Abstract
The RecQ helicases are guardians of the genome. Members of this conserved family of proteins have a key role in protecting and stabilizing the genome against deleterious changes. Deficiencies in RecQ helicases can lead to high levels of genomic instability and, in humans, to premature aging and increased susceptibility to cancer. Their diverse roles in DNA metabolism, which include a role in telomere maintenance, reflect interactions with multiple cellular proteins, some of which are multifunctional and also have very diverse functions. The results of in vitro cellular and biochemical studies have been complimented by recent in vivo studies using genetically modified mouse strains. Together, these approaches are helping to unravel the mechanism(s) of action and biological functions of the RecQ helicases.
RecQ helicases: guardians of the genome
The RecQ protein family is a highly conserved group of DNA helicases with diverse roles in multiple DNA metabolic processes, including DNA recombination, replication and repair, and a possible role in transcription. Only one RecQ homolog has been identified in Escherichia coli and the lower eukaryotes Saccharomyces cerevisiae and Schizosaccharomyces pombe, but five RecQ homologs have been identified in mammals, including humans. The human RecQ helicases include WRN (Werner), BLM (Bloom), RECQ4, RECQ1 and RECQ5. WRN and BLM helicases are relatively well studied, but comparatively little is known about human RECQ1, RECQ4 and RECQ5, and it is not yet clear whether the five human RecQ homologs provide partially redundant, complementary or independent cellular functions.
Three human RecQ helicases are associated with rare recessive genetic disorders: Werner syndrome (WS) is associated with defects in WRN helicase and exonuclease, Bloom syndrome (BS) is associated with defects in BLM helicase and Rothmund Thomson syndrome (RTS) is associated with defects in RECQ4. WS and RTS are characterized by premature aging. Although BS patients have a less pronounced, premature-aging phenotype, they have a strong predisposition to several types of cancer [1]. These phenotypes reflect the prominent role that human RecQ helicases have in maintaining genome stability and the response to cellular stress and/or DNA damage.
Succesful cloning, expression and purification of the human RecQ helicases has permitted detailed biochemical, molecular and cellular studies on these enzymes. Cellular studies, in a broad range of biological systems, show that deficiencies in the RecQ helicases lead to genome instability [2], a hallmark of both cancer and aging. Mouse models of RecQ syndromes have been developed with complete or partial defects in several RecQ helicases; however, only some of the mouse models recapitulate the clinical features of WS, BS or RTS. Regardless, these animal-model systems have provided valuable insights into the roles of RecQ helicases in cancer susceptibility and premature aging. In many cases, the molecular basis of the phenotypes of mice with RecQ helicase defects remains poorly understood. Nevertheless, these are invaluable experimental models that will eventually help to unravel the molecular and cellular functions of the RecQ helicases. Here, I focus on recent advances in molecular, biochemical and cellular studies of the RecQ helicases involved in the inherited human genetic disorders WS, BS and RTS, with emphasis on the role of RecQ helicases in the response to DNA damage and in other aspects of DNA metabolism.
The RecQ family
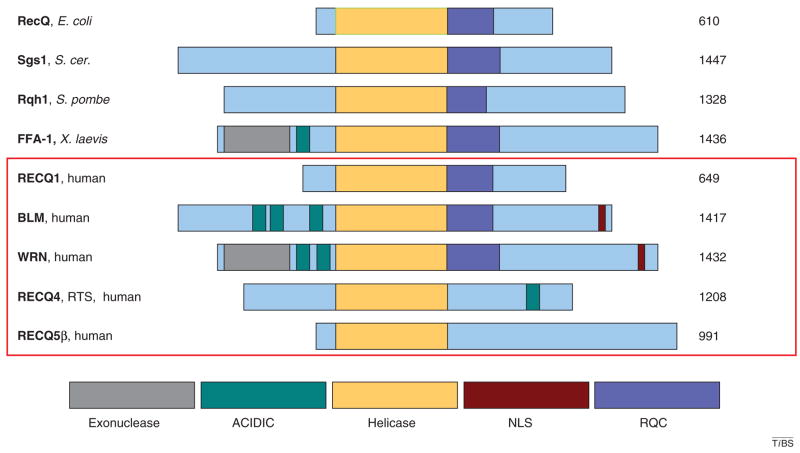
E. coli RecQ is the prototypical member of the RecQ helicase family. Alignment of E. coli RecQ with other RecQ family members reveals strong conservation of the helicase domain (Figure 1). This amino acid sequence conservation is reflected in the functional DNA helicase activity found in all RecQ homologs with the exception of RECQ4 [3]. The RQC (RecQ conserved) domain is less well conserved than the helicase domain but is present in most RecQ family members, including E. coli RecQ. However, the nuclear localization signal (NLS) and the exonuclease and acidic domains of the RecQ proteins are unique to mammalian RecQ family members. The precise biological functions of these distinct domains are not yet fully understood.
Figure 1.
The RecQ helicase family. Schematic diagram of RecQ helicases from diverse species. Conserved domains are aligned and color-coded, as indicated at the bottom of the figure. The size of each protein species in amino acids is indicated on the right. Protein name and species of origin is indicated on the left. The five human RecQ helicases are indicated by the red box. RECQ5 exists in three isoforms, the β form is shown.
WRN and BLM
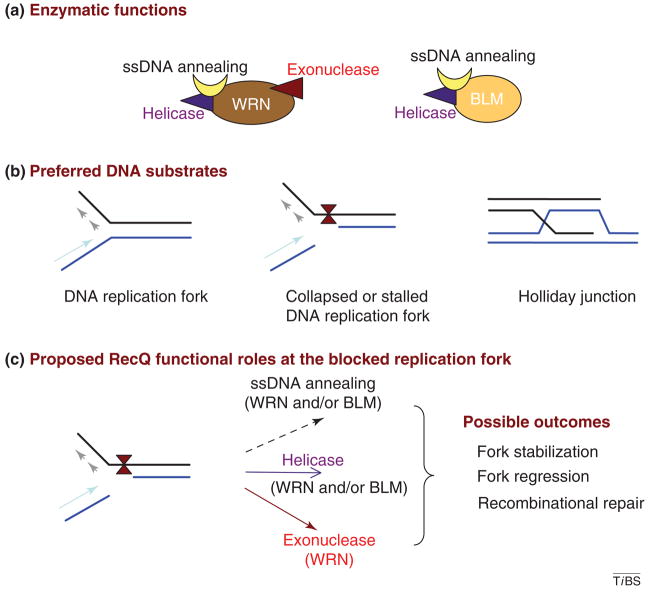
In addition to their helicase activity, WRN and BLM also have intrinsic DNA-dependent ATPase activity and single-strand (ss)DNA-annealing activity [3]. Although the biological importance of their ssDNA-annealing activity remains to be determined, it has been proposed that this activity might facilitate strand migration during recombination or replication fork movement at the site of DNA damage in vivo [3].
WRN and BLM (Figure 2a) both unwind various DNA helicase substrates with a 3′ to 5′ directionality. The preferred substrates of these enzymes are DNA structures that resemble homologous recombination (HR) intermediates such as Holliday junctions and G-rich quadruplex structures [4]. Other structures for which RecQ helicases have a high affinity include normal and blocked or collapsed replication forks, telomeric structures and recombination intermediates (Figure 2b). Using a wide range of DNA substrates, the specificities of WRN and BLM helicases are similar [4]. Because WRN and BLM interact with each other physically and functionally [5], they might function synergistically on some DNA substrates. However, to date, there is no evidence for such collaboration between these or other human RecQ helicases.
