Preface
The ubiquitin-proteasome system degrades an enormous variety of proteins, which are targeted by specific degradation signals (degrons). Besides the degradation of regulatory proteins, virtually every protein suffers from sporadic biosynthetic errors or misfolding, and cells can recognize such aberrant proteins and rapidly degrade them. Structural and functional data on a handful of degrons allows some generalizations about their mechanism of action. We focus on different strategies of degron recognition by the ubiquitin system, and contrast regulatory degrons subject to signalling-dependent modification and those controlled by protein folding or assembly, as frequently occurs during protein quality control.
Introduction
Intracellular protein degradation has been studied for more than half a century, and it became clear early on that such degradation is highly selective, with individual protein half-lives ranging from minutes to years (for reviews of the early literature, see refs. 1-2). Moreover, much of this degradation was found to be energy-dependent despite the exergonic nature of peptide-bond cleavage. This energy dependence derives from the dual requirements of high substrate specificity and substrate protein unfolding to make the polypeptide backbone fully accessible for proteolytic cleavage. The vast majority of regulated protein degradation in eukaryotes is executed by the ubiquitin-proteasome system 3-5. Polyubiquitin tagging of substrates by specific enzymes provides the major source of selectivity in the system (Box 1), whereas the 26S proteasome complex performs the protein unfolding necessary for processive cleavage of the tagged proteins into short peptides (Box 2). In addition, ubiquitin ligation can function independently of the proteasome by directing certain -usually membrane- proteins to the lysosome/vacuole for proteolysis. Conversely, proteasomes can degrade some proteins without their prior modification by ubiquitin.
Box 1. The ubiquitin-conjugation machinery.
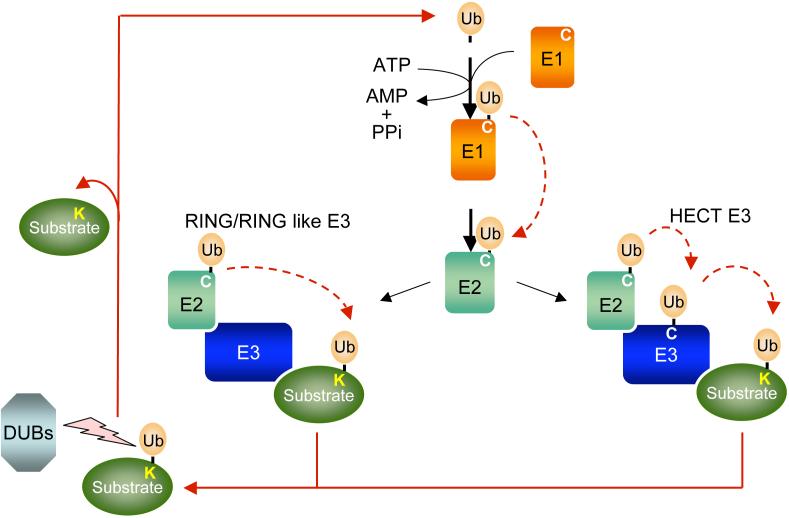
Substrate proteins destined for elimination are initially attached to polymers of the highly conserved ubiquitin (Ub) protein (see figure). This covalent modification of the substrate targets the conjugated protein to a multicatalytic protease complex, the 26S proteasome 5. The ubiquitin attachment site(s) in substrate proteins is commonly a lysine (K) side chain. A well-defined series of enzymes orchestrates polyubiquitin attachment to proteins. Ubiquitin is first activated in an ATP-consuming reaction by an E1 ubiquitin-activating enzyme, to which it becomes attached by a high-energy thioester bond. Subsequently, the activated ubiquitin is transferred to the active site cysteine (C) of a second protein, an E2 ubiquitin-conjugating enzyme. With the aid of a third enzyme, called E3 or ubiquitin-protein ligase, the E2 catalyzes transfer of (poly)ubiquitin onto the protein destined for degradation.
The E3 is the most important factor in determining the specificity of substrate ubiquitination. There are two major classes of mechanistically distinct E3s, characterized by the RING (or RING-like) and HECT domains. Both types of E3s are alike in their ability to establish selective substrate binding. The RING finger uses cysteine and histidine residues to coordinate a pair of zinc ions in a characteristic arrangement. A smaller set of E3s bear a domain called the U box, which is a degenerate version of the RING-finger that achieves the same general fold as the RING finger without coordinating any metal ions 143. RING and RING-like E3s bind to both E2 and substrate, and catalyze the transfer of ubiquitin directly from the E2. Unlike RING and U-box E3s, the HECT E3s have a more direct catalytic role in substrate ubiquitylation. The activated ubiquitin of the ubiquitin-E2 thioester is transferred to a conserved cysteine residue in the HECT domain of the E3 before finally being transferred to a substrate.
Ubiquitylation is reversed by deubiquitylating enzymes (DUBs) that remove ubiquitin from proteins and disassemble multi-ubiquitin chains. DUBs provide additional regulatory control prior to protein degradation, and they are also fundamental for maintaining a sufficient pool of free ubiquitin molecules in the cell.
Box 2. Substrate targeting to the 26S proteasome.
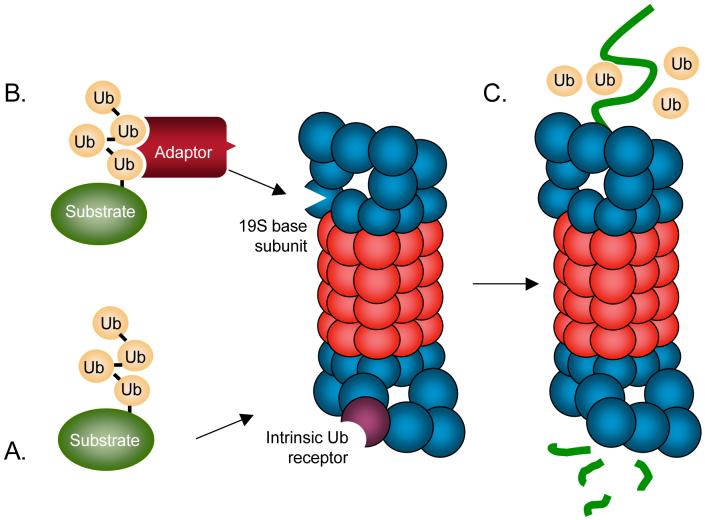
Once modified by a polyubiquitin chain of at least four ubiquitins (Ub), the substrate protein is able to bind either directly to intrinsic Ub receptors in the 19S regulatory complex of the 26S proteasome (see figure panel a) or to adaptor proteins that bear both polyubiquitin-binding and proteasome-binding domains (see figure panel b) 144. Exactly why certain polyubiquitin-modified substrates must be shuttled to the proteasome by adaptor proteins and others can associate directly with polyubiquitin-binding subunits in the regulatory complex of the proteasome is not fully understood. Binding of the substrate protein to the proteasome is followed by protein unfolding by the half-dozen ATPases that encircle the pore of the proteasome catalytic core, removal of the polyubiquitin chain by proteasome-associated deubiquitylating enzymes (DUBs), and translocation of the unfolded protein into the central proteolytic chamber, where it is cleaved into short peptides (see figure panel c).
A fundamental question about intracellular proteolysis is how specific proteins are recognized by the proteolytic machinery, resulting in proteins being degraded only under specific conditions with highly characteristic degradation rates. Early work had suggested that global structural features determine the metabolic stability of individual proteins. For instance, mutant proteins or proteins that had incorporated amino acid analogues during their synthesis were found to have shorter half-lives in vivo than their wild-type counterparts 6, 7. Moreover, protein degradation rates appeared to correlate with gross protein physicochemical properties such as molecular mass or isoelectric point 8, 9. However, later analyses revealed that correlations with gross protein properties did not generally hold true, and though abnormal proteins were frequently short-lived, this need not reflect a global change in their structure.
As will be discussed in detail below, most short-lived proteins are distinguished by localized structure determinants (‘signals’) that target them to the ubiquitin ligase machinery or to the proteasome (or lysosome in some cases). A degradation signal or ‘degron’ 10, is usually defined as a minimal element within a protein that is sufficient for recognition and degradation by a proteolytic apparatus. An important property of degrons is that they are transferable. That is, genetically engineered attachment of such sequences confers metabolic instability (a short half-life) on otherwise long-lived proteins 3. Degrons can be defined for distinct proteolytic pathways, but we will confine this review to the description of degrons that target proteins to the ubiquitin-proteasome pathway. At this point, generalizations in this field are limited to some degree by the sheer diversity of substrates of the system. For example, many regulatory proteins are degraded in a temporally and spatially specific manner. These proteins are often tightly controlled by other post-translational modifications that are dependent on cell-signalling events. On the other hand, quality control of newly synthesized proteins and removal of misfolded proteins is under a very different set of constraints. The recognition and destruction of such aberrant proteins is expected to depend on their folding or assembly state. If folding or assembly goes awry, proteins would be expected to expose normally cryptic degrons that exist in many different proteins.
