Summary
Understanding the DNA recognition and binding by the AbrB-like family of transcriptional regulators is of significant interest since these proteins enable bacteria to elicit the appropriate response to diverse environmental stimuli. Although these ‘transition-state regulator’ proteins have been well characterized at the genetic level, the general and specific mechanisms of DNA binding remain elusive. We present RDC-refined NMR solution structures and dynamic properties of the DNA-binding domains of three Bacillus subtilis transition-state regulators AbrB, Abh, and SpoVT. We combined previously investigated DNase I footprinting, DNA methylation, gel shift assays, mutagenic and NMR studies to generate a structural model of the complex between AbrBN55 and its cognate promoter, abrB8. These investigations have enabled us to generate the first model for the specific nature of the transition-state regulator-DNA interaction.
Introduction
Transcription factors are pivotal to gene regulation and elucidating their modes of action is essential for understanding gene expression. The adaptability and endurance of bacteria derives from their ability to initiate the suitable response at the appropriate time under a specific circumstance (Aertsen and Michiels, 2004; Sonenshein et al., 2002). Such circumstances include changes in chemical concentration, temperature, osmolarity, viscosity, light, pH, density, and exposure to anti-infectives. Substantial changes in transcriptional regulation occur upon sensing that environments become hostile. Moving from exponential growth phase to a more resistant stationary phase prepares a cell for survival by expression of bacterial protection genes. Correspondingly, bacterial responses also take various forms including biofilm formation, toxin secretion, production of polymer-degrading enzymes or antibiotics, and complete physiological transformations.
Like many bacteria, Bacillus subtilis responds to a multitude of environmental stimuli by using transcription factors called transition state regulators (TSRs) to orchestrate gene expression patterns (Sonenshein et al., 2002). In B. subtilis, TSRs play an essential role in cell survival by regulating spore formation, competence, and biofilm development (Strauch and Hoch, 1993). TSRs are generally referred to as ‘AbrB-like’, as the most widely studied TSR is the transcription factor AbrB. Numerous known or projected AbrB-like TSRs are found in an array of important human health-related organisms, including Staphylococcus, Streptococcus, Listeria and Clostridia (Bobay et al., 2004). However, while there is a surfeit of biochemical and genetic data on AbrB, there is no model for how this protein, or indeed how any other AbrB-like TSR, performs its biological role.
AbrB does not recognize a well-defined DNA base-pairing sequence. Instead, it appears to target a very weak pseudo consensus nucleotide sequence, TGGNA-5bp-TGGNA, which allows it to be rather promiscuous in binding. AbrB regulates the transcription of more than 60 different genes (Xu and Strauch, 1996). Then again, AbrB has high binding affinity to a finite subset of DNA targets (Bobay et al., 2004). The DNA-binding properties of AbrB have been described as three-fold (Bobay et al., 2006): (i) non-specific interactions arising from the recognition of general DNA features; (ii) limited promiscuity allowing for interactions with a subset of structurally related DNA sequences; and (iii) high affinity interactions with specific DNA sequences. It is possible to obtain a better understanding of the three-fold nature of DNA-binding by the AbrB-like TSRs using comparative structural studies as these are structurally homologous proteins that display different binding properties (structural differences) while also possessing general recognition characteristics (structural similarities).
This study extends our previous work on AbrB (Bobay et al., 2005) and Abh (Bobay et al., 2006) to include a third AbrB-like TSR paralog, SpoVT from B. subtilis. SpoVT is a transcription factor responsible for stimulating/repressing forespore-specific σG-dependent transcription (Bagyan et al., 1996; Dong et al., 2004; Shcheptov et al., 1997). For these three TSRs there is no known DNA target that they share in common, no overlapping DNA targets (Bobay et al., 2005; Bobay et al., 2004; Bobay et al., 2006), and furthermore these proteins are not expressed in the cell at the same time (Bagyan et al., 1996; Shcheptov et al., 1997; Strauch and Hoch, 1993). Given the high degree of similarity in both their primary sequences (36% overall and 78% within the N-terminal DNA-binding domain) and secondary structure within their N-terminal DNA-binding domains, structural similarities among these three proteins should lead to general TSR recognition properties, whereas structural differences should lead to protein specific structural requirements for selecting the correct DNA target.
Here we present RDC-refined NMR solution structures and backbone dynamics analyses of the N-terminal DNA-binding domains of three AbrB-like transcription factors from B. subtilis: AbrB, Abh, and SpoVT. This study represents the first structural characterization for SpoVT. Consequently, comparative analyses provide a unique opportunity to identify essential contributions to the DNA recognition and specificity of this class of proteins. We also incorporate a number of biochemical studies, including gel shift assays, DNase I footprinting, DNA methylation, as well as mutational and knockout data (unpublished, and (Bobay et al., 2005; Bobay et al., 2004; Cavanagh et al., 2002; Strauch, 1995c; Strauch and Ayazifar, 1995; Xu et al., 1996)) to develop a structural model of AbrB bound to the target promoter abrB8. The model reveals that AbrB binds to one side of the cognate promoters in both the major and minor grooves. Our comparative structural and dynamic studies, in conjunction with a model of the interaction between AbrB and abrB8, support the importance of conformational flexibility in the function of AbrB-like TSRs.
Results
Refined NMR Structures of AbrBN53, AbhN, and SpoVTN
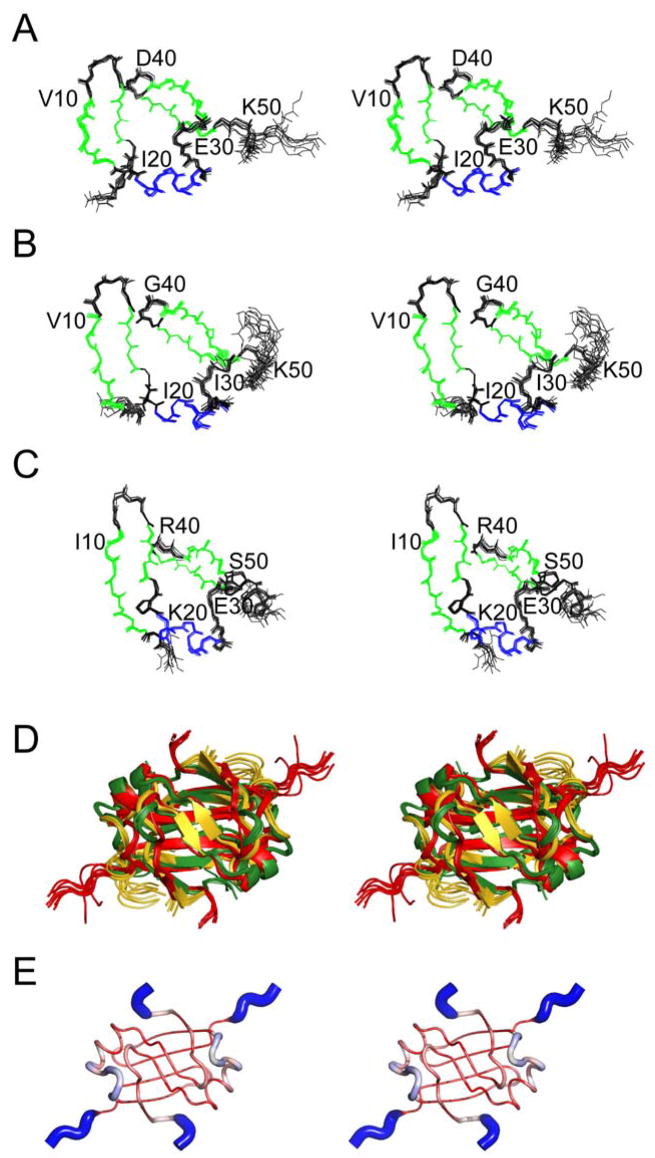
Chemical shift assignments for SpoVTN (residues 1–55) were obtained for 94% of backbone HN, N, Cα, and C′ nuclei and near complete assignments for side-chain nuclei. The 1H–15N HSQC spectrum of SpoVTN displays good peak dispersion, and size exclusion chromatography reveals a dimeric state in solution (not shown). Refined structures of AbrBN53 (residues 1–53; Figure 1A), AbhN (residues 1–54; Figure 1B), and SpoVTN (Figure 1C) were determined using nuclear Overhauser effect (NOE) distance, dihedral angle, hydrogen bond, and 1DNH residual dipolar coupling (RDC) restraints (Table 1). The average pairwise backbone root mean square deviation (rmsd) of AbrBN53, AbhN, and SpoVTN is 0.832±0.340Å, 0.742±0.227Å, and 0.603±0.199Å, respectively.