Figure 2.
RecQ enzymatic functions and possible functional roles. (a) Schematic summary of the enzymatic properties of WRN and BLM. (b) Preferred DNA substrates of WRN and BLM are shown. (c) Proposed functional roles of WRN and BLM at a stalled DNA replication fork. Possible outcomes are indicated. DNA damage is present at the replication fork.
Despite biochemical similarities between BLM and WRN, WRN is unique in the RecQ helicase family in having an intrinsic 3′ to 5′ exonuclease activity [6] (Figure 2a). WRN exonuclease degrades DNA substrates with a 5′ overhang, and this substrate has been widely used to assay for its activity; but WRN 3′ exonuclease can also degrade blunt-end and various other normal DNA structures in addition to those that contain bubbles and mismatches [7,8].
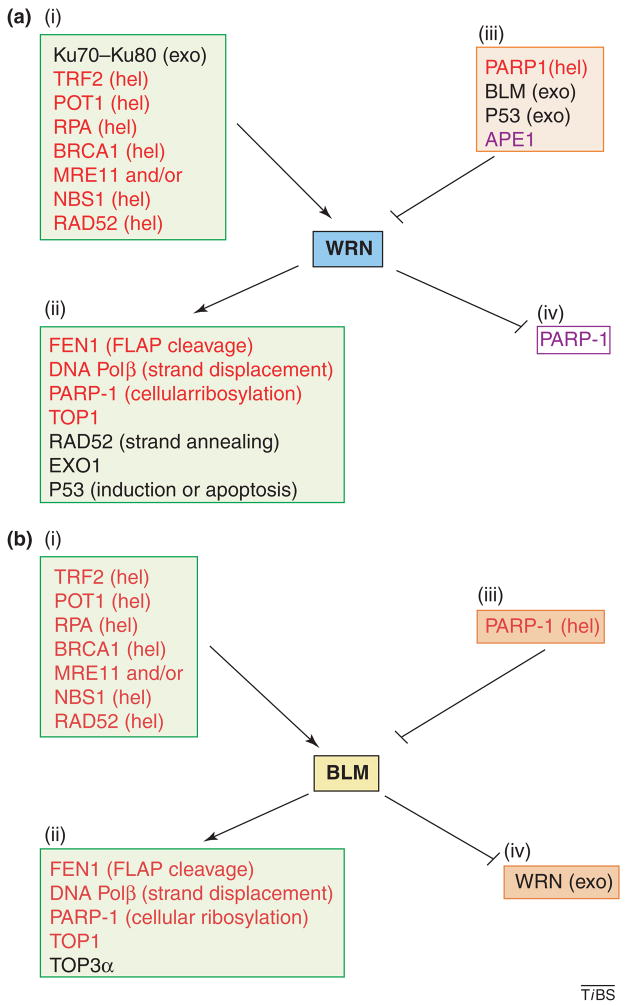
BLM and WRN have several protein interacting partners in common, but each protein also has unique protein interactions (Figure 3). Such differential interactions might contribute to the different roles of BLM and WRN in specific DNA metabolic pathways and to the distinct clinical and cellular phenotypes of BS and WS patients (Figure 4). The interacting proteins are discussed further here in the context of the DNA metabolic pathways in which they are involved. Many WRN protein interactions are mediated by the RQC domain. This non-catalytic region in WRN is also the primary DNA-binding domain [9] and the nucleolar targeting domain, which facilitates the localization of WRN to nucleoli in unstressed cells [10].
Figure 3.
WRN and BLM functional protein interactions. (a) Protein interacting partners of WRN are indicated according to functional effect. Note that this list only includes functional protein interactions (i.e. interactions that modulate the function of one or both of the protein partners (see elsewhere for further discussion). The target WRN activity is shown in parentheses. (i) Proteins that stimulate WRN. (ii) Proteins that are stimulated by WRN. (iii) Proteins that inhibit WRN. (iv) Proteins that are inhibited by WRN. Proteins that have analogous interactions with both WRN and BLM are shown in red. (b) Same as part (a), except for BLM. Abbreviations: exo, exonuclease; FLAP, DNA flap; hel, helicase; TOP1, topoisomerase I.
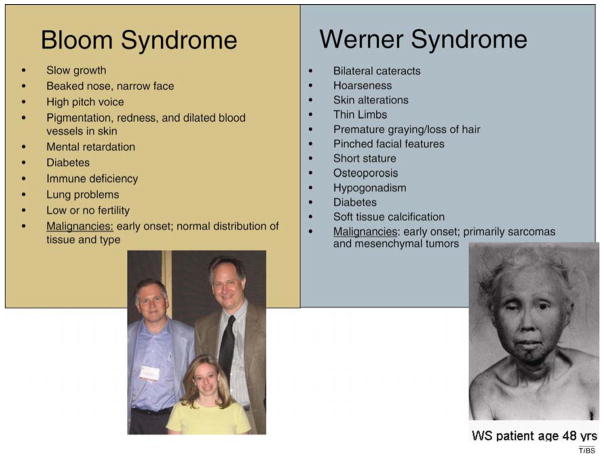
Figure 4.
Clinical features of BS and WS.
RECQ4
The native RECQ4 protein has proven to be difficult to purify and characterize. Recent studies of homogeneous recombinant human RecQ4 purified from E. coli revealed that the recombinant protein has DNA-annealing activity but no detectable helicase activity in vitro [11]. This is despite the presence of the conserved helicase domain in the primary sequence of the protein. Therefore, additional studies are required to determine whether native and/or recombinant RecQ4 can act as a helicase, for example under certain conditions of post-translational modification or in association with another protein.
RECQ1 and RECQ5
Studies of the molecular functions of RECQ1 and RECQ5 are at an early stage, and no disease has been associated with defects in either of these two human RecQ helicases. RECQ1 seems to be the most abundant RecQ helicase in human cells. Both RECQ1 and RECQ5 have helicase- and ssDNA-annealing activities in vitro [12,13]. A recent study indicated that higher-order oligomers of RECQ1 catalyze ssDNA annealing, whereas lower-order oligomers catalyze DNA unwinding [14]. Little is known about the protein interacting partners or biological function(s) of RECQ5. RECQ5 interacts with the DNA polymerase processivity factor PCNA (proliferating cell nuclear antigen) [15], indicating that PCNA might recruit RECQ5 to stalled replication forks. Very recently, RECQ5 has been shown to interact with RNA polymerase II [16]. RECQ5 exists as three splice variants, and the most studied form is RECQ5β (Figure 1), which is discussed in the following.
For further details on the biochemical characteristics of RecQ helicases, interested readers should consult Refs [3,8,17,18].
RecQ helicase syndromes
Several of the clinical features of WS and BS are identical, whereas other clinical features are unique to one or the other syndrome (Figure 4). Clinical features common to WS and BS include slow growth, abnormal facial features, infertility and high incidence and/or early onset of aging-related diseases. WS patients have a more pronounced premature-aging phenotype and, unlike normal individuals and BS patients, WS patients are highly susceptible to early onset of mesenchymal tumors such as sarcomas [1,19]. Microarray analysis of WS primary fibroblasts revealed a gene-expression profile more similar to normal fibroblasts from old individuals than to normal fibroblasts from young individuals [20]. Although WS and BS patients show early onset of many features of normal aging, not all features of normal aging are accelerated in these patients; thus, WS and BS are known as segmental progerias.