In this review, we will focus on how different physiological requirements for the degradation of specific proteins dictate the design of different degrons. Distinct determinants comprise a degron, and they have different roles in the degradation pathway. Specifically, we consider primary recognition determinants as those sequences or structures within the degron that bind directly to the E3-E2 ubiquitin-ligase complex or its ancillary factors. Another determinant in ubiquitin-dependent degrons is the presence of an appropriate acceptor site(s) for attachment of the polyubiquitin chain, such as a lysine residue. The (polyubiquitin-modified) substrate must also be able to interact with the proteasome or shuttling factors that deliver it to the proteasome. Finally, the degron must be situated within the substrate such that the proteasome can initiate its unfolding and translocate it into the proteasome core. Our emphasis here will be on primary recognition determinants.
Not all regulation of protein ubiquitylation occurs through substrate changes that activate a degron. Although we do not have space to cover this area, E3 and E2 enzymes can themselves be regulated by post-translational modification, such as the phosphorylation of the anaphase-promoting complex E3 11, or by binding to small molecules. For instance, dipeptide binding can allosterically modulate the N-recognin E3 (see next section) 12, and the growth-regulating plant indole auxin binds to a specific E3 ligase and forms part of an enlarged protein-binding interface that allows high-affinity interaction with specific protein substrates 13.
N-degrons and the N-end rule pathway
The notion that covalent ubiquitin conjugation commits proteins for degradation led the discoverers of ubiquitin-mediated proteolysis to propose that substrate selection takes place mainly at the stage of ubiquitin ligation 14, 15. By adding a variety of proteins to a rabbit reticulocyte lysate, Hershko and colleagues noted an apparent correlation between the presence of a free α-amino group in the proteins and their ubiquitin-dependent degradation 16. They subsequently isolated a 180 kD protein with E3 ubiquitin ligase activity that appeared to have higher affinity for proteins with a free α-amino group than those with a blocked N terminus 15.
This particular E3 distinguishes proteins not only by a free N-terminal α-amino group but also the side chain of the N-terminal residue. Varshavsky and co-workers systematically changed the N-terminal residue in an otherwise identical series of Escherichia coli β-galactosidase test substrates and expressed them in yeast, where they displayed a remarkable range of degradation rates 17. Half-lives ranged from a few minutes to greater than 20 hours. Thus, an E3 is able to bind protein substrates with very high selectivity, in this case being able to distinguish substrates by recognizing a specific residue at the N terminus of a protein 18. This degradation pathway, termed the ‘N-end rule pathway,’ states that the half-life of a protein is determined by the nature of its N-terminal residue. Peptide sequences within the N-terminal region of the substrate that are sufficient for ubiquitin-dependent turnover constitute the ‘N-degron.’ (Figure 1A).
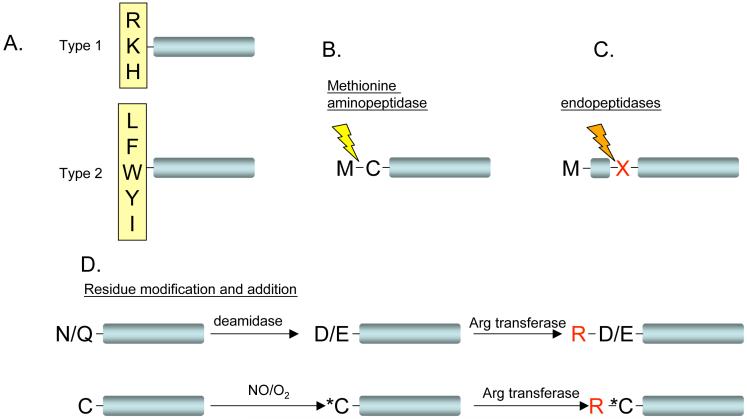
Figure 1. Mechanisms for activation of N-end rule pathway substrates.
a| Type 1 (basic) and type 2 (hydrophobic) destabilizing residues of the N-end rule pathway are shown in a single letter code. b| Cleavage by endopeptidases can lead to the positioning of a destabilizing residue (X) at the N terminus of the truncated protein. c| Cleavage between Met and Cys by methionine aminopeptidase (indicated by a lightning bolt) can lead to protein destabilization when followed by addition of Arg, a type 1 destabilizing residue (see panel d). d| Modification of Asn to Asp (or Gln to Glu) by specific deamidases can lead to the addition of Arg by arginyltransferase (upper panel). Oxidation of the N-terminal Cys residue (marked by Asterisk) can similarly lead to protein arginylation and degradation (lower panel).
The prokaryotic N-end rule pathway
Interestingly, prokaryotes also have an N-end rule pathway, even though prokaryotes lack a ubiquitin-conjugation system (reviewed in ref. 19). Whereas the 26S proteasome is the protease for N-end rule substrates in eukaryotes, the ATP-dependent Clp protease complex (ClpAP) is the principal protease that degrades such substrates in bacteria. Several findings suggest that both proteolytic systems share common principles of recognition for at least a subset of “destabilizing” N-terminal residues. In eukaryotes, a RING-class ubiquitin ligase, now called E3α or N-recognin, mediates ubiquitin transfer from the E2 onto specific lysines within the N-end rule substrate, allowing substrate delivery to the 26S proteasome. In bacteria, rather than an E3 ligase recognizing the N-terminal residue, an adaptor protein called ClpS binds to the N-terminal region of substrates and also to specific domains of the ClpA ATPase subunits of the ClpAP protease. Delivery to the ClpA hexameric ring leads to substrate unfolding and translocation into the ClpP serine protease core (reviewed in ref. 19). Limited homology was found between one of the substrate binding sites of eukaryotic N-recognin (the “type 2” site) and a conserved sequence at the surface of ClpS 20. This region of homology is enriched in acidic and hydrophobic residues, providing a potential complementary surface for interaction with N-degrons. Erbse and colleagues found that strong binding was favoured by a net positive charge in the substrate peptide, which is complementary to the postulated ClpS binding motif 21. Notably, acidic residues in a conserved position also exist in the eukaryotic type-2 substrate-binding site of N-recognin homologues. The function of ClpS in bacterial N-end rule degradation and its (limited) homology to eukaryotic N-recognin suggest that, despite obvious differences in the downstream proteolytic systems, recognition of N-end rule substrates shares certain features between bacteria and eukaryotes.
The eukaryotic N-end rule pathway
In Saccharomyces cerevisiae, where the N-end rule was first described, the physiological significance of the pathway was at first difficult to discern because mutants lacking N-recognin (Ubr1) have a phenotype indistinguishable from wild-type cells under most conditions. The first identified yeast substrate with a physiologically important N-degron was a proteolytic fragment of Scc1, a subunit of the cohesin complex that helps maintain sister chromatid cohesion during mitosis and meiosis 22. At the metaphase-to-anaphase transition, Scc1 is cleaved by an endoprotease called separase, allowing daughter chromosomes to separate. The resulting C-terminal fragment of Scc1 bears an arginine at its N terminus. This protein fragment is recognized by Ubr1 and targeted for proteasomal degradation. Failure to degrade the C-terminal Scc1 fragment results in increased rates of chromosome loss, and overexpression of the stabilized fragment is lethal. Therefore, an internal cleavage of a protein can provide the initiating event for protein degradation by the N-end rule pathway (Figure 1B).
Endoproteolytic cleavage is just one of many N-terminal modifications that can channel a protein to the N-end rule pathway. For instance, newly synthesized proteins contain N-terminal methionine, which is a stabilizing residue. Therefore, an N-terminal degron of the N-end rule pathway can be produced only from a pre-N-degron by co translational removal of the methionine (Figure 1C). Furthermore, mature proteins initiating with an acidic residue or (in mammalian cells) a cysteine are first modified with an arginine by arginyl-tRNA-protein transferase (Ate1) before they can be bound by N-recognin and ubiquitylated (Figure 1D) 23, 24. The regulation of short-lived proteins with an N-terminal cysteine is particularly intricate. In this case, the cysteine must first be oxidized to a sulfinic or sulfonic acid form through the action of molecular oxygen and nitric oxide (NO) before it is enzymatically arginylated 25-27. Degradation of several Regulator of G-protein Signalling (RGS) proteins depends on this NO-dependent mechanism. Recently, it has been shown that Ate1 is capable of arginylating many proteins, but this does not necessarily create an N-degron 28. This implies that multiple determinants are necessary for creating an N-degron, not just a destabilizing N-terminus (see below).
