Abstract
Separation of the two strands of DNA with heat (melting) is a fundamental property of DNA that is conveniently monitored with fluorescence. Conventional melting is performed after PCR on any real-time instrument to monitor product purity (dsDNA dyes) and sequence (hybridization probes). Recent advances include high-resolution instruments and saturating DNA dyes that distinguish many different species. For example, mutation scanning (identifying heterozygotes) by melting is closed-tube and has similar or superior sensitivity and specificity compared to methods that require physical separation. With high resolution melting, SNPs can be genotyped without probes and more complex regions can be typed with unlabeled hybridization probes. Highly polymorphic HLA loci can be melted to establish sequence identity for transplantation matching. Simultaneous genotyping with one or more unlabeled probes and mutation scanning of the entire amplicon can be performed at the same time in the same tube, vastly decreasing or eliminating the need for re-sequencing in genetic analysis. High-resolution PCR product melting is homogeneous, closed-tube, rapid (1–5 min), non-destructive and does not require covalently-labeled fluorescent probes. In the clinical laboratory, it is an ideal format for in-house testing, with minimal cost and time requirements for new assay development.
INTRODUCTION
The annealing and melting properties of DNA have enabled the development of many clinical, genetic, and forensic tests. Well established technologies using various nucleic acid and signal amplification methods for DNA detection and identification all depend on DNA hybridization (Wittwer and Kusukawa, 2005). The binding of labeled DNA probes to identify unique sequences is used in real time PCR, FISH, and array analysis. Recent advances in DNA melting techniques include instrumentation that allows for highly controlled temperature transitions and data acquisition (Gundry et al., 2003), and the development of fluorescent DNA binding dyes with improved saturation properties (Wittwer et al., 2003). These advances allow a more precise assessment of sequence variations based on melting analysis, without the need for labeled probes and have the potential to greatly decrease the burden of sequencing. Fluorescence instrumentation that focuses on high resolution melting has recently been introduced, and high resolution methods have also been adopted to real-time PCR instruments with variable success (Herrmann et al., 2006; Herrmann et al., 2007a; Herrmann et al., 2007b).
In addition to improved instrumentation, new DNA binding dyes have been developed that saturate available PCR products and allow detection of heterozygous DNA for genotyping and variant scanning. The newer dyes exhibit minimal redistribution during melting and do not inhibit PCR (Wittwer et al., 2003), two difficulties common to the use of earlier DNA binding dyes in PCR.
The fluorescence data generated during DNA melting can be analyzed based on the melting temperature (Tm) or on the shape of the melting curve. Tm data is best calculated following the mathematical removal of background and normalization of the melting curve. Tm differences can allow the discrimination of many genotypes but may not distinguish all homozygotes. Additional information is available if the entire melting curve is analyzed. Specifically, differences in the melting curve shape can easily identify heterozygotes. Shape differences in melting curves can be conveniently displayed by superimposing normalized curves and plotting the fluorescence differences between samples.
High resolution melting analysis may be applied to amplicon generated during PCR or to unlabeled probes to interrogate a short segment. Furthermore, amplicon and probe melting can both be analyzed from the same melting curve. This review will discuss clinical laboratory applications of high resolution melting using both amplicon and unlabeled probe methods.
GENOTYPING BY AMPLICON MELTING
High resolution melting analysis of amplicons depends on DNA melting in the presence of saturating DNA binding dyes. As the temperature of the solution is increased, the specific sequence of the amplicon (primarily the GC content and the length) determine the melting behavior. When the fluorescence signal is plotted against the temperature, the fluorescence intensity decreases as the double stranded DNA becomes single stranded and the dye is released. The melting temperature (Tm) at which 50% of the DNA is in the double stranded state may be approximated by taking the derivative of the melting curve. The unique pattern of the melting curve, the derivative plot, or the difference plot may be used for amplicon analysis. Examples of these plots are presented in Figure 1.
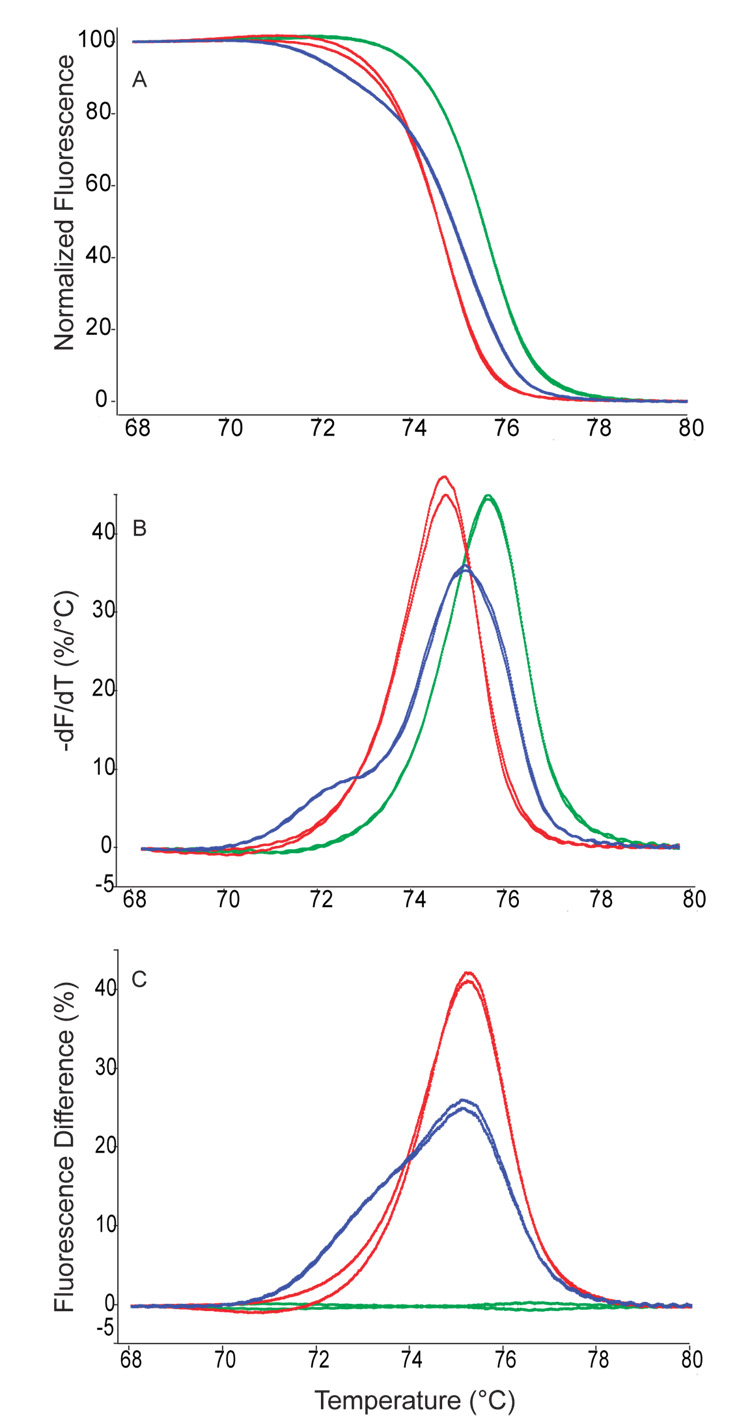
Figure 1.
Amplicon melting analyses for duplicate samples of factor V (Leiden) 1691 G>A wild-type (green), heterozygous (blue) and homozygous mutant (red) samples. (A) normalized melting curves, (B) derivative plots, (C) difference plots.