BS cells are characterized by higher levels of sister chromatid exchange (SCE) than wild-type cells, and both BS and WS cells exhibit higher-than-normal levels of chromosomal aberrations and hypersensitivity to inhibitors of topoisomerase I and DNA interstrand crosslinking agents [21,22]. BS and WS cells also progress more slowly through S-phase than wild-type cells, indicating that cell-cycle or replication checkpoints might be activated in these cells, even in the absence of induced DNA damage [18,23].
Mutations in RECQ4 cause three distinct clinical diseases: RTS, RAPIDILINO and Baller-Gerold syndromes. RTS is an autosomal recessive condition with skin manifestations (poikiloderma), juvenile cataracts, growth deficiency, premature aging and a predisposition for malignancies, particularly osteosarcoma. The other two syndromes are not as strongly associated with premature aging or cancer [24].
Recent studies indicate that expression of WRN and other premature-aging-associated proteins might be regulated epigenetically [25,26]. For example, the WRN gene is silenced by promoter hypermethylation in many tumors of mesenchymal and epithelial origin, including those commonly observed in WS patients [25]. In addition, when cancer cells are treated with DNA demethylating agents to reverse hypermethylation of the WRN promoter, the defect in WRN exonuclease and the cancer phenotype are rescued [25]. Epigenetic regulation of genes involved in premature aging has been observed by others; for example, the lamin A/C genes, mutations in which lead to premature aging, are regulated by hypermethylation [25]. Other studies also indicate that global changes in DNA methylation have a role in cancer and aging [27]. Epigenetic regulation is a newly recognized mechanism for modulating genes involved in aging; clearly, this is an area of considerable interest for further study.
Mouse models for the RecQ syndromes
The phenotype of RecQ-helicase-deficient mammalian cells typically includes genomic instability and sensitivity to DNA-damaging agents, which is consistent with the idea that RecQ helicases are ‘guardians of the genome’. In contrast to the cellular phenotype, the phenotypes of some RecQ-deficient mice are difficult to interpret. Those with homozygous deletion of BLM die during embryogenesis (for a review, see Ref. [28]). Heterozygous BLM mice display some features of human BS patients including increased frequency of SCE; however, increased frequency of SCE is only observed in humans that are homozygous for a BLM mutation. Mice expressing varying levels of BLM have also been generated; in these animals, the level of BLM correlates inversely with the frequency of SCE, the extent of genomic instability and cancer susceptibility [28]. Whether a similar correlation exists in humans remains to be determined.
Curiously, mice carrying a homozygous deletion of WRN are phenotypically normal. However, mice in which both WRN and Tert (which encodes telomerase) have been mutated have a complex WS-like phenotype characterized by premature death, hair graying, alopecia, osteoporosis, type II diabetes, cataracts and increased incidence of nonepithelial malignancies [29]. The cellular phenotype of WRN-deficient Tert-deficient double mutants includes accelerated replicative senescence and increased chromosomal instability [29]. These data indicate that the latent clinical features of WS might be caused by telomere shortening [29] and that WRN might play an important part in telomere maintenance.
RECQ1-deficient mice have been constructed and they seem to be normal, but mouse embryonic fibroblasts from these mice display genomic instability [27]. Mice deficient for the coding portion of RECQ4 show some typical features of RTS, such as genomic instability, abnormal skin pigmentation, skeletal defects and kyphosis, indicating that it might be a useful model of the human disease [30]. Moreover, cells from these mutant mice have a high frequency of premature centromere separation and aneuploidy, indicating a role for RECQ4 in sister chromatid cohesion. Additional genetic and biochemical studies are needed to elucidate the primary biological roles of RECQ4. Lastly, a recently developed mouse model for RecQ5 displayed an increased cancer susceptibility but normal development and lifespan [31]. Interestingly, the RECQ5-gene knockout mice cells aberrantly accumulated double-strand (ds)DNA breaks (DSBs) and have increased DSB repair (DSBR) by HR repair.
Additional RecQ-homolog-deficient mouse strains are currently being constructed and characterized, and it is hoped that the phenotypes of these mice will provide novel insights into the biological functions of mammalian RecQ proteins. In particular, it will be interesting to determine the phenotype of mice deficient in two or more RecQ helicases to understand whether or not they cooperate in specific pathways of genome maintenance.
RecQ helicases and DNA repair
Cellular DNA is continuously damaged by exogenous and endogenous agents and chemicals, which together generate many types of lesions throughout the genome. Many of the oxidative DNA modifications that accumulate in nucleic acids over time are thought to be caused by endogenous reactive oxygen species (ROS), which are produced as normal byproducts of oxidative phosphorylation in mitochondria and other metabolic processes. It has been estimated that 50 000–100 000 oxidative DNA lesions are generated per mammalian genome per day [32]. If DNA damage persists, it can cause errors in DNA replication or transcription leading to point mutations or chromosomal rearrangements or induce a stress response via various signaling pathways. Prokaryotic and eukaryotic cells express multiple DNA-repair pathways, which provide distinct but overlapping mechanisms to remove many of the DNA lesions that accumulate in cellular DNA (Figure 5). Recent studies indicate that the cellular response to DNA damage is mediated by a complex network of DNA-repair proteins, many of which form multifunctional multiprotein complexes that participate in more than one DNA-repair pathway [33].
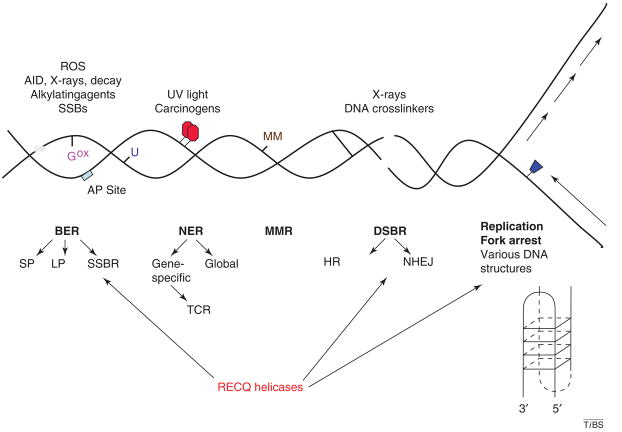
Figure 5.
Eukaryotic DNA repair. Schematic diagram summarizing eukaryotic DNA-repair pathways and their target DNA lesions. Abbreviations: AID, activation-induced cytidine deaminase; AP, apurinic or apyrimidinic; Gox, 8-oxoG; MM, mismatch; NHEJ, non-homologous end-joining; SSBR, single-strand break repair; TCR, transcription-coupled repair; U, uracil. See text for discussion of RecQ helicase roles in DNA repair.