Figure 1.
RDC-refined structure ensembles of AbrBN53, AbhN, and SpoVTN. Cartoon stereo-view diagrams of the refined structure ensemble of (A) AbrBN53, (B) AbhN, (C) SpoVTN, (D) superposition of the refined structure ensembles of AbrBN53, AbhN, and SpoVTN, and (E) putty/sausage PyMOL diagram illustrating regions of structural variation in the maximum likelihood (ML) superposition of AbrBN53, AbhN, and SpoVTN plotted on the structure of AbrBN53 as reported by THESEUS. The color scheme is red-white-blue, small-to-large structural deviation in the ML structural superposition. Note the N- and C-termini and LP2 regions show the largest structural divergence.
Table 1.
NMR and refinement statistics
| NMR distance and dihedral constraints | AbrBN53 | AbhN | SpoVTN |
|---|---|---|---|
| Distance constraints | |||
| Total NOE | 3083 | 2180 | 3282 |
| Ambiguous | 1392 | 767 | 1147 |
| Unambiguous | 1691 | 1413 | 2135 |
| Intra-residue | 901 | 329 | 918 |
| Sequential (|i–j|=1) | 933 | 730 | 808 |
| Medium range (|i–j|<4) | 461 | 478 | 621 |
| Long range (|i–j|>5) | 788 | 643 | 935 |
| Hydrogen bonds | 48 | 70 | 54 |
| Total dihedral angles | |||
| φ | 69 | 47 | 38 |
| ψ | 69 | 47 | 38 |
| RDC constraints | 46 | 46 | 38 |
|
| |||
| Structural statistics | |||
| Energies (kcal mol −1) | |||
| van der Waals | −1018.6 ± 32.0 | −1016.6 ± 29.8 | −1001.7 ± 43.5 |
| Electrostatic | −4044.3 ± 53.0 | −4051.1 ± 79.7 | −4238.8 ± 79.5 |
| Average violations per structure | |||
| NOEs and/or hydrogen bondsb | 0 | 0 | 0.1 |
| Dihedralsb | 0 | 0 | 0 |
| Violations (mean and S.D) | |||
| Distance constraints (Å) | 0.032 ± 0.001 | 0.030 ± 0.001 | 0.039 ± 0.001 |
| Hydrogen bonds (Å) | 0.055 ± 0.004 | 0.063 ± 0.004 | 0.091 ± 0.006 |
| Dihedral angle constraints (°) | 0.404 ± 0.134 | 0.316 ± 0.060 | 0.783 ± 0.089 |
| Deviations from idealized geometry | |||
| Bond lengths (Å) | 0.0042 ± 0.0001 | 0.0047 ± 0.0001 | 0.0063 ± 0.0001 |
| Bond angles (°) | 0.622 ± 0.014 | 0.693 ± 0.010 | 0.813 ± 0.020 |
| Impropers (°) | 1.66 ± 0.11 | 1.91 ± 0.10 | 1.93 ± 0.07 |
| RDC constraints | |||
| R-factor | 0.289 | 0.213 | 0.197 |
| Q-factor | 0.224 | 0.207 | 0.166 |
| R.m.s deviation (Hz) | 0.745 | 0.928 | 2.217 |
| Correlation (experimental to calculated) | 0.999 | 0.978 | 0.988 |
| Average pairwise r.m.s (Å)c | |||
| Secondary structure (backbone) | 0.223 ± 0.049 | 0.206 ± 0.048 | 0.221 ± 0.048 |
| Secondary structure (heavy) | 0.721 ± 0.096 | 0.564 ± 0.063 | 0.632 ± 0.066 |
| Backbone | 0.832 ± 0.340 | 0.742 ± 0.227 | 0.603 ± 0.199 |
| Heavy | 1.29 ± 0.281 | 1.03 ± 0.205 | 0.986 ± 0.156 |
| Ramachandran analysis | |||
| Most favored | 74.9 | 71.7 | 61.5 |
| Additionally allowed | 14.1 | 18.5 | 26.1 |
| Generously allowed | 6.4 | 7.6 | 8.7 |
| Disallowed | 4.6 | 2.2 | 3.7 |
Structural statistics for non-RDC refined SpoVTN.
0.5 Å Violations for the ensemble of 10 lowest energy structures.
Pairwise r.m.s. deviation was calculated among the 10 lowest energy structures for residues 1–53 (AbrBN53), 1–54 (AbhN) and 1–55 (SpoVTN).
The structures of AbrBN53, AbhN, and SpoVTN have similar secondary structure elements and global folds. The monomeric subunits have, with respect to SpoVTN, four β-strands (β1, residues 5–9; β2, residues 15–17; β3, residues 34–39; and β4, residues 43–47), one α-helix (α1, residues 20–26), and three loop regions (LP1, residues 11–14; LP2, residues 28–32; and LP3, residues 40–42). Superposition of the TSR structures (Figure 1D) reveals that the proteins overlay quite well, with Cα rmsd values (including residues 4–47) of 1.408Å between AbrBN53 and AbhN, 2.096Å between AbrBN53 and SpoVTN, and 1.988Å between AbhN and SpoVTN. The regions with the largest degree of structural variation in the THESEUS ML superposition include the N- and C-termini, as well as the LP2 region (Figure 1E).
The dimer quaternary structure involves extensive interactions between monomeric subunits making up a domain swap fold (Figure 1). An extensive β-sheet scaffold composed of β1, β3′, β4′, β4, β3, and β1′ on one face, and β2 and β2′ on the other, defines the core and dimer interface of these proteins. β1 and β2 from monomeric subunit A interacts with β3 and β4 from monomeric subunit B (residues in this monomer will be identified by notation from this point onward, i.e. R15 and R15′). A number of hydrogen bonds are involved in the dimerization interface, including hydrogen bonds between residues 44 and 46 in β4 and residues 46′ and 44′ in β4′, respectively, as well as residue 16 in β2 and residue 16′ in β2′. Short NOE distance interactions between residues 44 and 46 in β4 to residues 16′ and 18′ in β2′ further define the dimerization interface. Sequence alignment of AbrBN53, AbhN, and SpoVTN (Figure S1) reveals a glycine residue at position 42 in SpoVTN that is not present in AbrBN53 or AbhN, which extends the length of LP3 and shifts the register of β4 with respect to AbrBN53 and AbhN (shown diagrammatically at the top of Figure 3A).
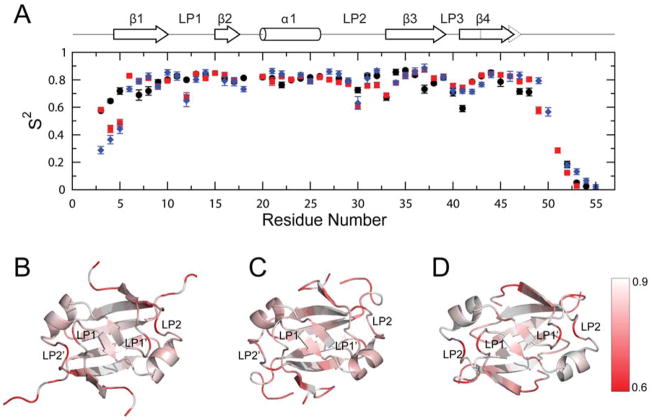
Figure 3.
