Abstract
Ovarian epithelial carcinomas (OEC) frequently exhibit amplifications at the 20q13 locus which is the site of several oncogenes, including the eukaryotic elongation factor EEF1A2 and the transcription factor ZNF217. We reported previously that overexpressed ZNF217 induces neoplastic characteristics in precursor cells of OEC. Unexpectedly, ZNF217, which is a transcriptional repressor, enhanced expression of eEF1A2. In this study, array comparative genomic hybridization, single nucleotide polymorphism and Affymetrix analysis of ZNF217-overexpressing cell lines confirmed consistently increased expression of eEF1A2 but not of other oncogenes, and revealed early changes in EEF1A2 gene copy numbers and increased expression at crisis during immortalization. We defined the influence of eEF1A2 overexpression on immortalized ovarian surface epithelial cells, and investigated interrelationships between effects of ZNF217 and eEF1A2 on cellular phenotypes. Lentivirally induced eEF1A2 overexpression caused delayed crisis, apoptosis resistance and increases in serum-independence, saturation densities, and anchorage independence. siRNA to eEF1A2 reversed apoptosis resistance and reduced anchorage independence in eEF1A2-overexpressing lines. Remarkably, siRNA to eEF1A2 was equally efficient in inhibiting both anchorage independence and resistance to apoptosis conferred by ZNF217 overexpression. Our data define neoplastic properties that are caused by eEF1A2 in nontumorigenic ovarian cancer precursor cells, and suggest that eEF1A2 plays a role in mediating ZNF217-induced neoplastic progression.
Keywords: Ovarian cancer, ZNF217, EEF1A2, oncogene, ovarian epithelial cells, neoplastic progression
Introduction
The transcription factor ZNF217 and the eukaryotic translation elongation factor EEF1A2 are both located on chromosome 20q13, a locus which is frequently amplified in ovarian epithelial carcinomas. There is both clinical and experimental evidence that these two proteins act as oncogenes in ovarian carcinogenesis as well as in other forms of cancer. However, because of the coexistence of multiple putative oncogenes on chromosome 20q, it has been difficult to define their individual specific influence on malignant progression.
20q13 amplification and overexpression of ZNF217 have been described in many types of human cancers, including ovarian carcinomas (1, 2). Recent evidence indicates that ZNF217 is a transcriptional repressor (3, 4) and that it may contribute to neoplastic progression by attenuating apoptotic signals (5, 6). In experimental systems, overexpression of ZNF217 has been shown to immortalize cell lines with limited life spans, such as human mammary epithelial cell lines and SV40 Tag/tag transduced ovarian surface epithelial cells (IOSE cells). In both of these cell types, ZNF217 increased telomerase activity and stabilize telomere lengths (7, 8). ZNF217-transduced IOSE cells also acquired anchorage independence and reduced serum dependence.
The translation elongation factor eEF1A2 is one of two isoforms of eEF1A, eEF1A-1 and eEF1A-2. In contrast to eEF1A-1, which is expressed ubiquitously, eEF1A2 is normally present only in brain and muscle. The classical role of these proteins is the regulation of ribosomal polypeptide elongation, but eEF1A2 also has noncanonical functions such as cytoskeletal modifications (9, 10), targeting proteins for degradation (11) involvement in heat shock responses (12), phosphatidylinositol signaling (9) and apoptosis (13). There is increasing evidence that it also exerts oncogenic functions. eEF1A2 is amplified and overexpressed in ovarian cancers (14, 15), and is overexpressed in cancers of the breast and lung (16, 17, 18, 19). It also induces neoplastic properties in NIH3T3 cells (14).
eEF1A2 has been reported to be antiapoptotic (20) and to regulate filopodia formation and migration through Akt/PI3-dependent cytoskeletal remodeling (9). The precise mode of action through which this elongation factor exerts its oncogenic properties remains unclear (21), though its effects on cytoskeletal modifications, apoptosis and AKT signaling likely play a role.
We recently reported that virally transduced, overexpressed ZNF217 induces neoplastic characteristics in nontumorigenic precursor cells of ovarian carcinomas (IOSE cells), derived from ovarian surface epithelial (OSE) cells (8). As in other systems, the effects of ZNF217 on IOSE cells were predominantly gene-suppressive. However, among the few genes that were upregulated, EEF1A2 stood out as the only oncogene that was consistently overexpressed, over long periods of time and many passages, in two IOSE cell lines (lines I-144RZ and I-80RZ) that were derived from two different women.
The present study was undertaken to further examine the relationship between ZNF217 and eEF1A2, and, in particular, to determine whether ZNF217-induced neoplastic changes require interactions with EEF1A2. Importantly, Krieg et al. (3) recently carried out a comprehensive identification of all genes that are directly regulated by the gene suppressor ZNF217, using ChiP-chip assays. There was negligible interaction between ZNF217 and EEF1A2, indicating that the effects of ZNF217 on EEF1A2 expression, as observed by Li et al. (8), were likely indirect, perhaps through inhibition by ZNF217 of negative regulator(s) of eEF1A2 expression (13, 22).
In the present study, we created the first stable preneoplastic ovarian cell lines that overexpress eEF1A2. These lines allowed us to provide the first direct evidence showing that eEF1A2 overexpression contributes neoplastic characteristics to precursor cells of ovarian carcinomas, and thus support previous evidence pointing to an oncogenic role of this protein in ovarian carcinogenesis. The results suggest further that eEF1A2 may play an important role in mediating and/or enhancing the oncogenic effects of ZNF217.
Material and methods
Cell culture and infection
Normal ovarian surface epithelial cells (OSE) were obtained at surgery for nonmalignant gynecological disorders. Ethical permits were obtained as required by the University of B.C. OSE cells in low passage were immortalized with SV40 large T and small t antigen to give rise to IOSE (“immortalized OSE”) lines (23). To obtain IOSE lines constitutively overexpressing eEF1A2, two IOSE lines that were derived from two different women, IOSE-80 and IOSE-144, were infected with the ViraPower Lentivial Expression System (Invitrogen, Carlsbad, CA), using constructs encoding lentiviral EEF1A2-V5 and β-galactosidase as control that were amplified in STBL3 competent cells (Invitrogen). High titer stocks of EEF1A2-V5 and control lentivirus were prepared using a transient packaging kit (Invitrogen). Starting at 48 hours after infection, cells were selected with 2 µg/ml blasticidin, and infection efficiency was evaluated cytochemically in cultures with the recombinant lentivirus encoding β-galactosidase.
As described previously (8), two ZNF217-overexpressing IOSE-derived cell lines, I-80RZ and I- 144RZ, were obtained from different women, and two isogenic sublines were derived from each of these two lines at the time of crisis: one subline was maintained with, and one without 10 ng/ml of EGF. All cells were grown under the conditions described previously (8).