The four main DNA-repair pathways are: base excision repair (BER), which repairs oxidative DNA base modifications such as 8-oxoguanine (8-oxoG), alkylation base damage and ssDNA breaks (SSBs); nucleotide excision repair (NER), which repairs bulky helix-distorting DNA lesions; mismatch repair (MMR), which repairs single-nucleotide mismatches and small insertion–deletion mispairs; and DSBR, which repairs DSBs. Several DNA-repair subpathways have been characterized, including long patch (LP-BER) and short patch (SP-BER) subpathways of BER, the global genome repair (GGR) and transcription-coupled repair (TCR) subpathways of NER, and HR and nonhomologous endjoining (NHEJ), which are subpathways of DSBR (Figure 5). Recent molecular studies show that RecQ helicases interact with DNA-repair proteins and might participate in specific DNA-repair pathways. This is consistent with the observation that RecQ helicases demonstrate structure-specific DNA binding and have higher affinity for DNA susbstrates that resemble DNA-repair intermediates (Figure 2b) than for simple duplex DNA [3,4,34].
BER
BER involves five important steps: removal of a damaged base by DNA glycosylase, incision of the phosphodiester backbone by an abasic (AP) site endonuclease, diesterase- or lyase-mediated modification of DNA termini, DNA synthesis to fill in gapped DNA, and ligation of nicked DNA by DNA ligase. Several DNA glycosylases have been identified in mammals (including TDG, MBD4, UDG, MPG, MYH1, OGG1, NEIL1 and NTH1). These proteins recognize a specific subset of base modifications and hydrolyze the N-glycosylic bond, which releases the damaged base and initiates BER. Collectively, the DNA glycosylases can remove a wide range of damaged bases, and there is considerable overlap in their substrate specificities. The next enzymatic step in BER is catalyzed by DNA polymerase β (POLβ) [35]. POLβ can fill short gaps in DNA and remove residues at a ssDNA break. BER proceeds either via single-nucleotide replacement (i.e. SP-BER) or multiple-nucleotide strand displacement (i.e. LP-BER). SP-BER most commonly involves POLβ, whereas LP-BER typically engages the replicative PCNA-dependent polymerases, although POLβ can also be involved. The LP-BER pathway also involves the flap endonuclease FEN1, which removes the flap structure generated after the polymerase progresses.
Several enzymes exist to deal with specific intermediates that arise in the context of SSB processing. These proteins are more commonly considered to be components of SSB repair (SSBR), a pathway associated with BER.
WRN and BLM interact functionally with many proteins involved in BER and SSBR (Figure 3). These protein interactions have been demonstrated in vitro and in vivo. In particular, APE1, the endonuclease that incises abasic sites during BER, inhibits WRN; this interaction could possibly prevent promiscuous unwinding of DNA-repair intermediates [36]. WRN and BLM can also stimulate DNA POLβ, enhancing base incorporation and facilitating strand displacement [37]. WRN and BLM strongly stimulate FEN1 [38,39]. In vitro evidence also indicates that WRN exonuclease can act as an autonomous proofreading enzyme for DNA POLβ during LP-BER [40]. Collectively, these data indicate that WRN could participate in BER, specifically in LP-BER [37]. Although WRN is not essential for BER, its role in this process is supported by the in vivo finding that WRN-deficient cells accumulate 8-oxoG [41] and are sensitive to some DNA-damaging agents that generate BER substrates [37,42]. Although WRN does not seem to interact with human OGG1, the most important glycosylase for 8-oxoG in human cells, it does interact in vivo and in vitro with NEIL1 [41], a human glycosylase for formamidopyrimidine lesions [41,43]. These lesions are common [44] but not well characterized [45], and they accumulate in cells deficient in WRN [41]. WRN also interacts functionally and reciprocally with polyADP ribose polymerase (PARP-1), a protein with a key role at various steps during BER and SSBR. PARP-1 ribosylates many cellular proteins but it does so at a lower level in WRN-deficient cells, indicating that PARP-1 is activated or stimulated by WRN [46]. PARP-1 also colocalizes with RECQ4 [24]. Thus, WRN has several roles in BER and SSBR, and RECQ4 might also function in SSBR [24].
WRN deficiency leads to hyper-oxidation [47] and rapid and premature accumulation of protein carbonyls [48], which normally accumulate primarily in older individuals. Increased protein oxidation might explain high frequency and early onset of cataracts in WS patients, possibly due to oxidation of lens proteins. The increased incidence of cataracts in WS patients might also be mediated by a direct association between WRN and NBS1 [49], a protein involved in DSBR. Mice deficient in NBS1 develop cataracts at an early age and demonstrate aberrant lens-fiber differentiation [50]. High levels of oxidative stress and oxidative DNA damage correlates with increased risk for sarcomas; thus, hyper-oxidation and dysfunctional or decreased BER could potentially explain the high incidence of sarcomas in WS patients [51].
DSBR and recombination
DSBs are common DNA lesions induced by many types of stress and exposure, including ionizing radiation. DSBs can be visualized by immunochemical staining for foci enriched in phosphorylated histone H2AX (γ-H2AX) or by biochemical and cell biological methods that directly identify DSBs. Two main pathways repair DSBs: HR and NHEJ. Our current understanding of these pathways has been summarized in recent reviews [52,53].
RecQ helicases seem to have an important role in DSBR (Figure 6). For example, RecQ helicases interact with several proteins that have essential roles in DSBR. RAD51, which is a key player in the strand-invasion event during HR, interacts with WRN [31,54], BLM [55], RECQ4 [56] and RECQ5β [31]. The physical interaction between BLM and RAD51D stimulates branch migration by BLM on Holliday junctions [57]. RAD52 both inhibits and enhances WRN helicase activity in a DNA structure-dependent manner, whereas WRN increases the efficiency of RAD52-mediated strand annealing [58], indicating that RAD52 and WRN might cooperatively facilitate rescue of stalled or blocked DNA replication forks. RAD54, another key protein in this pathway, co-localizes with WRN in response to replicative stress [54]. WRN also associates with the MRE11–RAD50–NBS1 complex via NBS1 [49] and the tumor suppressor BRCA1 [59]. Some of these protein interactions are functional; for example, BRCA1 stimulates WRN helicase [59], which is required for HR in cell extracts [60]. Furthermore, WS cells are deficient in the removal of DNA interstrand crosslinks, a process that requires recombination [59,61].
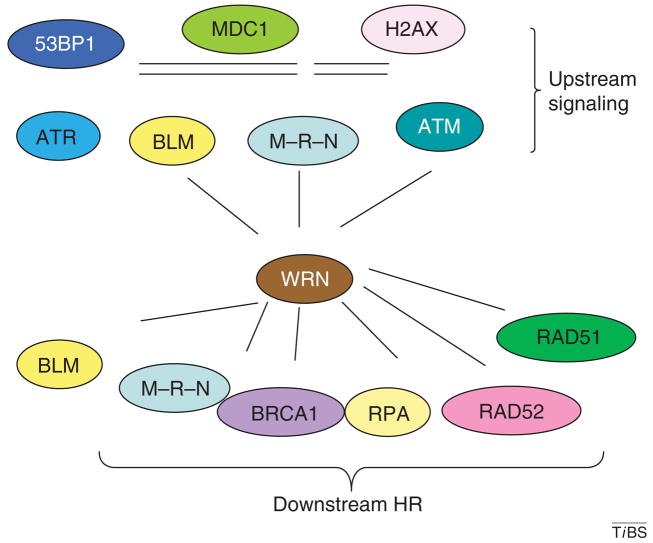
Figure 6.
WRN in the DSBR DNA-damage response. WRN protein interactions indicate possible roles in early and late steps of the response to replication stress. Several of the proteins involved in the formation of the DNA double-strand breaks repair formation are shown under upstream signaling, and several of the proteins involved in the DNA-repair process of homologous recombination are shown. Abbreviations: 53BP1, 53 binding protein; MDC1, mediator of DNA damage checkpoint 1; M–R–N, MRE11–RAD50–NBS1 complex.