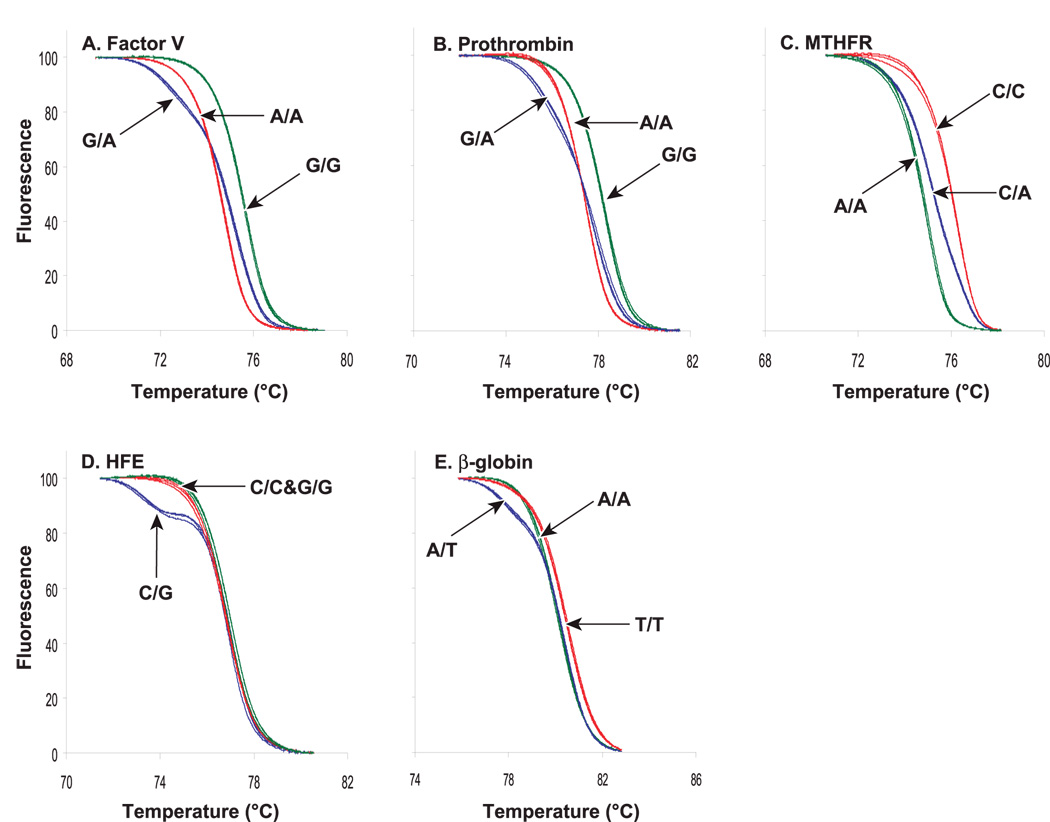
Shorter amplicons generally allow better discrimination of small sequence variations such as single base differences. Liew et al. (Liew et al., 2004) demonstrated the power of high resolution melting analysis for SNP genotyping using small amplicons for factor V (Leiden) 1691G>A, prothrombin 20210G>A, methylenetetrahydrofolate reductase (MTHFR)1298A>C, hemochromatosis (HFE) 187C>G, and beta-globin (hemoglobin S) 17A>T. The PCR products were from 38 to 50 bp in length and provided good differentiation of genotypes. (Figure 2)
Figure 2.
Normalized, high-resolution melting curves from (A) factor V (Leiden) 1691G>A, (B) prothrombin 20210G>A, (C) MTHFR 1298A>C, (D) HFE 187C>G, and beta-globin 17A>T SNPs. Three individuals of each genotype were analyzed and are displayed for each SNP. Wild-type (green), heterozygous (blue) and homozygous mutant (red). Reprinted with permission from Clin Chem 50, 1156-1146 (2004).
The use of small amplicons for genotyping simplifies assay design since the primers are chosen as close to the SNP as possible. As the size of the amplicons is decreased, the Tm differences among the genotypes are increased, thus allowing better differentiation. Cycling times for small amplicons can be very short because lower melting temperatures are used for denaturation and no holds are necessary during amplification.
Additional studies on samples with heterozygous sequence variants near the standard expected mutations for factor V (Leiden) 1691G>A, prothrombin 20210G>A, HFE 187C>G, further demonstrated that unexpected heterzygotes can be distinguished from the typical targeted ones using high resolution melting analysis of small amplicons (Graham et al., 2005). In this study, the use of difference plots revealed unique melting curve shapes for each genotype. The factor V 1691G>A was clearly separated from 3 rare heterozygotes, (1690delC, 1690C>T, and compound 1696A>G/1690G>A). Prothrombin 20210G>A was distinguished from 2 heterozygotes (20209C>T and 20218A>G) and HFE 187C>G was distinguished from 5 rare variants (187C>G/189T>C, 187C>G/193A>T, 189T>C, 193A>T, and 197G>A).
SNP genotyping using high resolution melting analysis has also been applied to larger amplicons for human platelet antigens 1 to 5 and 15 (amplicons 160 to 218 bps) (Liew et al., 2006; Liew et al., 2007). Homozygous wild type and mutant human platelet antigen samples generated clearly defined single peaks separated by 0.3 to 1 °C, while heterozygotes were easily identified by curve shape. Human platelet antigen genotyping by high resolution melting analysis was compared to genotyping using conventional hybridization probes and performed comparably.
High resolution amplicon melting analysis was used to differentiate mycobacteria species within the Mycobacterium chelonae-abscessus group. Odell et al. (Odell et al., 2005) used 105 bp amplicons from the heat shock protein 65 (hsp65) gene to differentiate the M. chelonae-abscessus group. The target region selected contained 4 T>C and one G>T base changes that could differentiate M. chelone and M. abscessus. The assay also identified and differentiated M. chelone and M. abscessus sequence variants as well as M. immunogenum. The assay could be performed in 20 minutes in a closed tube system with no separation steps and no requirement for labeled probes.
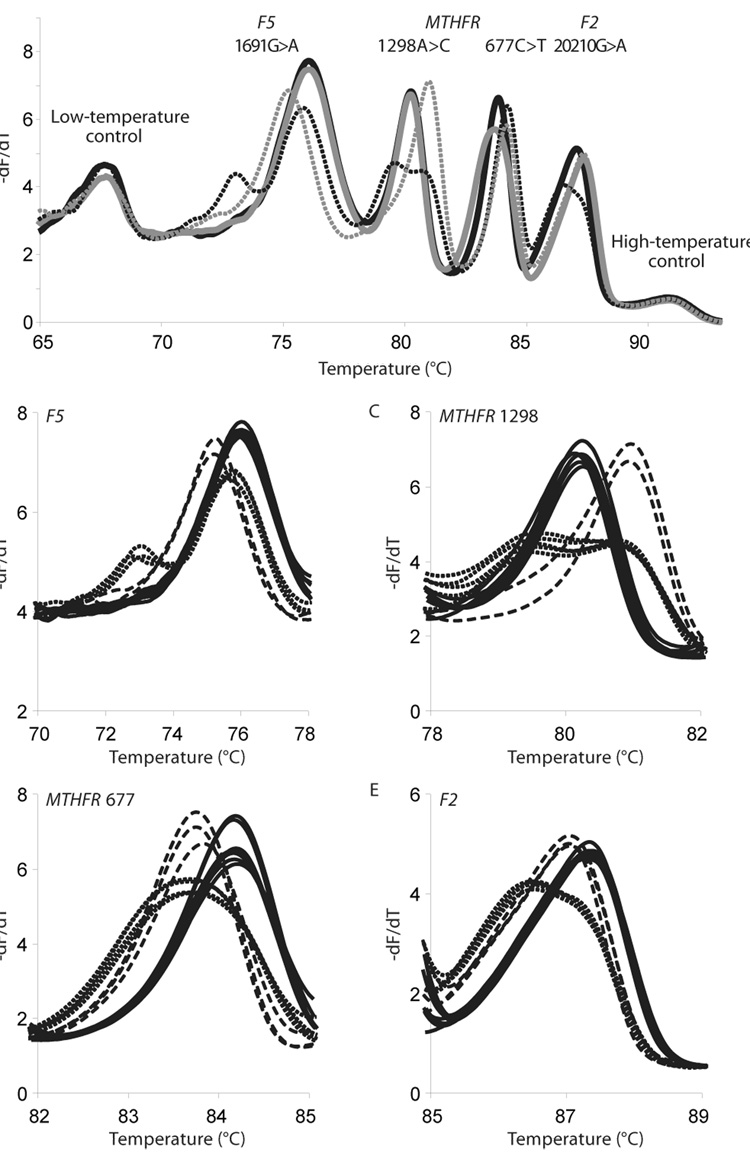
For discrimination of closely related sequences or multiplex reactions, modifications to the primers can be made to improve the differences in the melting patterns of the amplicons. In sequences with very similar Tms, additional differentiation can be obtained using GC-rich tailed primers, locked nucleic acids, or modified bases. Seipp et al. (Seipp et al., 2007b; Seipp et al., 2008) demonstrated the use of primers with GC- or AT-rich 5’ tails to generate amplicons with varying lengths and Tms in a quadraplex assay that differentiated factor V (Leiden) 1691G>A, MTHFR 1298A>C, MTHFR 677C>T, and prothrombin 20210G>A (figure 4). A similar use of tailed primers was applied to the detection and differentiation of three Aspergillus species in a multiplex assay (Erali et al., 2006).
