Abstract
Two genetically engineered, conditional mouse models of lung tumor formation, K-rasLSL-G12D and K-rasLSL-G12D/p53LSL-R270H, are commonly used to model human lung cancer. Developed by Tyler Jacks and colleagues, these models have been invaluable to study in vivo lung cancer initiation and progression in a genetically and physiologically relevant context. However, heterogeneity, multiplicity and complexity of tumor formation in these models make it challenging to monitor tumor growth in vivo and have limited the application of these models in oncology drug discovery. Here, we describe a novel analytical method to quantitatively measure total lung tumor burden in live animals using micro-computed tomography imaging. Applying this methodology, we studied the kinetics of tumor development and response to targeted therapy in vivo in K-ras and K-ras/p53 mice. Consistent with previous reports, lung tumors in both models developed in a time- and dose (Cre recombinase)-dependent manner. Furthermore, the compound K-rasLSL-G12D/p53LSL-R270H mice developed tumors faster and more robustly than mice harboring a single K-rasLSL-G12D oncogene, as expected. Erlotinib, a small molecule inhibitor of the epidermal growth factor receptor, significantly inhibited tumor growth in K-rasLSL-G12D/p53LSL-R270H mice. These results demonstrate that this novel imaging technique can be used to monitor both tumor progression and response to treatment and therefore supports a broader application of these genetically engineered mouse models in oncology drug discovery and development.
Introduction
Lung cancer is the leading cause of cancer deaths worldwide [1]. Mutations in K-ras proto-oncogene are closely associated with approximately 30% of non-small cell lung cancers. Expression of an oncogenic mutant K-rasG12D gene in lung tissues of mice resulted in the development of lung adenocarcinoma, resembling human lung cancers [2,3]. Alterations in the p53 tumor suppressor gene are also found in 50% human non-small cell lung cancer patients [4]. Conditional expression of a p53R270H dominant-negative mutant gene using the Cre-lox recombinase system promotes K-rasG12D-initiated lung adenocarcinomas in a compound knock-in mouse model [5,6]. This conditional compound knock-in mouse line contains a Lox-Stop-Lox (LSL) K-rasG12D (referred to as K-rasLSL-G12D, or K-ras) and a Lox-Stop-Lox p53R270H (referred to as p53LSL-R270H, or p53) transgenes. The oncogenic K-rasG12D and the dominant-negative tumor suppressor gene p53R270H are simultaneously expressed by the removal of transcriptional termination Stop element in the LSL cassette when the Cre recombinase is expressed [7,8]. Intranasal introduction of the Cre recombinase by Adeno-Cre viral infection activates expression of these two genes in lung tissue, leading to the development of lung adenocarcinoma.
Application of lung cancer animal models for drug discovery has been challenging. Tumor progression and phenotype is determined by grading tumor stages with histologic examination [6]. Tumor burden is currently measured by examining the percentage of total lung area occupied by tumor in a histologic slice. These time-consuming assessments lack accuracy because of the limited areas of lung tissue analyzed. Computed tomography (CT) imaging has been used for the detection of lung cancer in human clinical settings and in preclinical studies using laboratory animals. However, lung tumor measurement has been limited in mouse models to quantify discrete tumors that are large and located away from major vascular structures [9,10]. Micro-CT has also been applied in the K-ras knock-in model by counting the number of tumors and measuring three to five individual tumors in each animal [11,12]. However, small tumors are hard to distinguish from vasculature and difficult to measure. It is particularly challenging to quantify individual tumor volumes in these lung models because intranasal adenoviral infection leads to formation of multifocal small tumors with irregular shapes. Although attempts have been made to monitor protease activity during tumor initiation and progression in this model using the fluorescence molecular tomography imaging technique [13], it has not been applied to quantify tumor burden in drug discovery. To enhance incorporation of these lung tumor genetic mouse models into drug discovery programs, we developed a high-throughput analytical method using micro-CT images to quantitatively measure total lung tumor burden. This novel analytical method was evaluated using the well-characterized tumor progression in K-ras and K-ras/p53 models [6,14]. K-ras-driven mouse lung tumors have been reported to be sensitive to epidermal growth factor receptor tyrosine kinase inhibitors [15,16], which have not been shown to be effective in human lung tumors [17,18]. However, to test our methodology in a therapy setting, tumor-bearing K-ras/p53 mice were treated with erlotinib.