In mammals, functions for several of the enzymes that contribute to the N-end rule pathway have been determined by mouse knockout analyses and human disease gene mapping (reviewed in ref. 29). Beside E3α, the mammalian genome encodes at least five more UBR box-containing proteins with specific signatures unique to E3 ubiquitin ligases 30. Several of these putative E3s, along with enzymes known to modify N-terminal residues for potential targeting by the N-end rule pathway, have been shown to contribute to cardiovascular development, spermatogenesis and viral defences, among other roles 31-33.
Phosphodegron recognition by SCFCdc4
F-Box proteins (FBPs) are the substrate-specificity subunits of the multisubunit SKP1-CUL1-F-box (SCF) family of E3 ligases (Box 3). Genetic and structural approaches have provided many insights into the composition of degrons of SCF substrates and their recognition 34-40. Degradation of most SCF substrates requires phosphorylation on specific serine or threonine residue(s), and the short phosphorylated peptide motif can then bind to a specific FBP (Figure 2).
Box 3. The SCF complex.
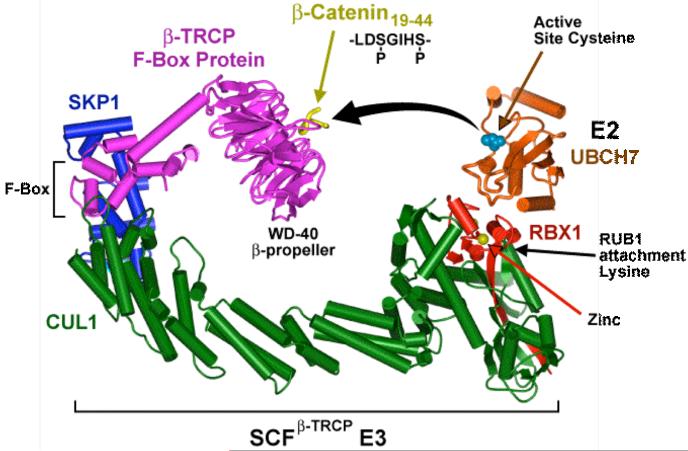
Multisubunit ubiquitin ligases that are assembled on a type of scaffolding subunit called a cullin represent the largest subclass of E3s (for review, see 145). The most thoroughly characterized cullin-dependent ligases belong to the SCF (Skp1-Cul1-F-box protein) family of ligases, which frequently target proteins bearing a specific phosphorylated sequence element referred to as a phosphodegron. In the SCF ligases, the Cul1 cullin subunit functions as a molecular scaffold that interacts simultaneously with S-phase-kinase-associated protein 1 (Skp1) and with the RING-box 1 protein Rbx1 (see figure panel a). Skp1 functions as an adaptor between Cul1 and a variety of F-box proteins (FBPs). The F-box element binds directly to Skp1 whereas the C-terminal domain of the FBP supplies a substrate-recognition platform, most often composed of WD40- or leucine-rich repeats 36, 146, 147. SCFFBP complexes participate in cell cycle regulation as well as other processes such transcription, cellular signalling, and endoplasmic reticulum-associated protein degradation (ERAD). A model for the orientation of the SCFβTrCP complex together with an E2 enzyme and the phosphodegron of the substrate β-catenin (panel b) 36. The model places the E2 active site cysteine ∼50 Å away from the β-catenin peptide, suggesting that the active form of SCFβTrCP either has a distinct conformation or functions as part of a larger multimeric unit.
Figure 2. Structure of CycE phosphodegron bound to the F-box protein Fbw7.
a| Overall architecture of the Skp1-Fbw7-CycE C-terminal phosphodegron (CycEdegC) complex, with the secondary structure elements of Skp1, F-box, and linker domains labelled. b| CycEdegC binds across the narrow face of the Fbw7 β-propeller structure. The two phosphorylated residues Trp380 and Ser384 are shown. The eight Fbw7 blades and the strands for one blade are labelled by numbers and letters, respectively.
The best-characterized of such phosphodegrons are the ones involved in the ordered elimination of specific cyclins and cyclin-dependent kinase (CDK) inhibitors by the ubiquitin system (for reviews, see refs. 41, 42). In yeast, commitment to cell division requires a threshold level of G1 cyclins (Cln1/2/3), which gradually accumulate during G1 and lead to activation of the Cdc28 kinase (Cdk1) in late G1 phase. The primary function of Cln-Cdc28 kinase is to phosphorylate Sic1, an inhibitor of cyclin B-regulated kinase, thereby targeting Sic1 for degradation and enabling entry into S phase 43. Phosphorylated Sic1 is specifically recognized by the FBP Cdc4, which is part of the SCFCdc4 ubiquitin ligase. In the mammalian cell cycle, similar SCF complexes target phosphorylated forms of cyclin E and the CDK inhibitor p27Kip1 44, 45. Abnormal persistence of the latter proteins causes cell-cycle misregulation and genome instability 46, 47.
The mechanisms by which phosphorylation drives substrate binding to the SCF and subsequent ubiquitin conjugation are under active investigation. Studies of the phosphorylation of Sic1 and its binding to SCFCdc4 provide a good example of the complexity of this coordinated control 34, 37, 40. Sic1 is phosphorylated at nine different sites, and phosphorylation of a minimum of six of them enhances its interaction with the WD40-repeat domain of Cdc4. Importantly, the nine Sic1 phosphorylation sites share very little similarity among themselves or with other degrons of SCFCdc4 substrates 34. Testing the optimal binding affinity of synthetic phosphopeptides to Cdc4 yielded a consensus binding site for the Cdc4 phosphodegron (CPD; ref. 34). Surprisingly, none of the Sic1 CPDs exactly matches the consensus sequence. One rationalization for the multiple weak CPD sites in Sic1 is that they allow an ultrasensitive response to Cln-Cdc28 kinase activity that both builds in a time delay for Sic1 turnover and prevents its premature degradation from small fluctuations in phosphorylation status. Sic1 turnover and entry into S phase will then require a relatively high level of active kinase, which in turn depends on the appropriate nutritional signals to the cell 34, 48.
Analysis of the crystal structure of Cdc4 in complex with a nine-residue optimal phosphodegron yielded insight into the mechanistic basis for Cdc4 binding preferences 37. The crystallographic data showed that binding of the CPD to Cdc4 is governed by three principal features of the interface: 1) Peptide phosphorylation is absolutely necessary for binding to the WD40 domain of Cdc4 because of favourable electrostatic interactions and hydrogen bonding with the phosphate group. 2) A deep hydrophobic pocket on Cdc4 selects for a conserved hydrophobic residue in the phosphodegron. 3) Charged and polar residues on Cdc4 select against basic residues that are present in suboptimal phosphodegrons. This latter feature could explain why multiple phosphorylation events are required for Sic1-Cdc4 interaction 49.
In contrast to the cooperative binding of multiple, single phosphorylated sites in Sic1 to Cdc4, crystal structures of two related SCF complexes, SCFβTrCP1 and SCFFbw7 bound to their respective doubly phosphorylated peptide substrates revealed a binding mode where both of the phosphorylated residues in the peptide bind to two phosphate-binding sites on the WD40 domain of the FBPs 36, 40 (Figure 2). The two phosphate-binding sites in Fbw7 are conserved in the Cdc4 structure 37, suggesting that Cdc4, similar to Fbw7, can bind a doubly phosphorylated peptide with greater affinity than a singly phosphorylated one. Indeed, doubly phosphorylated Sic1 CPDs showed up to 19-fold higher affinity for Cdc4 than did singly phosphorylated versions 40. According to the emerging model, Sic1 contains three separate CPDs, each with two essential phosphorylation sites. Each of these degrons is sufficient for tight binding to Cdc4 in vitro, with similar affinity to those of the optimal CPD. These findings have implications for the proposed cooperative model of Cdc4-Sic1 interaction since it does not support a simple threshold mechanism 34.