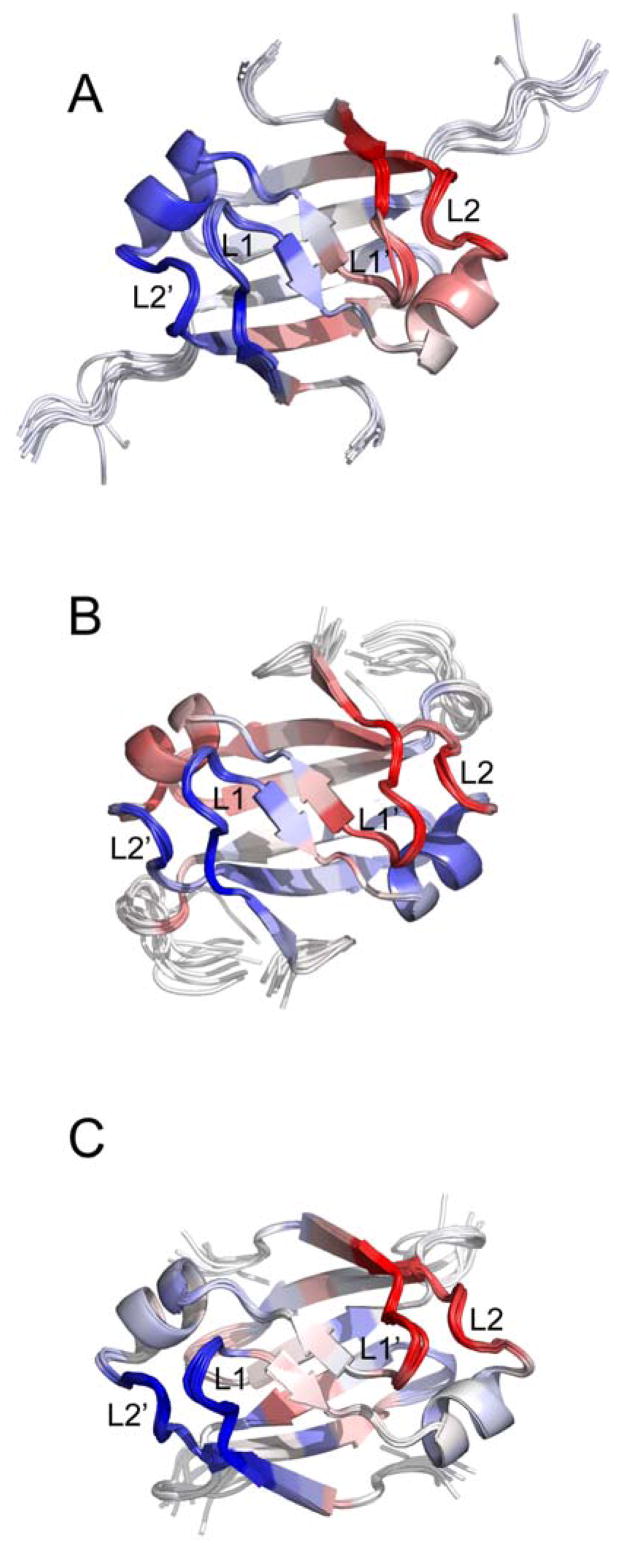
Model-free backbone order parameters for AbrBN53, AbhN and SpoVTN. (A) Backbone order parameter (S2) vs. residue number for AbhN (black circles), AbrBN53 (red squares) and SpoVTN (blue diamonds). The positions of the α-helical and β-strand secondary structure, as well as loop regions defined in the text, are indicated schematically at the top. Error bars indicate the fitting errors. S2 varies between 0 and 1, with a value of 1 represents completely restricted motion. (B–D) S2 values plotted on the solution structures of (B) AbrBN53, (C) AbhN and (D) SpoVTN most similar to the average structure in the ensemble reported by THESEUS. LP1 and LP2 (chain A) and LP1′ and LP2′ (chain B) are noted, as well as the color scale of S2 order parameters ranging from >=0.9 (white) to <=0.6 (red). Proteins are oriented as depicted in Figure 1.
The α-helix in each monomer contains residues critical to the DNA-binding properties of TSR proteins. The refined structures presented here reveal a difference in the angle of the α-helix with respect to β2, with values of 9.0° between AbrBN53 and AbhN, 21.2° between AbrBN53 and SpoVTN, and 12.2° between AbhN and SpoVTN. However, as noted in our previous study (Bobay et al., 2006), the α-helical pitch (i.e. the angle of the helix with respect to the plane of β2) among these proteins is very similar (~52° in AbrBN53 and SpoVTN and ~56° in AbhN). The similarity in pitch may have functional relevance to the general recognition properties of these DNA binding proteins, whereas the dissimilarity in angle may have implications for target specificity. Because the β-sheet scaffold is located on the surface opposite to the putative DNA-binding surface, the slight difference noted in the register of β4/β4′ of SpoVTN most likely does not play a role in directing DNA specificity (see the modeling section below).
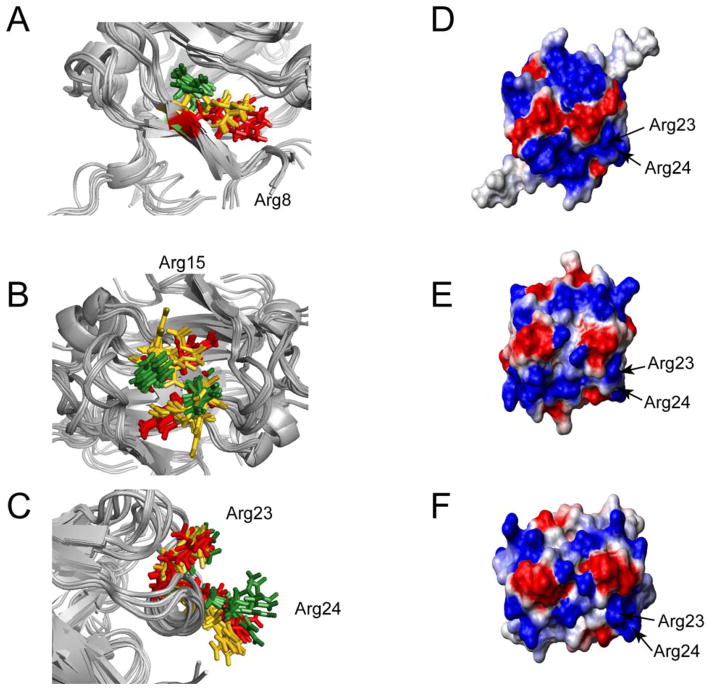
Mutagenic analysis of AbrB identified residues R8, R15, R23, and R24 as critical to AbrB’s ability to bind DNA (Bobay et al., 2005; Vaughn et al., 2000; Xu et al., 1996). Slight differences in position and orientation of these residues in the refined structures of AbrBN53, AbhN and SpoVTN are noted. An ensemble superposition reveals that R8 (Figure 2A) has a similar position and orientation in AbrBN53 and AbhN, whereas R8 in SpoVTN is oriented towards LP1. The position and orientation of R15 is similar between AbhN and SpoVTN, whereas R15 in AbrBN53 crosses the dimerization interface between β2 and β2′ (Figure 2B). Finally, the position and orientation of R23 and R24 are similar among the three proteins (Figure 2C). The similarity in the spatial arrangement of R23 and R24 may have functional relevance to the general recognition properties of these DNA binding proteins, whereas the dissimilarity in the spatial arrangement of R8 and R15 between the structures of AbrBN53, AbhN, and SpoVTN may have implications for target specificity.
Figure 2.
Structural similarities and differences among AbrBN53, AbhN, and SpoVTN. Critical arginine residues involved in DNA binding – orientations for (A) R8, (B) R15 and (C) R23 and R24 from AbrBN53 (red), AbhN (gold), and SpoVTN (green). Comparison of the electrostatic surface potential between (D) AbrBN53, (E) AbhN, and (F) SpoVTN. Blue regions indicated positive charge, whereas red regions indicate negative charge. Proteins are oriented as depicted in Figure 1.
The electrostatic surface potential of the putative DNA-binding surface of AbrBN53, AbhN, and SpoVTN (Figure 2D, 2E, and 2F, respectively) reveal that the three proteins have a considerable amount of positive charge resulting from conserved arginine and lysine residues (R8, R9/K9, R15, R23, R24 and K31) mutagenically identified as critical for AbrB to bind DNA (unpublished results, (Bobay et al., 2005; Vaughn et al., 2000)). R23 and R24 display similar electrostatic surfaces in the refined structures of AbrBN53, AbhN, and SpoVTN. We note that the electrostatic character provided by R8, R15, and LP1 is very similar between AbhN and SpoVTN (Figure 2E and 2F, respectively), whereas a slight difference in the electrostatic surface provided by R15 in AbrBN53 (Figure 2D) is noted. These similarities and differences in electrostatic characteristics may contribute to the general and specific mechanisms of DNA binding.
15N Backbone Relaxation Measurements and Model-Free Analysis of Backbone Motions
Spin-lattice relaxation (R1), spin-spin relaxation (R2), and steady-state {1H}-15N NOE data were collected at a magnetic field strength of 9.4T for 44, 47, and 43 residues of AbrBN53, AbhN, and SpoVTN out of a possible 51, 52, and 52 total residues, respectively. Residues at the N- and C-terminal regions have relaxation values lower than the average of core residues, indicating considerable motion (very fast internal motions; τe ≪ τm) on the picosecond-nanosecond (ps-ns) time scale. A few residues of AbhN and AbrBN53, but not SpoVTN, have elevated R2/R1 or R1·R2 values compared to the average core of residues, indicating a chemical exchange broadening (Rex) contribution on the microsecond-millisecond (μs-ms) time scale (i.e., lower frequency motions) (Jarymowycz and Stone, 2006; Kneller et al., 2002). For AbhN, these residues include I4, G5, D11, I34, and V38. For AbrBN53, they include Y37 and D41.