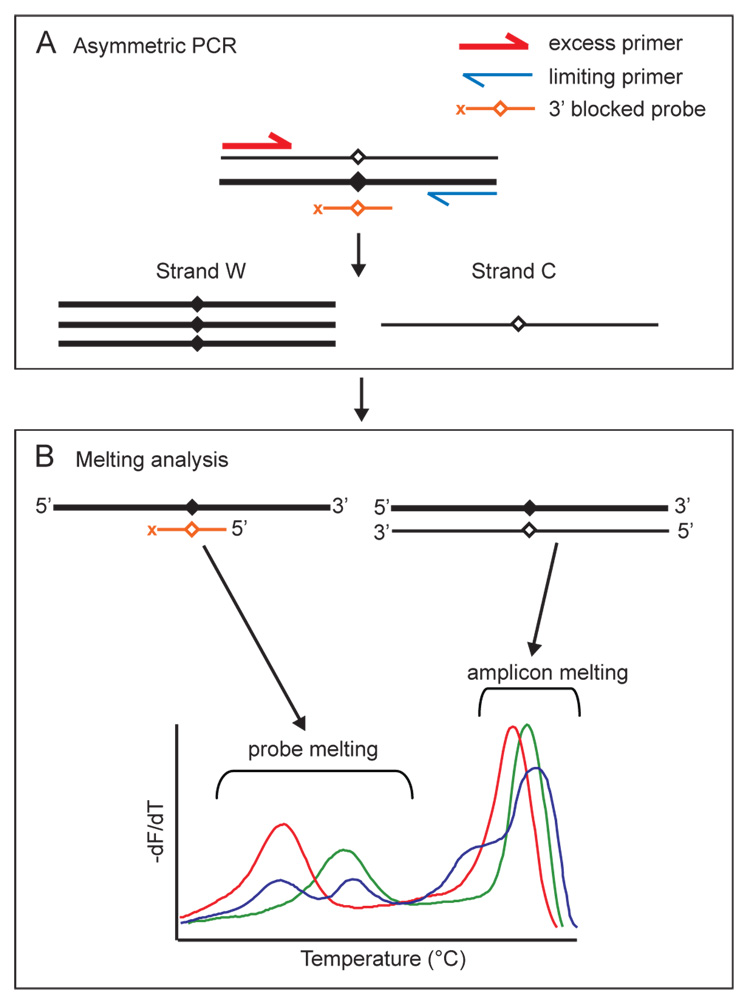
Figure 4.
Combined unlabeled probe and amplicon melting analysis. (A) Asymmetric PCR produces excess copies of strand W and limiting copies of strand C. Excess copies of strand W are available for probe binding, revealing expected and variant sequences under the probe as melting peaks. Sufficient copies of strand C are present so that the full length amplicon duplexes allow variant scanning anywhere within the amplicon. (B) Both product/probe melting transitions and product/product melting transitions are visible on a single negative derivative plot of normalized background corrected fluorescence. Examples of wild-type (green), mutant (red) and heterozygous (blue) melting peaks are illustrated.
Additional discrimination of target Tms can be accomplished using internal temperature standards that bracket the target Tms. This can be helpful in highly multiplexed assays or for larger amplicons (>200 bp). The temperature standards are small synthetic oligonucleotides that serve as reference points for any adjustments needed to compensate for sample differences or well-to-well variations in a plate. Locked nucleic acids can also be incorporated into the standards to adjust melting temperatures. (Liew et al., 2007; Seipp et al., 2007a)
Temperature controls were used in an amplicon melting assay for MTHFR 1298A>C and 677C>T variants (Liew et al., 2007). In this study, the Tm variations resulting from different extraction methods were reduced when two temperature controls that bracketed the target Tms were used. The derivative melting curves for samples were shifted and scaled through linear expansion or compression of the temperature axis based on aligning the positions of the Tm controls. Temperature controls also improved the genotyping of the lactose gene (LCT) 13910C>T SNP associated with lactose intolerance. Genotyping accuracy from melting the 206 bp amplicon on a 96-well plate was improved by incorporating internal temperature controls.
In some cases, homozygous SNPs may have melting characteristics identical to that of the wild-type. Palais et al. (Palais et al., 2005) described the addition of a known reference genotype to samples before PCR to introduce heteroduplex formation, thus allowing discrimination among the three genotypes. Theoretical and experimental data were presented to show that addition of the reference DNA at one seventh of the total DNA provided the best discrimination.
High resolution melting analysis has been used for the rapid identification of bacteria. Broad range PCR of the 16S rRNA gene followed by high resolution melting was applied to the identification of 25 bacterial species (Cheng et al., 2006). Examination of difference plots allowed the direct identification of 9 bacterial species. The remaining 16 species clustered into 4 melting groups. For one group of bacteria, heteroduplexing with the PCR amplicon of P. mirabilis allowed differentiation of P. mirabilis, K. pneumoniae, and S. marcescens. A second group of bacteria, A. baumannii and H. influenzae was identified by heteroduplexing with PCR amplicon of A. baumannii and a third group, S. pneumoniae, E. faecalis, Bacillus spp., and S. pyogenes/S. agalactiae, was heteroduplexed with S. pneumoniae amplicon for differentiation. The fourth group, E. coli, S. typhimurium, S. enteritidis, and S.flexneri included bacterial species with no sequence variations in the targeted amplicon and required an additional primer set for differentiation.
Other applications of high resolution melting analysis to infectious disease targets have been described for determining Staphylococcus aureus genotypes (Stephens et al., 2008), influenza A subtypes (Lin et al., 2008), Bacillus anthracis strains (Fortini et al., 2007), and Mycoplasma synoviae strains (Jeffery et al., 2007). Other recent applications of high resolution melting analysis to human genotyping include the detection of the alpha-thalassemia-1 Southeast Asian allele (Pornprasert et al., 2008), the G11338A mutation in the fibroblast growth factor receptor 3 gene (Hung et al., 2008), and SNPs in the the ryanodine receptor gene associated with malignant hyperthermia and/or central core disease (Grievink and Stowell, 2008).
DNA VARIANT SCANNING
High resolution melting analysis of amplicons has also been used to scan for heterozygote sequence variants. Unlike other scanning methods, high resolution melting analysis provides a closed tube system that reduces the risk of contamination, decreases analysis time, and requires no sample processing or separations after PCR.