Materials and Methods
Animals
All animal procedures were reviewed and approved by the Merck-Boston Institutional Animal Care and Use Committee. K-rasLSL-G12D/p53LSL-R270H (K-ras/p53 or KP) and K-rasLSL-G12D (K-ras or K) mouse models were licensed form Dr. Tyler Jacks' laboratory at MIT, Cambridge, MA, and mouse colonies were maintained at Taconic, Germantown, NY. Mixed sexes of mice approximately 2 months old were received from Taconic. At Taconic, mice were housed in groups in a barrier rodent facility using the isolated barrier unit system on wood chip bedding and fed standard rodent chow ad libitum. Mice were tested for routine mouse pathogens and were found to be negative. Mice were acclimated for 1 week in Merck Research Laboratories, Boston animal facility, before initiating imaging studies.
Animal Treatments
For the tumor development study, mice were intranasally infected with either a low dose (1 x 107 plaque-forming units (PFUs)), medium dose (2.5 x 107 PFU), or high dose (5 x 107 PFU) of adenovirus containing Cre recombinase (Adeno-Cre) purchased from Vector BioLabs, Philadelphia, PA. Control adenovirus containing an empty vector was administered at the high dose (5 x 107 PFU). Mice were then scanned using micro-CT at weeks 8, 14, 18, and 20 after infection. Group sizes varied from 2 to 6 animals for control viral-infected groups and from 6 to 17 animals for Cre viral-infected groups at different time points. At week 8, groups include control KP (n = 2), high-dose KP (n = 4), control K (n = 2), and high-dose K (n = 3). At week 14, groups include control KP (n = 5), high-dose KP (n = 16), medium-dose KP (n = 17), control K (n = 6), high-dose K (n = 7), and medium-dose KP (n = 14). At weeks 18 and 20, groups include control KP (n = 3), high-dose KP (n = 8), medium-dose KP (n = 9), control K (n = 4), high-dose K (n = 8), and medium-dose KP (n = 7). To validate imaging method for therapy studies, erlotinib therapy was evaluated in K-ras/p53 knock-in mice (n = 12–15) 16 weeks after Adeno-Cre viral infection, and animals were orally treated once daily for 4 weeks with erlotinib at 75 or 150 mg/kg or with vehicle. Erlotinib was dissolved in vehicle containing 1% methylcellulose and 0.1% Tween-80 and was orally administered once daily. Micro-CT was performed before and after drug treatments.
Micro-CT Imaging
Mice were anesthetized with a continuous flow of 2% to 4% isoflurane/oxygen mixture (2.5 L/min), and the whole body was able to be imaged at one time using the GE eXplore Locus Ultra Pre-Clinical CTscanner (GE Healthcare, London, Canada). The isotropic resolution of this instrument is 185 µm. The micro-CT image acquisition consisted of 1000 projections collected in one full rotation of the gantry in approximately 16 seconds. The image acquisition was not respiratory-gated. The X-ray tube settings were 80 kV and 70 mA. The resulting raw data were reconstructed to a final image volume of 512 x 512 x 1000 slices at 100-µm3 voxel dimensions. The reconstructed slices were output in the CT manufacturer's raw format and were not scaled to Hounsfield units. The reconstructed images were viewed and analyzed using the Amira 4.1.1 software (Mercury Computer Systems, Inc., Chelmsford, MA).
Volumetric Measurement of Tumor Volume
The image analysis method was based on using threshold and region-grow algorithms to segment the image data and define the separate anatomic structures of interest. As a first step, images were cropped to display the lung region. Then, the functional lung volume was determined by using a threshold-based region grow algorithm with voxel value thresholds set from 450 to 800 to segment air space in the lung. Next, the total chest space volume, excluding the heart, was selected using a combination of manual segmentation and semiautomated contouring. Vascular structures around the heart were manually included in the total chest space. The combined tumor and vasculature (T & V) volume was then determined by subtracting the functional lung volume from the total chest space volume. Because tumor tissue and vascular tissue have similar grayscale values, they cannot be separated using this analysis method. However, nontumor vasculature volume should remain relatively constant as tumor volume increases over time. Thus, this method can provide a relative measure of tumor burden. All data are presented as mean ± SE.