Initial estimates for isotropic, axially symmetric and anisotropic diffusion tensors were obtained using the 15N relaxation rates and RDC-refined solution structures. Isotropic or prolate tensor parameter estimates were subsequently used for model-free analysis of internal motion parameters (Figure 3; Tables S1–S3). The refined NMR structures presented here afford a more accurate description of NH bond vector orientations, which is important for model-free analysis assuming a non-isotropic tensor. The initial global rotational correlation time (τc) estimates and D||/D⊥ values can be found in Supplemental Data. An isotropic tensor was found to be most appropriate for AbhN (τc=6.38 ns), whereas a prolate tensor was most appropriate for AbrBN53 (τc=6.36 ns; D||/D⊥= 1.14; φ=172.10°; ϕ=72.06°) and SpoVTN (τc=7.01 ns; D||/D⊥ = 1.21; φ=170.73°; ϕ=2.14°).
A detailed look at the S2 general order parameter (Figure 3) reveals some trends in mobility of backbone amide groups among the three proteins. All three proteins have increased mobility in the regions of the GD-box and LP1 and LP2, in particular around residues 30 and 41. A closer look at the S2 values for several regions of AbrBN53, AbhN, and SpoVTN reveals propensities for motion that may play a role in the specificity exhibited by each protein. AbrBN53 and SpoVTN have increased mobility (lower S2) in the LP1 region (residue 12) compared to AbhN. AbhN is more restricted in LP2 and less restricted in LP3 compared to AbrBN53 and SpoVTN, although all three proteins have lower S2 values for these regions compared to the average core of residues. AbhN is less restricted in β1 surrounding residue 8 compared to AbrBN53 or SpoVTN, indicating a greater ability for AbhN to reorient this portion of the protein. Finally, the region of LP3 in SpoVTN (residues 42 and 43) is slightly less restricted compared to AbrBN53 or AbhN, perhaps a result of the glycine that is not present in AbrBN53 or AbhN.
A few residues have significant chemical exchange contributions (Rex) to the 15N R2 relaxation rates indicative of motions on the μs-ms timescale (Tables S1–S3). Two residues in LP3 of AbrBN53, Y37 and E41, have significant Rex values (3.88±0.34 s−1 and 10.10±0.40 s−1, respectively). In AbhN, only a few residues have significant Rex values, including G5 (3.99±0.37 s−1; β1), D11 (3.20±0.23 s−1; LP1), and V38 (2.00±0.15 s−1; LP3). SpoVTN does not have any residues with significant Rex contributions. The primary location for residues with significant Rex contributions is at the surface opposite that containing residues involved in DNA binding. These residues may be involved in domain-domain interactions (N- and C- or C- and C′-terminal interactions) with functional significance in determining the quaternary structure. This is supported by the observation that the HSQC peak corresponding to AbrBN53 residue Y37 either disappears or experiences a significant change in chemical shift upon multimerization (Figure S2).
Model of the AbrBN55:abrB8 complex
Previous studies showed: (i) full length AbrB functions as a tetramer; (ii) dimeric AbrBN55 (residues 1–55) can be oxidized to form a tetramer (AbrBN55-tetramer; via a C54–C54′ disulfide bond); and (iii) that the AbrBN55-tetramer binds to cognate promoters of full length AbrB with near wild-type affinity (Phillips and Strauch, 2001; Xu and Strauch, 2001). This suggests that the binding mechanism of the AbrBN55-tetramer is analogous to that of full length AbrB. 1H-15N HSQC spectra of AbrBN53 (AbrBN53 and AbrBN55 are used throughout to describe the N-terminal domain of AbrB) and the AbrBN55-tetramer (Figure S2) overlay well, confirming that no major structural differences exist between dimeric AbrBN53 and the AbrBN55-tetramer (apart from multimerization).
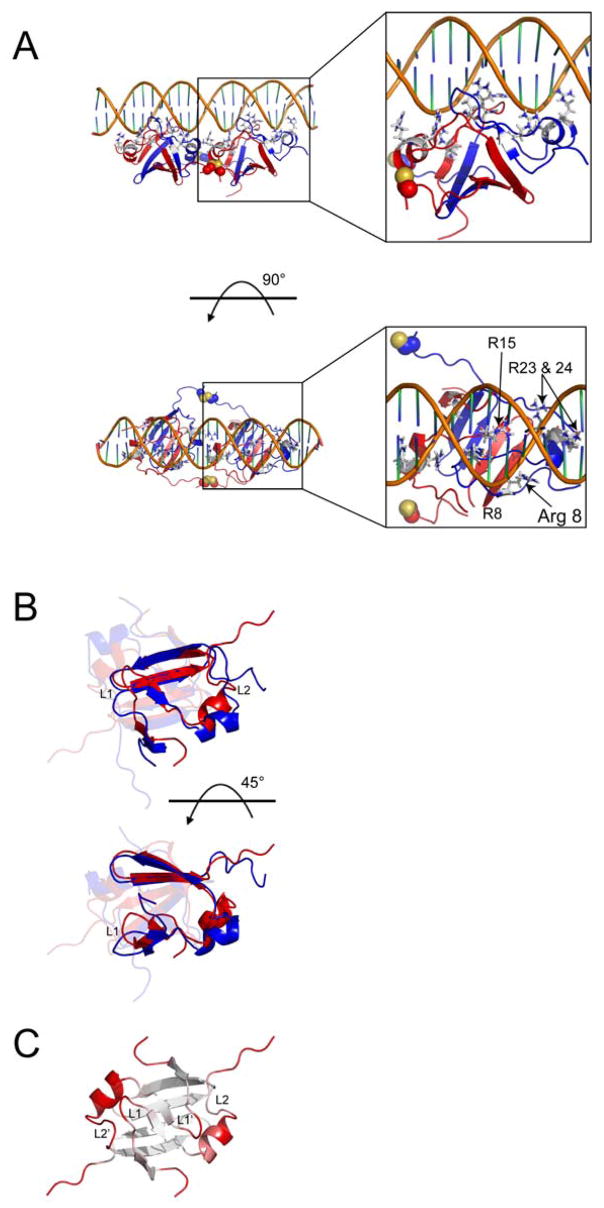
HADDOCK was used to develop a structural model of the AbrBN55-tetramer (Figure 4) using the NMR structure of AbrBN53, previously published biochemical data (Strauch, 1995c; Xu et al., 1996; Xu and Strauch, 1996, 2001), and new mutational data (unpublished results; see Supplemental Data). A plot of the Einter (sum of restraint, van der Waals, and electrostatic energy terms) as a function of backbone rmsd (Figure S3) from the lowest energy model reveals that the models converge to a Cα (protein) and P (DNA phosphate backbone) rmsd of 0.49±0.07Å at the defined protein-DNA interface with an average buried surface area of 3721±360Å2. One cluster of structures with low rmsd and energy was obtained for all calculated models (Table 2 and Figure S3) based on a minimum cluster size of four models and a Cα+P rmsd of 1.5Å. PROCHECK analysis (Laskowski et al., 1996) of the resulting models in this cluster revealed that 96.4% were in allowed regions of Ramachandran space. Comparison of the AbrBN55-tetramer-abrB8 modeled complex and the NMR structure of unbound AbrBN53 indicates that the monomeric subunits in the AbrBN55-tetramer maintain a similar global fold upon binding (Figure 4B and C). The Cα rmsd between a dimer of the modeled AbrBN55 tetramer and the refined, unbound NMR structure of AbrBN53 is 2.84Å measured over the protein for residues 5–47.
Figure 4.