An oxygen-dependent degron
Another interesting example of protein ubiquitylation regulated by signal-dependent post-translational substrate modification occurs with hypoxia-inducible factor-1 (HIF-1), a heterodimeric transcriptional complex that mediates the transcriptional response to oxygen availability 50. Under hypoxic conditions, dimerization of HIF-1 activates the transcription of genes involved in the adaptation of cells to low oxygen tension, such as those encoding vascular endothelial growth factor and erythropoietin, which are important for formation of new blood vessels and red blood cells. The HIF-1 complex is stable under hypoxia, but the HIF-1α subunit is rapidly degraded by the proteasome under normoxic conditions. This proteolytic regulation depends on a distinct cullin-RING ubiquitin ligase composed of von Hippel-Lindau protein (VHL), elongins B and C, the cullin Cul2A and Rbx1 51, 52. VHL is the substrate recognition subunit in the complex, and it binds to HIF-1α via an oxygen-dependent degron (ODD) 53 54. In well-oxygenated cells, a HIF-1α-specific prolyl hydroxylase uses molecular oxygen to hydroxylate one or two specific prolyl residues 54, 55.
Crystal structures of the hydroxylated degron of human HIF-1α bound to VHL revealed a crucial role for the hydroxyl group on proline 564 35, 56. The ODD binds VHL primarily via a six-residue segment in the ODD that is centred on hydroxyproline (Hyp) 564. This residue is nearly entirely buried in a hydrophobic pocket of the VHL binding surface, forming hydrogen and van der Waals interactions with surrounding VHL residues. The backbone of HIF-1α in the vicinity of Hyp564 is held in place through an extensive network of hydrogen bonds with VHL. The affinity of VHL for the hydroxylated HIF-1α peptide is ∼1,000-fold higher than for the nonhydroxylated form 56. In patients with von Hippel Lindau disease, an hereditary cancer-susceptibility syndrome, each of the five VHL residues that make direct contact with Hyp564 is subject to missense mutations 57. Thus, high affinity binding of Hyp564 to VHL is maintained by multiple interactions and is essential for abrogating HIF-1α-dependent tumorigenesis.
In general, post-translational modifications allow protein ubiquitylation rates to be coupled to specific stimuli. Such substrate alterations also displace the major point of regulation to steps prior to ubiquitin attachment. In addition to the examples of N-terminal residue processing, phosphorylation and prolyl hydroxylation discussed above, protein glycosylation (see below) and even prior modification by other ubiquitin-like proteins such as the small ubiquitin-related modifier (SUMO) can modulate the probability that a specific protein will be ubiquitylated and degraded 58.
Degrons in the ER
All the degrons discussed so far can be classified as conditional signals for which post-translational modifications at specific sites are necessary to create a functional degron. The modified residues, along with neighbouring regions in the polypeptide, comprise the basic structural elements for recognition by a specific ubiquitin-ligase complex. However, not all substrate proteins are recognized through prior covalent modifications. Structural features that are revealed when a protein assumes a specific conformation or assembly state can serve as recognition elements in a wide range of degrons. Polypeptides that fail to assume their native tertiary or quaternary structures, collectively referred to as protein quality control (PQC) substrates, are often subject to this mode of substrate recognition (see refs. 59, 60). A major site for ubiquitin-dependent PQC is the endoplasmic reticulum (ER), where most secretory and integral membrane proteins are folded and assembled before being trafficked to their site of action. Proteins unable to fold or assemble properly usually never make it from the ER to the Golgi but instead are extracted back across the bilayer into the cytosol, ubiquitylated and degraded by the cytosolic proteasome (reviewed in refs. 61-64). Components of this ER-associated degradation (ERAD) system can also recognize native proteins undergoing transient or induced structural changes, allowing regulation of the levels of specific ER-resident proteins.
What are the crucial features that characterize degrons of misfolded or misassembled proteins? Much still needs to be learned in this area, and there are no degrons (or degron-E3 complexes) of this type that have been visualized by high-resolution structural methods (but see next section). However, some hints have emerged, including, surprisingly, from studies of certain naturally short-lived regulatory proteins.
The hydrophobic degron of yeast Matα2
In budding yeast, cell identity is controlled by the MAT mating-type locus. This locus can carry two distinct DNA sequences, MATa and MATα, which direct expression of different transcriptional regulators. In certain haploid strains, rapid switching between these two states occurs, with the associated phenotypic changes manifested within a single cell division. For such a rapid change in cellular phenotype, the proteins encoded by the discarded MAT locus must be quickly degraded 65. Matα2 is a MATα-encoded transcriptional repressor. One pathway for α2 ubiquitylation requires two E2s, Ubc6 and Ubc7, which function together with a RING-type E3 ligase, Doa10 66, 67. Unexpectedly, these E2 and E3 enzymes form part of a large complex embedded in the ER membrane, where they also act on specific ERAD substrates 67-70.
A ∼60-residue N-terminal region in α2 contains a degron, called Deg1, which is targeted by the Doa10 complex; a ∼19-residue determinant within Deg1 is the most critical element 71. This determinant appears to form an amphipathic helix, and almost all the degron-inactivating mutations cluster on the hydrophobic face of this helix (Figure 3A). Deg1 also functions when appended to an integral ER membrane protein, suggesting that membrane substrates of the Doa10 pathway could utilize related degrons 68. Similar hydrophobic elements were also found in degrons of the mammalian serum- and glucocorticoid-regulated kinase 1 (Sgk1) and of a set of artificial peptide sequences fused to a reporter protein, all substrates of the Doa10 pathway in yeast 72-74. Together, these observations suggest that a surface-exposed hydrophobic (helical) structure could also serve as a recognition motif within ERAD substrates of the Doa10 pathway, although this remains to be examined by structural studies.
Figure 3. Degrons in ER-associated degradation.
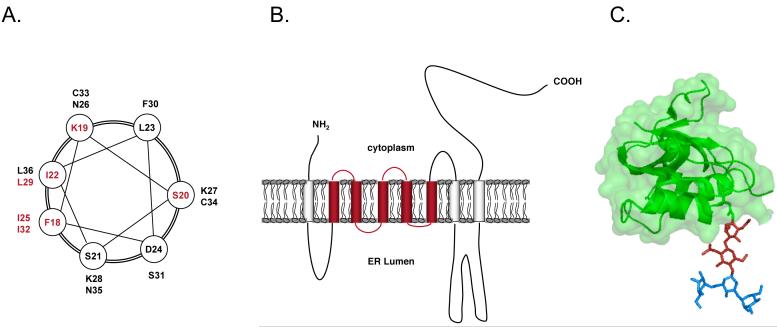
a| Helical-wheel representation of a region (residues 18-36) of the yeast α2 Deg1 degron that is predicted to form part of a coiled-coil structure. Residues whose mutation inhibited Deg1-mediated proteolysis are marked in bold. b| A model for 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) organization in the endoplasmic reticulum (ER) membrane. The sterol-sensing domain is marked in orange. c| 3D structure of the Man3GlcNAc2 oligosaccharide attached to Asn34 of RNase B. The β-1,4-linked dimer of glucosamine (chitobiose), which functions as a binding site for SCFFbs1, is marked in red.
Proteolytic targeting of most aberrant ER proteins in yeast is carried out by two ubiquitin-ligase complexes, the Doa10 complex discussed above and the Hrd1 complex (for review, see refs. 61, 64, 75). Mammals have orthologues of both these E3s as well as additional ER-associated E3s 76. Doa10 and Hrd1 recognize distinct sets of ER membrane and luminal substrates. Doa10 can also target α2 and other nuclear substrates by virtue of its ability to reach the inner nuclear envelope, an ability not shared by Hrd1 77. Most PQC substrates of Doa10 in the ER are integral membrane proteins with cytosolically disposed lesions 68, 78. Hrd1, in contrast, seems to act primarily on soluble ER luminal substrates or membrane proteins with luminal lesions 78. Because the number of substrates examined is still low, these generalizations should be regarded with caution. Interestingly, the ability of Hrd1 to target specific substrates is influenced by both N-linked carbohydrates and polypeptide determinants in the substrate (reviewed in ref. 79).
The distributed degron of HMG-CoA reductase
In addition to aberrant proteins, the Hrd1 pathway also targets naturally short-lived substrates. Best studied is 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR), a 97-kDa integral membrane glycoprotein of the ER. HMGR is a key enzyme in the mevalonate pathway, from which sterols are synthesized, and is subject to feedback regulation as part of the cellular control of sterol synthesis (for review, see ref. 80). HMGR consists of a C-terminal catalytic domain that faces the cytoplasm and a non-catalytic N-terminal membrane domain that anchors the enzyme in the ER 81, 82. This hydrophobic domain, with its eight membrane spans, constitutes a cis-acting element that is necessary and sufficient to regulate the enzyme's stability (Figure 3B) 83-85. A similar stretch of membrane-spanning sequences has been identified in several other proteins, each of which is influenced by sterols; this region has therefore been termed the sterol-sensing domain 86-88.