Gene scanning depends on the recognition of changes in the shape of the amplicon melting curve that result from heterozygous sequence alterations. The c-kit activating mutations in exons 9, 11, 13 and 17, associated with human malignancies, have been detected using high resolution melting analysis. Mutation screening for c-kit activating mutations was demonstrated for gastrointestinal stroma tumors (GIST) with follow-up confirmation by sequence analysis (Willmore et al., 2004). This application was further extended to include screening for platelet-derived growth factor receptor α (PDGFRA) gene activating mutations in exons 12 and 18, also associated with GISTs (Holden et al., 2007). High resolution melting analysis of PCR amplicons was shown to detect mutations in 91% of tumors diagnosed as GIST and all cases with abnormal melting curves were confirmed by sequence analysis.
Epidermal Growth Factor Receptor (EGFR) mutation status was assessed using high resolution melting analysis with fine needle aspirate samples as the DNA source (Smith et al., 2007). The identification of EGFR mutation status in non-small cell lung carcinoma patients was described for 4 EGFR exons. Deletions, missense mutations and silent mutations were detected with high resolution melting analysis for both heterozygous and homozygous cases.
Scanning with high resolution melting analysis was described for mutations in 9 exons of the cystic fibrosis transmembrane conductance regulator gene (CFTR) and the melting analysis method was compared to denaturing high-performance liquid chromatography (dHPLC) (Chou et al., 2005a). Melting analysis correctly identified 20/20 samples containing heterozyogous mutations, while dHPLC identified 19/20 samples. Homozygous mutations G542X and F508del could not be detected by dHPLC or by the analysis of melting curve shapes. However, when temperature shifting was omitted from the melting analysis and the absolute temperature was analyzed, the G542A, but not the F508del homozygote, could be identified.
In a more recent application to CFTR scanning (Montgomery et al., 2007) all 27 exons of CFTR, including all ACMG mutations were amplified. Analysis of 96 blood donor samples identified 22 different sequence variants. These variants included the F508del and 4 novel variants. In a blinded study with samples chosen to contain disease causing variants, 40/40 expected heterozygotes were detected, most of which produced unique melting patterns when analyzed using difference plots. To further distinguish heterozygotes, specific genotyping using unlabeled probes was described. If common heterozygote melting profiles are considered while analyzing unknown samples, false positive calls were greatly decreased. Some homozygous variants could be distinguished from wild type when analyzed with melting curve difference plots, however, there were some, including the F508del homozygote described above, that required unlabeled probes for genotyping.
Scanning for mutations associated with genetic conditions for which no common causative mutation has been identified can be an effective tool in clinical management. Mutations associated with primary carnitine deficiency in the OCTN2 carnitine transporter encoded by the SLC22A5 gene were evaluated using high resolution melting analysis (Dobrowolski et al., 2005). Both melting curves and difference plots were used to correctly identify all known patients with compound hererozygotes and to identify novel mutations in new patients. Of continued concern was the detection of homozygous variants. In this study, a second amplification was performed in which the patient DNA was mixed with normal control DNA. This allows formation of heteroduplexes that can be distinguished from wild type when the sample is homozygous. This process was also described previously (Liew et al., 2004; Palais et al., 2005).
High resolution melting analysis was used by Margarf et al. (Margraf et al., 2006a) to develop a screen for RET protooncogene mutations associated with multiple endocrine neoplasia type 2 (MEN2) syndromes. This assay was designed to amplify 6 RET exons to include all known pathogenic mutations in a total of 20 codons. The assay distinguished the presence of pathogenic mutations by comparing the derivative or difference plots for wild type and sample melting curves. Additionally, both Tm and the shape of the derivative melting curve could be used to determine the genotype for some samples and sequence variations that were not pathogenic could also be identified.
A interpretive challenge of heteroduplex scanning by high resolution melting analysis is that all heterozygotes are detected, including variants that are not of clinical interest. This challenge was addressed in a study that targeted 24 exons of the ACVRL1 and ENG genes implicated in hereditary hemorrhagic telangiectasia (HHT) (Vandersteen et al., 2007). By including the analysis of DNA from unaffected individuals, a reference panel of normal genetic variations was generated. Comparison of unknown heterozygous melting profiles with normal population profiles, allowed common benign variants to be eliminated. Further discrimination of disease causing variants was provided by evaluating data from both the normalized melting curves and the difference plots. Hierarchical clustering of melting curves aided analysis.
The use of high resolution melting to establish HLA identity between individuals as a screen or possible alternative to current molecular and serologic HLA typing has been described (Zhou et al., 2004). In this report, genotypic HLA matching of polymorphic exons of HLA-A was demonstrated using high resolution melting analysis of PCR products. Melting curves from single samples and 1:1 mixtures of samples were compared to determine if the samples matched. If the samples were identical, the melting curves were the same. If the samples were different, the melting curves varied for either the single samples or for the mixed samples.
Several reports have described the application of high resolution melting to the assessment of DNA methylation (Dahl and Guldberg, 2007). Bisulfite treatment converts unmethylated cytosines into uracil and subsequent PCR incorporates thymine into the product. Methylated cytosines are protected from bisulfite treatment and PCR incorporates cytosine into the product. This results in PCR product with a higher GC content when the target is methylated and allows resolution of DNA methylation status using high resolution melting analysis. White et al. described this application for a diagnostic screening method for Prader-Willi and Angelman syndromes (White et al., 2007) and Wojdacz and Dobrovic reported the determination of methylation status of the O6-methylguanine-DNA methyltransferase (MGMT) gene promoter (Wojdacz and Dobrovic, 2007). Snell et al. (Snell et al., 2008) recently described the use of high resolution melting as a scanning method for the detection of methylation in the BRCA1 gene promoter that may be associated with development of breast cancer.
High resolution melting analysis has also been reported for detecting epidermal growth factor receptor (EGFR) mutations associated with non-small cell lung cancer (Fukui et al., 2007; Nomoto et al., 2006; Takano et al., 2007), KRAS mutations (Krypuy et al., 2006), and TP53 mutations (Krypuy et al., 2007). Mutations in the BRCA1 and BRCA2 genes (Takano et al., 2008), the IGF1 gene (Palles et al., 2008), the factor VIII gene (Laurie et al., 2007), the ornithine transcarbamylase gene (Dobrowolski et al., 2007a), and the phenylalanine hydroxylase gene (Dobrowolski et al., 2007b) have also been described. Application of high resolution melting to scan RNA transcripts for editing sites in a plant model has been reported (Chateigner-Boutin and Small, 2007) as well as detection of IgH gene rearrangements using high resolution melting analysis (Uemura et al., 2007).
UNLABELED PROBES
High resolution DNA melting of PCR products is a simple method for SNP analysis and mutation scanning. However, the type of base change, the presence of a homozygous variant, or the presence of common non-disease causing variants, may complicate the interpretation of melting analysis. Additional information for interpretation in these cases can be obtained with the use of unlabeled probes. When unlabeled probes are included, the melting patterns of both the amplicon and the probe can be considered. Unlabeled probes are small oligonucleotides blocked at the 3’-end to prevent polymerase extension. They are included in the PCR master mix which also contains asymmetric ratios of primers. Asymmetric PCR is optimized so that sufficient signal is produced for both amplicon melting and unlabeled probe melting. A diagram of asymmetric PCR with unlabeled probes is presented in Figure 4.
Unlabeled probes can be designed to complement either the wild-type or variant sequence and the best signals are obtained with probes of 20–30 base pairs. The placement of mismatches in the central portion of the probe allows for more destabilization and better differentiation of mismatches than if the mismatches are at the ends of the probe. The 3’ end of the probe must be blocked to prevent extension of the probe itself. This is commonly accomplished with 3’-phosphorylation, however a recent report indicates that 3’-phosphorylation may be incomplete or unstable and can lead to aberrant melting profiles (Dames et al., 2007a). Improved probe blocking and stability was reported using amino-modified C6, inverted dT, or C3 spacer as blocking agents.