Lowest energy DNA-bound AbrBN55 model from semi-flexible docking and comparison to unbound AbrBN53. (A) Two views of the lowest energy HADDOCK structure from the semi-flexible docking studies. Disulfide linkages are shown as spheres in the image. Insets show a detailed look at the positioning of the arginine residues involved in binding. (B) Overlay of unbound AbrBN53 (red) and the lowest HADDOCK score model of AbrBN55 bound to abrB8 (blue). One monomer is highlighted for clarity. (C) The degree of structural variation between the unbound AbrBN53 NMR structure and the modeled AbrBN55 bound to abrB8, colored from white (little variation) to red (large variation) as calculated in the Cα alignment by THESEUS plotted on the refined AbrBN53 solution structure. The unbound and bound AbrBN53 dimer structures overlay with a Cα r.m.s.d. of 2.84Å. LP 1 and 2 (chain A) and 1′ and 2′ (chain B) are noted. Proteins are oriented as depicted in Figure 1 on structures most similar to the average structure in the ensemble reported by THESEUS.
Table 2.
HADDOCK refinement statistics
| Violations (mean and S.D.) | |
| Distance constraints (Å) | 0.23 ± 0.04 |
| Deviations from idealized geometry | |
| Bond Lengths (Å) | 0.0031 ± 0.0001 |
| Bond Angles (°) | 0.64 ± 0.01 |
| Impropers (°) | 0.51 ± 0.01 |
| Average pairwise r.m.sa (Å) | |
| Interface | 2.84 ± 0.64 |
| Buried surface area (Å2) | 3721.98 ± 359.8 |
| Ramachandran analysis | |
| Favored | 71.5 % |
| Allowed | 24.8 % |
| Generously allowed | 1.1 % |
| Disallowedb | 2.6 % |
Pairwise r.m.s deviation was calculated (10 structures) based on all heavy atoms at the interface of binding.
Those residues that are in the disallowed regions are either in loop 3 (residue Asp40) and/or at the C-terminus of the protein (residues 51–55).
The interface between the AbrBN55-tetramer and abrB8 (Figure 4A) reveals an extended network of intermolecular hydrogen bonds that stabilizes the complex. These hydrogen bonds involve both charged and polar amino acids at the interface and within the protein structure. The structures show the positioning of the AbrBN55-tetramer in consecutive major grooves (left panels of Figure 4A) with the loop regions extending into the major groove to make base-pair specific interactions, primarily targeting guanines. Analysis of the most populated hydrogen-bond interactions between the protein and the DNA show extensive hydrogen bonds between the δ–guanidino groups of R15 (β2) and N7 and O6 of guanine residues (in the loose conserved sequence – right panels of Figure 4A), as well as with atoms on the DNA phosphate backbone (O#P, O5′ and O3′). In all models, the δ–guanidino groups of R8 (β1), R23 (α1) and R24 (α1), as well as the amino groups of K9 (β1) and K31 (LP2), move toward the DNA phosphate backbone and only contact atoms on the DNA phosphate backbone (O#P, O5′ and O3′) (~ 4 base pairs in length). In some cases, the terminal oxygen (OE1) of E12 (LP1) was seen to hydrogen bond to the NH2 groups of adenine and cytosine (H6# and H4#, respectively), whereas the terminal oxygens (OD1 and OD2) of D11 (LP1) hydrogen bond to the NH2 groups of cytosine (H4#).
In the modeled AbrBN55-tetramer-abrB8 structures β1 moves slightly, allowing R8 and K9 to bind to the DNA backbone while still providing an appropriate dimerization interface. LP1 and LP2 undergo significant movement upon complex formation and move to facilitate limited contacts between K31 (LP2) and the DNA backbone. Additionally, α1 moves towards the DNA backbone, allowing R23 and R24 to make contacts with the phosphate backbone. Several hydrogen bonds are observed between the δ–guanidino groups of R23 and R24 and the oxygen atoms of the phosphate backbone. R24 appears to undergo the most significant conformational change of the residues directly responsible for binding DNA.
In summary, the dimerization interface is preserved, with minimal changes observed in the positions of β3 and β4. However, upon complex formation α1, LP1, and LP2 of the AbrBN55-tetramer are displaced compared to AbrBN53 (Figure 4B and C). These regions cooperatively migrate towards the DNA phosphate backbone making limited base specific interactions. This cooperative migration complements the backbone dynamics data and structural PCA plots (discussed below), which suggest the ability to change conformation in these regions. The model also suggests local deformation of the DNA upon binding, as the model DNA base pair parameter values deviate from normal values (Table S4).
Discussion
To gain further insight into DNA recognition and binding by the AbrB-like TSRs, in particular the three-fold nature of DNA-binding, we extended previous structural studies of AbrBN53 and AbhN. We conducted structural and dynamic comparisons among the refined structures of the DNA-binding domains of AbrB, Abh, and SpoVT, and developed a structural model for the mode of interaction between AbrBN55 and the DNA target abrB8 (Bobay et al., 2005; Bobay et al., 2006). To assess the correlation between looped-hinge/DNA recognition helix mobility and protein function among AbrBN53, AbhN, and SpoVTN, 15N NMR relaxation experiments were performed. Motion on similar timescales can define residue networks within proteins that are important for function. The clustered patterns of backbone mobility identified in all three TSR proteins suggests that the ability of this class of proteins to recognize DNA sequences with no apparent consensus sequence may be attributed to a synchronization of motion between structurally independent regions. Increased mobility of backbone amide groups was observed for residues in LP1, LP2, and LP3 in all proteins, and β1 in AbhN (Figure 3). These residues provide a convenient hinge mechanism that allows LP1 and LP2 to reorient and make hydrogen bonds with the phosphate backbone. The motions of LP1 and LP2 also allow α1 to alter its conformation and migrate towards the DNA backbone, as shown in the model of AbrBN55-tetramer bound to abrB8 (Figure 4). The propensity of these regions to readily adopt multiple conformations enables the straightforward re-positioning of residues critical to the binding interaction.
Principal components analysis (PCA) is a standard tool in the field of multivariate analysis for extracting from a set of inter-related variables a much smaller set that retains most of the variation contained in the full set. By setting up a correlation matrix whose elements are the ensemble average of the pairwise products of displacements from their average position of landmarks (such as the Cα positions in a protein) PCA can be very helpful in identifying, from an NMR structural ensemble, correlations in conformational rearrangements within a protein (Theobald and Wuttke, 2008). Structural PCA plots of the refined NMR structures of AbrBN53, AbhN, and SpoVTN (Figure 5A, 5B, and 5C, respectively) reveal the following: on average, within the NMR structure ensemble, (i) LP1′ and LP2 move in the same direction; (ii) LP1 and LP2′ move in the same direction but in an opposite sense to LP1′/LP2. This suggests the propensity for a general concerted motion involving LP1, LP1′, LP2, and LP2′. For example, as LP1′ and LP2 move together towards the dimer interface, symmetry related LP1 and LP2′ also move towards the dimer interface. This flexibility, as evidenced by lower S2 values (Figure 3), and correlated conformational changes observed via PCA for LP1, LP1′, LP2, and LP2′, is likely central to the general DNA-recognition properties for TSRs. These concerted motions allow the proteins to appropriately reorient during complex formation with target DNA sequences, enabling Arg residues critical to DNA-recognition and binding (R8, R15, R23, R24) to make suitable contacts. Interestingly, we also note that the α1 regions in each of the TSR proteins studied show different degrees of motional correlation with the loop regions. For example, α1 from AbrBN53 moves in the same direction as the proximal LP2′ and LP1, whereas α1 from AbhN moves in the opposite direction as the proximal LP2′ and LP1. The differences suggest that these regions may experience complicated movements as a result of DNA binding and contribute to DNA recognition.
Figure 5.
THESEUS PCA analysis of the TSR structure ensembles. PCA plots of the maximum likelihood correlation matrix for the structure ensembles of (A) AbrBN53, second principal component; (B) AbhN, first principal component; and (C) SpoVTN, second principal component. Regions colored similarly (red or blue) are self-correlated, whereas regions colored differently (red vs. blue) are anti-correlated.