To monitor the timing of changes in EEF1A2 at the genomic and transcriptional level, IOSE-144 cells, infected with HA-tagged ZNF217 as described above, were examined by array CGH and Affymetrix array analysis as described previously (8), both prior to and after crisis. Furthermore, ZNF217-HA-infected IOSE-144 cultures entering crisis were monitored for the appearance of growing colonies. When such colonies had reached a size representing approximately 20,000 to 40,000 cells, they were scraped off, lysed and analysed for eEF1A2 expression by CGH–SNP arrays.
CGH–Single nucleotide polymorphism (SNP) arrays
DNA was extracted from cells using a Puregene protocol (Gentra Systems, Minneapolis, MN). Genomic DNA was amplified with phi29 enzyme using the REPLI-g Midi kit (Qiagen, Valencia, CA). The DNA concentration was determined using a spectrophotometer and diluted to a concentration of 50 ng/µL. Preparation of DNA for hybridization to the GeneChip® Human Mapping 50K Array Xba 240 was done according to the manufacturer's instructions (100K Mapping Assay Manual; Affymetrix, Santa Clara, CA). Briefly, 250 ng of genomic DNA was digested with XbaI and a common adapter oligo ligated to the ends of the digestion fragments and amplified using a universal primer set. The amplified product was fragmented and labeled with biotin and hybridized to the Mapping 50K Xba array overnight. After removal of the hybridization cocktail, the chip was stained with streptavidin-R-phycoerythrin conjugate and scanned using a GeneChip® Scanner 3000 7G, and genotyped and signals were detected by Affymetrix GeneChip® Operating Software (GCOS). DNA copy number and LOH calls were generated using dSNPChip software from Harvard (http://biosun1.harvard.edu/complab/dchip/). The SNPs are positioned using the May 2004 assembly (NCBI Build 35) from the UCSC genome browser.
Reverse Transcription PCR
Total RNA was extracted and purified from cells grown to 70–80% confluency with the RNeasy kit from Qiagen by following manufacturer’s instructions (Qiagen, Mississauga, ON, Canada), while 1st-strand cDNA synthesis was performed with a first-strand cDNA synthesis kit (Amersham Biosciences, Baie d’ Urfe, Quebec). RNA denaturation, reaction setup and sample treatment were performed as previously reported (8). A pair of primers spanning the 3’ terminal of EEF1A2 gene and 5’ terminal of V5 tag (5’—CCGGCGGGGCAACGTGTGTGGGGAC—3’ and 5’—CGAGGAGAGGGTTAGGGATAGGCTTAC—3’, respectively) was used to amplify the transgene EEF1A2-V5. PCR amplification was carried out as reported previously (8), with PCR products of EEF1A2-V5 analyzed by agarose gel electrophoresis.
Western blot analysis
Cells were lysed as described previously (8) with protein concentrations measured by Bio-Rad protein assay kit (Bio-Rad Laboratories, Missisauga, ON, Canada). To analyse the expression of recombinant EEF1A2-V5, total proteins (30ug) were separated on 8% SDS-PAGE gels followed by transfer onto nitrocellulose membranes. After blocking for 30 min with 5% nonfat dried milk, blots were probed at 4 ° C for overnight with mouse monoclonal anti-EEF1A2 (Cell Signaling, Danvers, MA) at 1:10000 Upon extensive washing with TBST, the immune complexes were detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibody at 1:3000 (Bio-Rad Laboratories, Hercules, CA) for 1 hr followed by enhanced chemiluminescence (ECL) detection system (BioLynx, Brockville, ON, Canada).
Anchorage independence assay
Anchorage independent cell growth was determined by analyzing colony formation of cells in soft agar. Cells (1.3 × 104) were suspended in 5 ml 0.3% agarose in M199:105 medium containing 10% FBS and plated on pre-solidified agarose (0.5%) in 60 mm dishes with grids for colony quantification. Five milliliters of medium were added on top of the agarose and changed once per week. After 3 weeks, colonies of different sizes were counted microscopically, with data obtained from triplicate experiments.
Cell growth and serum dependence assays
Cells in the exponential growth phase were trypsinized and plated in 96-well strip plates (Corning Inc., Corning, NY) at 2 × 103 cells/well. For proliferation assays, cells were incubated in 199:105 medium (5% FBS). Six replicates were collected on days 0, 1, 3, 5, 7, 9, 11, 13, respectively. For the serum dependence assays, cells were incubated in 199:105 medium with serum concentrations ranging from 0 to 5%, and collected after 7–9 days. For both assays, at the end of the growth period, cells were washed with serum-free medium and fixed with −20 ° C methanol 20 min followed by 3 washes with PBS. Cells were then stained with 5.0 ug/ml Hoechst 33258 (Sigma) for 2 minutes, washed to remove unbound dye, and analyzed on a fluorescence plate reader with filters of wavelength 360/40 and 460/40.
Apoptosis assays
Cells were either treated with 1 µM camptothecin for 16–20 hr at 37°C to induce apoptosis or left untreated as controls. Both induced and control cells were then harvested, lysed and assayed as follows:
(1) EnzChek caspase-3 assay
Cells were harvested after being incubated with apoptosis inducer for optimized lengths of time and washed in phosphate-buffered saline (PBS). Each cell sample was resuspended in 50 µl of the 1× lysis buffer, and cells were subject to lysis on ice for 30 min. At the same time, a 2× reaction buffer was prepared by adding 400 µl of the 5× reaction buffer and 10 µl of 1M DTT to 590 µl of dH2O, and a 2× substrate working solution was made by mixing 10 µ of the 5mM Z-DEVD-R110 substrate with 990 µl of the 2× reaction buffer. The lysed cells were centrifuged at 500 rpm for 5 min at room temperature to pellet the cellular debris. Fifty microliters of 2× substrate working solution was added to each sample. The microplate was covered and samples incubated at room temperature for 30 min. Fluorescence was measured in a microplate reader with the excitation filter at 485±20nm and emission detection at 530±20nm after the indicated amount of time.
(2) Cell death detection ELISA
Quantification of apoptotic cell death was determined by an ELISA assay that measures cytoplasmic histone-complexed DNA fragments, using the one-step sandwich immunoassay to detect nucleosomes released from the nuclei (Roche Diagnostics, Indianapolis, IN). After dilution to 1×105 cells/ml, 100 µl of suspended cells were added into individual wells of a 96-well plate and incubated for 4 hr at 37°C with 5% CO2. The 96-well plate was then centrifuged, cell pellets resuspended and mixtures incubated according to the manufacturer’s instructions (Roche Diagnostics)‥ The solutions were removed thoroughly by tapping or suction, and each well was given 100 µl ABTS solution. The reactions were incubated on a shaker at 250 rpm until the color development was sufficient for photometric analysis. Absorbance was measured at 405 nm against ABTS solution as a blank.