Histology Examination of Lung Tissues
For histologic diagnosis, formalin-fixed, paraffin-embedded blocks of the left side of the lung were cut in a transverse manner. Serial slices, 5 to 8 µm thick, of lung tissue were prepared by hematoxylin and eosin (H&E) staining. Slides were scanned with an Ariol SL-50 automated microscope and image analyses software (Applied Imaging, San Jose, CA) at 5x magnification. Total lung and tumor areas were manually demarcated by Ariol Review software, and data were presented as percent of tumor area.
Statistical Analysis
A repeated-measures analysis of variance (ANOVA) was used to assess statistically significant differences. Post hoc one-sided tests were performed with Bonferroni adjustment. For the tumor development experiment (Figures 2A and 4A), 24 statistical contrasts were estimated: for each time point, these were the differences in total T & V volume between each dose (high or low) for each animal model versus the corresponding control for that time point and animal model, as well as the within-group differences of each time point with the earliest available time point (8 weeks for high dose, 14 weeks for low dose). Similar analyses were done for the lung and chest volumes but with only the within-group contrasts calculated (Figure 4A). For the erlotinib therapeutic study (Figure 5), five statistical contrasts were estimated. These include three tests for change from baseline of total T & V volume within each group. These also include two tests for the difference between both doses versus vehicle at the postdosing time point, subtracting the appropriate predose difference. These statistical results were consistent with those from a one-way ANOVA on the percent change from baseline scores (with Bonferroni adjustment; Figure 5B). The repeated-measures ANOVA analysis is equivalent to that when the end point is made to be total T & V volume minus the overall baseline vasculature volume of 128 mm3. Separate t tests were done comparing the groups at baseline, simply to illustrate the heterogeneity of the sample before treatment.
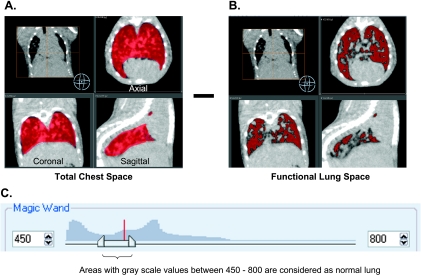
Figure 1.
Method of micro-CT measurement of total tumor burden in lung GEMMs. Tumor and vasculature were measured to represent total tumor burden. The image was adjusted to 450 to 800 grayscale to define boundaries between functional lung area, vasculature, tumor, and chest. (A) Total chest space volume in red includes vessels that come from the heart. (B) Lung area in red with lower CT density was then quantified. Tumor and vasculature were calculated by subtracting the functional lung volume from the total chest space. (C) The total image histogram is shown with the region grow threshold set between 450 and 800.
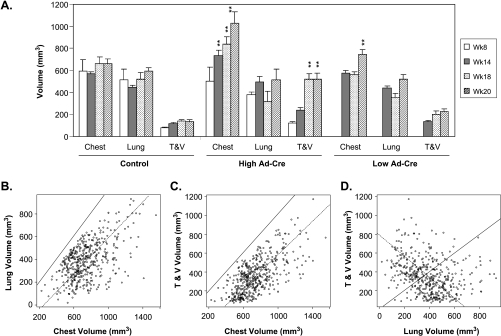
Figure 4.
Enlargement of chest space associated with tumor growth. (A) Chest space and lung volume were measured. The tumor and vasculature (T & V) volume was calculated. Data from KP mice induced by control or Adeno-Cre virus were depicted. (B) Relationship between chest space and lung volume from 458 individual measurements. (C) Relationship between chest space and calculated T & V volume. (D) Relationship between T & V and lung volumes. **P = .01, statistical significant compared to the values of the week 8 time point (for KP high) or the week 14 time point (for KP low).
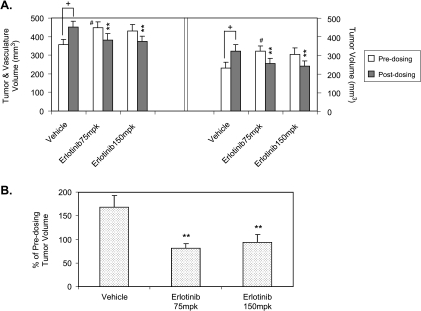
Figure 5.
Effect of erlotinib on lung tumor growth in the compound knock-in mouse model. K-rasLSL-G12D/p53LSL-R270H (KP) mice were infected with Adeno-Cre virus. Mice were randomly assigned into study groups 16 weeks after infection and were treated. (A) Effect on T & V volume and calculated tumor volumes by subtracting the baseline vasculature, 128 mm3. (B) Percentage changes in calculated tumor volume over predosing. **P < .01, statistical significant compared to vehicle controls. + P < .05, statistical significant compared to predosing values within the same group. #P < .05, statistically significant difference (t test) between groups at the predosing time point.