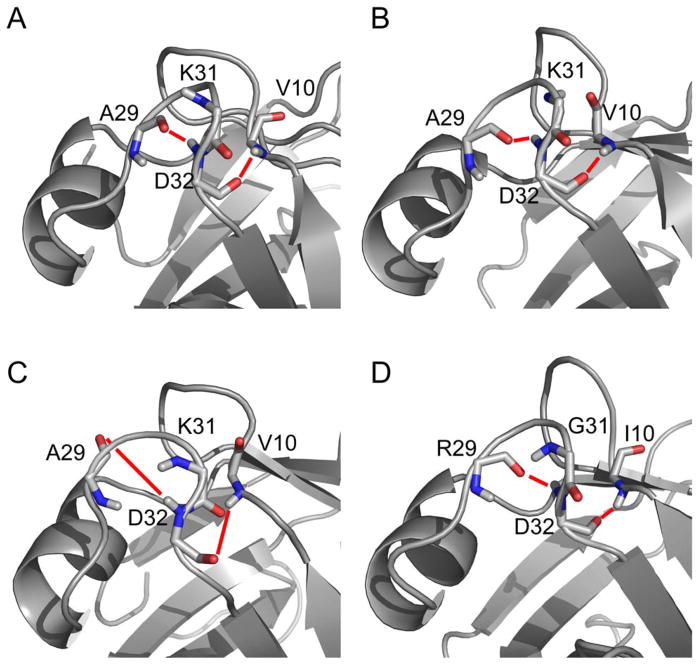
Previous studies suggested that LP1 and LP2 play an integral part in DNA recognition for TSRs as a hinge mechanism for β2 and α1, as well as the formation of a structural feature known as a GD box (Bobay et al., 2005; Bobay et al., 2006; Coles et al., 2005; Vaughn et al., 2001). The GD box is an evolutionarily conserved sequence motif described as a hallmark of the βαβ element of the cradle-loop metafold forming β hairpin turns that are anchored to the hydrophobic core of the protein by flanking hydrophobic residues. This motif was observed in a recent structural analysis of AbrBN53 (Coles et al., 2005). However, we failed to find any significant hydrogen bonds from hydrogen exchange experiments that would facilitate formation of a robust GD box in AbrBN53 or AbhN (Figure S4A and S4B) (Bobay et al., 2005; Bobay et al., 2006). Despite these previous observations, the refined structure of AbrBN53 presented here supports the possible formation of the characteristic conserved hydrogen bonds (Figure 6A). The hydrogen bond distances observed by Coles et al. are: A29 to D32, 2.14Å; D32 to V10, 2.16Å. In our refined AbrBN53 structure, these distances are: A29 to D32, 1.84Å; D32 to V10, 2.35Å. Fast hydrogen exchange (Figure S4A) and low S2 values (Figure 3A and 3B) suggest that this region has significant flexiblity. Taken together, the data suggest that there is a possibility for this region of the protein to form a GD box, although it is likely to fluctuate between several conformations.
Figure 6.
GD-Box motif of TSRs. The residues involved in the characteristic hydrogen bond network of the GD box of loop 2 for (A) refined AbrBN53 structure, (B) AbrBN53 structure by Coles et al., (C) refined structure of AbhN, and (D) refined structure of SpoVTN. Key residues involved in the GD box hydrogen-bonding network are depicted. Thick red lines indicate hydrogen bonds necessary for GD-Box formation.
Analysis of the GD box region of the refined AbhN structure indicates that this protein does not contain the characteristic hydrogen bond network, as backbone hydrogen bond donors and acceptor groups in the refined structure are neither close in proximity to support the characteristic hydrogen bonds (A29 to D32, 6.01Å; D32 to V10, 4.83Å), nor are they properly oriented to allow productive interactions (Figure 6C and Figure S4B). Finally, the LP2 region of the refined SpoVTN structure contains the classical GD box motif. Slow hydrogen exchange (Figure S4C) and hydrogen bond donor/acceptor positioning (R29 to D32, 2.15Å; D32 to I10 2.05Å) and orientation (Figure 6D) support the classical GD box hydrogen bond network, forming a β hairpin turn and linking LP2 to β1 via a hydrogen bond between D32 and I10.
In summary, the data suggest that the ability or inability to form a classical GD box motif may have far reaching implications in the specific mechanisms of DNA binding for these three proteins given that the data presented here suggests that AbhN does not form a GD box, AbrBN53 has the propensity to form a GD box and SpoVT has the classic GD box motif. This is solidified by the fact that these three proteins share no known DNA target in common, no overlapping DNA target sequences and regulate the same synthetic DNA target with drastically different affinity (Bobay et al., 2005; Bobay et al., 2004; Bobay et al., 2006).
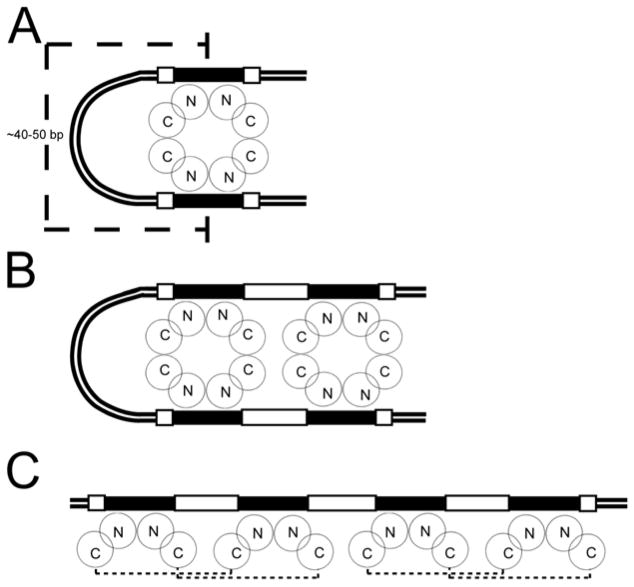
The data presented thus far when extended to our model of the AbrBN55-tetramer-abrB8 complex – the first of such kind for any AbrB-like TSR bound to a DNA target – reveal that structural and dynamic similarities and differences may contribute to the general and specific mechanisms of binding. Furthermore, the specific amino acid-nucleotide interactions described above can be accommodated within three broader binding models in vivo (Figure 7). Electrophoretic mobility assays demonstrate that AbrB can bind bent and linear DNA (Strauch and Ayazifar, 1995). DNA loops generally have helical repeats of 10–11 base pairs (i.e., n repeats of 10–11 base pairs) (Schleif, 1992) and are typically 40–50 base pairs long (Nakano and Zuber, 1993; Ptashne, 1986; Schleif, 1987, 1992). A schematic of tetrameric AbrB binding to 40–50 base pair looped DNA (Figure 7A) requires that the approximate coverage protected from DNase I cleavage be 60–70 base pairs. Although some AbrB-target interactions have been shown to result in protection regions of this length (Strauch, 1995b, 1996; Xu et al., 1996; Xu and Strauch, 1996), the universality of this model is not supported by other studies which show that the base pair regions typically protected from DNase I cleavage in many AbrB DNA targets are between 25–45 base pairs (Furbass et al., 1991; Strauch, 1995a, b, c, 1996; Strauch et al., 1989; Xu and Strauch, 1996) (M.A. Strauch, unpublished). A schematic representation of two tetrameric AbrB molecules binding to 40–50 base pair looped DNA in consecutive major grooves (Figure 7B) requires that the approximate coverage protected from DNase I cleavage would be 80–90 base pairs. Again, while there are some examples of interactions giving this extent of protection from DNase I cleavage, the model is not universally supported by all previous DNase I footprinting studies (Strauch, 1995c; Strauch et al., 1989). A schematic of two tetrameric AbrB molecules binding to ~40 base pair linear DNA in pairs of consecutive major grooves (Figure 7C) is supported by DNase I footprinting studies, which show 25–45 base pairs of AbrB-afforded protection (Strauch, 1995b, 1996; Xu et al., 1996; Xu and Strauch, 1996). Previous μESI-MS studies revealed that the largest detected protein-DNA interaction was that of a single AbrB tetramer with a single 30 base-pair DNA fragment, affording a molecular weight of 60 kDa (Bobay et al., 2004; Cavanagh et al., 2002). Although this molecular weight precludes the binding of multiple DNA fragments, model C could also apply to interactions giving longer regions of DNase I protection if more than two tetramers of AbrB were binding side-by-side. Although in vitro observations indicate that most AbrB-DNA interactions can be accommodated by model C, none of the three models can be ruled out entirely and may reflect different forms of AbrB interactions that occur in a target-specified manner.
Figure 7.
Schematic models for AbrB-DNA binding. Binding models for (A) one tetramer bound to two target regions separated by a DNA loop (typical length of DNA loops shown), (B) two AbrB tetramers bound to four target regions separated by a DNA loop, and (C) two AbrB tetramers bound to four target regions not separated by a DNA loop. Solid black boxes denote the loosely conserved recognition sequence of TGGNA; white boxes denote 4 base pairs immediately adjacent to this sequence, “N” denotes the N-terminal domain of AbrB while the “C” represents the C-terminal domain. Small dashed lines in panel “C” define the AbrB tetramer.