Migration assays
Cells were grown to confluency (95%) in 60-mm cell culture dishes. A scratch was made through the cell monolayer using a 200 µl pipette tip. After two washes to remove floating cells and debris, fresh culture medium with 5% FBS was added. Distances between the edges of the wounds were measured at specific time points at 0, 12 and 24 hr after wounding, and the changes from time 0 were halved to calculate the distance that the cells had migrated.
siRNA assays
Exponentially growing cells were seeded at 70% density for 24h. Then, the cells were transfected with EEF1A2-specific siRNA and negative control (NC) siRNA (Qiagen) at varying concentrations using HiPerFect (Giagen). The transfected cells were incubated overnight before being trypsinized, counted, and assayed for anchorage independent growth as described above. After 7 days of incubation, colonies were counted. Changes in growth and morphology associated with the presence of EEF1A2 siRNA assay were also examined with cells that were transfected with siRNA and maintained under standard conditions for 1–4 days posttransfection.
Results
Generation of IOSE cell lines overexpressing eEF1A2
IOSE-144 cells (see Materials and Methods for definition) at passage 6~7 were lentivirally infected with EEF1A2-V5 or a LacZ-V5 control vector. Thirteen independent cell lines infected with EEF1A2-V5 (LE-lines) and 2 lines infected with LacZ-V5 control vector (LL lines) were established. Expression of β-galactosidase from the control construct confirmed that infection efficiency was near 80% (not shown). Infection with either lentiviral EEF1A2-V5 or vector control caused no obvious phenotypic changes other than nonspecific transient damage such as detachment and vacuolation, which was similar in both groups.
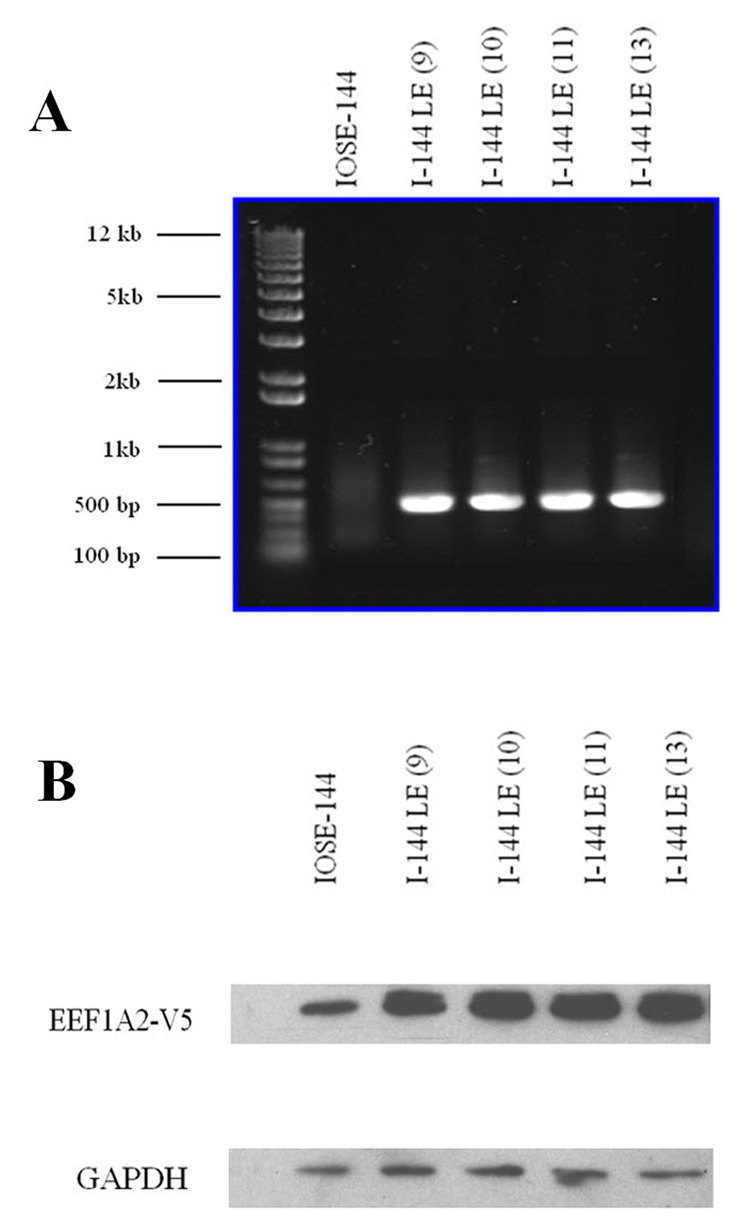
The EEF1A2-V5 infected cells were resistant to blasticidin, were positive for the construct RNA by RT-PCR and showed enhanced expression of eEF1A2 by Western blots with anti-eEF1A2 antibody (Fig. 1A, B). The overexpressed eEF1A2 appeared as a double band with a slower upper band, representing the tagged form of the molecule.
Figure 1.
A, RT-PCR, using a primer spanning the 3’ terminal of EEF1A2 gene and 5’ terminal of V5 tag, demonstrates EEF1A2-V5 expression in I-144LE cell lines, but not in the parent line. From the left to right: GeneRuler DNA ladder mix, IOSE-144 (parent line), I-144LE(9) 9, I-144LE (10), I-144LE (11), I-144LE (13) all at passage 9. The numbers in parentheses indicate independently created lines. B, Western blot: monoclonal anti-eEF1A2 reveals eEF1A2 overexpression in EEF1A2-V5-infected cultures of the 4 LE lines shown in A.
Phenotypic changes in lines overexpressing eEF1A2
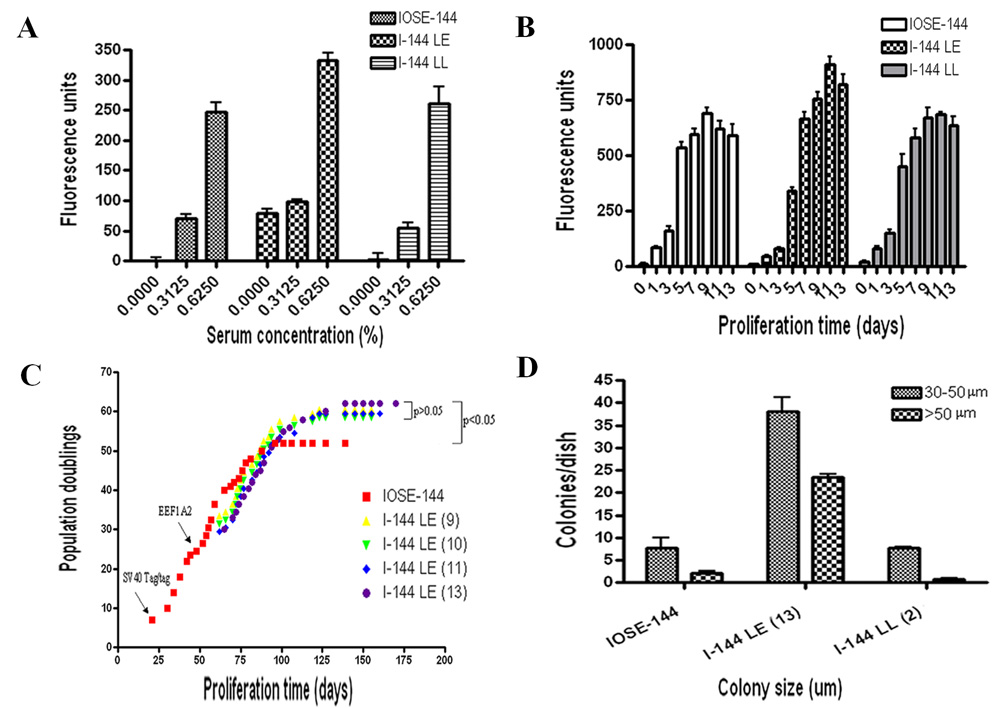
eEF1A2 overexpressing lines were significantly less serum dependent (p<0.05) than the parent line, and cells proliferated even in the complete absence of serum (Fig. 2). In contrast, the control β-Gal overexpressing line I-144LL did not exhibit enhanced serum-independence.