Results
Quantitative Measurement of Total Lung Tumor Burden in K-ras/p53 and K-ras Model
We developed a micro-CT image analysis method to quantitatively measure the sum of T & V volume as a readout of total tumor burden in lung. Noncontrast micro-CT imaging cannot readily distinguish tumor tissue from vasculature because the two tissues have similar X-ray densities. However, one would expect that total tumor burden would be proportional to the combined T & V volume in the lung. Voxel grayscale values between 450 and 800 were considered as functional lung volume, whereas grayscale values above 800 within the chest space except for heart were calculated as T & V. The T & V volume was then calculated by subtracting the functional lung volume from the total chest space (Figure 1).
Using this method, we studied the time course of tumor development in both the single K-ras and compound K-ras/p53 knock-in mouse models. Mice from each model were infected with either control adenovirus lacking the Cre gene (Adeno-Null) or high (5 x 107 PFU) or low (1 x 107 PFU) doses of Adeno-Cre virus. Mice were scanned using micro-CT at 8, 14, 18, and 20 weeks after viral infection. Eight weeks after infection there was no significant increase in T & V volume in either GEM model, although a trend increase was noted in the K-ras/p53 mice treated with the high dose of Adeno-Cre virus compared to control animals (Figure 2A). At week 8, the total vasculature volume in lung space infected with control Adenovirus was between 72 and 122 mm3. The average of T & V volume in KP and K mice infected with the high dose of Adeno-Cre virus was 122 and 95 mm3, respectively, suggesting that mice did not develop detectable tumor burden 8 weeks after infection. With time, animals infected with the high dose of Adeno-Cre virus in both models showed significant increases in T & V volume compared to control animals, whereas a trend of increase was noted in both models with a low dose of infection (Figure 2A). In addition, these data suggest that the K-ras/p53 compound knock-in mice developed tumors faster and more robustly than the K-ras mice, which is consistent with literature reports [6]. The average of total T & V volume in KP group with a high dose of Adeno-Cre increases from 122 ± 12 mm3 at week 8 to 240 ± 22, 518 ± 49, and 520 ± 51 mm3 at 14, 18, and 20 weeks after infection, respectively. The average vasculature volume calculated from all the mice (n = 29) that were infected with control Adeno-null virus at all time points was 128 ± 7 mm3. This baseline vasculature volume was used to approximate the actual tumor volume by subtracting it from the total T & V volume.
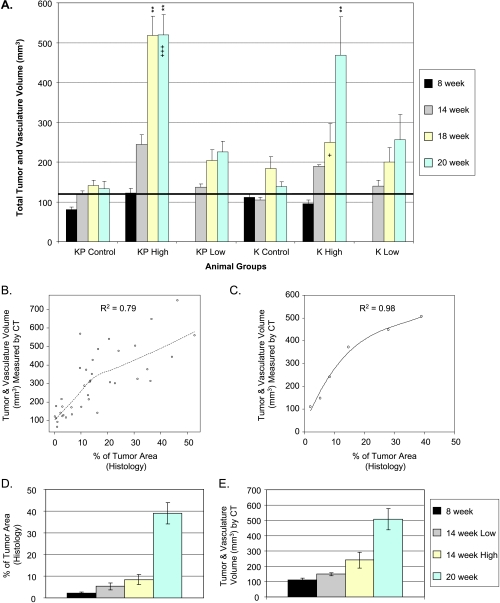
Figure 2.
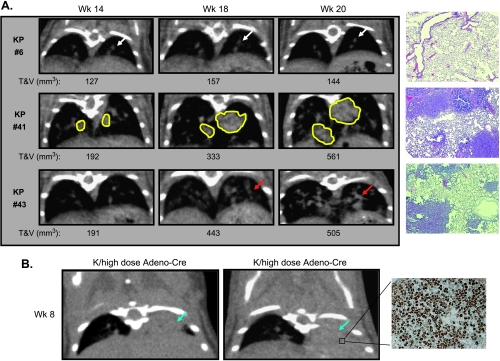
Validation of CT image analysis method and detection of tumor development in K-ras/p53 and K-ras transgenic models. K-rasLSL-G12D/p53LSL-R270H (KP) or K-rasLSL-G12D (K) mice were infected with control adenovirus or with a high or low dose of Adeno-Cre virus. Mice were imaged at 8, 14, 18, and 20 weeks after infection. (A) Tumor development was depicted as total T & V volume over time. The bold black line indicates the average baseline vasculature, 128 mm3. (B) Spearman's rank correlation between CT measurement and histologic assessment of individual animals. (C) Trend correlation approximation using averages of group data from B. (D and E) Time and dose dependence of tumor development in the same animals assessed by histology and CT analysis. **P < .01, statistically significant compared to their vehicle controls. “+” and “+++” indicate that one and three animals with highest tumor burden in their groups were found dead before or during CT scan, respectively.