Conclusion
Elucidating the mechanism by which TSRs interact with DNA targets is particularly important, since these proteins allow bacteria to adapt to changing environments in various ways. This family of proteins is of particular interest because as of yet, no TSR has been found to have overlapping regulation of exactly the same DNA target in the same location. This study determined the structure and dynamics of three such TSRs, representing the first structural attempt to determine how a TSR binds its DNA target and what structural changes occur during complex formation. The modeled AbrBN55-tetramer-abrB8 structures are especially helpful in identifying the role of residues at the protein-DNA interface.
Experimental Procedures
Cloning, Expression, and Purification of AbrBN53, AbrBN55, AbhN, and SpoVTN
Uniformly labeled 15N samples of AbrBN53, AbrBN55, and AbhN were expressed and purified as previously described (Bobay et al., 2005; Bobay et al., 2006; Phillips and Strauch, 2001). DNA fragments coding the N-terminal domain SpoVT were obtained through PCR utilizing the Stratagene QuikChange Site-Directed Mutagenesis Kit (inserting a TAA stop codon at residue 56 from a construct containing the DNA sequence of full length SpoVT). SpoVTN was purified in a similar manner to AbrBN53 and AbhN with a few differences. E. coli cell growth lasted 2 hours post induction. Solid (NH4)2SO4 (35%) was added to the supernatant, which was loaded onto a Phenyl Sepharose (Sigma) column and eluted using a 35% - 0% (NH4)2SO4 gradient. SpoVTN fractions were dialyzed into 10mM Tris-HCl (pH 7.9), 10mM KCl, 1mM EDTA, 1mM DTT, and 0.02% NaN3, loaded onto a Q-Sepharose column (GE Healthcare). Pure SpoVTN fractions were dialyzed into 10mM Tris-HCl (pH 5.8), 150mM KCl, 1mM EDTA, 1mM DTT, and 0.02% NaN3 for NMR experiments. All purification was performed at 4°C.
NMR Spectroscopy and Structure Calculations
AbrBN53 and AbrBN55 1H-15N HSQC NMR experiments carried out on 1–2 mM samples in 90:10% H2O:D2O 10mM KH2PO4 (pH 5.8), 15mM KCl, 1mM EDTA, 1mM DTT (not present in AbrBN55 sample), and 0.02% NaN3. NMR data for SpoVTN were collected in a similar manner as for AbrBN53 and AbhN (Bobay et al., 2005; Bobay et al., 2006) with a few differences. Experiments were carried out on 1–2 mM samples in 90:10% or 1:99% H2O:D2O 10mM Tris-HCl (pH 5.8), 150mM KCl, 1mM EDTA, 1mM DTT, and 0.02% NaN3. The 13C NOESY-HSQC experiments were carried out on a 600 MHz Varian INOVA equipped with a Varian cryogenic probe. All data was collected at 305K, processed with NMRPipe (Delaglio et al., 1995), and analyzed with NMRView (Johnson and Blevins, 1994). 1DNH RDCs were measured on 1.5–3 mM uniformly labeled 15N samples of AbrBN53, AbhN, and SpoVTN using HSQC and IPAP-HSQC experiments recorded on a Varian INOVA spectrometer at 500 MHz from 6.0 mm to 4.2 mm (AbrBN53 and SpoVTN) or 5.0 mm to 4.2 mm (AbhN) radially compressed 7% polyacrylamide gels (Chou et al., 2001).
Structure calculations used previously reported NMR restraints for AbrBN53 (Bobay et al., 2005) and AbhN (Bobay et al., 2006) and were performed in a similar manner to that described except the initial and final susceptibility anisotropy (SANI) force constants for the RDC restraints were 0.2 and 1.0 kcal Hz−2, respectively, adjusted to give average rmsd values within a reasonable range of error (Lipsitz and Tjandra, 2004). RDC Da and R values within the ARIA protocol were as follows: AbrBN53 (−13.241 and 0.289), AbhN (−4.781 and 0.213), and SpoVTN (−12.130 and 0.197). RDC restraints for the dimer structures were fit and analyzed with the RDCA program (L.E. Kay, University of Toronto) using SVD, where the measured RDC value was assigned to the N-H vectors of a given residue in each of the two monomeric subunits. Structural images were created using PyMOL or MOLMOL.
15N Backbone Relaxation Measurements and Model-Free Analysis of Backbone Motions
Backbone 15N R1, R2, and {1H}-15N NOE relaxation measurements were acquired on a Varian INOVA spectrometer at 9.4T and 305K on uniformly 15N-labeled samples of AbhN, AbrBN53, and SpoVTN (1–2mM). 15N relaxation data for some residues were not obtained due to unobserved or overlapped resonances (listed in Supplemental Data). Measurements made on ½ or ⅓ diluted samples indicated that there were no concentration-dependent effects on the 15N R2 relaxation rates. For all analyses, 15N relaxation data was applied to both monomers within the dimer structures. The program THESEUS (Theobald and Wuttke, 2006) was used to determine the structure most similar to the average structure in the refined AbhN, AbrBN53, and SpoVTN NMR structure ensembles that were subsequently used for relaxation and model-free analysis. The program relax (d’Auvergne and Gooley, 2008) was used to fit the experimental 15N relaxation data to five motional models of the Lipari-Szabo formalism (Clore et al., 1990; Lipari, 1982a, b): (1) S2, (2) S2 and τe, (3) S2 and Rex, (4) S2, τe, and Rex, and (5) Sf2, S2, and τe. Final selection of the appropriate diffusion tensor was determined using the χ2 and AIC statistics describing the fit of the tensor after convergence to the optimal diffusion tensor parameters, as well as agreement between experimental and back-calculated 15N relaxation parameters. Principle components analysis (PCA) was performed on the NMR structure ensembles using THESEUS.
HADDOCK Docking Procedure
Default HADDOCK (Dominguez et al., 2003) parameters were used throughout the docking procedure. Active and passive residues (Table S5) with solvent accessibility >40% calculated by NACCESS (Hubbard, 1993) were assigned from AbrB mutational studies. 1000 structures were generated per iteration, and 200 lowest energy structures were water refined. Each docking attempt was performed 10 times and the solution with the lowest HADDOCK score was retained. The rmsd values of the complexes were calculated using ProFit (A. Martin, UCL). Values for base pair and base pair step parameters, as well as torsion angles for the sugar-phosphate backbone and sugar pucker (Table S4) were obtained using the program 3DNA (Lu and Olson, 2003). A cluster analysis was performed on the final docking solutions using a minimum cluster size of 4. The cut-off for clustering was manually determined for each docking run. The rmsd matrix was calculated over the backbone atoms of the interface residues of the DNA after fitting on the interface residues of the protein.
Supplementary Material
Additional methodological details, four supplemental figures, and five supplemental tables can be found in Supplemental Data.
Acknowledgments
Funding
This work was supported in part by National Institutes of Health grants (GM55769 to J.C., GM46700 to MAS, GM063855 to MR), NIEHS training grant ES07250 (DK), and a grant from the Kenan Institute for Engineering, Technology and Science (JC).
We would like to thank Dr. Ronald A. Venters (Duke) for SpoVTN 13C NOESY data, Dr. Geoffrey A. Mueller (NIEHS) for assistance with RDC experiments and HADDOCK, Dr. Edward J. d’Auvergne for assistance with the program relax, Dr. Walter Chazin (Vanderbilt) for helpful discussions, and Cincinnati Children’s Hospital Medical Center, Division of Biomedical Informatics, for computational resources.