Figure 2.
A. Serum dependence. At low serum concentrations the eEF1A2 overexpressing line I-144LE was significantly less serum dependent (p<0.05) than either the parent line or the β-Gal overexpressing line I-144LL, and proliferated even in the complete absence of serum. B. Short term growth assay. In medium with 5% FBS I-144LE cells grew slowly at low densities but reached saturation densities ranging from 10% to 33% above the control lines IOSE-144 and I-144LL by 13 days. C. Proliferation potential. I-144LE cell lines gained a prolonged lifespan, with the population doublings increased significantly as compared to controls. The times when Tag/tag and EEF1A2 were introduced into the cultures are indicated on the graph. Crisis occurred as proliferation ceased. D. Anchorage independence assay. Upon 3-week incubation in soft agar, the I-144LE line had 6 to 7 times more colonies per 60 mm dish, as compared with either the IOSE-144 parent line or the I-144LL line, though the absolute number of colonies was small, suggesting that it represented a minor subpopulation.
Short term proliferation assays showed that I-144LE cells grew relatively slowly at low densities as compared with the IOSE-144 and I-144LL cells, but they overtook the latter by 7 days of continued growth and reached saturation densities ranging from 10% to 33% higher, as compared with the parent IOSE-144 line and the I-144LL line (Fig. 2B). Proliferation over long periods demonstrated a significant increase in population doubling potential, increased lifespan and delayed crisis of eEF1A2-overexpressing LE cell lines, when compared with the parent IOSE line (Fig. 2C). The control cells ceased proliferating and entered crisis at approximately 80 days (52 PDs), while I-144LE cells reached 120–130 days (60 PDs) before undergoing crisis. Delayed crisis of a cell line is indicative of an increase in population doubling potential, which leads to an increased lifespan of precrisis cells. This change usually results from an increase in proliferative capacity and/or enhanced resistance to apoptosis. However, overexpression of eEF1A2 alone was insufficient to induce further immortalization and circumvention of crisis
Upon 3 weeks incubation in soft agar, anchorage independence assays demonstrated that the I-144LE line had 6 to7 times more cell colonies (Fig. 2D), as compared with either the IOSE-144 parent line or I-144LL line, indicating decreased anchorage dependence with increased eEF1A2 expression. However, the total number of colonies was small, suggesting that the change took place in a minor subpopulation of cells.
Scratch assays indicated that the migration capability of I-144LE cells into wounds in cell monolayers was different, which is in contrast to the parent line-IOSE144 cells (Fig. 3 a–f), with the migration rate of the former increased significantly (p<0.05) as compared to the latter (Fig. 3g).
Figure 3.
Cell migration. A–C: IOSE144 cells at 0 hrs (A), 12 hrs (B) and 24 hrs (C). after the monolayers were scratched. D–F: I-144LE cells at 0 hrs (D) 12 hrs (E) and 24 hrs (F) after the monolayers were scratched. G. Migration distances of two sets of IOSE-144 and I-144LE cells during the wound healing assay, measured at different time points shows significant differences (p< 0.05).
EEF1A2 gene expression and gene copy numbers in ZNF217-transformed lines
We examined two ZNF217-transformed isogenic sublines derived from I-144RZ and two ZNF217-transformed isogenic sublines derived from I-80RZ (8): medium in one of each pair of sublines was supplemented with 10ng EGF/ml which had been shown previously to be required transiently for immortalization (8). As shown in the present report, EGF had no influence on gene expression or other properties of the cell lines. eEF1A2 expression was consistently amplified in all 4 sublines (Table 1). Importantly, in line I–80RZ, where the 20q13 locus is amplified, eEF1A2 was significantly more overexpressed (23–27- fold) than in line I-144RZ (4.8- 6.6 fold) where 20q13 is not amplified. Of 11 established or putative ovarian epithelial oncogenes, the only ones that were sporadically overexpressed were STAT1, STAT3 and PI3KCA in the two I-144RZ lines and STAT1, STAT3 and RAB6B in the I-80RZ lines; all other oncogenes were either downregulated or unchanged, and none besides eEF1A2 were consistently overexpressed in all 4 sublines.
TABLE 1. CHANGES IN GENE EXPRESSION IN 2 ISOGENIC LINES I-144RZ AND 2 ISOGENIC LINES I-80RZ, INFECTED WITH RETROVIRAL ZNF217 IN PASSAGE 9, AND MAINTAINED WITH (+EGF) OR WITHOUT (−EGF) 10 ng EGF/ML MEDIUM.
Crisis occurred around passages 14–20. The genes listed are known or putative ovarian oncogenes and related genes, followed by HOXA genes known to influence ovarian carcinogenesis. EEF1A2 is the only gene consistently overexpressed after crisis.
