed by immunoblot over time after glucose withdrawal for expression of Bcl-2 family proteins. Statistical significance is shown by an asterisk to indicate Students t test p < 0.01 for death curves (A) in which p < 0.01 for multiple time points for test samples relative to control samples and for glucose uptake in cells with (+) and without (-) glucose (B).
FIGURE 4.
Puma and Bim are essential for apoptosis following glucose withdrawal. A and B, control cells were transfected with control, Bim, Puma, or both Bim and Puma shRNAi and cell death after glucose withdrawal was observed over time by PI exclusion (A) and after 15 h by flow cytometry (B) for active conformation Bax (top row) and DNA fragmentation (bottom row). The percentages of cells in areas indicated by the bars are provided within each plot. The values are the means for cell survival from triplicate samples; the error bars indicate standard deviations. Statistical significance is shown by an asterisk to indicate Student's t test p < 0.01 for death curves with multiple time points for test samples relative to control samples.
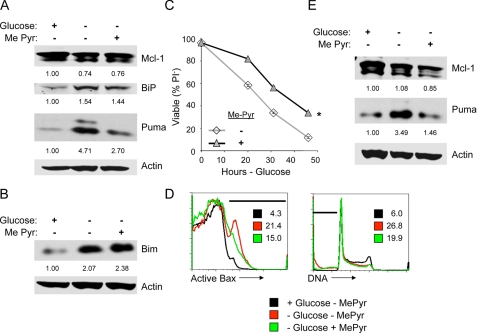
Glucose-mediated regulation of Puma and Mcl-1 may occur through a number of mechanisms, such as altered lipid pathways (16), endoplasmic reticulum (ER) stress because of deficient glycosylation, or an inability to maintain mitochondrial fuel. To test the role of ER stress and mitochondrial fuel in the regulation of Mcl-1 and Puma by glucose, cells were glucose-deprived and untreated or provided a cell-permeable form of pyruvate, methyl-pyruvate (Me-Pyr), as a mitochondrial fuel to replace glucose for 10 h. Mcl-1 and Puma levels were analyzed along with the ER stress marker and chaperone, BiP, by immunoblot. Glucose deprivation led to induction of both Puma and BiP (Fig. 5A). Consistent with previous analyses (Fig. 3C), Mcl-1 was only marginally reduced by glucose deprivation at this time point. Importantly, whereas addition of Me-Pyr did not relieve ER stress to suppress BiP induction, Me-Pyr did inhibit Puma up-regulation. Mcl-1 levels, however, were not detectably altered, and Bim induction was not affected by addition of Me-Pyr (Fig. 5, A and B). Consistent with regulation of Puma playing a role in cell death upon glucose withdrawal, treatment with Me-Pyr was sufficient to delay the induction of death of glucose-deprived cells, with delayed membrane permeability to PI (Fig. 5C), Bax activation, and DNA fragmentation (Fig. 5D). This metabolic regulation of Puma was not unique to IL-3-dependent cells because activated primary T lymphocytes showed similar regulation of Puma by glucose deprivation and Me-Pyr addition (Fig. 5E). Puma, therefore, appears to be regulated at least in part by the availability of glucose-derived mitochondrial fuels.
FIGURE 5.
Puma induction upon glucose withdrawal is suppressed by methyl-pyruvate. A–D, cells were cultured in glucose or withdrawn from glucose without or with addition of Me-Pyr. Puma (A) and Bim (B) expression were determined by immunoblot after 10 h, and cell survival was measured by propidium iodide exclusion over time (C) and after 12 h by flow cytometry (D) for active conformation Bax (left) and DNA fragmentation (right). The percentage of cells in areas indicated by bars are provided within each plot. E, primary T cells were isolated and stimulated in vitro on anti-CD3- and anti-CD28-coated plates for 2 days. Activated T cells were cultured in IL-2 in the presence of glucose or after glucose withdrawal without or with Me-Pyr for 1 day and analyzed by immunoblot. The values are the means for cell survival from triplicate samples, and error bars indicate standard deviations. Statistical significance is shown by an asterisk to indicate Student's t test p < 0.01 for death curves with multiple time points for test samples relative to control samples.