As already mentioned, WRN and BLM helicases bind preferentially in vitro to DNA substrates that resemble recombination intermediates such as Holliday junctions (Figure 2b). BLM is the only RecQ helicase that, in cooperation with topoisomerase IIIα [62] and BLAP75 [63], can resolve double Holliday-junction substrates to exclusively generate non-crossover products; this observation is consistent with the fact that BLM-defective cells are deficient in SCE.
WRN and BLM both play a part in assembly of DSBR complexes at γ-H2AX foci, an early step in DSBR. However, WRN and BLM also have downstream roles in NHEJ and DSBR (Figure 6). For example, WRN interacts with the Ku70–Ku80 heterodimer, a primary mediator of NHEJ [64], and this interaction strongly stimulates WRN exonuclease activity in vitro [65]. WRN also interacts with the catalytic subunit of DNA-dependent protein kinase (DNA-PK) [64], indicating that WRN might participate in NHEJ and/or telomere repair. It has been proposed that WRN might be a partner in the end-joining process, at least in part because both WRN exonuclease and helicase are required for NHEJ. However, WS cells are not particularly sensitive to γ-irradiation, which generates DSBs. It remains possible that WRN participates in a NHEJ subpathway or in end-joining in a sub-genomic region such as telomeres or ribosomal (r)DNA (which encodes pre-rRNA transcripts).
WRN might also play a part in DNA mismatch repair. For example, like other proofreading nucleases with which it shares homology [66], WRN can remove a mismatched nucleotide incorporated by a DNA polymerase during DNA replication [67] or DNA repair [40]. Although WRN interacts with human DNA polymerase δ (Polδ) [68] and stimulates the extension activity of yeast DNA Polδ and human translesion DNA polymerases [69], interaction with WRN reduces the fidelity of Polδ [69]. Also, WRN exonuclease fails to bypass some DNA lesions [70]. Further work is needed to understand how WRN interacts with DNA mismatches and many other types of DNA lesions.
In summary, a substantial body of biochemical, genetic and cell biological data strongly indicates that RecQ helicases are key regulators and/or modulators of genetic recombination and DSBR, especially in response to replicative stress (Figures 2c and 6).
RecQ helicases, replication, transcription and chromatin structure
Replication defects in WS and BS cells were first described many years ago. In BS cells, the rate of fork progression is slower than in wild-type cells [71], whereas, in WS cells, progression through S-phase is delayed [23,72]. Decreased proliferation in mouse embryonic fibroblasts was also reported after silencing of RECQ4 [73] and in UV-treated RTS human cells [74].
Subsequent studies indicate that RecQ helicases are recruited to collapsed or arrested DNA replication forks, where they stabilize the replication complex, facilitate DNA repair and promote replication re-start (Figure 2b,c). Recent developments indicate that this could be one of the most important functions of the RecQ helicases [3,18,75]. Insight into the role of RecQ helicases in replication comes from biochemical studies (including analysis of protein–protein interactions) and cell biological studies. RecQ helicases interact physically and functionally with several key proteins involved in replication, such as replication protein A (RPA), PCNA, DNA Polδ and FEN1 [76]. RPA has essential and integral roles in DNA replication and repair. RPA interacts with WRN and BLM and strongly stimulates their respective helicase activities. Thus, interaction between RPA and RecQ helicases might be required for efficient unwinding of large regions of duplex DNA [77,78].
Numerous in vitro studies have examined the effect of induced replication arrest on the interactions between specific DNA substrates and purified RecQ helicases. The results of these studies indicate that RecQ helicases, in conjunction with DNA polymerases and other proteins, play an important part in resolving stalled or collapsed replication forks (for examples, see Refs [3,8,17]).
It is also possible that the DNA-strand-annealing activity of RecQ helicases, including WRN and BLM, might be important for their function at blocked DNA replication forks (Figure 2a,c). For example, BLM, but not E. coli RecQ, can facilitate DNA-replication-fork regression in vitro [79]. The BLM helicase can promote the regression of a model replication fork [3]. Furthermore, WRN and BLM (but not other helicases including UvrD, Rep, PriA, E. coli RecQ) can regress a model replication-fork substrate in vitro [8]. Several detailed studies indicate that uncoupling of leading and lagging strand DNA synthesis is required to enable RecQ helicases to promote replication-fork regression at DNA replication forks blocked by DNA lesions [3]. The helicases might have several functions at replication forks that are blocked by DNA damage. They unwind unusual structures in the vicinity, they promote replication and repair proteins via physical interactions, thus facilitating lesion removal, and they promote unwinding and strand exchange to facilitate the progression of replication. More studies are needed to understand the molecular details of such reactions and how they promote restart of replication by a non-recombinogenic mechanism.
RecQ helicases might also play a part in transcription. For example, biochemical and cellular evidence indicates that WRN modulates RNA polymerase II transcription [80], which could explain differences between gene-expression patterns in normal and WS cells [81]. BLM and WRN might also play a part in regulating chromatin structure via interactions with chromatin accessibility factor 1 [82,83]. This notion is supported by a recent demonstration that WRN-dependent changes in chromatin structure can reduce the incidence of DNA-strand breaks [22]. However, the molecular basis of this effect is not yet clear. Some, but not all, oxidative and bulky DNA base modifications inhibit WRN exonuclease activity [70]; thus, it is possible that RecQ helicases have a role in the early response to and recognition of DNA damage before initiation of DNA repair [70].
RecQ helicases and telomere maintenance
Early studies of WS fibroblasts revealed that WRN deficiency is associated with a defect in telomere maintenance [84]. Furthermore, in vitro experiments with oligomeric telomere substrates demonstrated that WRN processes telomeric DNA and activates a DNA damage response [85]. Previous studies showed that WRN and BLM both interact with telomere proteins TRF1 and TRF2 [86–87], both of which are components of shelterin, a protein complex involved in telomere maintenance. WRN is enriched at the telomeres only during S-phase of the cell cycle, as revealed by live-cell imaging and direct chromatin immunoprecipitation of synchronized cells [88]. In vitro, both BLM and WRN unwind an artificial D-loop substrate in coordination with TRF1 and TRF2 [88], and a tetromeric G-quadruplex is an excellent substrate for WRN and BLM helicases [89]. TRF1 and TRF2 bind with high affinity to telomere repeat sequences but, because they have no known catalytic activity, one likely function of TRF1 and TRF2 is to recruit proteins that are essential for telomere maintenance and/or repair. The binding of TRF1 and TRF2 to telomere repeat sequences is disrupted by oxidative DNA damage in vitro [90], leading to the accumulation of oxidative lesions in telomere sequences that might hamper the early stages of telomere maintenance. Such lesions will, therefore, need to be rapidly repaired to maintain genome stability. POT1 (protection of telomeres 1) is a ssDNA-binding protein that binds with high specificity to telomere repeats and strongly stimulates WRN and BLM helicases [87]. Together with these helicases, POT1 plays an important part in telomere maintenance. Thus, WRN might help to resolve aberrant DNA structures that tend to form as the replication fork progresses through telomeric repeats. This model is consistent with the observation that the frequency of chromosome fusions is higher in WS fibroblasts than in normal fibroblasts and that overexpression of telomerase reduces the frequency of chromosomal aberrations in cells lacking WRN [91]. In this way, genomic instability, one of the hallmarks of WS cells, might be linked to the defect in telomere maintenance in these cells. As mentioned previously, although WRN-deficient mice seem to be normal, WRN-deficient Tert-deficient double-mutant mice have a complex WS-like phenotype [29]. This result and many other recent biochemical and cell biological studies (for a review, see Ref. [92]) imply that WRN might have a particularly important role in telomere maintenance.