Budding yeast express two isozymes of HMGR, Hmg1 and Hmg2, of which only the second is normally regulated by ERAD. An extensive series of sequence swaps between the transmembrane domains of Hmg1 and Hmg2 revealed that information necessary for degradation is dispersed throughout the >500 residues of the Hmg2 transmembrane domain, suggesting the existence of a “distributed degron.” 89 Analysis of Hmg2 folding in cell-free extracts further demonstrated that certain lipids induce structural changes of the Hmg2 membrane domain, which can be counteracted by chemical chaperones in vitro and in vivo 90, 91. Thus, Hmg2 can undergo significant, reversible structural changes that stimulate its Hrd1-dependent degradation, potentially by mimicking structures seen in misfolded proteins subject to PQC in the ER.
How is such a “distributed degron” recognized by the ubiquitin-proteasome system? Earlier work demonstrated that inhibition of protein translation abrogated sterol-stimulated degradation of mammalian HMGR concomitant with a marked reduction in polyubiquitylation of the enzyme 92. This suggested that a short-lived protein might have an essential role in the regulated turnover of HMGR. More recently, a short-lived protein that stimulates HMGR turnover was identified 93. This protein, called Insig-1, interacts with the mammalian RING E3 ligase gp78, which is related to yeast Hrd194. In sterol-depleted cells, gp78 associates with Insig-1, which leads to the ubiquitylation and proteasomal degradation of the latter protein. Conversely, ubiquitylation of Insig-1 is blocked, and the protein is stabilized, when intracellular sterols accumulate. High sterol levels also facilitate binding of the gp78-Insig-1 complex to HMGR, resulting in the ubiquitylation and proteasomal degradation of the enzyme 95. Therefore, the Insig proteins can be regarded as adaptor proteins that recognize disordered sterol sensing domain-containing client proteins and help shepherd them to the appropriate ubiquitin ligase for ubiquitylation and down regulation.
The chitodegrons of N-glycosylated proteins
As noted earlier , many membrane and soluble proteins in the ER are glycosylated, and specific N-linked glycans provide signals by which misfolded proteins in the ER are recognized for eventual retrotranslocation to the cytosol (for review, see ref. 96). In other cases, most obviously for proteins that are not glycosylated, the polypeptide itself carries the primary recognition determinants, and there are examples where both sugar and protein moieties participate in substrate recognition 97. Importantly, the original recognition of these carbohydrate structures is carried out by specialized lectins acting on the luminal side of the ER membrane 98-100.
In mammals, a distinct mode of cytosolic recognition of misfolded glycosylated proteins is utilized by Fbs1 101. Fbs1 is the F-box protein of an SCF complex, SCFFbs1, which catalyzes ubiquitylation of misfolded N-glycosylated proteins. Several high-mannose oligosaccharide-recognition molecules are involved in quality control assurance in the ER lumen, which prevents improperly folded, glycosylated proteins from leaving the ER 102, 103. Instead, these eventually enter the ERAD pathway and are retrotranslocated to the cytosol. Notably, SCFFbs1 specifically binds to proteins bearing high-mannose N-linked oligosaccharides in the cytosolic face of the ER membrane and promotes their ubiquitylation and degradation 104. Structural studies identified a sugar-binding domain (SBD) in Fbs1, composed of a 10-stranded β-sandwich with two α-helices 38, 101. Whereas Man8GlcNAc2 is thought to be the major N-glycan on unfolded glycoproteins that are translocated to the cytosol in ERAD, the SBD domain primarily recognizes the disaccharide GlcNAc2 (chitobiose) in the base of this high mannose structure (Figure 3C). Only limited contacts exist between the protein portions of the glycoprotein and the SBD, suggesting that the sugar component of the glycoprotein defines the interaction with the E3 ligase. In native glycoproteins, however, the sugar-polypeptide junction is shielded by protein residues surrounding the glycosylation site, so the chitobiose at the base of the sugar is unlikely to make contacts with Fbs1. Both Fbs1 and its isozyme Fbs2 interact more efficiently with denatured glycoproteins 105. Therefore, solvent exposure of the sugar-protein junction in the misfolded glycoprotein is likely to be the initial step for recognition by these ubiquitin ligases.
Degron recognition by chaperones
The notion that adaptor proteins mediate PQC substrate recognition by their cognate E3s is not unique to HMGR degradation. Genetic analysis in yeast has implicated specific molecular chaperones in the degradation of nearly all PQC substrates analyzed so far (reviewed in ref. 106). Different chaperones have distinct functions in the pathway of substrate recognition, ubiquitylation and degradation, but in most cases their exact roles are not well defined. An exception is the relatively well-characterized mechanism of PQC involving the HSC70/HSP70/HSP90 cytosolic chaperones and the mammalian U-box protein called CHIP (carboxyl terminus of Hsc70-interacting protein) 107, 108. In addition to its U-box, CHIP possesses a tetratricopeptide repeat motif (TPR), which binds to Hsp70 and Hsp90. This association drives the ubiquitin-dependent degradation of various chaperone client proteins 107, 109-115. In vitro reconstitution of CHIP-dependent ubiquitylation of a firefly luciferase directly demonstrated that CHIP is a chaperone-dependent E3 ligase that selectively ubiquitylates proteins in their unfolded state 107. CHIP essentially uses Hsp70 or Hsp90 as a substrate-recognition subunit that binds unstructured regions of client proteins and positions them for CHIP-mediated ubiquitin ligation. Nevertheless, CHIP can also bind non-native proteins in the absence of Hsp70 or Hsp90, suggesting that there are multiple ways by which substrates can be recognized by this E3 116.
Ubiquitin ligation-site determinants
In most proteins, the preferred acceptor site for polyubiquitin chain addition is a lysine side chain. The majority of ubiquitylated polypeptides have multiple lysines, but in some only one or a few can be efficiently ubiquitylated 117-121. This implies that for these substrates, the position of the ubiquitin acceptor site or the local structure surrounding it serves as a determinant for degron function. For example, the yeast Sic1 cell cycle regulator is polyubiquitylated only on the six most N-terminal of its 20 lysines in vivo, and any one of these is sufficient to support normal Sic1 degradation kinetics 122. However, in vitro, polyubiquitin chain position within this subset of sites contributes significantly to the rate of Sic1 proteolysis by the proteasome. In contrast, many proteins can be ubiquitylated on different lysines with degradation efficiency depending little, if at all, on which particular lysine(s) is chosen 123-126. Structural flexibility in the substrate or E3 complex (or both) might allow various substrate lysine residues to be brought near the ubiquitin thioester linkage in the E2 (or E3) active site to allow efficient ubiquitin transfer.
A special case where lysine position is important for ubiquitin transfer efficiency is during auto-ubiquitylation of E2s. A number of E2s bear a lysine in close proximity to the catalytic cysteine, and this lysine is a primary target for their auto-ubiquitylation. The ubiquitin molecule that anchors the chain is transferred to this lysine from the active site of the same E2 enzyme molecule 127-129. Conservation of the positioning of this lysine among different E2s in multiple organisms suggests its functional importance for auto-regulation. Attachment of ubiquitin molecule(s) close to the E2 active site may hinder the E2 enzyme activity, as shown for UBE2T, the E2 enzyme in the ubiquitin-dependent DNA repair pathway that is defective in Fanconi anemia patients 127. UBET2 auto-ubiquitylation might be essential for negative regulation of this pathway.
The importance of the specific localization of the E2 acceptor lysine was highlighted by studies of the regulated turnover of the yeast E2 enzyme Ubc7 130. When Ubc7 is overexpressed, it forms a polyubiquitin chain on its active site, Cys89, the sole acceptor site for ubiquitin in the Ubc7 protein, and it is rapidly degraded by the proteasome. Unlike most E2s, Ubc7 does not contain a lysine residue near its catalytic core, and sequence alignments revealed that a histidine is at the position usually occupied by a lysine. Substitution of this histidine with a lysine seems to enable polyubiquitin chain transfer from the active site cysteine to the lysine side chain, suggesting a mechanism for polyubiquitin chain assembly that precedes substrate modification.
There are now multiple examples of amino acids other than lysine serving as ubiquitin acceptor sites. The first identified was the α-amino group of the substrate N-terminal residue (αN). Attachment of a polyubiquitin chain in this way requires the formation of a standard peptide bond between the basal ubiquitin C-terminus and the substrate protein (reviewed in ref. 131). The most compelling evidence for this mode of post-translational ubiquitylation came from mass spectrometric sequencing of tryptic peptides derived from ubiquitylated forms of certain substrates. Sequenced peptides included fragments that spanned the C-terminal diglycine of ubiquitin and the N-terminal segment of the substrate 132, 133.