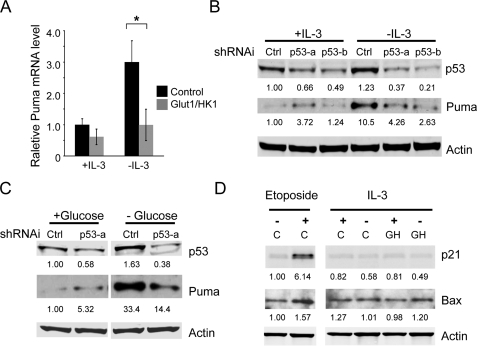
Glucose Metabolism Inhibits Puma Induction by p53 in Growth Factor Withdrawal—Puma is well described to be transcriptionally induced in stressed cells (28, 29). To determine whether the levels of Puma mRNA were increased in growth factor withdrawal and whether maintenance of glucose metabolism could prevent this increase, control and Glut1/HK1 cells were cultured in medium with IL-3 or withdrawn from IL-3 for 10 h. Total RNA was extracted to generate cDNA, and the Puma message level was determined by quantitative real time PCR (Fig. 6A). Puma RNA levels increased in control cells after IL-3 withdrawal, correlating with the increase in its protein level. This increase in Puma mRNA was inhibited, however, in Glut1/HK1 cells, suggesting that glucose metabolism may inhibit transcriptional activation of Puma in growth factor withdrawal.
FIGURE 6.
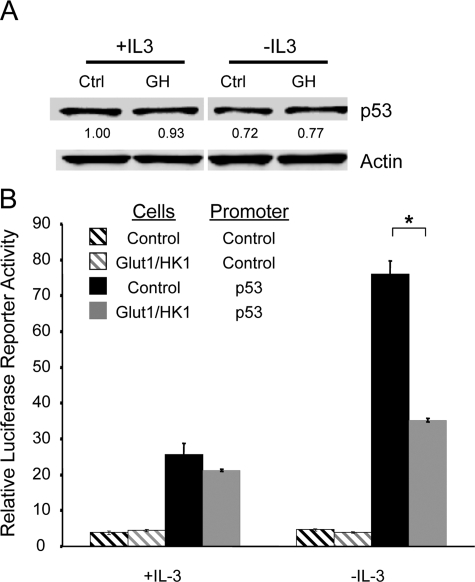
Puma is transcriptionally regulated by p53 in response to growth factor or glucose withdrawal. A, control and Glut1/HK1 cells were cultured in the presence or absence of IL-3 for 10 h. Total RNA was extracted, and levels of Puma mRNA were determined by quantitative real time PCR. B-D, cells were transfected with control (Ctrl) or independent p53 shRNAi plasmids (p53-a or p53-b), and p53 and Puma protein levels were determined after IL-3 (B) or glucose withdrawal (C). D, cells were untreated (vehicle only) or etoposide-treated or cultured in the presence or absence of IL-3 for 10 h, and p21 and Bax proteins were analyzed by immunoblot. Lanes C, control; lanes GH, Glut1/HK1. The values are the means from triplicate independent experiments, and error bars indicate standard deviations. Statistical significance is shown by an asterisk to indicate Students t test p < 0.01.
Given the described role of p53 in Puma transcriptional induction (28–31), it is possible that p53 mediated the metabolic regulation of Puma. To determine whether Puma was up-regulated by p53 upon IL-3 withdrawal or glucose deprivation, p53 levels were decreased using two independent shRNAis, and Puma induction was observed after IL-3 (Fig. 6B) or glucose withdrawal (Fig. 6C). In both cases, p53 deficiency strongly attenuated Puma induction, indicating a critical role for p53. Importantly, growth factor withdrawal did not lead to induction of p53 target genes in general, because unlike after treatment with DNA damaging agents, growth factor deprivation did not lead to induction of the p53 target genes p21 or Bax (Fig. 6D). Puma, therefore, is selectively induced upon growth factor or glucose withdrawal in a p53-dependent manner.
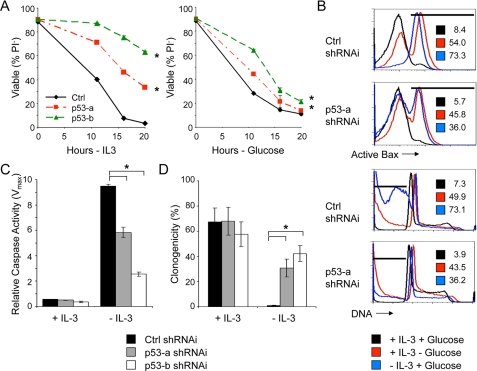
The requirement for p53 to achieve maximal Puma induction suggested that p53 deficiency may attenuate or delay cell death after growth factor or glucose withdrawal. To test this, cells were transfected with control shRNAi or two independent p53 shRNAi, and cells were analyzed after growth factor or glucose withdrawal. Consistent with an important role for Puma and a role for p53 in metabolic induction of Puma, p53 deficiency delayed plasma membrane permeability and PI uptake (Fig. 7A) as well as Bax activation and DNA fragmentation (Fig. 7B) following both growth factor and glucose withdrawal. In addition, p53 deficiency attenuated caspase activation (Fig. 7C) and allowed increased clonogenic survival (Fig. 7D) of growth factor-deprived cells. In each case, however, p53 deficiency protected against apoptosis less well after glucose withdrawal than growth factor withdrawal.