| Growth stage: PrecrisisCrisis | Postcrisis | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cell Line | I-144RZ | I-80RZ | |||||||
| EGF* | − EGF | +EGF | −EGF | +EGF | |||||
| Passage | 14 | 15 | 20 | 29 | 51 | 29 | 62 | 59 | 48 |
| Gene | |||||||||
| ANXA10 | 3.57 | 3.34 | 0.35 | 0.42 | 0.29 | 0.41 | 0.39 | 0.02 | 0.06 |
| THBS1 | 1.22 | 0.42 | 0.11 | 0.13 | 0.12 | 0.12 | 0.26 | 0.05 | 0.12 |
| CDH11 | 6.93 | 4.28 | 0.97 | 0.35 | 0.41 | 0.34 | 0.36 | 0.10 | 0.29 |
| DSC3 | 3.31 | 1.90 | 1.00 | 0.27 | 0.32 | 0.29 | 0.31 | 0.09 | 0.09 |
| CDH2 | 3.81 | 3.19 | 1.31 | 0.31 | 0.44 | 0.32 | 0.36 | 0.23 | 0.24 |
| KRA 2 | 4.05 | 3.88 | 3.88 | 0.63 | 0.79 | 0.69 | 0.76 | 0.42 | 0.35 |
| ERBB2 | 1.59 | 1.93 | 2.51 | 1.13 | 1.25 | 1.19 | 1.17 | 1.32 | 0.54 |
| BRAF | 0.57 | 0.65 | 0.57 | 0.88 | 1.17 | 1.45 | 0.92 | 0.57 | 1.00 |
| EVI1 | 1.39 | 1.39 | 1.52 | 1.09 | 1.37 | 1.06 | 1.15 | 1.28 | 1.05 |
| RAB25 | 1.80 | 1.90 | 1.90 | 1.35 | 1.28 | 1.05 | 1.24 | 0.95 | 1.14 |
| MYC | 6.77 | 7.33 | 10.71 | 1.30 | 0.90 | 1.43 | 1.42 | 0.92 | 1.51 |
| MYB | 0.76 | 0.79 | 0.71 | 1.09 | 1.07 | 1.00 | 1.03 | 0.79 | 0.68 |
| ETS1 | 0.75 | 0.75 | 0.62 | 1.00 | 0.73 | 1.06 | 0.96 | 0.81 | 0.87 |
| EGFR | 1.96 | 1.44 | 2.06 | 1.55 | 1.63 | 1.56 | 1.55 | 0.67 | 1.23 |
| MET | 1.77 | 1.15 | 0.94 | 1.05 | 1.09 | 0.96 | 1.26 | 0.53 | 0.88 |
| PIK3CA | 2.10 | 0.49 | 0.81 | 1.71 | 2.07 | 1.41 | 1.47 | 0.94 | 0.88 |
| RAB6B | 0.57 | 0.57 | 0.57 | 0.87 | 0.80 | 0.80 | 1.04 | 2.37 | 2.55 |
| STAT1 | 2.35 | 3.32 | 10.02 | 3.32 | 1.40 | 3.33 | 0.45 | 5.13 | 2.20 |
| STAT3 | 2.00 | 2.56 | 4.03 | 1.02 | 1.14 | 1.05 | 1.12 | 0.79 | 2.00 |
| EEF1A2 | 1.67 | 2.31 | 33.14 | 6.57 | 6.57 | 5.23 | 4.82 | 23.18 | 36.99 |
| HOXA4 | 4.62 | 7.27 | 4.16 | 0.88 | 0.75 | 0.88 | 0.70 | 0.59 | 0.36 |
| HOXA7 | 1.27 | 1.32 | 1.31 | 1.11 | 1.18 | 1.08 | 0.98 | 0.64 | 0.48 |
| HOXA9 | 0.37 | 0.40 | 0.36 | 0.22 | 0.21 | 0.20 | 0.20 | 0.25 | 0.16 |
| HOX10 | 0.45 | 0.52 | 0.45 | 1.12 | 0.85 | 1.15 | 0.70 | 0.26 | 0.31 |
| HOX11 | 0.66 | 0.67 | 0.66 | 0.78 | 0.80 | 0.72 | 0.83 | 0.53 | 0.59 |
Bold = overexpressed genes (2.0 or more), underlined = underexpressed genes (0.5 or less); EGF*, parallel lines were maintained with and without 10 ng/ml EGF (Li et al., 2007)
Note: ANXA10, DSC3 and CDH2 are tumor suppressors.
Early stages of ZNF217-induced immortalization in line IOSE-144 were studied in colonies that appeared in ZNF217-infected cultures at the time of crisis. As shown in Table 1, eEF1A2 expression increased dramatically at the time of crisis and remained continuously upregulated thereafter. To determine whether EEF1A2 gene copy changes could be detected very early in the immortalization process, IOSE-144 cultures were infected with ZNF217 in passage 15, i.e. just prior to crisis, and when growing colonies, estimated to contain 20,000 to 40,000 cells, appeared during crisis, they were scraped off the culture dishes, lysed and analysed by CGH by Affymetrix SNP chip.
Of 9 such colonies analysed, one EEF1A2 gene copy was lost in five of the colonies, and the gene was amplified to 3–4 copies in 3 other colonies (Table 2). In Table 2, we included SNPs surrounding SNP_A-1757091 rs1406 and SNP_A-1698559 rs6089982, where eEF1A2 genetically resides, to emphasize the point that the observed variability in copy numbers is not limited to EEF1A2 but is a generalized phenomenon. Multiple other factors determine which variants will have a selective advantage that will allow the cells to overcome crisis. Cells with EEF1A2 amplification appear to be such variants. The most consistent feature among these colonies and fully transformed I-144RZ cell lines was gene loss at chromosome 18 (Table 3), which has also been reported as a frequent change in benign and neoplastic ovarian tumors (Tibiletti et al., 2003)(23).
TABLE II. GENE COPY NUMBER VARIATION ON CHROMOSOME 20 AMONG IOSE-144B COLONIES DURING CRISIS (20,000 to 40,000 cells per colony).
| COLONY NO. | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | ||||||
| SNP_A-1655213 | rs6061845 | 20 | 61.0842 | 61.0842 | NA | 2 | 3 | 2 | 3 | 2 | 2 | 3 | 3 | 3 |
| SNP_A-1655949 | rs10485827 | 20 | 61.0852 | 61.0852 | NA | 2 | 3 | 2 | 3 | 2 | 2 | 3 | 3 | 3 |
| SNP_A-1674859 | rs4308178 | 20 | 61.0862 | 61.0862 | NA | 2 | 3 | 2 | 3 | 2 | 2 | 3 | 3 | 3 |
| SNP_A-1748779 | rs6121489 | 20 | 61.1719 | 61.1719 | NA | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 3 | 3 |
| SNP_A-1692074 | rs708629 | 20 | 62.1332 | 62.1332 | NA | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 3 | 3 |
| SNP_A-1702295 | rs3746765 | 20 | 62.2471 | 62.2471 | NA | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 3 | 3 |
| SNP_A-1749637 | rs2295001 | 20 | 62.2674 | 62.2674 | NA | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 3 | 3 |
| SNP_A-1660585 | rs2427462 | 20 | 62.3174 | 62.3174 | NA | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 3 | 3 |
| SNP_A-1659348 | rs910933 | 20 | 62.3177 | 62.3177 | NA | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 3 | 3 |
| SNP_A-1757091 | rs1406961* | 20 | 62.6223 | 62.6223 | NA | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 3 | 3 |
| SNP_A-1698559 | rs6089982* | 20 | 63.1275 | 63.1275 | NA | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 3 | 3 |
| SNP_A-1758391 | rs2427625 | 20 | 63.5986 | 63.5986 | NA | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 3 | 3 |
location of EEF1A2
TABLE III. aCGH: COMMON FEATURES OF INDIVIDUAL COLONIES APPEARING DURING CRISIS, AND ESTABLISHED LINES.