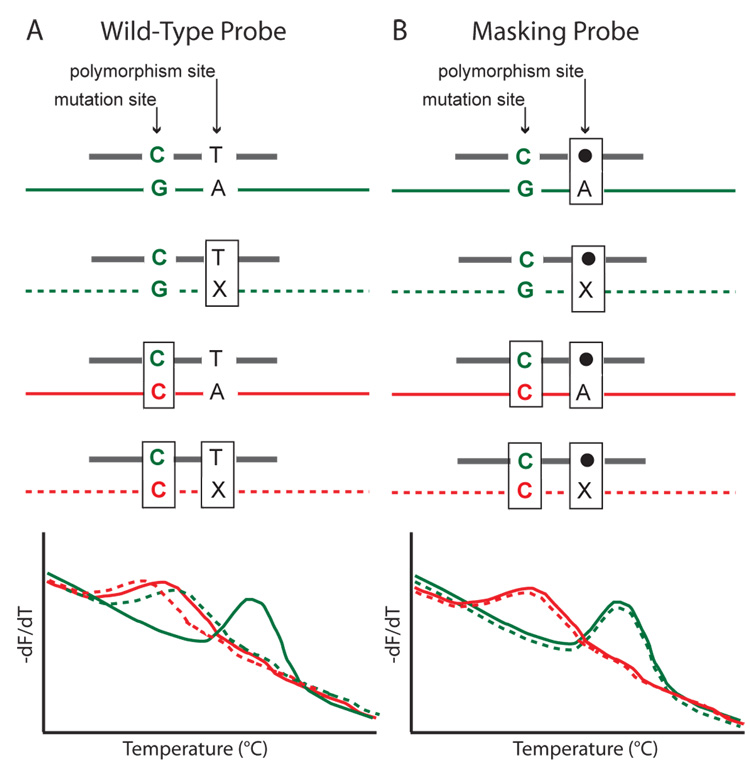
The analysis of probe melting profiles can be complicated if there are common, benign sequence variations near the targeted mutations of interest. Margraf et al. (Margraf et al., 2006b) described the use of “masking” probes in which additional mismatches were incorporated in the probe at the site of known sequence variations.. These mismatches included deletions, unmatched nucleotides, or universal bases in the probe at the site of the variant. An overview of the masking technique is presented in Figure 5.
Figure 5.
Demonstration of mismatched “masking” probes to conceal polymorphisms in probe melting analysis. Two single base variant loci are considered. The locus of interest is a “mutation site”, while the other is a benign variant at the “polymorphism site”. The four different possible genotypes (green/red, solid/dotted lines) are shown hybridized to two different probes (thick grey lines). The probes are either, A) perfectly-matched to wild type (wild-type probe) or B) mismatched at the polymorphism site (masking probe). The filled circles indicate the masking position in the probe that can be a deletion, a mismatched base, or a universal base. The X represents the polymorphic variant base. Mismatched bases are enclosed by rectangles. With a wild-type probe as shown in (A), it is difficult to distinguish the mutation from the polymorphism on a derivative plot (bottom left), because both result in one mismatch. However, the masking probe shown in (B) is always mismatched at the polymorphism site so that the presence (2 mismatches) or absence (1 mismatch) of the mutation is always clear on derivative plots (bottom right).
The use of masking probes allowed all targeted mutations to be discriminated from polymorphisms in the RET proto-oncogene, as long as the mutation and variation were at least one base pair apart. This technique was further applied to develop a two-stage RET genotyping assay that included mutation scanning and genotyping without the time and cost requirements of sequencing (Margraf et al., 2007).
Although not necessary with high resolution melting, improved discrimination of variants was addressed by including locked nucleic acids (LNAs) in the unlabeled probe (Chou et al., 2005b). In an unlabeled probe assay described for the Factor V Leiden (1691G>A) mutation, an increase in the ΔTm (difference in melting temperature) between the peaks generated by a heterozygous sample was achieved with the use of LNAs in the probe. An increased ΔTm was also noted with heterozygous samples with unexpected sequence variants near the targeted mutation allowing discrimination of the targeted mutation from the variants on standard real-time instruments.
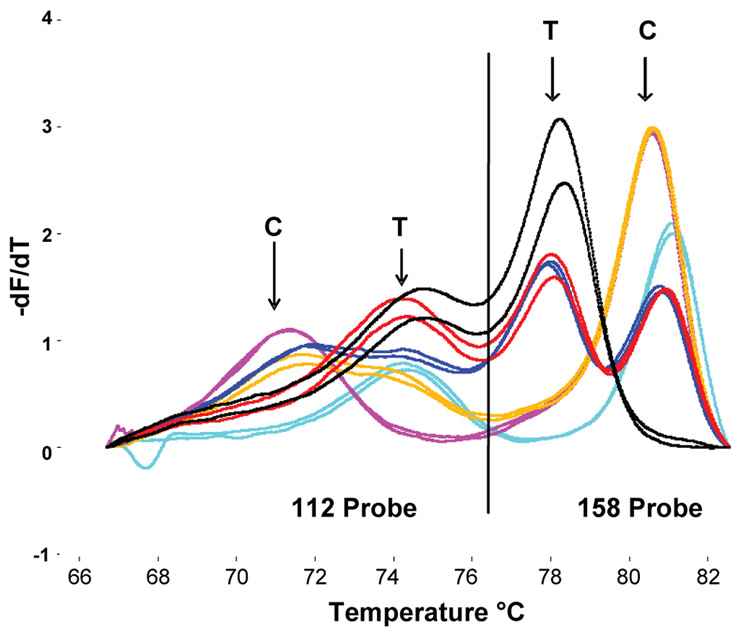
A multiplex combination of two unlabeled probes in a single reaction was used to identify 2 SNPs located 139 base pairs apart in the apolipoprotein E (APOE) gene (Poulson and Wittwer, 2007). A single 277 base pair PCR product was generated and the probes were designed to have well separated Tms. The SNP 112 probe had Tm peaks at 72 °C and 75 °C for the mismatched and matched targets, respectively. The SNP 158 probe had Tm peaks at 78 °C and 81.5 °C for the mismatched and matched targets, respectively. Representative melting peaks are shown in Figure 6. This report addressed the challenges of detecting SNPs in high-GC rich targets using DMSO and adding the probe after PCR while maintaining a closed-tube system.
Figure 6.
Derivative melting plots for APOE genotyping using two unlabeled probes added after asymmetric PCR. The vertical black line separates data generated by probes that target the 112 (left) and 158 (right) codons. All six common genotypes: e2/e2 (black), e2/e3 (red), e2/e4 (dark blue), e3/e3 (light blue), e3/e4 (orange), and e4/e4 (pink) are shown in duplicate. Reprinted with permission from BioTechniques 43, 87–91 (2007).
Simultaneous mutation scanning and genotyping using both amplicon and unlabeled probe melting analysis has been described for factor V Leiden and CFTR mutations (Zhou et al., 2005). For factor V, the PCR product melted at 78–82 °C and the probe melted at 58–68 °C. Identification of the genotype could be made using analysis of either the PCR product or the unlabeled probe melting transitions. The melting curves for PCR product generated from wild-type samples were more stable than product from homozygous mutants and heterozygous samples generated a skewed curve shape. Clearer identification of genotype could be obtained when the amplicon melting curves were plotted as difference curves using the wild-type sample as reference. Data from the melting curve in the region of the unlabeled probe was easiest to visualize as a derivative plot. The wild-type samples matched the unlabeled probe and were most stable. The melting peaks for homozygous mutant samples were shifted ~ 6°C lower, and heterozygous samples showed both peaks.