FIGURE 7.
p53 is required for efficient growth factor or glucose withdrawal-induced apoptosis. Cells were transfected with control shRNAi or one of two independent p53 shRNAis. The cells were withdrawn from IL-3 or glucose, and cell viability was determined by propidium iodide exclusion over time (A) or flow cytometry (B) after 15 h for active conformation Bax (top two panels) and DNA fragmentation (bottom two panels). The percentages of cells in areas indicated by bars are provided within each plot. C and D, cells transfected with control (Ctrl) shRNAi or p53 shRNAis were cultured in IL-3 or withdrawn from IL-3 for 12 h, and caspase activity (C) and clonogenic survival (D) were determined. The values are the means (n = 3 for A and C; n = 6 for D), and error bars indicate standard deviations. Statistical significance is shown by an asterisk to indicate Student's t test p < 0.01 for cells with (+) and without (-) IL-3 (C and D) and for death curves (A) in which p < 0.01 for multiple time points for test samples relative to control samples.
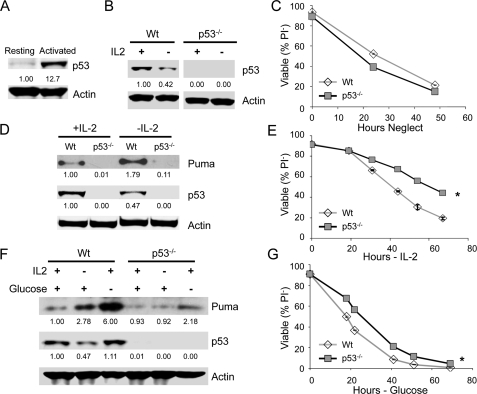
p53 Is Required to Induce Puma after Growth Factor or Glucose Withdrawal in Activated T Cells—Primary T cells become highly glycolytic and glucose-dependent after activation by T cell receptor stimulation with CD28-mediated co-stimulation (10, 11). To determine whether p53 was also required for up-regulation of Puma and cell death upon growth factor withdrawal or glucose deprivation in primary lymphocytes, we analyzed the effect of p53 deficiency on cell death of resting and activated T lymphocytes. Peripheral T cells were purified from wild type and p53-/- mice and cultured in vitro in the absence of growth factors (neglect). Resting T cells expressed little p53, but T cell activation led to robust p53 induction (Fig. 8A). Maintenance of maximal p53 protein levels required continued T cell stimulation because culture of activated T cells in the T cell growth factor, IL-2, maintained p53 but p53 levels decreased by approximately half following withdrawal from IL-2 (Fig. 8B; 0.45 ± 0.03-fold change of p53 levels after 1 day IL-2 deprivation, n = 3 independent experiments). Accordingly, unstimulated T cells from both wild type and p53-/- mice died at a similar rate when cultured in vitro in the absence of growth factors (neglect; Fig. 8C), indicating that p53 does not play an important role in death by neglect of resting T cells. To confirm the role of p53 in Puma induction (26) and to determine whether p53 was necessary for efficient cell death of activated T cells, wild type and p53-/- T cells were activated by anti-CD3 and anti-CD28 for 2 days and cultured in the presence or absence of IL-2. Consistent with the results from cell lines, Puma protein was up-regulated in IL-2-deprived wild type T cells, whereas p53-/- cells showed very modest and barely detectable Puma induction (Fig. 8D). Importantly, cell death upon IL-2 deprivation as determined by PI exclusion was delayed in p53-/- cells (Fig. 8E). In addition, glucose deprivation of activated T cells led to strong Puma induction (Fig. 8F) and cell death (Fig. 8G) that was partially prevented with p53 deficiency. p53-dependent Puma induction and cell death upon growth factor or glucose withdrawal, therefore, appear to occur in multiple types of stimulated cells, such as cell lines and activated T cells.