I-144RZ and I-144RZ-B are lines obtained from independently ZNF217-infected IOSE-144. Colonies of 20,000 to 40,000 cells, appearing during crisis, were scraped off and lysed. Only changes that occurred in 4 or more colonies are listed.↑ and ↓ indicate gene gains and losses.
| I-144RZ-B colonies | Chromosomes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | ↓: 1 | 7 | 18 | ↑: | 5 | 8 | 15 | 20 | |||
| 8 | ↓: 1q | 7 | 18 | ↑: | 5 | 8 | 14 | 15 | |||
| 20 | |||||||||||
| 9 | ↓: 1 | 11 | 18 | ↑: | 5 | 8 | 14 | 15 | 20 | ||
| 10 | ↓: 1 | 18 | ↑: | 5 | 20 | ||||||
| 11 | ↓: | 3 | 11 | 18 | ↑: | 5 | 14 | 20 | |||
| 12 | ↓: 1 | 18 | ↑: | 5 | 8 | 14 | 15 | 20 | |||
| 13 | ↓: | 3 | 11 | 18 | ↑: 1p | ||||||
| 14 | ↓: 1 | 11 | 18 | ↑: 1p | 5 | ||||||
| 15 | ↓: 1 | 11 | 18 | ↑: 1p | |||||||
| Postcrisis and long-term changes, respectively | |||||||||||
| I-144RZ passage 20: | ↓: 1q | 3p | 7q | 18 X | ↑: 3q | 8q | 14 | ||||
| I-144RZ passage 61: | ↓: | 3p | 7q | 18 X | ↑: 3q | 5 | 8q | 14 | |||
Phenotypic characteristics
To determine whether the phenotypic changes in I-144LE lines were caused specifically by eEF1A2, and whether eEF1A2 mediated effects of ZNF217, we transiently transfected I-144LE and I-144RZ cells with siRNA against eEF1A2 and studied the effects on protein expression, anchorage independence and apoptosis.
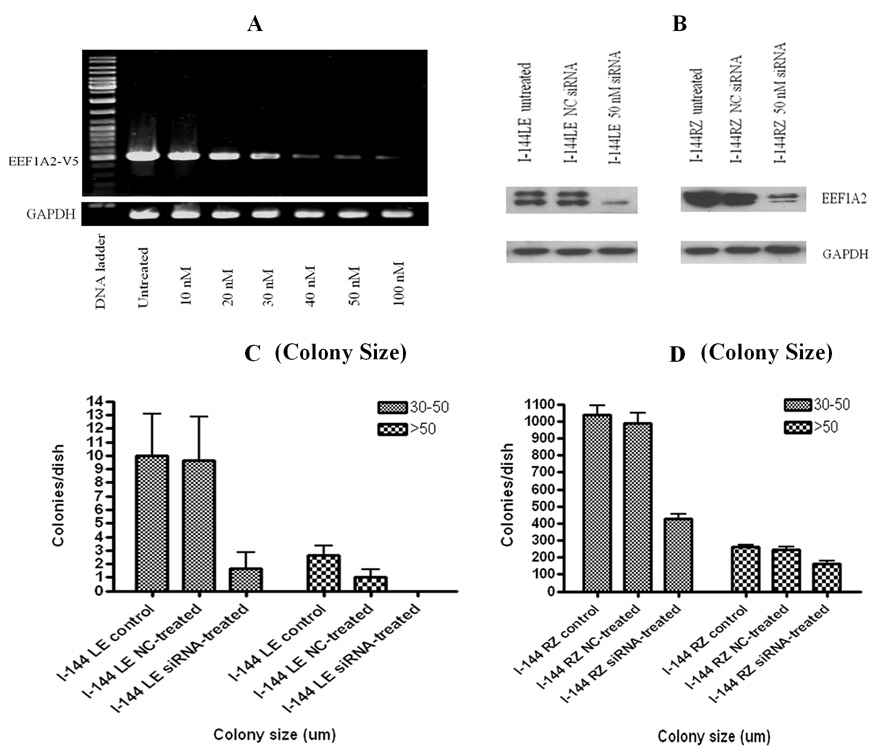
Upon siRNA transfections, RT-PCR revealed diminishing eEF1A2 mRNA signals in eEF1A2 overexpressing cell lines (Fig. 4A), and Western blots indicated reduced eEF1A2 protein expression, in both I-144LE cells and I-144RZ cells (Fig. 4B).
Figure 4.
Effects of siRNA against eEF1A2 on protein expression and anchorage independence. NC is control siRNA. A, RT-PCR demonstrated reduced eEF1A2 mRNA levels in the eEF1A2- overexpressing cell line I-144LE upon transfection with increasing concentrations of siRNA. B, Western blot: siRNA significantly reduced eEF1A2 expression in both I-144LE and I-144RZ cells. GAPDH was a loading control. C, D. Effects of eEF1A2-siRNA on anchorage independence, measured as average colony numbers per dish according to the colony size (30–50 µm, and >50 µm. In both the I-144LE line and the I-144RZ line, siRNA significantly inhibited colony formation in agarose compared to controls and NC siRNA (p<0.01). Note that the scale bars differ between the two cell lines: I-144RZ cells form approximately 100 –fold more colonies than I-144LE cells.
Anchorage independence assays showed that I-144RZ cells formed approximately 100 times more colonies than I-144LE cells. However, importantly, not only the I-144LE cells but also the I- 144RZ cells formed significantly fewer colonies in the presence of siRNA to eEF1A2 (Fig. 4C,D).
The susceptibility to apoptosis of lines I-144LE and I-144RZ was examined by treatment with the DNA topoisomerase I inhibitor camptothecin (CAM). After treatment for 16–20 hr, dramatic cell death was observed in the parent line IOSE-144 (data not shown). In contrast, the damaging effects were alleviated significantly in I-144LE cells, although both I-144RZ cells and SKOV3 cells, representing a camptothecin resistant established ovarian cancer line, exhibited relatively more robust resistance against this cytotoxic drug (Fig. 5A,B). In parallel, the EnzChek Caspase-3 Assay and Cell Death Detection ELISA also demonstrated greatly reduced apoptosis in lines I- 144LE and I-144RZ compared with the parent line IOSE-144 (Fig.5A,B)‥ These assays measure caspase-3 activity and increased histone-associated DNA fragmentation into mono- and oligonucleosomes, respectively, as markers of these responses to apoptosis-inducing drugs among known drug-resistant cell lines. Caspase-3 activity and DNA fragmentation were significantly increased upon apoptosis induction in the presence of eEF1A2-siRNA in both I-144LE and I- 144RZ cells (Figs.5C, D). Importantly, the anti-apoptotic effects of both eEF1A2 and ZNF217 were reduced to a similar degree by siRNA to eEF1A2. These results confirmed the specific role of eEF1A2 in regulating anchorage independence and apoptosis resistance in I-144LE cells, and support the hypothesis that eEF1A2 mediates proneoplastic features resulting from overexpression of ZNF217.
Figure 5.
A, B, apoptosis resistance of lines IOSE-144, I-144LE, I-144RZ and SKOV3 prior to and after camptothecin (CAM) treatment. A EnzCheck caspase-3 activity assay of the above cell lines. B. Cell death detection ELISA assay for histone-associated DNA fragmentation into monoand oligonucleosomes. C, D, Effects of eEF1A2-siRNA on apoptosis resistance in lines I-144LE and I-144RZ before and after CAM treatment. C. EnzChek caspase-3 activity assay. D. Cell death detection ELISA.