Footnotes
Accession Numbers
Coordinates have been deposited in the Protein Data Bank with the following codes: RDC-refined structures of AbrBN53 (2ro4), AbhN (2ro3), SpoVTN (2ro5), and the AbrBN55-tetramer/abrB8 modeled complex (2k1n).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aertsen A, Michiels CW. Stress and how bacteria cope with death and survival. Crit Rev Microbiol. 2004;30:263–273. doi: 10.1080/10408410490884757. [DOI] [PubMed] [Google Scholar]
- Bagyan I, Hobot J, Cutting S. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J Bacteriol. 1996;178:4500–4507. doi: 10.1128/jb.178.15.4500-4507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay BG, Andreeva A, Mueller GA, Cavanagh J, Murzin AG. Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins. FEBS Lett. 2005;579:5669–5674. doi: 10.1016/j.febslet.2005.09.045. [DOI] [PubMed] [Google Scholar]
- Bobay BG, Benson L, Naylor S, Feeney B, Clark AC, Goshe MB, Strauch MA, Thompson R, Cavanagh J. Evaluation of the DNA binding tendencies of the transition state regulator AbrB. Biochemistry. 2004;43:16106–16118. doi: 10.1021/bi048399h. [DOI] [PubMed] [Google Scholar]
- Bobay BG, Mueller GA, Thompson RJ, Murzin AG, Venters RA, Strauch MA, Cavanagh J. NMR structure of AbhN and comparison with AbrBN: FIRST insights into the DNA binding promiscuity and specificity of AbrB-like transition state regulator proteins. J Biol Chem. 2006;281:21399–21409. doi: 10.1074/jbc.M601963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh J, Thompson R, Bobay B, Benson LM, Naylor S. Stoichiometries of protein-protein/DNA binding and conformational changes for the transition-state regulator AbrB measured by pseudo cell-size exclusion chromatography-mass spectrometry. Biochemistry. 2002;41:7859–7865. doi: 10.1021/bi0202225. [DOI] [PubMed] [Google Scholar]
- Chou JJ, Gaemers S, Howder B, Louis JM, Bax A. A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J Biomol NMR. 2001;21:377–382. doi: 10.1023/a:1013336502594. [DOI] [PubMed] [Google Scholar]
- Clore GM, Driscoll PC, Wingfield PT, Gronenborn AM. Analysis of the backbone dynamics of interleukin-1 beta using two-dimensional inverse detected heteronuclear 15N-1H NMR spectroscopy. Biochemistry. 1990;29:7387–7401. doi: 10.1021/bi00484a006. [DOI] [PubMed] [Google Scholar]
- Coles M, Djuranovic S, Soding J, Frickey T, Koretke K, Truffault V, Martin J, Lupas AN. AbrB-like transcription factors assume a swapped hairpin fold that is evolutionarily related to double-psi beta barrels. Structure. 2005;13:919–928. doi: 10.1016/j.str.2005.03.017. [DOI] [PubMed] [Google Scholar]
- d’Auvergne EJ, Gooley PR. Optimisation of NMR dynamic models I. Minimisation algorithms and their performance within the model-free and Brownian rotational diffusion spaces. J Biomol NMR. 2008;40:107–119. doi: 10.1007/s10858-007-9214-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Dong TC, Cutting SM, Lewis RJ. DNA-binding studies on the Bacillus subtilis transcriptional regulator and AbrB homologue, SpoVT. FEMS Microbiol Lett. 2004;233:247–256. doi: 10.1016/j.femsle.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Furbass R, Gocht M, Zuber P, Marahiel MA. Interaction of AbrB, a transcriptional regulator from Bacillus subtilis with the promoters of the transition state-activated genes tycA and spoVG. Mol Gen Genet. 1991;225:347–354. doi: 10.1007/BF00261673. [DOI] [PubMed] [Google Scholar]
- Hubbard SJTJM. NACCESS Computer Program. Department of Biochemistry and Molecular Biology; University College London: 1993. [Google Scholar]
- Jarymowycz VA, Stone MJ. Fast time scale dynamics of protein backbones: NMR relaxation methods, applications, and functional consequences. Chem Rev. 2006;106:1624–1671. doi: 10.1021/cr040421p. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kneller JM, Lu M, Bracken C. An effective method for the discrimination of motional anisotropy and chemical exchange. J Am Chem Soc. 2002;124:1852–1853. doi: 10.1021/ja017461k. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Lipari GaSA. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1 Theory and range of validity. Journal of the American Chemistry Society. 1982a;104 [Google Scholar]
- Lipari Ga SA. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. Journal of the American Chemistry Society. 1982b;104:4559–4570. [Google Scholar]
- Lipsitz RS, Tjandra N. Residual dipolar couplings in NMR structure analysis. Annu Rev Biophys Biomol Struct. 2004;33:387–413. doi: 10.1146/annurev.biophys.33.110502.140306. [DOI] [PubMed] [Google Scholar]
- Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Zuber P. Mutational analysis of the regulatory region of the srfA operon in Bacillus subtilis. J Bacteriol. 1993;175:3188–3191. doi: 10.1128/jb.175.10.3188-3191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips ZE, Strauch MA. Role of Cys54 in AbrB multimerization and DNA-binding activity. FEMS Microbiol Lett. 2001;203:207–210. doi: 10.1111/j.1574-6968.2001.tb10842.x. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986;322:697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Schleif R. Gene regulation: why should DNA loop? Nature. 1987;327:369–370. doi: 10.1038/327369a0. [DOI] [PubMed] [Google Scholar]
- Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- Shcheptov M, Chyu G, Bagyan I, Cutting S. Characterization of csgA, a new member of the forespore-expressed sigmaG-regulon from Bacillus subtilis. Gene. 1997;184:133–140. doi: 10.1016/s0378-1119(96)00603-8. [DOI] [PubMed] [Google Scholar]
- Sonenshein AL, Hoch JA, Losick R. Bacillus subtilis and its closest relatives: from genes to cells. Washington, D.C: ASM Press; 2002. [Google Scholar]
- Strauch MA. AbrB modulates expression and catabolite repression of a Bacillus subtilis ribose transport operon. J Bacteriol. 1995a;177:6727–6731. doi: 10.1128/jb.177.23.6727-6731.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch MA. Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promote. J Bacteriol. 1995b;177:6999–7002. doi: 10.1128/jb.177.23.6999-7002.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch MA. In vitro binding affinity of the Bacillus subtilis AbrB protein to six different DNA target regions. J Bacteriol. 1995c;177:4532–4536. doi: 10.1128/jb.177.15.4532-4536.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch MA. Dissection of the Bacillus subtilis spoOE binding site for the global regulator AbrB reveals smaller recognition elements. Mol Gen Genet. 1996;250:742–749. doi: 10.1007/BF02172986. [DOI] [PubMed] [Google Scholar]
- Strauch MA, Ayazifar M. Bent DNA is found in some, but not all, regions recognized by the Bacillus subtilis AbrB protein. Mol Gen Genet. 1995;246:756–760. doi: 10.1007/BF00290723. [DOI] [PubMed] [Google Scholar]
- Strauch MA, Hoch JA. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol Microbiol. 1993;7:337–342. doi: 10.1111/j.1365-2958.1993.tb01125.x. [DOI] [PubMed] [Google Scholar]
- Strauch MA, Perego M, Burbulys D, Hoch JA. The transition state transcription regulator AbrB of Bacillus subtilis is autoregulated during vegetative growth. Mol Microbiol. 1989;3:1203–1209. doi: 10.1111/j.1365-2958.1989.tb00270.x. [DOI] [PubMed] [Google Scholar]
- Theobald DL, Wuttke DS. THESEUS: maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006;22:2171–2172. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Wuttke DS. Accurate structural correlations from maximum likelihood superpositions. PLoS Comput Biol. 2008;4:e43. doi: 10.1371/journal.pcbi.0040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn JL, Feher V, Naylor S, Strauch MA, Cavanagh J. Novel DNA binding domain and genetic regulation model of Bacillus subtilis transition state regulator abrB. Nat Struct Biol. 2000;7:1139–1146. doi: 10.1038/81999. [DOI] [PubMed] [Google Scholar]
- Vaughn JL, Feher VA, Bracken C, Cavanagh J. The DNA-binding domain in the Bacillus subtilis transition-state regulator AbrB employs significant motion for promiscuous DNA recognition. J Mol Biol. 2001;305:429–439. doi: 10.1006/jmbi.2000.4305. [DOI] [PubMed] [Google Scholar]
- Xu K, Clark D, Strauch MA. Analysis of abrB mutations, mutant proteins, and why abrB does not utilize a perfect consensus in the −35 region of its sigma A promoter. J Biol Chem. 1996;271:2621–2626. doi: 10.1074/jbc.271.5.2621. [DOI] [PubMed] [Google Scholar]
- Xu K, Strauch MA. In vitro selection of optimal AbrB-binding sites: comparison to known in vivo sites indicates flexibility in AbrB binding and recognition of three-dimensional DNA structures. Mol Microbiol. 1996;19:145–158. doi: 10.1046/j.1365-2958.1996.358882.x. [DOI] [PubMed] [Google Scholar]
- Xu K, Strauch MA. DNA-binding activity of amino-terminal domains of the Bacillus subtilis AbrB protein. J Bacteriol. 2001;183:4094–4098. doi: 10.1128/JB.183.13.4094-4098.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional methodological details, four supplemental figures, and five supplemental tables can be found in Supplemental Data.