FIGURE 8.
p53 is required to induce Puma and promote efficient cell death after growth factor or glucose withdrawal in stimulated, but not resting, T cells. A, primary T cells were isolated, and p53 protein levels determined by immunoblot in resting T cells or T cells stimulated with anti-CD3 and anti-CD28 for 1 day. B, T cells from wild type (Wt) or p53-/- mice were stimulated as described in A; were cultured in IL-2 or deprived IL-2 and p53 levels determined by immunoblot. C, Wt and p53-/- T cells were purified and cultured without stimulation (neglect), and cell death was measured by propidium iodide exclusion over time. D and E, purified T cells from wild type and p53-/- mice were activated by CD3/CD28 for 2 days and cultured in the presence of IL-2 for additional 2 days. T cells were then washed and cultured in the presence or absence of IL-2. D, levels of Puma and p53 protein were determined after 24 h of additional culture. E, cell viability was analyzed by propidium iodide exclusion over time. F and G, primary T cells were stimulated as described above and cultured with or without IL2 or glucose. (F), Puma and p53 levels were observed by immunoblot after 1 day. (G), cell viability was observed by propidium iodide exclusion over time. The values are the means of triplicates, and error bars indicate standard deviations. Statistical significance is shown by an asterisk to indicate Student's t test p < 0.01 for death curves with multiple time points for test samples relative to control samples.
Glut1 and HK1 Signal to Suppress p53 Activity without Affecting p53 Protein Level—Activation of p53 to allow new gene transcription is mediated through accumulation of p53 protein and/or post-translational p53 modification (32). We sought, therefore, to determine whether IL-3 withdrawal activated p53 and whether increased glucose metabolism affected p53 level or activity. Control and Glut1/HK1 cells were cultured in the presence of IL-3 or withdrawn from IL-3 for 10 h, and levels of p53 were analyzed by immunoblot (Fig. 9A). Just as IL-2 deprivation of activated T cells did not cause p53 accumulation (Fig. 8, B, D, and F), IL-3 deprivation did not increase p53 expression level. In addition, increased glucose metabolism in Glut1/HK1 expressing cells did not have a significant affect on p53 protein levels compared with p53 levels in control cells. Next, p53 transcriptional activity was analyzed using a luciferase reporter construct driven by a series of tandem p53 binding elements. Control and Glut1/HK1 cells were transfected with control promoter or p53 response element luciferase reporter plasmids and luciferase activity was measured in cells cultured in the presence of absence of IL-3 for 10 h (Fig. 9B). IL-3 withdrawal led to a 3-fold increase in p53 reporter activity in control cells, whereas only a modest increase in p53 reporter activity was observed in Glut/HK1 cells. Therefore, p53 appeared to be activated by growth factor deprivation without an increase in protein level, and maintenance of glucose metabolism attenuated this p53 activation.
FIGURE 9.
Increased glucose metabolism suppresses p53 activity in growth factor withdrawal. A, levels of p53 protein were determined by immunoblot in control (Ctrl) and Glut1/HK1 (GH) cells in the presence or absence of IL-3. B, activity of control luciferase reporter or p53 response element-driven luciferase reporter was determined in control and Glut1/HK1 cells in the presence of IL-3 or after 10 h withdrawal from IL-3. The values are the means of triplicates, and error bars indicate standard deviations.
DISCUSSION
Highly proliferative cells such as growth factor-stimulated or cancerous cells often have high rates of glucose metabolism and are resistant to cell death. The contribution of metabolic signaling to the regulation of cell death pathways, however, has been unclear. Here we show that Puma induction by p53 is metabolically regulated in growth factor or glucose withdrawal in both cell lines and primary T cells. Increased glucose metabolism suppressed p53-mediated Puma induction and attenuated cell death after growth factor withdrawal, whereas decreased glucose promoted p53-mediated Puma induction. Importantly, the cell-permeable metabolite methyl-pyruvate was sufficient to suppress Puma induction in cell lines and primary T cells and protect glucose-deprived cells from apoptosis. Thus regulation of Puma by glucose metabolism appears to be mediated directly through availability of mitochondrial fuels and cellular bioenergetics. Although p53 can be regulated by changes in expression levels, levels of p53 protein did not increase to support p53-dependent Puma induction. Nevertheless, p53 activity increased after growth factor withdrawal, and this increase was suppressed by elevated glucose metabolism. Together these data show that p53 activity can be metabolically regulated and that glucose stimulates anti-apoptotic signaling pathways to attenuate growth factor withdrawal-induced apoptosis through both inhibition of GSK-3 to maintain the anti-apoptotic protein Mcl-1 (16) and inhibition p53-dependent induction of the pro-apoptotic BH3-only protein Puma.