Whereas both the lysine and αN determinants are linked to ubiquitin via primary amines, ubiquitin is also capable of covalent coupling to other nucleophilic protein side chains. Cysteine residues can form a thioester bond to ubiquitin, and this of course occurs as a standard enzyme intermediate with E1, E2 and HECT-class E3 enzymes. As noted earlier, the catalytic cysteine of the yeast Ubc7 E2 can serve as a polyubiquitin acceptor site 130, and in vitro data demonstrated the assembly of a polyubiquitin chain on the active site cysteine of the mammalian ortholog of yeast Ubc7 134. Ubiquitin can also be conjugated to cysteines in proteins that are not ubiquitinconjugation enzymes. For instance, this occurs on the cytosolic tail of major histocompatibility complex (MHC) class I molecules at the cell surface, resulting in their endocytosis and lysosomal degradation 135. Finally, recent evidence strongly suggests that serine and threonine residues of the MHC-I tail can also be ubiquitylated in vivo 136. Thus, there might be considerably more flexibility in protein-ubiquitin modification than had been suspected previously.
Concluding remarks
As is apparent from this limited survey, degrons are fundamental elements of protein degradation by the ubiquitin system. The foregoing discussion focused on elements within ubiquitin-dependent degrons that function in ubiquitin-ligase binding and ubiquitin-substrate conjugation. Once a polyubiquitin chain has been attached to a protein, the protein must still be properly routed to the proteasome, unfolded and then degraded. These steps depend on additional features of the degron or the proteolytic substrate. For example, several Lys48-linked polyubiquitin chain-modified cellular proteins have also been shown to bind the proteasome without concomitant degradation 137-139. It is possible in these cases that the protein substrate lacks an appropriate degradation initiation site. Several studies have indicated that in addition to tethering the substrate to the proteasome, the degron must also have a more loosely structured peptide segment that initiates unfolding and insertion into the proteasome 140-142.
The general principles of substrate recognition summarized in this review should be useful guides for addressing many unanswered questions. We still have limited knowledge of the structures of E3/E2 complexes bound to substrate proteins that are in a state competent for ubiquitin transfer. The true range and variety of protein quality-control degrons are poorly defined, including the question of whether soluble and membrane PQC substrates use related or distinct degrons. The mechanisms of polyubiquitin chain assembly on substrates may vary among different E3/E2 enzymes, and current chain assembly models remain to be tested rigorously. Other vexing questions include whether attachment of polyubiquitin chains of different linkage results in differences in proteasomal degradation efficacy, why different substrates are targeted directly to the proteasome and others through mobile adaptor proteins, and what exact series of molecular events occur once a polyubiquitylated substrate reaches the proteasome. Answering these questions will allow us to understand much more fully how the ubiquitin-proteasome system is deployed in the myriad physiological pathways with which it has been connected.
Acknowledgements
We thank J. Bloom, Y. Reiss and Y. Xie for helpful comments on the manuscript. Work from the M.H.'s laboratory was supported by grants from the U.S. National Institutes of Health (GM046904, GM053756 and GM083050). T. Ravid is supported by the European Union (grant IRG-205425) and by the Lejwa Fund for Biochemistry.
Glossary
- TETRATRICOPEPTIDE REPEAT (TPR) MOTIF
Tandem repeats of a degenerate 34-amino acid sequence, which mediates protein-protein interactions
- RING FINGER DOMAIN
“Really Interesting New Gene” motif consisting of a defined pattern of cysteine and histidine residues that coordinate two zinc ions. This motif is engaged in ubiquitin ligation through recruitment and positioning of the E2 enzyme
- HECT DOMAIN
(Homologous to E6-AP C Terminus domain). HECT- and RING-domain-containing proteins represent the two main classes of E3 ubiquitin ligases. In contrast to RING ligases, HECT-domain ligases form an essential thioester intermediate with ubiquitin as it is being transferred from the E2 enzyme to the substrate
- F-BOX DOMAIN
A protein motif of ∼50 residues that functions as a binding site for the Skp1 adaptor protein. F-box proteins contain additional protein-protein interaction motifs, such as WD40 or leucine-rich repeats, and are the substrate recognition subunits of SCF ligases
- UBR-BOX
A ∼70-residue zinc-finger-like motif in E3 ubiquitin ligases that serves as a substrate recognition domain for N-end rule substrates
- WD40 REPEAT
A repeat of 40 amino acids with a characteristic Trp-Asp motif, first found in the β subunit of heterotrimeric G proteins and involved in protein-protein interactions. F-box motif-containing proteins often also have these repeats
References
- 1.Schimke RT, Doyle D. Control of enzyme levels in animal tissues. Annu Rev Biochem. 1970;39:929–976. doi: 10.1146/annurev.bi.39.070170.004433. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AL, Dice JF. Intracellular protein degradation in mammalian and bacterial cells. Annu Rev Biochem. 1974;43:835–869. doi: 10.1146/annurev.bi.43.070174.004155. [DOI] [PubMed] [Google Scholar]
- 3.Hochstrasser M. Ubiquitin-dependent protein degradation. Ann Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- References 3-5 review the older literature on intracellular proteolysis and various features of the ubiquitin-proteasome system
- 6.Platt T, Miller JH, Weber K. In vivo degradation of mutant lac repressor. Nature. 1970;228:1154–1156. doi: 10.1038/2281154a0. [DOI] [PubMed] [Google Scholar]
- 7.Rabinovitz M. Translational repression in the control of globin chain initiation by hemin. Ann N Y Acad Sci. 1974;241:322–333. doi: 10.1111/j.1749-6632.1974.tb21890.x. [DOI] [PubMed] [Google Scholar]
- 8.Dice JF, Goldberg AL. Relationship between in vivo degradative rates and isoelectric points of proteins. Proc Natl Acad Sci U S A. 1975;72:3893–3897. doi: 10.1073/pnas.72.10.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dice JF, Hess EJ, Goldberg AL. Studies on the relationship between the degradative rates of proteins in vivo and their isoelectric points. Biochem J. 1979;178:305–312. doi: 10.1042/bj1780305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varshavsky A. Naming a targeting signal. Cell. 1991;64:13–15. doi: 10.1016/0092-8674(91)90202-a. [DOI] [PubMed] [Google Scholar]
- 11.Lahav-Baratz S, Sudakin V, Ruderman JV, Hershko A. Reversible phosphorylation controls the activity of cyclosome-associated cyclin-ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 1995;92:9303–9307. doi: 10.1073/pnas.92.20.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner GC, Du F, Varshavsky A. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature. 2000;405:579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- 13.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 14.Hershko A, Ciechanover A, Heller H, Haas AL, Rose IA. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci U S A. 1980;77:1783–1786. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershko A, Heller H, Eytan E, Reiss Y. The protein substrate binding site of the ubiquitin-protein ligase system. J Biol Chem. 1986;261:11992–11999. [PubMed] [Google Scholar]
- 16.Hershko A, Leshinsky E, Ganoth D, Heller H. ATP-dependent degradation of ubiquitin-protein conjugates. Proc. Natl. Acad. Sci. U.S.A. 1984;81:1619–1623. doi: 10.1073/pnas.81.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Shows the importance of the N-terminal amino acid in targeting specific substrates for ubiquitin-dependent turnover
- 18.Bartel B, Wunning I, Varshavsky A. The recognition component of the N-end rule pathway. Embo J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mogk A, Schmidt R, Bukau B. The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Lupas AN, Koretke KK. Bioinformatic analysis of ClpS, a protein module involved in prokaryotic and eukaryotic protein degradation. J Struct Biol. 2003;141:77–83. doi: 10.1016/s1047-8477(02)00582-8. [DOI] [PubMed] [Google Scholar]
- 21.Erbse A, et al. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature. 2006;439:753–756. doi: 10.1038/nature04412. [DOI] [PubMed] [Google Scholar]
- 22.Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410:955–959. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 23.Balzi E, Choder M, Chen WN, Varshavsky A, Goffeau A. Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J Biol Chem. 1990;265:7464–7471. [PubMed] [Google Scholar]
- 24.Ciechanover A, et al. Purification and characterization of arginyl-tRNA-protein transferase from rabbit reticulocytes. Its involvement in post-translational modification and degradation of acidic NH2 termini substrates of the ubiquitin pathway. J Biol Chem. 1988;263:11155–11167. [PubMed] [Google Scholar]
- 25.Davydov IV, Varshavsky A. RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J Biol Chem. 2000;275:22931–22941. doi: 10.1074/jbc.M001605200. [DOI] [PubMed] [Google Scholar]
- 26.Hu RG, et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature. 2005;437:981–986. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 27.Lee MJ, et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci U S A. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong CC, et al. Global analysis of posttranslational protein arginylation. PLoS Biol. 2007;5:e258. doi: 10.1371/journal.pbio.0050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tasaki T, Kwon YT. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem Sci. 2007;32:520–528. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Tasaki T, et al. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Groot RJ, Rumenapf T, Kuhn RJ, Strauss EG, Strauss JH. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc Natl Acad Sci U S A. 1991;88:8967–8971. doi: 10.1073/pnas.88.20.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon YT, et al. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 33.Zenker M, et al. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome) Nat Genet. 2005;37:1345–1350. doi: 10.1038/ng1681. [DOI] [PubMed] [Google Scholar]
- 34.Nash P, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Identifies the optimal degradation signal for SCFCdc4 and gives a model for how multisite phosphorylation controls SCF substrate ubiquitylation.