It is interesting to consider whether, how and under what circumstances WRN helicase and exonuclease activities might be coordinated or compartmentalized. In vitro evidence indicates that both exonuclease and helicase functions are required for some biological functions, for example, for the unwinding of a telomeric D-loop substrate [88] (see also later).
Post-translational modification of RecQ helicases
Early studies identified post-translational modifications of RecQ helicases, some of which might have a regulatory role (Figure 7). For example, WRN is phosphorylated on serine, threonine and tyrosine residues in vitro and in vivo. Phosphorylation of WRN increases in cells exposed to bleomycin or other types of replication stress [64,93]. Bleomycin-induced serine/threonine phosphorylation of WRN requires ataxia telangiectasia mutated protein (ATM) and the DNA-PK complex, as it is not observed in MO59J cells that are deficient in these two proteins [64]. At present, it remains unclear how ATM modulates the phosphorylation status of WRN and whether ATM activity affects downstream WRN activity in cells with DNA damage or other types of replication stress. However, WRN phosphorylation clearly depends on the heterotrimeric DNA-PK complex in vivo and in vitro [64,94] and, importantly, serine/threonine phosphorylation of WRN by the DNA-PK complex inhibits the WRN helicase and exonuclease activities [64]. This negative regulatory mechanism might have a role in facilitating NHEJ-mediated repair of DSBs.
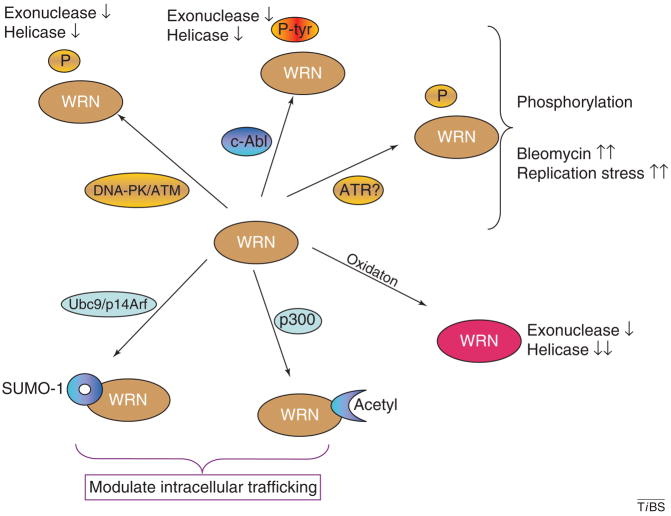
Figure 7.
WRN post-translational modification. Schematic diagram summarizing post-translational modifications of WRN including phosphoryation on Ser/Thr or Tyr (upper half of figure), sumoylation (bottom left), acetylation (bottom middle) and oxidation (bottom right). Probable mediators and modifiers of WRN post-translational modifications are indicated. Functional effects of post-translational modifications are indicated, where known. The ‘?’ indicates uncertainty. Arrows up or down: two arrows indicate stronger reaction than one arrow.
WRN is also phosphorylated by c-Abl, a protein tyrosine kinase, with which it is constitutively associated [93]. c-Abl is activated by and dissociates from WRN in response to bleomycin, indicating that c-Abl-mediated tyrosine phosphorylation might destabilize the interaction between c-Abl and WRN. This cAbl-mediated tyrosine phosphorylation, like the serine/threonine phosphorylation mediated by the DNA-PK complex, inhibits WRN exonuclease and helicase activities [93]. It has been reported, but not confirmed, that WRN is also phosphorylated by ATR during activation of the S-phase checkpoint [95].
WRN is also sumoylated in vitro and in vivo. The small ubiquitin-related modifier (SUMO)-1 conjugating system requires a SUMO-activating (E1) enzyme, a SUMO-conjugating (Ubc9) enzyme, and a SUMO-ligating enzyme. WRN interacts with Ubc9 [96], which is required for conjugation of SUMO-1 to WRN. However, the functional consequences of sumoylation of WRN remain to be established. In addition to sumoylation, acetylation and tyrosine phosphorylation of WRN (or of a putative ‘WRN-nucleolar carrier’) have been proposed to modulate nucleolar trafficking of WRN [93]. Indeed, acetylation of WRN by the transcriptional co-activator and acetylase p300 results in translocation of WRN from the nucleolus to nuclear foci [97]. The precise role of WRN or other RecQ helicases in the nucleoli is not yet understood.
It will also be interesting to determine whether and how post-translational modification of WRN is coordinated and how one post-translational modification might promote or inhibit additional modifications of WRN or other RecQ helicases. Initial studies indicate that BLM is phosphorylated and sumoylated in vitro and in vivo [28,98,99].
Post-translational modification of RecQ helicases might facilitate DNA repair or otherwise regulate RecQ-associated enzymatic functions. As such, post-translational modification of RecQ helicases could ultimately modulate multiple pathways, including HR, NHEJ, BER and DNA replication.
Conclusions
RecQ helicases have an important role in maintaining genome stability. Defects in RecQ helicases are associated with susceptibility to cancer and premature aging in humans and genome instability and hypersensitivity to DNA damaging agents in cultured cells. Purified RecQ helicases from higher organisms bind preferentially to DNA substrates that resemble intermediates in DNA repair, replication or recombination. Extensive characterization of the human RecQ helicases indicates that they have important roles in DNA replication, telomere maintenance and DNA repair.
More sophisticated protein-interaction technologies will be needed to further investigate the interactions between RecQ helicases and other proteins. The relative affinity of different RecQ helicase protein partners needs to be determined and their protein-interaction domains mapped by site-directed mutagenesis. Additionally, RecQ helicase protein–protein interactions need to be evaluated based on tissue specificity and/or developmental stages. The results of such studies might indicate whether WRN and BLM interact simultaneously with their various protein partners and which protein interactions are most important. Rigorous biophysical studies of such interactions could also identify their functional effects and ultimately help to define the emerging roles of RecQ helicases in different DNA metabolic pathways.
Eventually, it is hoped that experimental studies will reveal the role of RecQ helicases in vivo; however, this research goal might not be easily achieved, especially because mouse models have been less revealing than expected in this regard. Fortunately, new mouse models are emerging with targeted or non-targeted deficiencies in RecQ helicases, some of which seem to mimic human WS, BS or RTS, indicating that there could be more benefit from pursuit of this avenue than the initial mouse models provided.
Individuals who lack RecQ family proteins suffer from high rates of cancer and/or premature aging. Further studies are needed to clarify the molecular mechanisms by which the RecQ helicases exert their effects on these processes in eukaryotic cells and in living organisms, including humans. Lastly, it will be a challenge to understand how the activity of multiple RecQ helicases are coordinated and regulated in human cells, which express five RecQ homologues that might have overlapping, but distinct, functional roles.
Acknowledgments
I would like to thank M. Sander, D. Wilson and D. Croteau for suggestions. I acknowledge support from the Intramural Research Program of the National Institute on Aging, National Institutes of Health.