Discussion
While increased eEF1A2 expression has previously been demonstrated in ovarian carcinomas (14), our study defines, for the first time, direct proneoplastic effects of eEF1A2 on the nontumorigenic precursors of these neoplasms. Furthermore, our results suggest that the two known ovarian oncogenes, ZNF217 and EEF1A2, may act interdependently in ovarian neoplastic progression, thus revealing a possible mechanism by which ZNF217, a gene suppressor protein, may exert its oncogenic effects. Krig et al. (3) recently carried out a comprehensive identification of all genes that are directly regulated by the gene suppressor ZNF217, using ChiP-chip assays (3). They found that there is negligible direct interaction between ZNF217 and EEF1A2. This conclusion is consistent with our present and previous results which indicate that, in contrast to a large number of genes eEF1A2 expression is not downregulated by overexpressed ZNF217. The results of the present study, which show that eEF1A2 is in fact overexpressed in the presence of overexpressed ZNF217, suggests that the interrelationships between ZNF217 and EEF1A2 must be indirect, perhaps through inhibition by ZNF217 of physiologic negative regulator(s) of eEF1A2 expression (13, 20).
Overexpressed eEF1A2 rendered IOSE cells resistant to apoptosis and resulted in increased anchorage- and serum independence, suggestive of altered production of growth factors and/or extracellular matrix components. It also resulted in increased saturation densities, which indicate potential transition from normalcy toward malignancy. Normal cell populations become growth arrested when they reach specific levels of confluency (saturation density) through inhibition of further proliferation by intercellular contact. This regulatory process, named contact inhibition of growth, is compromised in neoplastic cells, thus allowing the cells to continue growing to increased saturation densities. Interestingly, the changes induced by eEF1A2 resemble the neoplastic characteristics that were induced in IOSE by ZNF217 (8), but in eEF1A2-overexpressing cells, they were subtle compared to the more robust effects of ZNF217. Both of these genes exert some of their oncogenic effects by activating Akt2 and thereby increasing its antiapoptotic and anchorage independence-enhancing action (5, 9). Most importantly, siRNA to eEF1A2 reversed the anchorage independence and the resistance to apoptosis induced by overexpression of either eEF1A2 or of ZNF217.
Our results indicate that eEF1A2 may mediate the proneoplastic changes caused by ZNF217, and/or that ZNF217 may enhance the proneoplastic effects of eEF1A2 by upregulating its expression. In addition, eEF1A2 may complex with ZNF217, since this protein has been reported to function through other protein-protein interactions: it forms complexes with Prdx-1 which protect cells from apoptosis more efficiently than either protein alone (25) and it forms complexes with the zinc finger protein ZRP1, which translocate to the nucleus to maintain cell cycling (26). However, at present there is no evidence for such an interaction between eEF1A2 and ZNF217.
In our studies of ZNF217-transduced IOSE cells, most genes were downregulated (8), as would be expected from a transcriptional gene suppressor (3). In our past and present studies, EEF1A2 stood out as the only oncogene that was consistently overexpressed, independently of source, duration in culture or EGF treatment. Although eEF1A2 was consistently overexpressed in all four I-144RZ and I-80RZ lines, the q20.13 locus where the EEF1A2 gene is located was amplified only in the I-80RZ lines but not in the I-144RZ lines (8). It has been reported previously that copy numbers of EEF1A2 do not always correlate with expression levels of the gene, indicating that eEF1A2 expression and/or protein levels must also be regulated at other levels (15). The fact that, in our studies, eEF1A2 was overexpressed in lines with and without 20q13 amplification indicates that, similar to Tomlinson’s observations (15, 16), mechanisms besides gene dosage were responsible for its overexpression. Furthermore, eEF1A2 overexpressing IOSE cells appeared to have a selective advantage regardless of the means by which overexpression was achieved, since they were selected for in every line. Interestingly, the degree of overexpression was approximately 5-fold greater in the lines with 20q13 gene amplification compared to the lines with normal gene dosage. This observation suggests that 20q13 amplification may have enhanced the overexpression of eEF1A2 and thus the selective advantage of these cell populations.
Using SNP analysis of very small colonies consisting of approximately 20,000–40,000 cells, which appeared during senescence and crisis of ZNF217-transfected IOSE cultures, we observed considerable variation in gene copy numbers, indicative of genetic instability. The number of EEF1A2 copies ranged from 1–4 in various colonies. Since the colonies had to be lysed for analysis, and recovery from crisis is a rare event, it was impossible to determine whether this variability had any relationship to the establishment of permanent immortal lines – however, the results do show that the genetic instability at the time of crisis affects EEF1A2 gene expression which may be a factor influencing the success of crisis survival (27).
In summary, this study further defines the role of eEF1A2 in ovarian carcinogenesis and shows that this protein confers neoplastic characteristics on preneoplastic, nontumorigenic precursors of ovarian cancer. eEF1A2 thus has the potential to exert some of its transforming effects early in ovarian neoplastic progression. Another important point raised by the results of this study is the possibility that at least some of the proneoplastic effects of the transcriptional gene suppression protein ZNF217 may be conveyed indirectly by upregulated EEF1A2. In view of the complexity of the transcriptional regulation of eukaryotic ribosomal protein genes (22), the detailed molecular mechanisms of such an interaction remain to be determined.
Acknowledgments
Supported by grants to N.A. by the National Cancer Institute of Canada through the Canadian Cancer Society to N.A. and by a fellowship to Y.S. from OvCaRe Canada. J.W. Gray’s work was supported by the NIH (CA58207 and CA112970), the Office of Health and Environmental Research of the U.S. Department of Energy (contract DE-AC03-76SF00098), and the Avon Foundation. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. For full disclaimer, see http://www-library.lbl.gov/public/tmRco/howto/RcoBerkeleyLabDisclaimer.htm. We thank Sarah Maines-Bandiera for expert assistance.
Footnotes
Novely and impact: (1) First report on direct oncogenic effects of eEF1A2 on ovarian cancer precursors; (2) evidence that eEF1A2 mediates or facilitates oncogenic effects of ZNF217 – a new example of oncogene interaction.