- 35.Min JH, et al. Structure of an HIF-1alpha -pVHL complex: hydroxyproline recognition in signaling. Science. Online. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 36.Wu G, et al. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 37.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural Basis for Phosphodependent Substrate Selection and Orientation by the SCF(Cdc4) Ubiquitin Ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 38.Mizushima T, et al. Structural basis of sugar-recognizing ubiquitin ligase. Nat Struct Mol Biol. 2004;11:365–370. doi: 10.1038/nsmb732. [DOI] [PubMed] [Google Scholar]
- 39.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 40.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Structural study that provides an alternative view of multisite phosphorylation and SCF substrate recognition.
- 41.Tyers M, Jorgensen P. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr Opin Genet Dev. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]
- 42.Ang XL, Harper JW. Interwoven ubiquitination oscillators and control of cell cycle transitions. Sci STKE. 2004;2004:pe31. doi: 10.1126/stke.2422004pe31. [DOI] [PubMed] [Google Scholar]
- 43.Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 44.Winston JT, et al. The SCFbetaTrCP ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999 doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amati B, Vlach J. Kip1 meets SKP2: new links in cell-cycle control. Nat Cell Biol. 1999;1:E91–3. doi: 10.1038/12087. [DOI] [PubMed] [Google Scholar]
- 46.Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 47.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 48.Klein P, Pawson T, Tyers M. Mathematical modeling suggests cooperative interactions between a disordered polyvalent ligand and a single receptor site. Curr Biol. 2003;13:1669–1678. doi: 10.1016/j.cub.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 49.Willems AR, Schwab M, Tyers M. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 50.Semenza GL, Nejfelt MK, Chi SM, Antonarakis SE. Hypoxia-inducible nuclear factors bind to an enhancer element located 3′ to the human erythropoietin gene. Proc Natl Acad Sci U S A. 1991;88:5680–5684. doi: 10.1073/pnas.88.13.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamura T, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 53.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 54.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 55.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 56.Hon WC, et al. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- Provides detailed structural data on the interaction between hydroxylated HIF-1alpha and VHL.
- 57.Beroud C, et al. Software and database for the analysis of mutations in the VHL gene. Nucleic Acids Res. 1998;26:256–258. doi: 10.1093/nar/26.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Hatakeyama S, Nakayama KI. Ubiquitylation as a Quality Control System for Intracellular Proteins. J Biochem. 2003;134:1–8. doi: 10.1093/jb/mvg106. [DOI] [PubMed] [Google Scholar]
- 60.Laney JD, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 61.Hampton RY. ER-associated degradation in protein quality control and cellular regulation. Curr Opin Cell Biol. 2002;14:476–482. doi: 10.1016/s0955-0674(02)00358-7. [DOI] [PubMed] [Google Scholar]
- 62.Kostova Z, Wolf DH. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO Journal. 2003;22:2309–2317. doi: 10.1093/emboj/cdg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sayeed A, Ng DT. Search and destroy: ER quality control and ER-associated protein degradation. Crit Rev Biochem Mol Biol. 2005;40:75–91. doi: 10.1080/10409230590918685. [DOI] [PubMed] [Google Scholar]
- 64.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- References 61-64 review the mechanisms of endoplasmic-reticulum associated degradation (ERAD).
- 65.Laney JD, Hochstrasser M. Ubiquitin-dependent degradation of the yeast Matα2 repressor enables a switch in developmental state. Genes Dev. 2003;17:2259–2270. doi: 10.1101/gad.1115703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 67.Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 70.Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- 71.Johnson PR, Swanson R, Rakhilina L, Hochstrasser M. Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell. 1998;94:217–227. doi: 10.1016/s0092-8674(00)81421-x. [DOI] [PubMed] [Google Scholar]
- Characterizes the Deg1 degron of Matα2 and physiological regulation of degron recognition by changes in protein quaternary structure.
- 72.Arteaga MF, Wang L, Ravid T, Hochstrasser M, Canessa CM. An amphipathic helix targets serum and glucocorticoid-induced kinase 1 to the endoplasmic reticulum-associated ubiquitin-conjugation machinery. Proc Natl Acad Sci U S A. 2006;103:11178–11183. doi: 10.1073/pnas.0604816103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gilon T, Chomsky O, Kulka RG. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. Embo J. 1998;17:2759–2766. doi: 10.1093/emboj/17.10.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilon T, Chomsky O, Kulka RG. Degradation signals recognized by the Ubc6p-Ubc7p ubiquitin-conjugating enzyme pair. Mol Cell Biol. 2000;20:7214–7219. doi: 10.1128/mcb.20.19.7214-7219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ismail N, Ng DT. Have you HRD? Understanding ERAD is DOAble! Cell. 2006;126:237–239. doi: 10.1016/j.cell.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 76.Kostova Z, Tsai YC, Weissman AM. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin Cell Dev Biol. 2007;18:770–779. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443:827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- 78.Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang S, Ng DTW. Lectins sweet-talk proteins into ERAD. Nat Cell Biol. 2008;10:251–253. doi: 10.1038/ncb0308-251. [DOI] [PubMed] [Google Scholar]
- 80.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 81.Liscum L, et al. 3-Hydroxy-3-methylglutaryl-CoA reductase: a transmembrane glycoprotein of the endoplasmic reticulum with N-linked “high-mannose” oligosaccharides. Proc Natl Acad Sci U S A. 1983;80:7165–7169. doi: 10.1073/pnas.80.23.7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liscum L, et al. Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum. J Biol Chem. 1985;260:522–530. [PubMed] [Google Scholar]
- 83.Chin DJ, et al. Nucleotide sequence of 3-hydroxy-3-methyl-glutaryl coenzyme A reductase, a glycoprotein of endoplasmic reticulum. Nature. 1984;308:613–617. doi: 10.1038/308613a0. [DOI] [PubMed] [Google Scholar]
- 84.Gil G, Faust JR, Chin DJ, Goldstein JL, Brown MS. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 1985;41:249–258. doi: 10.1016/0092-8674(85)90078-9. [DOI] [PubMed] [Google Scholar]
- 85.Skalnik DG, Narita H, Kent C, Simoni RD. The membrane domain of 3-hydroxy-3-methylglutaryl-coenzyme A reductase confers endoplasmic reticulum localization and sterol-regulated degradation onto beta-galactosidase. J Biol Chem. 1988;263:6836–6841. [PubMed] [Google Scholar]
- 86.Hua X, Sakai J, Brown MS, Goldstein JL. Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane. J Biol Chem. 1996;271:10379–10384. doi: 10.1074/jbc.271.17.10379. [DOI] [PubMed] [Google Scholar]
- 87.Loftus SK, et al. Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science. 1997;277:232–235. doi: 10.1126/science.277.5323.232. [DOI] [PubMed] [Google Scholar]
- 88.Burke R, et al. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99:803–815. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- References 86-88 identify sterol sensing domains in the transmembrane regions of several integral membrane proteins.
- 89.Gardner RG, Hampton RY. A ‘distributed degron’ allows regulated entry into the ER degradation pathway. Embo J. 1999;18:5994–6004. doi: 10.1093/emboj/18.21.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shearer AG, Hampton RY. Structural control of endoplasmic reticulum-associated degradation: effect of chemical chaperones on 3-hydroxy-3-methylglutaryl-CoA reductase. J Biol Chem. 2004;279:188–196. doi: 10.1074/jbc.M307734200. [DOI] [PubMed] [Google Scholar]
- 91.Shearer AG, Hampton RY. Lipid-mediated, reversible misfolding of a sterol-sensing domain protein. Embo J. 2005;24:149–159. doi: 10.1038/sj.emboj.7600498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ravid T, Doolman R, Avner R, Harats D, Roitelman J. The ubiquitin-proteasome pathway mediates the regulated degradation of mammalian 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 2000;275:35840–35847. doi: 10.1074/jbc.M004793200. [DOI] [PubMed] [Google Scholar]
- 93.Yang T, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Reports the discovery of Insig1, a sterol sensing protein.