The finding here that Puma is regulated by glucose metabolism is another example of metabolic regulation of apoptosis through Bcl-2 family members. We have previously shown that the anti-apoptotic Bcl-2 protein Mcl-1 is stabilized by increased glucose metabolism (16). Importantly, pro-apoptotic Puma and anti-apoptotic Mcl-1 can interact with and antagonize each other to exert opposite effects on cell death (33). Glucose-stimulated maintenance of Mcl-1 and inhibition of Puma, therefore, both result in an elevated ratio of anti-apoptotic to pro-apoptotic proteins to set a high threshold for cell death upon growth factor withdrawal. It is currently unclear whether the nutrient-regulated signaling pathway that regulates Mcl-1 is equivalent or distinct from that which regulates p53-dependent Puma induction. Mcl-1 stability is regulated by GSK-3-mediated phosphorylation (16, 19, 20). GSK-3, in turn, was shown to be subject to inhibitory phosphorylation by protein kinase Cs in glycolytic cells (16). GSK-3 activity has also been implicated as necessary for efficient p53-mediated Puma up-regulation after DNA damage (34), suggesting that GSK-3 may connect Mcl-1 and Puma regulation by glucose metabolism. In addition, because other Bcl-2 family members, such as Bax, have been shown to be regulated by GSK-3 (35), it will be interesting to further analyze connections between nutrient signaling pathways that regulate Bcl-2 family members and cell fate.
In addition to a well described role for p53 in DNA damage-induced cell cycle arrest and cell death, p53 has also been previously shown to play a role in growth factor withdrawal-induced death (36–45). The mechanisms by which p53 affects growth factor withdrawal-induced cell death and how p53 activity is regulated by growth factor signaling, however, have been unclear. Our study demonstrates that p53 transcriptional activity is increased upon growth factor withdrawal to up-regulate Puma, and this is both necessary and sufficient to efficiently initiate apoptosis. In addition to Puma, Bim also contributed significantly to cell death but did not appear to be either glucose- or p53-regulated. Bim and Puma are both involved in the initiation of cell death in response to a variety of cell stresses, and combined deficiency of these proteins provide synergistic protection from apoptosis in many instances (25), indicating the importance of understanding the roles and regulation of both Bim and Puma.
It is unclear precisely how p53 activity may be regulated by glucose metabolism. Typically, pathways leading to activation of p53 cause p53 protein stabilization and accumulation. In addition to protein levels, however, p53 activity can also be regulated by a wide variety of modifications, including phosphorylation, acetylation, and sumolyation (32). Regulation of p53 associating proteins and co-activators can also impact p53 activity. Activation of p53 to induce Puma after glucose or growth factor withdrawal therefore likely results from p53 post-translational modification or alterations in proteins associations, because levels of p53 did not increase and instead decreased in growth factor withdrawal. Because p53 levels were not increased upon growth factor or glucose withdrawal, this pathway for regulation of Puma and cell death by p53 is likely to play a role only in cells that already express p53. Indeed, resting T cells do not express significant p53, and p53 deficiency did not affect cell death upon neglect. Activated T cells, however, expressed p53 protein in the absence of apparent DNA damage and resisted growth factor withdrawal-induced cell death when deficient for p53. These data suggest that p53 may preferentially regulate cell death in proliferating cells, which are glycolytic (7, 10) and may up-regulate p53 protein levels but suppress p53 activity to prevent cell death and allow cell growth because elevated glucose metabolism was provided by growth factor signaling. If nutrients become limiting, however, p53 could cause cell cycle arrest (46) or cell death.
The pro-apoptotic proteins Bax and Noxa can also be induced by p53 (28, 30, 31, 47) and may participate in the initiation of cell death. We have shown, however, that levels of Bax do not change in response to growth factor withdrawal (Fig. 6D). Noxa is also a p53 target and often cooperates with Puma to mediate a p53-dependent death pathway (31). An important feature of Noxa is that it selectively interacts with Mcl-1 to facilitate the action of other toxic proteins (48). It has also been reported that Noxa is necessary for the death of activated T cells upon glucose limitation (21), suggesting that Noxa may also be glucose-regulated. We were, however, unable to detect Noxa in the experiments described here (data not shown), leaving the significance of Noxa in growth factor withdrawal uncertain. p53 can also function as a BH3-only protein itself and induce apoptosis independent of its transcriptional activity through direct inhibitory interaction with the anti-apoptotic proteins Bcl-2 and Bcl-xL (49). The clear role of Puma in the cell death system described here, however, suggests that the role of p53 may be primarily as a transcription factor and that Puma is an essential target for p53-mediated apoptosis.