The same report also described the use of 2 probes to simultaneously scan and genotype mutations in the CFTR gene. In one assay, 2 probes were designed to detect 3 SNPs in two regions of exon 11, and in a second assay, 2 probes were designed to detect 3 SNPs and 2 deletions in exon 10. The exon 11 PCR product melted at 80–83 °C and the probe melting region was 56–74 °C. The exon 10 PCR product melted at 80–83 °C with a low Tm probe melting region at 56–67 °C and a high Tm probe melting region at 67–75 °C. In both assays, optimum discrimination was achieved by analyzing the difference plots of the PCR products referenced to the wild-type samples.
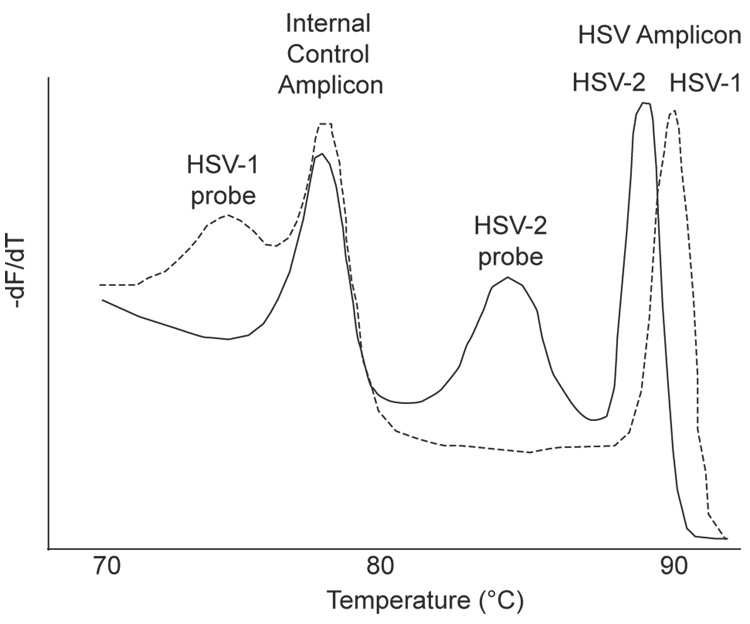
An infectious disease application for simultaneous detection and typing of herpes simplex virus (HSV) was described in which HSV-1 and HSV-2 were identified and differentiated based on analysis of the amplicon and probe melting profiles (Dames et al., 2007b). Universal primers to the HSV glycoprotein D gene were designed to amplify both HSV-1 and HSV-2 in a region where there were 8 known sequence variations between the 2 HSV types. These sequence differences resulted in a 1 °C difference in the Tms of the HSV-1 and HSV-2 PCR products. An unlabeled probe with complete homology to HSV-2 and 5 mismatches to HSV-1 was used that produced a probe Tm of 83.0 °C with HSV-2 and 71.6 °C with HSV-1. An internal control to monitor inhibitors and extraction efficiency was designed with an amplicon Tm of 76.0 °C, between the HSV-1 and HSV-2 Tms. Melting profiles were analyzed as derivative plots as shown in Figure 7. Of concern in infectious disease testing is whether the use of asymmetric PCR impacts the sensitivity of detection. In this study, sensitivity was stochastically limited, and the presence of as few as 10 copies of HSV DNA per reaction was sufficient for detection and genotyping by amplicon and unlabeled probe melting analysis.
Figure 7.
High resolution amplicon and unlabeled probe melting analysis of herpes simplex virus (HSV) type 1 and type 2. Dashed line is HSV-1 and solid line is HSV-2. Universal primers amplified both HSV types in a region with 8 sequence variations producing a 1 °C difference in the amplicon melting peaks. The unlabeled probe was homologous to HSV-2. Primers to an internal control produced an amplicon used to monitor inhibitors. Adapted and reprinted with permission from Clin Chem 53, 1847–1854 (2007).
High-resolution melting analysis is conceptually a simple tool for genotyping, variant scanning and sequence matching that does not require real-time PCR, labeled probes, processing or separations after PCR. Unlabeled probes can be used when increased specificity is needed and both scanning and genotyping can be performed simultaneously. However, the accuracy of the technique depends on appropriate instrumentation, saturation dyes, and software for analysis. This review updates prior reviews (Dujols et al., 2006; Erali et al., 2007; Reed et al., 2007) with a focus on implementation in the clinical laboratory.
Figure 3.
Derivative melting plots for the multiplex thrombophilia melting assay. (A): Four representative melting profiles containing examples of all of the genotypes for each locus. Melting plots are shown as a solid black line (F5 1691GG, MTHFR 1298AA and 677TT, and F2 20210AA), a solid gray line (F5 1691GG, MTHFR 1298AA and 677CT, and F2 20210GG), a dotted black line (F5 1691GA, MTHFR 1298AC and 677CC, and F2 20210GA), and a dotted gray line (F5 1691AA, MTHFR 1298CC and 677CC, and F2 20210GG). The derivative melting plot includes all 4 thrombophilia loci and 50-bp complementary oligonucleotide temperature-correction controls for high and low temperature. (B–E): Representative derivative melting plots for the F5 1691, MTHFR 1298, MTHFR 677, and F2 20210 loci, with homozygous wild-type, homozygous variant, and heterozygous genotypes indicated by solid black lines, dashed black lines, and dotted black lines, respectively. Reprinted with permission from Clin Chem 54, 108–115 (2008).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Chateigner-Boutin AL, Small I. A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 2007;35:e114. doi: 10.1093/nar/gkm640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JC, Huang CL, Lin CC, Chen CC, Chang YC, Chang SS, Tseng CP. Rapid detection and identification of clinically important bacteria by high-resolution melting analysis after broad-range ribosomal RNA real-time PCR. Clin Chem. 2006;52:1997–2004. doi: 10.1373/clinchem.2006.069286. [DOI] [PubMed] [Google Scholar]
- Chou LS, Lyon E, Wittwer CT. A comparison of high-resolution melting analysis with denaturing high-performance liquid chromatography for mutation scanning: cystic fibrosis transmembrane conductance regulator gene as a model. Am J Clin Pathol. 2005a;124:330–338. doi: 10.1309/BF3M-LJN8-J527-MWQY. [DOI] [PubMed] [Google Scholar]
- Chou LS, Meadows C, Wittwer CT, Lyon E. Unlabeled oligonucleotide probes modified with locked nucleic acids for improved mismatch discrimination in genotyping by melting analysis. Biotechniques. 2005b;39:644–646. doi: 10.2144/000112050. 648 passim. [DOI] [PubMed] [Google Scholar]
- Dahl C, Guldberg P. High-Resolution Melting for Accurate Assessment of DNA Methylation. Clin Chem. 2007;53:1877–1878. doi: 10.1373/clinchem.2007.094854. [DOI] [PubMed] [Google Scholar]
- Dames S, Margraf RL, Pattison DC, Wittwer CT, Voelkerding KV. Characterization of aberrant melting peaks in unlabeled probe assays. J Mol Diagn. 2007a;9:290–296. doi: 10.2353/jmoldx.2007.060139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dames S, Pattison DC, Bromley LK, Wittwer CT, Voelkerding KV. Unlabeled probes for the detection and typing of herpes simplex virus. Clin Chem. 2007b;53:1847–1854. doi: 10.1373/clinchem.2007.090761. [DOI] [PubMed] [Google Scholar]
- Dobrowolski SF, C EE, Caldovic L, Tuchman M. Streamlined assessment of gene variants by high resolution melt profiling utilizing the ornithine transcarbamylase gene as a model system. Hum Mutat. 2007a doi: 10.1002/humu.20558. [DOI] [PubMed] [Google Scholar]
- Dobrowolski SF, Ellingson C, Coyne T, Grey J, Martin R, Naylor EW, Koch R, Levy HL. Mutations in the phenylalanine hydroxylase gene identified in 95 patients with phenylketonuria using novel systems of mutation scanning and specific genotyping based upon thermal melt profiles. Molecular Genetics and Metabolism. 2007b;91:218–227. doi: 10.1016/j.ymgme.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Dobrowolski SF, McKinney JT, Amat di San Filippo C, Giak Sim K, Wilcken B, Longo N. Validation of dye-binding/high-resolution thermal denaturation for the identification of mutations in the SLC22A5 gene. Hum Mutat. 2005;25:306–313. doi: 10.1002/humu.20137. [DOI] [PubMed] [Google Scholar]