References
- 1.German J. Bloom’s syndrome. XX The first 100 cancers. Cancer Genet Cytogenet. 1997;93:100–106. doi: 10.1016/s0165-4608(96)00336-6. [DOI] [PubMed] [Google Scholar]
- 2.Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 3.Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu Rev Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- 4.Mohaghegh P, et al. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Kobbe C, et al. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J Biol Chem. 2002;277:22035–22044. doi: 10.1074/jbc.M200914200. [DOI] [PubMed] [Google Scholar]
- 6.Huang S, et al. The premature ageing syndrome protein, WRN, is a 3′ → 5′ exonuclease. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen JC, Loeb LA. Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res. 2000;28:3260–3268. doi: 10.1093/nar/28.17.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma S, et al. Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Kobbe C, et al. Werner syndrome protein contains three structure-specific DNA binding domains. J Biol Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 10.von Kobbe C, Bohr VA. A nucleolar targeting sequence in the Werner syndrome protein resides within residues 949–1092. J Cell Sci. 2002;115:3901–3907. doi: 10.1242/jcs.00076. [DOI] [PubMed] [Google Scholar]
- 11.Macris MA, et al. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Garcia PL, et al. Human RECQ5β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma S, et al. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 14.Muzzolini L, et al. Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol. 2007;5:e20. doi: 10.1371/journal.pbio.0050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanagaraj R, et al. Human RECQ5β helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aygun O, et al. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci U S A. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cobb JA, Bjergbaek L. RecQ helicases: lessons from model organisms. Nucleic Acids Res. 2006;34:4106–4114. doi: 10.1093/nar/gkl557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu L. Role of the BLM helicase in replication fork management. DNA Repair (Amst) 2007;6:936–944. doi: 10.1016/j.dnarep.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Huang S, et al. The spectrum of WRN mutations in Werner syndrome patients. Hum Mutat. 2006;27:558–567. doi: 10.1002/humu.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kyng KJ, et al. Gene expression profiling in Werner syndrome closely resembles that of normal aging. Proc Natl Acad Sci U S A. 2003;100:12259–12264. doi: 10.1073/pnas.2130723100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laine JP, et al. Werner protein stimulates topoisomerase I DNA relaxation activity. Cancer Res. 2003;63:7136–7146. [PubMed] [Google Scholar]
- 22.Turaga RV, et al. Werner syndrome protein prevents DNA breaks upon chromatin structure alteration. Aging Cell. 2007;6:471–481. doi: 10.1111/j.1474-9726.2007.00301.x. [DOI] [PubMed] [Google Scholar]
- 23.Poot M, et al. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cells. Exp Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- 24.Dietschy T, et al. The molecular role of the Rothmund-Thomson-, RAPADILINO- and Baller-Gerold-gene product, RECQL4: recent progress. Cell Mol Life Sci. 2007;64:796–802. doi: 10.1007/s00018-007-6468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agrelo R, et al. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proc Natl Acad Sci U S A. 2006;103:8822–8827. doi: 10.1073/pnas.0600645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Opresko PL, et al. Role for the Werner syndrome protein in the promotion of tumor cell growth. Mech Ageing Dev. 2007;128:423–436. doi: 10.1016/j.mad.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16:R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 28.Woo LL, et al. The broken genome: genetic and pharmacologic approaches to breaking DNA. Ann Med. 2007;39:208–218. doi: 10.1080/08035250601167136. [DOI] [PubMed] [Google Scholar]
- 29.Chang S, et al. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 30.Mann MB, et al. Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Hum Mol Genet. 2005;14:813–825. doi: 10.1093/hmg/ddi075. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shigenaga MK, Ames BN. Oxidants and mitogenesis as causes of mutation and cancer: the influence of diet. Basic Life Sci. 1993;61:419–436. doi: 10.1007/978-1-4615-2984-2_37. [DOI] [PubMed] [Google Scholar]
- 33.Hoeijmakers JH. DNA repair mechanisms. Maturitas. 2001;38:17–22. doi: 10.1016/s0378-5122(00)00188-2. [DOI] [PubMed] [Google Scholar]
- 34.Opresko PL, et al. Junction of RecQ helicase biochemistry and human disease. J Biol Chem. 2004;279:18099–18102. doi: 10.1074/jbc.R300034200. [DOI] [PubMed] [Google Scholar]
- 35.Wilson SH. Mammalian base excision repair and DNA polymerase β. Mutat Res. 1998;407:203–215. doi: 10.1016/s0921-8777(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 36.Ahn B, et al. Regulation of WRN helicase activity in human base excision repair. J Biol Chem. 2004;279:53465–53474. doi: 10.1074/jbc.M409624200. [DOI] [PubMed] [Google Scholar]
- 37.Harrigan JA, et al. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase β. Nucleic Acids Res. 2006;34:745–754. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brosh RM, Jr, et al. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma S, et al. Stimulation of flap endonuclease-1 by the Bloom’s syndrome protein. J Biol Chem. 2004;279:9847–9856. doi: 10.1074/jbc.M309898200. [DOI] [PubMed] [Google Scholar]
- 40.Harrigan JA, et al. WRN exonuclease activity is blocked by DNA termini harboring 3′ obstructive groups. Mech Ageing Dev. 2007;128:259–266. doi: 10.1016/j.mad.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das A, et al. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase Neil1. J Biol Chem. 2007;282:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 42.Blank A, et al. The Werner syndrome protein confers resistance to the DNA lesions N3-methyladenine and O6-methylguanine: implications for WRN function. DNA Repair (Amst) 2004;3:629–638. doi: 10.1016/j.dnarep.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Imoto S, et al. Synthesis, DNA polymerase incorporation, and enzymatic phosphate hydrolysis of formamidopyrimidine nucleoside triphosphates. J Am Chem Soc. 2006;128:14606–14611. doi: 10.1021/ja065525r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu J, et al. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J Biol Chem. 2005;280:40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 45.Jaruga P, et al. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 46.von Kobbe C, et al. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol Cell Biol. 2003;23:8601–8613. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szekely AM, et al. Werner protein protects nonproliferating cells from oxidative DNA damage. Mol Cell Biol. 2005;25:10492–10506. doi: 10.1128/MCB.25.23.10492-10506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oliver CN, et al. Age-related changes in oxidized proteins. J Biol Chem. 1987;262:5488–5491. [PubMed] [Google Scholar]
- 49.Cheng WH, et al. Linkage between Werner syndrome protein and the Mre11 complex via Nbs1. J Biol Chem. 2004;279:21169–21176. doi: 10.1074/jbc.M312770200. [DOI] [PubMed] [Google Scholar]
- 50.Yang YG, et al. A novel function of DNA repair molecule Nbs1 in terminal differentiation of the lens fibre cells and cataractogenesis. DNA Repair (Amst) 2006;5:885–893. doi: 10.1016/j.dnarep.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Kansara M, Thomas DM. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007;26:1–18. doi: 10.1089/dna.2006.0505. [DOI] [PubMed] [Google Scholar]