Deficiency for p53 protected cells against both glucose and growth factor withdrawal but more potently inhibited cell death following growth factor withdrawal. In each case glucose uptake decreased, leading to possible metabolic activation of p53. This apparent discrepancy in p53 dependence for cell death in these stress pathways is likely because of the more severe and rapid loss of glucose availability upon glucose withdrawal than occurs in growth factor withdrawal, in which glucose uptake decreases slowly over hours. In particular, glucose but not growth factor withdrawal can lead to a pro-apoptotic ER stress (Fig. 5A and data not shown) that p53 deficiency would not inhibit. In addition, p53 has been shown to be necessary to promote oxidative metabolism through regulation of SCO2 and cytochrome c oxidase (50). In the complete absence of glucose, the cells must rapidly switch from glycolysis to an aerobic metabolism that is highly dependent on mitochondrial respiration. Cells lacking p53, therefore, may be unable to adequately utilize alternative metabolic fuels and suffer from acute bioenergetic stress upon glucose withdrawal that may partially offset the reduced induction of Puma.
It is well documented that increased glucose metabolism is characteristic of cancer cells and cells stimulated by growth factors (1, 51, 52); yet the impact of this altered glucose metabolism on cell physiology, particularly cell death, is poorly understood. Here we identified an additional novel anti-apoptotic signaling pathway stimulated by glucose that suppressed Puma up-regulation upon growth factor withdrawal through the inhibition of p53 activity. We have shown here that Puma, in addition to GSK-3 regulation of Mcl-1 (16), is a target of glucose metabolism, and our results again highlight the critical role of glucose metabolism in regulation of Bcl-2 family members. This study further supports the notion that metabolic pathways interact with apoptotic pathways to regulate cell fate and that glucose metabolism can act as a direct survival mechanism in highly proliferating cells, such as cancer cells and activated lymphocytes.
Acknowledgments
We thank Brian Altman for constructive comments on the manuscript. We also thank M. Datto (Duke University), D. Plas (University of Cincinnati), and R. Ray (St. Louis University) for providing reagents. We also thank P. A. Wolthusen and T. Van Dyke (University of North Carolina, Chapel Hill, NC) and David Kirsch (Duke University) for providing p53null splenocytes for primary T cell experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 CA123350 (to J. C. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Glut, glucose transporter; HK, hexokinase; IL, interleukin; Me-Pyr, methyl-pyruvate; shRNAi, small hairpin RNA interference; PI, propidium iodide; ER, endoplasmic reticulum.
References
- 1.Plas, D. R., Rathmell, J. C., and Thompson, C. B. (2002) Nat. Immunol. 3 515-521 [DOI] [PubMed] [Google Scholar]
- 2.Raff, M. C. (1992) Nature 356 397-400 [DOI] [PubMed] [Google Scholar]
- 3.Gottlob, K., Majewski, N., Kennedy, S., Kandel, E., Robey, R. B., and Hay, N. (2001) Genes Dev. 15 1406-1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plas, D. R., Talapatra, S., Edinger, A. L., Rathmell, J. C., and Thompson, C. B. (2001) J. Biol. Chem. 276 12041-12048 [DOI] [PubMed] [Google Scholar]
- 5.Rathmell, J. C., Fox, C. J., Plas, D. R., Hammerman, P., Cinalli, R. M., and Thompson, C. B. (2003) Mol. Cell. Biol. 23 7315-7328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rathmell, J. C., Vander Heiden, M. G., Harris, M. H., Frauwirth, K. A., and Thompson, C. B. (2000) Mol. Cell 6 683-692 [DOI] [PubMed] [Google Scholar]