- Dujols V, Kusukawa N, McKinney JT, Dobrowolsky SF, Wittwer CT. High-resolution melting analysis for scanning and genotyping, Chapter 9. In: Tevfik D, editor. Real Time PCR. Abingdon: Taylor and Francis; 2006. pp. 155–169. [Google Scholar]
- Erali M, Palais R, Wittwer CT. SNP genotyping by unlabeled probe melting analysis. In: Seitz O, Marx A, editors. Molecular Beacons – Signalling Nucleic Acid Probes, Methods and Protocols. Vol. 429. Totowa, New Jersey: Humana Press; 2007. pp. 199–206. [Google Scholar]
- Erali M, Pounder JI, Woods GL, Petti CA, Wittwer CT. Multiplex single-color PCR with amplicon melting analysis for identification of Aspergillus species. Clin Chem. 2006;52:1443–1445. doi: 10.1373/clinchem.2006.068510. [DOI] [PubMed] [Google Scholar]
- Fortini D, Ciammaruconi A, De Santis R, Fasanella A, Battisti A, D'Amelio R, Lista F, Cassone A, Carattoli A. Optimization of high-resolution melting analysis for low-cost and rapid screening of allelic variants of Bacillus anthracis by multiple-locus variable-number tandem repeat analysis. Clin Chem. 2007;53:1377–1380. doi: 10.1373/clinchem.2007.085993. [DOI] [PubMed] [Google Scholar]
- Fukui T, Tsuta K, Furuta K, Watanabe SI, Asamura H, Ohe Y, Maeshima AM, Shibata T, Masuda N, Matsuno Y. Epidermal growth factor receptor mutation status and clinicopathological features of combined small cell carcinoma with adenocarcinoma of the lung. Cancer Sci. 2007;98:1714–1719. doi: 10.1111/j.1349-7006.2007.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R, Liew M, Meadows C, Lyon E, Wittwer CT. Distinguishing different DNA heterozygotes by high-resolution melting. Clin Chem. 2005;51:1295–1298. doi: 10.1373/clinchem.2005.051516. [DOI] [PubMed] [Google Scholar]
- Grievink H, Stowell KM. Identification of ryanodine receptor 1 single-nucleotide polymorphisms by high-resolution melting using the LightCycler 480 System. Analytical Biochemistry. 2008;374:396–404. doi: 10.1016/j.ab.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Gundry CN, Vandersteen JG, Reed GH, Pryor RJ, Chen J, Wittwer CT. Amplicon melting analysis with labeled primers: a closed-tube method for differentiating homozygotes and heterozygotes. Clin Chem. 2003;49:396–406. doi: 10.1373/49.3.396. [DOI] [PubMed] [Google Scholar]
- Herrmann MG, Durtschi JD, Bromley LK, Wittwer CT, Voelkerding KV. Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin Chem. 2006;52:494–503. doi: 10.1373/clinchem.2005.063438. [DOI] [PubMed] [Google Scholar]
- Herrmann MG, Durtschi JD, Bromley LK, Wittwer CT, Voelkerding KV. Instrument Comparison for Heterozygote Scanning of Single and Double Heterozygotes: A Correction and Extension of Herrmann et al., Clin Chem 2006;52:494–503. Clin Chem. 2007a;53:150–152. doi: 10.1373/clinchem.2006.081240. [DOI] [PubMed] [Google Scholar]
- Herrmann MG, Durtschi JD, Wittwer CT, Voelkerding KV. Expanded instrument comparison of amplicon DNA melting analysis for mutation scanning and genotyping. Clin Chem. 2007b;53:1544–1548. doi: 10.1373/clinchem.2007.088120. [DOI] [PubMed] [Google Scholar]
- Holden JA, Willmore-Payne C, Coppola D, Garrett CR, Layfield LJ. High-resolution melting amplicon analysis as a method to detect c-kit and platelet-derived growth factor receptor alpha activating mutations in gastrointestinal stromal tumors. Am J Clin Pathol. 2007;128:230–238. doi: 10.1309/7TEH56K6WWXENNQY. [DOI] [PubMed] [Google Scholar]
- Hung C-C, Lee C-N, Chang C-H, Jong Y-J, Chen C-P, Hsieh W-S, Su Y-N, Lin W-L. Genotyping of the G1138A mutation of the FGFR3 gene in patients with achondroplasia using high-resolution melting analysis. Clinical Biochemistry. 2008;41:162–166. doi: 10.1016/j.clinbiochem.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Jeffery N, Gasser RB, Steer PA, Noormohammadi AH. Classification of Mycoplasma synoviae strains using single-strand conformation polymorphism and high-resolution melting-curve analysis of the vlhA gene single-copy region. Microbiology. 2007;153:2679–2688. doi: 10.1099/mic.0.2006/005140-0. [DOI] [PubMed] [Google Scholar]
- Krypuy M, Ahmed AA, Etemadmoghadam D, Hyland SJ, Group AO, Brenton JD, Fox SB, Defazio A, Bowtell DD, Dobrovic A. High resolution melting for mutation scanning of TP53 exons 5–8. BMC Cancer. 2007;7:168. doi: 10.1186/1471-2407-7-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krypuy M, Newnham GM, Thomas DM, Conron M, Dobrovic A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer. 2006;6:295. doi: 10.1186/1471-2407-6-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie AD, Smith MP, George PM. Detection of Factor VIII Gene Mutations by High-Resolution Melting Analysis. Clin Chem. 2007 doi: 10.1373/clinchem.2007.093781. [DOI] [PubMed] [Google Scholar]
- Liew M, Nelson L, Margraf R, Mitchell S, Erali M, Mao R, Lyon E, Wittwer C. Genotyping of human platelet antigens 1 to 6 and 15 by high-resolution amplicon melting and conventional hybridization probes. J Mol Diagn. 2006;8:97–104. doi: 10.2353/jmoldx.2006.050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- Liew M, Seipp M, Durtschi J, Margraf RL, Dames S, Erali M, Voelkerding K, Wittwer C. Closed-tube SNP genotyping without labeled probes/a comparison between unlabeled probe and amplicon melting. Am J Clin Pathol. 2007;127:341–348. doi: 10.1309/N7RARXH3623AVKDV. [DOI] [PubMed] [Google Scholar]
- Lin J-H, Tseng C-P, Chen Y-J, Lin C-Y, Chang S-S, Wu H-S, Cheng J-C. Rapid differentiation of influenza A virus subtypes and genetic screening for virus variants by high-resolution melting analysis. J. Clin. Microbiol. 2008 doi: 10.1128/JCM.02015-07. JCM.02015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margraf RL, Mao R, Highsmith WE, Holtegaard LM, Wittwer CT. Mutation scanning of the RET protooncogene using high-resolution melting analysis. Clin Chem. 2006a;52:138–141. doi: 10.1373/clinchem.2005.052951. [DOI] [PubMed] [Google Scholar]
- Margraf RL, Mao R, Highsmith WE, Holtegaard LM, Wittwer CT. RET proto-oncogene genotyping using unlabeled probes, the masking technique, and amplicon high-resolution melting analysis. J Mol Diagn. 2007;9:184–196. doi: 10.2353/jmoldx.2007.060091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margraf RL, Mao R, Wittwer CT. Masking selected sequence variation by incorporating mismatches into melting analysis probes. Hum Mutat. 2006b;27:269–278. doi: 10.1002/humu.20290. [DOI] [PubMed] [Google Scholar]