- 52.Lobrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 53.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 54.Otterlei M, et al. Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J Cell Sci. 2006;119:5137–5146. doi: 10.1242/jcs.03291. [DOI] [PubMed] [Google Scholar]
- 55.Wu L, et al. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 56.Petkovic M, et al. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- 57.Braybrooke JP, et al. Functional interaction between the Bloom’s syndrome helicase and the RAD51 paralog, RAD51L3 (RAD51D) J Biol Chem. 2003;278:48357–48366. doi: 10.1074/jbc.M308838200. [DOI] [PubMed] [Google Scholar]
- 58.Baynton K, et al. WRN interacts physically and functionally with the recombination mediator protein RAD52. J Biol Chem. 2003;278:36476–36486. doi: 10.1074/jbc.M303885200. [DOI] [PubMed] [Google Scholar]
- 59.Cheng WH, et al. Collaboration of Werner syndrome protein and BRCA1 in cellular responses to DNA interstrand cross-links. Nucleic Acids Res. 2006;34:2751–2760. doi: 10.1093/nar/gkl362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang N, et al. The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J Biol Chem. 2005;280:40559–40567. doi: 10.1074/jbc.M508453200. [DOI] [PubMed] [Google Scholar]
- 61.Poot M, et al. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 2001;15:1224–1226. doi: 10.1096/fj.00-0611fje. [DOI] [PubMed] [Google Scholar]
- 62.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 63.Yin J, et al. BLAP75, an essential component of Bloom’s syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24:1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karmakar P, et al. Werner protein is a target of DNA-PK in vivo and in vitro and its catalytic activities are regulated by phosphorylation. J Biol Chem. 2002;277:18291–18302. doi: 10.1074/jbc.M111523200. [DOI] [PubMed] [Google Scholar]
- 65.Cooper MP, et al. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- 66.Brosh RM, Jr, et al. Enzymatic mechanism of the WRN helicase/nuclease. Methods Enzymol. 2006;409:52–85. doi: 10.1016/S0076-6879(05)09004-X. [DOI] [PubMed] [Google Scholar]
- 67.Kamath-Loeb AS, et al. Werner Syndrome Protein. Ii characterization of the integral 3′ → 5′ dna exonuclease. J Biol Chem. 1998;273:34145–34150. doi: 10.1074/jbc.273.51.34145. [DOI] [PubMed] [Google Scholar]
- 68.Kamath-Loeb AS, et al. Interactions between the Werner syndrome helicase and DNA polymerase δ specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J Biol Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- 69.Kamath-Loeb AS, et al. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc Natl Acad Sci U S A. 2007;104:10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Machwe A, et al. Selective blockage of the 3′ →5′ exonuclease activity of WRN protein by certain oxidative modifications and bulky lesions in DNA. Nucleic Acids Res. 2000;28:2762–2770. doi: 10.1093/nar/28.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hand R, German J. A retarded rate of DNA chain growth in Bloom’s syndrome. Proc Natl Acad Sci U S A. 1975;72:758–762. doi: 10.1073/pnas.72.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanaoka F, et al. Autoradiographic studies of DNA replication in Werner’s syndrome cells. Adv Exp Med Biol. 1985;190:439–457. doi: 10.1007/978-1-4684-7853-2_22. [DOI] [PubMed] [Google Scholar]
- 73.Sangrithi MN, et al. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 74.Park SJ, et al. A positive involvement of RecQL4 in UV-induced S-phase arrest. DNA Cell Biol. 2006;25:696–703. doi: 10.1089/dna.2006.25.696. [DOI] [PubMed] [Google Scholar]
- 75.Bachrati CZ, Hickson ID. Analysis of the DNA unwinding activity of RecQ family helicases. Methods Enzymol. 2006;409:86–100. doi: 10.1016/S0076-6879(05)09005-1. [DOI] [PubMed] [Google Scholar]
- 76.Ozgenc A, Loeb LA. Current advances in unraveling the function of the Werner syndrome protein. Mutat Res. 2005;577:237–251. doi: 10.1016/j.mrfmmm.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 77.Brosh RM, Jr, et al. Functional and physical interaction between WRN helicase and human replication protein A. J Biol Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- 78.Brosh RM, Jr, et al. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J Biol Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 79.Ralf C, et al. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 80.Balajee AS, et al. The Werner syndrome protein is involved in RNA polymerase II transcription. Mol Biol Cell. 1999;10:2655–2668. doi: 10.1091/mbc.10.8.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kyng KJ, Bohr VA. Gene expression and DNA repair in progeroid syndromes and human aging. Ageing Res Rev. 2005;4:579–602. doi: 10.1016/j.arr.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 82.Jiao R, et al. Physical and functional interaction between the Bloom’s syndrome gene product and the largest subunit of chromatin assembly factor 1. Mol Cell Biol. 2004;24:4710–4719. doi: 10.1128/MCB.24.11.4710-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiao R, et al. The Werner syndrome protein is required for recruitment of chromatin assembly factor 1 following DNA damage. Oncogene. 2006;26:3811–3822. doi: 10.1038/sj.onc.1210150. [DOI] [PubMed] [Google Scholar]
- 84.Kruk PA, et al. DNA damage and repair in telomeres: relation to aging. Proc Natl Acad Sci U S A. 1995;92:258–262. doi: 10.1073/pnas.92.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eller MS, et al. A role for WRN in telomere-based DNA damage responses. Proc Natl Acad Sci U S A. 2006;103:15073–15078. doi: 10.1073/pnas.0607332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Opresko PL, et al. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 87.Opresko PL, et al. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J Biol Chem. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- 88.Opresko PL, et al. The Werner Syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 89.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 90.Opresko PL, et al. Oxidative damage in telomeric DNA disrupts recognition by TRF1 and TRF2. Nucleic Acids Res. 2005;33:1230–1239. doi: 10.1093/nar/gki273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Crabbe L, et al. Telomere dysfunction as a cause of genomic instability in Werner syndrome. Proc Natl Acad Sci U S A. 2007;104:2205–2210. doi: 10.1073/pnas.0609410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Multani AS, Chang S. WRN at telomeres: implications for aging and cancer. J Cell Sci. 2007;120:713–721. doi: 10.1242/jcs.03397. [DOI] [PubMed] [Google Scholar]
- 93.Cheng WH, et al. Werner syndrome protein phosphorylation by abl tyrosine kinase regulates its activity and distribution. Mol Cell Biol. 2003;23:6385–6395. doi: 10.1128/MCB.23.18.6385-6395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yannone SM, et al. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J Biol Chem. 2001;276:38242–38248. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- 95.Pichierri P. Interplay between wrn and the checkpoint in s-phase. Ital J Biochem. 2007;56:130–140. [PubMed] [Google Scholar]
- 96.Woods YL, et al. p14 Arf promotes small ubiquitin-like modifier conjugation of Werners helicase. J Biol Chem. 2004;279:50157–50166. doi: 10.1074/jbc.M405414200. [DOI] [PubMed] [Google Scholar]
- 97.Blander G, et al. DNA damage-induced translocation of the Werner helicase is regulated by acetylation. J Biol Chem. 2002;277:50934–50940. doi: 10.1074/jbc.M210479200. [DOI] [PubMed] [Google Scholar]
- 98.Eladad S, et al. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum Mol Genet. 2005;14:1351–1365. doi: 10.1093/hmg/ddi145. [DOI] [PubMed] [Google Scholar]
- 99.Rao VA, et al. Phosphorylation of BLM, dissociation from topoisomerase IIIα, and colocalization with gamma-H2AX after topoisomerase I-induced replication damage. Mol Cell Biol. 2005;25:8925–8937. doi: 10.1128/MCB.25.20.8925-8937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]