- 7.Vander Heiden, M. G., Plas, D. R., Rathmell, J. C., Fox, C. J., Harris, M. H., and B., T. C. (2001) Mol. Cell. Biol. 21 5899-5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edinger, A. L., Cinalli, R. M., and Thompson, C. B. (2003) Dev. Cell 5 571-582 [DOI] [PubMed] [Google Scholar]
- 9.Wieman, H. L., Wofford, J. A., and Rathmell, J. C. (2007) Mol. Biol. Cell 18 1437-1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frauwirth, K. A., Riley, J. L., Harris, M. H., Parry, R. V., Rathmell, J. C., Plas, D. R., Elstrom, R. L., June, C. H., and Thompson, C. B. (2002) Immunity 16 769-777 [DOI] [PubMed] [Google Scholar]
- 11.Jacobs, S. R., Herman, C. E., Maciver, N. J., Wofford, J. A., Wieman, H. L., Hammen, J. J., and Rathmell, J. C. (2008) J. Immunol. 180 4476-4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatenby, R. A., and Gillies, R. J. (2004) Nat. Rev. Cancer 4 891-899 [DOI] [PubMed] [Google Scholar]
- 13.Bensaad, K., Tsuruta, A., Selak, M. A., Vidal, M. N., Nakano, K., Bartrons, R., Gottlieb, E., and Vousden, K. H. (2006) Cell 126 107-120 [DOI] [PubMed] [Google Scholar]
- 14.Chi, M. M., Pingsterhaus, J., Carayannopoulos, M., and Moley, K. H. (2000) J. Biol. Chem. 275 40252-40257 [DOI] [PubMed] [Google Scholar]
- 15.Nutt, L. K., Margolis, S. S., Jensen, M., Herman, C. E., Dunphy, W. G., Rathmell, J. C., and Kornbluth, S. (2005) Cell 123 89-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao, Y., Altman, B. J., Coloff, J. L., Herman, C. E., Jacobs, S. R., Wieman, H. L., Wofford, J. A., Dimascio, L. N., Ilkayeva, O., Kelekar, A., Reya, T., and Rathmell, J. C. (2007) Mol. Cell. Biol. 27 4328-4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Opferman, J. T., Iwasaki, H., Ong, C. C., Suh, H., Mizuno, S., Akashi, K., and Korsmeyer, S. J. (2005) Science 307 1101-1104 [DOI] [PubMed] [Google Scholar]
- 18.Opferman, J. T., Letai, A., Beard, C., Sorcinelli, M. D., Ong, C. C., and Korsmeyer, S. J. (2003) Nature 426 671-676 [DOI] [PubMed] [Google Scholar]
- 19.Ding, Q., He, X., Hsu, J. M., Xia, W., Chen, C. T., Li, L. Y., Lee, D. F., Liu, J. C., Zhong, Q., Wang, X., and Hung, M. C. (2007) Mol. Cell. Biol. 27 4006-4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurer, U., Charvet, C., Wagman, A. S., Dejardin, E., and Green, D. R. (2006) Mol. Cell 21 749-760 [DOI] [PubMed] [Google Scholar]
- 21.Alves, N. L., Derks, I. A., Berk, E., Spijker, R., van Lier, R. A., and Eldering, E. (2006) Immunity 24 703-716 [DOI] [PubMed] [Google Scholar]
- 22.Brunelle, J. K., Shroff, E. H., Perlman, H., Strasser, A., Moraes, C. T., Flavell, R. A., Danial, N. N., Keith, B., Thompson, C. B., and Chandel, N. S. (2007) Mol. Cell. Biol. 27 1222-1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouillet, P., Metcalf, D., Huang, D. C., Tarlinton, D. M., Kay, T. W., Kontgen, F., Adams, J. M., and Strasser, A. (1999) Science 286 1735-1738 [DOI] [PubMed] [Google Scholar]
- 24.Ekoff, M., Kaufmann, T., Engstrom, M., Motoyama, N., Villunger, A., Jonsson, J. I., Strasser, A., and Nilsson, G. (2007) Blood 110 3209-3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erlacher, M., Labi, V., Manzl, C., Bock, G., Tzankov, A., Hacker, G., Michalak, E., Strasser, A., and Villunger, A. (2006) J. Exp. Med. 203 2939-2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.You, H., Pellegrini, M., Tsuchihara, K., Yamamoto, K., Hacker, G., Erlacher, M., Villunger, A., and Mak, T. W. (2006) J. Exp. Med. 203, 1657-1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fox, C. J., Hammerman, P. S., Cinalli, R. M., Master, S. R., Chodosh, L. A., and Thompson, C. B. (2003) Genes Dev. 17 1841-1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano, K., and Vousden, K. H. (2001) Mol. Cell 7 683-694 [DOI] [PubMed] [Google Scholar]