- Montgomery J, Wittwer CT, Kent JO, Zhou L. Scanning the Cystic Fibrosis Transmembrane Conductance Regulator Gene Using High-Resolution DNA Melting Analysis. Clin Chem. 2007 doi: 10.1373/clinchem.2007.092361. [DOI] [PubMed] [Google Scholar]
- Nomoto K, Tsuta K, Takano T, Fukui T, Fukui T, Yokozawa K, Sakamoto H, Yoshida T, Maeshima AM, Shibata T, Furuta K, Ohe Y, Matsuno Y. Detection of EGFR mutations in archived cytologic specimens of non-small cell lung cancer using high-resolution melting analysis. Am J Clin Pathol. 2006;126:608–615. doi: 10.1309/N5PQNGW2QKMX09X7. [DOI] [PubMed] [Google Scholar]
- Odell ID, Cloud JL, Seipp M, Wittwer CT. Rapid species identification within the Mycobacterium chelonae-abscessus group by high-resolution melting analysis of hsp65 PCR products. Am J Clin Pathol. 2005;123:96–101. doi: 10.1309/wdr082x9ffjbqqgb. [DOI] [PubMed] [Google Scholar]
- Palais RA, Liew MA, Wittwer CT. Quantitative heteroduplex analysis for single nucleotide polymorphism genotyping. Anal Biochem. 2005;346:167–175. doi: 10.1016/j.ab.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Palles C, Johnson N, Coupland B, Taylor C, Carvajal J, Holly J, Fentiman IS, dos Santos Silva I, Ashworth A, Peto J, Fletcher O. Identification of genetic variants that influence circulating IGF1 levels: a targeted search strategy. Hum. Mol. Genet. 2008 doi: 10.1093/hmg/ddn034. ddn034. [DOI] [PubMed] [Google Scholar]
- Pornprasert S, Phusua A, Suanta S, Saetung R, Sanguansermsri T. Detection of alpha-thalassemia-1 Southeast Asian type using real-time gap-PCR with SYBR Green1 and high resolution melting analysis. Eur J Haematol. 2008 Feb;12 doi: 10.1111/j.1600-0609.2008.01055.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Poulson MD, Wittwer CT. Closed-tube genotyping of apolipoprotein E by isolated-probe PCR with multiple unlabeled probes and high-resolution DNA melting analysis. Biotechniques. 2007;43:87–91. doi: 10.2144/000112459. [DOI] [PubMed] [Google Scholar]
- Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;8:597–608. doi: 10.2217/14622416.8.6.597. [DOI] [PubMed] [Google Scholar]
- Seipp MT, Durtschi JD, Liew MA, Williams J, Damjanovich K, Pont-Kingdon G, Lyon E, Voelkerding KV, Wittwer CT. Unlabeled oligonucleotides as internal temperature controls for genotyping by amplicon melting. J Mol Diagn. 2007a;9:284–289. doi: 10.2353/jmoldx.2007.060136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipp MT, Pattison D, Durtschi JD, Jama M, Voelkerding KV, Wittwer CT. Quadruplex Genotyping of F5, F2, and MTHFR Variants in a Single Closed Tube by High-Resolution Amplicon Melting. Clin Chem. 2007b doi: 10.1373/clinchem.2007.097121. [DOI] [PubMed] [Google Scholar]
- Seipp MT, Pattison D, Durtschi JD, Jama M, Voelkerding KV, Wittwer CT. Quadruplex Genotyping of F5, F2, and MTHFR Variants in a Single Closed Tube by High-Resolution Amplicon Melting. Clin Chem. 2008;54:108–115. doi: 10.1373/clinchem.2007.097121. [DOI] [PubMed] [Google Scholar]
- Smith GD, Chadwick BE, Willmore-Payne C, Bentz JS. Detection of EGFR gene mutations in cytology specimens from patients with non-small cell lung cancer utilizing high-resolution melting amplicon analysis. J Clin Pathol. 2007 doi: 10.1136/jcp.2007.051425. [DOI] [PubMed] [Google Scholar]
- Snell C, Krypuy M, Wong E, kConFab i, Loughrey M, Dobrovic A. BRCA1 promoter methylation in peripheral blood DNA of mutation negative familial breast cancer patients with a BRCA1 tumour phenotype. Breast Cancer Research. 2008;10:R12. doi: 10.1186/bcr1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AJ, Inman-Bamber J, Giffard PM, Huygens F. High-Resolution Melting Analysis of the spa Repeat Region of Staphylococcus aureus. Clin Chem. 2008;54:432–436. doi: 10.1373/clinchem.2007.093658. [DOI] [PubMed] [Google Scholar]
- Takano EA, Mitchell G, Fox SB, Dobrovic A. Rapid detection of carriers with BRCA1 and BRCA2 mutations using high resolution melting analysis. BMC Cancer. 2008;8:59. doi: 10.1186/1471-2407-8-59. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Ohe Y, Tsuta K, Fukui T, Sakamoto H, Yoshida T, Tateishi U, Nokihara H, Yamamoto N, Sekine I, Kunitoh H, Matsuno Y, Furuta K, Tamura T. Epidermal growth factor receptor mutation detection using high-resolution melting analysis predicts outcomes in patients with advanced non small cell lung cancer treated with gefitinib. Clin Cancer Res. 2007;13:5385–5390. doi: 10.1158/1078-0432.CCR-07-0627. [DOI] [PubMed] [Google Scholar]
- Uemura A, Mori S, Sugahara K, Akamatsu N, Tsuruda K, Tsukasaki K, Hirakata Y, Atogami S, Hasegawa H, Yamada Y, Kamihira S. Rapid and high-resolution detection of IgH gene rearrangements using PCR and melting curve analysis. Int J Lab Hematol. 2007;29:200–207. doi: 10.1111/j.1751-553X.2006.00832.x. [DOI] [PubMed] [Google Scholar]
- Vandersteen JG, Bayrak-Toydemir P, Palais RA, Wittwer CT. Identifying common genetic variants by high-resolution melting. Clin Chem. 2007;53:1191–1198. doi: 10.1373/clinchem.2007.085407. [DOI] [PubMed] [Google Scholar]
- White HE, Hall VJ, Cross NCP. Methylation-Sensitive High-Resolution Melting-Curve Analysis of the SNRPN Gene as a Diagnostic Screen for Prader-Willi and Angelman Syndromes. Clin Chem. 2007;53:1960–1962. doi: 10.1373/clinchem.2007.093351. [DOI] [PubMed] [Google Scholar]
- Willmore C, Holden JA, Zhou L, Tripp S, Wittwer CT, Layfield LJ. Detection of c-kit-activating mutations in gastrointestinal stromal tumors by high-resolution amplicon melting analysis. Am J Clin Pathol. 2004;122:206–216. doi: 10.1309/4E6U-YBY6-2N2F-CA6N. [DOI] [PubMed] [Google Scholar]
- Wittwer CT, Kusukawa N. Nucleic Acid Techniques, Chapter 37. In: Burtis C, Ashwood ER, Bruns DE, editors. Tietz Textbook of Clinical Chemistry and Molecular Diagnostics. Philadelphia: Elsevier Science; 2005. pp. 1407–1449. [Google Scholar]
- Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007;35:e41. doi: 10.1093/nar/gkm013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Vandersteen J, Wang L, Fuller T, Taylor M, Palais B, Wittwer CT. High-resolution DNA melting curve analysis to establish HLA genotypic identity. Tissue Antigens. 2004;64:156–164. doi: 10.1111/j.1399-0039.2004.00248.x. [DOI] [PubMed] [Google Scholar]
- Zhou L, Wang L, Palais R, Pryor R, Wittwer CT. High-resolution DNA melting analysis for simultaneous mutation scanning and genotyping in solution. Clin Chem. 2005;51:1770–1777. doi: 10.1373/clinchem.2005.054924. [DOI] [PubMed] [Google Scholar]