- 94.Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005;19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 95.Lee JN, Song B, DeBose-Boyd RA, Ye J. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J Biol Chem. 2006;281:39308–39315. doi: 10.1074/jbc.M608999200. [DOI] [PubMed] [Google Scholar]
- 96.Yoshida Y. A novel role for N-glycans in the ERAD system. J Biochem. 2003;134:183–190. doi: 10.1093/jb/mvg128. [DOI] [PubMed] [Google Scholar]
- 97.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 98.Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 99.Kim W, Spear ED, Ng DT. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol Cell. 2005;19:753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 100.Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- References 97-100 identify Yos9 as a lectin that detects misfolded glycoproteins in the ER lumen.
- 101.Mizushima T, et al. Structural basis for the selection of glycosylated substrates by SCF(Fbs1) ubiquitin ligase. Proc Natl Acad Sci U S A. 2007;104:5777–5781. doi: 10.1073/pnas.0610312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shows that Fbs1 of SCF(Fbs1) in the cytosol recognizes the chitobiose domain in the oligosaccharide base of misfolded glycoproteins.
- 102.Helenius A. Quality control in the secretory assembly line. Philos Trans R Soc Lond B Biol Sci. 2001;356:147–150. doi: 10.1098/rstb.2000.0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Spiro RG. Role of N-linked polymannose oligosaccharides in targeting glycoproteins for endoplasmic reticulum-associated degradation. Cell Mol Life Sci. 2004;61:1025–1041. doi: 10.1007/s00018-004-4037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yoshida Y, et al. E3 ubiquitin ligase that recognizes sugar chains. Nature. 2002;418:438–442. doi: 10.1038/nature00890. [DOI] [PubMed] [Google Scholar]
- Identifies Fbs1 as a new sugar-binding F-box subunit of the SCF E3 ligase family.
- 105.Yoshida Y, Adachi E, Fukiya K, Iwai K, Tanaka K. Glycoprotein-specific ubiquitin ligases recognize N-glycans in unfolded substrates. EMBO Rep. 2005;6:239–244. doi: 10.1038/sj.embor.7400351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nakatsukasa K, Brodsky JL. The Recognition and Retrotranslocation of Misfolded Proteins from the Endoplasmic Reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Murata S, Minami Y, Minami M, Chiba T, Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Reports. 2001;2:1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shows that CHIP specifically ubiquitylates misfolded substrates in the presence of Hsp70 family members.
- 108.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang J, et al. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 110.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 111.Connell P, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 112.Sahara N, et al. In vivo evidence of CHIP up-regulation attenuating tau aggregation. J Neurochem. 2005;94:1254–1263. doi: 10.1111/j.1471-4159.2005.03272.x. [DOI] [PubMed] [Google Scholar]
- 113.Hatakeyama S, Matsumoto M, Yada M, Nakayama KI. Interaction of U-box-type ubiquitin-protein ligases (E3s) with molecular chaperones. Genes Cells. 2004;9:533–548. doi: 10.1111/j.1356-9597.2004.00742.x. [DOI] [PubMed] [Google Scholar]
- 114.Miller VM, et al. CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J Neurosci. 2005;25:9152–9161. doi: 10.1523/JNEUROSCI.3001-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Younger JM, et al. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 116.Rosser MF, Washburn E, Muchowski PJ, Patterson C, Cyr DM. Chaperone functions of the E3 ubiquitin ligase CHIP. J Biol Chem. 2007;282:22267–22277. doi: 10.1074/jbc.M700513200. [DOI] [PubMed] [Google Scholar]
- 117.Bachmair A, Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989;56:1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 118.Chau V, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 119.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115:565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 120.Scherer DC, Brockman JA, Chen Z, Maniatis T, Ballard DW. Signal-induced degradation of I kappa B alpha requires site-specific ubiquitination. Proc Natl Acad Sci U S A. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Baldi L, Brown K, Franzoso G, Siebenlist U. Critical role for lysines 21 and 22 in signal-induced, ubiquitin-mediated proteolysis of I kappa B-alpha. J Biol Chem. 1996;271:376–379. doi: 10.1074/jbc.271.1.376. [DOI] [PubMed] [Google Scholar]
- 122.Petroski MD, Deshaies RJ. Redundant degrons ensure the rapid destruction of Sic1 at the G1/S transition of the budding yeast cell cycle. Cell Cycle. 2003;2:410–1. [PubMed] [Google Scholar]
- 123.Banerjee A, Gregori L, Xu Y, Chau V. The bacterially expressed yeast CDC34 gene product can undergo autoubiquitination to form a multiubiquitin chain-linked protein. J. Biol. Chem. 1993;268:5668–5675. [PubMed] [Google Scholar]
- 124.Fung TK, Yam CH, Poon RY. The N-terminal regulatory domain of cyclin A contains redundant ubiquitination targeting sequences and acceptor sites. Cell Cycle. 2005;4:1411–1420. doi: 10.4161/cc.4.10.2046. [DOI] [PubMed] [Google Scholar]
- 125.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 126.King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and the structural characterization of ubiquitinated intermediates. Mol. Biol. Cell. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Machida YJ, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Shows that monoubiquitylation of UBE2T near its catalytic site hinders activity of the E2 enzyme.
- 128.Lin Y, Hwang WC, Basavappa R. Structural and functional analysis of the human mitotic-specific ubiquitin-conjugating enzyme, UbcH10. J Biol Chem. 2002;277:21913–21921. doi: 10.1074/jbc.M109398200. [DOI] [PubMed] [Google Scholar]
- 129.Hodgins R, Gwozd C, Arnason T, Cummings M, Ellison MJ. The tail of a ubiquitin-conjugating enzyme redirects multi-ubiquitin chain synthesis from the lysine 48-linked configuration to a novel nonlysine-linked form. J Biol Chem. 1996;271:28766–28771. doi: 10.1074/jbc.271.46.28766. [DOI] [PubMed] [Google Scholar]
- 130.Ravid T, Hochstrasser M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol. 2007;9:422–427. doi: 10.1038/ncb1558. [DOI] [PubMed] [Google Scholar]
- Demonstrates that a polyubiquitin chain attached to the catalytic cysteine residue of an E2 enzyme can act as a degron.
- 131.Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 132.Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell. 2003;115:71–82. doi: 10.1016/s0092-8674(03)00755-4. [DOI] [PubMed] [Google Scholar]
- 133.Ben-Saadon R, et al. The tumor suppressor protein p16(INK4a) and the human papillomavirus oncoprotein-58 E7 are naturally occurring lysine-less proteins that are degraded by the ubiquitin system. Direct evidence for ubiquitination at the N-terminal residue. J Biol Chem. 2004;279:41414–41421. doi: 10.1074/jbc.M407201200. [DOI] [PubMed] [Google Scholar]
- 134.Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- Shows that the attachment of a polyubiquitin chain on the catalytic cysteine of an E2 enzyme is an intermediate step in substrate ubiquitylation.
- 135.Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- Shows that ubiquitin covalently attached to a cysteine residue in a substrate can target it for degradation.
- 136.Wang X, et al. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J Cell Biol. 2007;177:613–624. doi: 10.1083/jcb.200611063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Elsasser S, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nature Cell Biology. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 138.Alberti S, et al. Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J Biol Chem. 2002;277:45920–45927. doi: 10.1074/jbc.M204196200. [DOI] [PubMed] [Google Scholar]
- 139.Rao H, Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J Biol Chem. 2002;277:11691–11695. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- 140.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Shows that the ability of a protein to be degraded depends on the position of the degron within the protein structure and its ability to initiate unfolding.
- 142.Takeuchi J, Chen H, Coffino P. Proteasome substrate degradation requires association plus extended peptide. EMBO J. 2007;26:123–131. doi: 10.1038/sj.emboj.7601476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Aravind L, Koonin EV. The U box is a modified RING finger - a common domain in ubiquitination. Curr Biol. 2000;10:R132–4. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- Reports the discovery of the U-box motif.
- 144.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 145.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 146.Schulman BA, et al. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 147.Hao B, et al. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]