- 29.Yu, J., Zhang, L., Hwang, P. M., Kinzler, K. W., and Vogelstein, B. (2001) Mol. Cell 7 673-682 [DOI] [PubMed] [Google Scholar]
- 30.Jeffers, J. R., Parganas, E., Lee, Y., Yang, C., Wang, J., Brennan, J., MacLean, K. H., Han, J., Chittenden, T., Ihle, J. N., McKinnon, P. J., Cleveland, J. L., and Zambetti, G. P. (2003) Cancer Cell 4 321-328 [DOI] [PubMed] [Google Scholar]
- 31.Villunger, A., Michalak, E. M., Coultas, L., Mullauer, F., Bock, G., Ausserlechner, M. J., Adams, J. M., and Strasser, A. (2003) Science 302 1036-1038 [DOI] [PubMed] [Google Scholar]
- 32.Liu, G., and Chen, X. (2006) J. Cell. Biochem. 97 448-458 [DOI] [PubMed] [Google Scholar]
- 33.Willis, S. N., and Adams, J. M. (2005) Curr. Opin. Cell Biol. 17 617-625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan, J., Zhuang, L., Leong, H. S., Iyer, N. G., Liu, E. T., and Yu, Q. (2005) Cancer Res. 65 9012-9020 [DOI] [PubMed] [Google Scholar]
- 35.Linseman, D. A., Butts, B. D., Precht, T. A., Phelps, R. A., Le, S. S., Laessig, T. A., Bouchard, R. J., Florez-McClure, M. L., and Heidenreich, K. A. (2004) J. Neurosci. 24 9993-10002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aloyz, R. S., Bamji, S. X., Pozniak, C. D., Toma, J. G., Atwal, J., Kaplan, D. R., and Miller, F. D. (1998) J. Cell Biol. 143 1691-1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blandino, G., Scardigli, R., Rizzo, M. G., Crescenzi, M., Soddu, S., and Sacchi, A. (1995) Oncogene 10 731-737 [PubMed] [Google Scholar]
- 38.Canman, C. E., Gilmer, T. M., Coutts, S. B., and Kastan, M. B. (1995) Genes Dev. 9 600-611 [DOI] [PubMed] [Google Scholar]
- 39.Goetz, A. W., van der Kuip, H., Maya, R., Oren, M., and Aulitzky, W. E. (2001) Cancer Res. 61 7635-7641 [PubMed] [Google Scholar]
- 40.Gottlieb, E., Lindner, S., and Oren, M. (1996) Cell Growth Differ. 7 301-310 [PubMed] [Google Scholar]
- 41.Grayson, J. M., Lanier, J. G., Altman, J. D., and Ahmed, R. (2001) J. Immunol. 167 1333-1337 [DOI] [PubMed] [Google Scholar]
- 42.Lotem, J., and Sachs, L. (1993) Blood 82 1092-1096 [PubMed] [Google Scholar]
- 43.Ohkusu-Tsukada, K., Tsukada, T., and Isobe, K. (1999) J. Immunol. 163 1966-1972 [PubMed] [Google Scholar]
- 44.Vaghefi, H., Hughes, A. L., and Neet, K. E. (2004) J. Biol. Chem. 279 15604-15614 [DOI] [PubMed] [Google Scholar]
- 45.Wang, P., Lushnikova, T., Odvody, J., Greiner, T. C., Jones, S. N., and Eischen, C. M. (2008) Oncogene 27 1590-1598 [DOI] [PubMed] [Google Scholar]
- 46.Jones, R. G., Plas, D. R., Kubek, S., Buzzai, M., Mu, J., Xu, Y., Birnbaum, M. J., and Thompson, C. B. (2005) Mol. Cell 18 283-293 [DOI] [PubMed] [Google Scholar]
- 47.Miyashita, T., Krajewski, S., Krajewska, M., Wang, H. G., Lin, H. K., Liebermann, D. A., Hoffman, B., and Reed, J. C. (1994) Oncogene 9 1799-1805 [PubMed] [Google Scholar]
- 48.Chen, L., Willis, S. N., Wei, A., Smith, B. J., Fletcher, J. I., Hinds, M. G., Colman, P. M., Day, C. L., Adams, J. M., and Huang, D. C. (2005) Mol. Cell 17 393-403 [DOI] [PubMed] [Google Scholar]
- 49.Chipuk, J. E., Kuwana, T., Bouchier-Hayes, L., Droin, N. M., Newmeyer, D. D., Schuler, M., and Green, D. R. (2004) Science 303 1010-1014 [DOI] [PubMed] [Google Scholar]
- 50.Matoba, S., Kang, J. G., Patino, W. D., Wragg, A., Boehm, M., Gavrilova, O., Hurley, P. J., Bunz, F., and Hwang, P. M. (2006) Science 312 1650-1653 [DOI] [PubMed] [Google Scholar]
- 51.Semenza, G. L., Artemov, D., Bedi, A., Bhujwalla, Z., Chiles, K., Feldser, D., Laughner, E., Ravi, R., Simons, J., Taghavi, P., and Zhong, H. (2001) Novartis Found. Symp. 240 251-260 [PubMed] [Google Scholar]
- 52.Warburg, O. (1956) Science 123 309-314 [DOI] [PubMed] [Google Scholar]