References
- 1.Tanner MM, Grenman S, Koul A, Johansson O, Meltzer P, Pejovic T, Borg A, Isola JJ. Frequent amplification of chromosomal region 20q12–q13 in ovarian cancer. Clin Cancer Res. 2000;6:1833–1839. [PubMed] [Google Scholar]
- 2.Watanabe T, Katahira II, Hirasawa A, Ishiwata I, Emi M, Takayama M. Differentially regulated genes as putative targets of amplifications at 20q in ovarian cancers. Jap J Cancer Res. 2002;93:1114–1122. doi: 10.1111/j.1349-7006.2002.tb01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krig SR, Jin VX, Bieda MC, O’Geen H, Yaswen P, Green R, Farnham PJ. Identification of genes directly regulated by the oncogene ZNF217 using chromatin immunoprecipitation (ChIP)-chip assays. J Biol Chem. 2007;282:9703–9712. doi: 10.1074/jbc.M611752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowger JJM, Zhao Q, Isovic M, Torchia J. Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: identification of a ZNF217 consensus recognition sequence. Oncogene. 2007;26:3378–3386. doi: 10.1038/sj.onc.1210126. [DOI] [PubMed] [Google Scholar]
- 5.Huang G, Krig S, Kowbel D, Xu H, Hyun B, Volik S, Feuerstain B, Mills GB, Stokoe D, Yaswen P, Colloins C. ZNF217 suppresses cell death associated with chemotherapy and telomere dysfunction. Hum Mol Genet. 2005;14:3219–3225. doi: 10.1093/hmg/ddi352. [DOI] [PubMed] [Google Scholar]
- 6.Quinlan KG, Verger A, Yaswen PK, Crossley M. Amplification of zinc finger gene 217 (ZNF217) and cancer: when good fingers go bad. Biochem Biophys Acta. 2007;1775:333–340. doi: 10.1016/j.bbcan.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Nonet GH, Stampfer MR, Chin K, Gray JW, Collins CC, Yaswen P. The ZNF217 gene amplified in breast cancers promotes immortalization of human mammary epithelial cells. Cancer Res. 2006;61:1250–1254. [PubMed] [Google Scholar]
- 8.Li P, Maines-Bandiera S, Kuo WL, Guan Y, Sun Y, Hills M, Huang G, Collins CC, Leung PC, Gray JW, Auersperg N. Multiple roles of the candidate oncogene ZNF217 in ovarian epithelial neoplastic progression. Int J Cancer. 2007;120:1863–1873. doi: 10.1002/ijc.22300. [DOI] [PubMed] [Google Scholar]
- 9.Amiri A, Noei F, Jeganathan S, Kulkarni G, Pinke DE, Lee JM. eEF1A2 activates Akt and stimulates Akt-dependent actin remodeling, invasion and migration. Oncogene. 2007;26:3027–3040. doi: 10.1038/sj.onc.1210101. [DOI] [PubMed] [Google Scholar]
- 10.Condeelis J. Elongation factor 1α, translation and the cytoskeleton. Trends in Biochemical Sciences. 1995;20:169–170. doi: 10.1016/s0968-0004(00)88998-7. [DOI] [PubMed] [Google Scholar]
- 11.Chung CM, Man C, Jin Y, Jin C, Guan XY, Wang Q, Wan TS, Cheung AL, Tsao SW. Amplification and overexpression of aurora kinase A (AURKA) in immortalized human ovarian epithelial (HOSE) cells. Mol Carcinog. 2005;43:165–174. doi: 10.1002/mc.20098. [DOI] [PubMed] [Google Scholar]
- 12.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 13.Ejiri S. Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1-associated nuclear localization. Biosci Biotechnol Biochem. 2002;66:1–21. doi: 10.1271/bbb.66.1. [DOI] [PubMed] [Google Scholar]
- 14.Anand N, Murthy S, Amann G, Wernick M, Porter LA, Cukier IH, Collins C, Gray JW, Diebold J, Demetrick DJ, Lee JM. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat Genet. 2002;31:301–305. doi: 10.1038/ng904. [DOI] [PubMed] [Google Scholar]
- 15.Tomlinson VA, Newbery HJ, Bergmann JH, Boyd J, Scott D, Wray NR, Sellar GC, Gabra H, Graham A, Williams AR, Abbott CM. Expression of eEF1A2 is associated with clear cell histology in ovarian carcinomas: overexpression of the gene is not dependent on modifications at the EEF1A2 locus. Brit J Can. 2007;96:1613–1620. doi: 10.1038/sj.bjc.6603748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomlinson VA, Newbery HJ, Wray NR, Jackson J, Larionov A, Miller WR, Dixon JM, Abbott CM. Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two-thirds of breast tumours. BMC Cancer. 2005;5:113. doi: 10.1186/1471-2407-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li R, Wang H, Belkele BN, Yin Z, Caraway NP, Katz RL, Staass SA, Jiang F. Identification of putative oncogenes in lung adenocarcinoma by a comprehensive functional genomic approach. Oncogene. 2006;25:2628–2635. doi: 10.1038/sj.onc.1209289. [DOI] [PubMed] [Google Scholar]
- 18.Kulkarni G, Turbin DA, Amiri A, Jeganathan S, Andrade-Navarro MA, Wu TD, Huntsman DG, Lee GM. Expression of protein elongation factor eEF1A2 predicts favorable outcome in breast cancer. Breast Cancer Research and Treatment. 2007;102:31–41. doi: 10.1007/s10549-006-9315-8. [DOI] [PubMed] [Google Scholar]
- 19.Zhu H, Lam DC, Han KC, Tin VP, Suen WS, Wang E, Lam WK, Cai WW, Chung LP, Wong MP. High resolution analysis of genomic aberrations by metaphase and array comparative genomic hybridization identifies candidate tumour genes in lung cancer cell lines. Cancer Lett. 2007;245:303–314. doi: 10.1016/j.canlet.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Ruest LB, Marcotte R, Wang E. Peptide elongation factor eEF1A-2/S1 expression in cultured differentiated myotubes and its protective effect against caspase-3-mediated apoptosis. J Biol Chem. 2007;277:5418–5425. doi: 10.1074/jbc.M110685200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goncalves J, Malta-Vacas J, Louis M, Brault L, Bagrel D, Monteiro C, Brito M. Modulation of translation factor’s gene expression by histone deacetylase inhibitors in breast cancer cells. Clin Chem Lab Med. 2005;43:151–156. doi: 10.1515/CCLM.2005.025. [DOI] [PubMed] [Google Scholar]
- 22.Hu H, Li X. Transcriptional regulation in eukaryotic ribosomal protein genes. Genomics. 2007;90:421–423. doi: 10.1016/j.ygeno.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Tibiletti MG, Bernasconi B, Taborelli M, Facco C, Riva C, Capella C, Franchi M, Binelli G, Acquati F, Taramelli R. Genetic and cytogenetic observations among different types of ovarian tumors are compatible with a progression model underlying ovarian tumorigenesis. Cancer Genet Cytogenet. 2003;146:145, 153. doi: 10.1016/s0165-4608(03)00134-1. [DOI] [PubMed] [Google Scholar]
- 24.Maines-Bandiera SL, Kruk PA, Auersperg N. Simian virus 40-transformed human ovarian surface epithelial cells escape normal growth controls but retain morphogenetic responses to extracellular matrix. Am J Obstet Gynecol. 1992;167:729–735. doi: 10.1016/s0002-9378(11)91579-8. [DOI] [PubMed] [Google Scholar]
- 25.Chang R, Wang E. Mouse translation elongation factor eEF1A-2 interacts with Prdx-1 to protect cells against apoptotic death induced by oxidative stress. J of Cell Biochem. 2007;100:267–278. doi: 10.1002/jcb.20969. [DOI] [PubMed] [Google Scholar]
- 26.Gangwani L, Mikrut M, Galcheva-Gargova Z, Davis RJ. Interaction of ZPR1 with translation elongation factor-1alpha in proliferating cells. J Cell Biol. 1998;143:1471–1484. doi: 10.1083/jcb.143.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung AL, Deng W. Telomere dysfunction, genome instability and cancer. Front Biosci. 2008;13:2075–2090. doi: 10.2741/2825. [DOI] [PubMed] [Google Scholar]