To determine whether the T & V volume imaging measurements represent the actual tumor burden, we measured percentage of tumor area from two-dimensional histologic slices and compared it with T & V volume measured by CT imaging from the same animals. Animals from the time course experiment and a drug test experiment were euthanized after the final micro-CT imaging session. The left lobe of the lung was collected for histologic examination and percentage of tumor area calculated from the histologic sections. Because the imaging measurement is a volume and the histologic measurement is a percentage of area, there should be a monotonically increasing, but nonlinear, relation between the two sets of measurements. Spearman's rank correlation, a nonparametric (model-free) assessment of the degree of monotonic association between two variables, was performed. Data from 38 animals from six different groups were pooled, and a nonparametric smooth curve (Figure 2B) was fit to the data using a loess (robust local linear smoother) with a bandwidth of 2/3. This correlation (R2 = 0.79) is statistically significantly higher than zero chance of correlation with P < .001. Considering that tumor burden measured from one slice of the left side of the lung by histologic diagnosis may not represent the total tumor burden, we compared the means from different groups. Total tumor burden measured by histologic diagnosis (Figure 2D) was increased in a time- and Cre dose-dependent manner, which is consistent with total T & V volume measurement by micro-CT (Figure 2E). The averages of tumor burden from all six groups of mice measured by histologic diagnosis and micro-CT showed an excellent correlation with R2 = 0.98 using a polynomial approximation (Figure 2C).
Micro-CT Imaging Reveals Complexity of the Lung Tumor Models
Micro-CT not only detected isolated large tumors that had distinct boundaries (Figure 3A, panel 2) but also revealed the multiplicity of tumor development spread throughout the entire lung (Figure 3A, panel 3). It was difficult to distinguish these small tumors from vascular structures and to quantify volumes of individual tumor nodules. Even for larger isolated tumors, lesions often fused or connected to each other in different areas and the irregular shapes of tumors made it even more difficult to define tumor boundaries for volumetric measurement. Histologic examination of lung slices at the end point confirmed the presence of tumors viewed by micro-CT images (Figure 3A). In addition, micro-CT images revealed that in some animals (2/13) 8 weeks after a high dose of viral infection, the left side of the lung was completely opaque and filled (Figure 3B). This increased cellularity in the left side of the lungs was caused by macrophage infiltration resulting from inflammation due to viral infection, which is a common feature of the model [6,12]. These imaging findings were confirmed by histologic examination stained with an anti-Mac-3 monoclonal antibody that detects Mac-3 antigen on tissue macrophages, thioglycollate-elicited peritoneal macrophages, and some myeloid cell lines, but not on lymphocytes or monocytes (Figure 3B). However, this phenomenon was rarely observed at later time points. Of more than 200 animals 15 to 17 weeks after infection with high doses of Adeno-Cre virus, only one mouse was found to have a complete macrophage-filled lung and was excluded from enrollment of the efficacy study.
Figure 3.
Representative micro-CT images of lung tumors from Figure 2. (A) Visualization of lung tumor development in K-rasLSL-G12D/p53LSL-R270H (KP) or K-rasLSL-G12D (K) models. Panel 1: Example of CT images of a control KP mouse with no tumor and H&E stain. Panel 2: Example of CT images of a KP mouse infected with a high dose of Adeno-Cre virus with isolated lung tumors and H&E stain. Panel 3: Example of CT images of a KP mouse infected with a high dose of Adeno-Cre virus with small multifocal tumors and H&E stain. (B) Lung images of macrophage infiltration and histologic staining with a purified rat antimouse Mac-3 antibody (BD Pharmingen, Mississauga, Ontario, Canada) in virus-infected mice. Blue arrow indicates microphage filled lung; white arrow, normal vasculature; yellow line, isolated tumor.
Interestingly, we noted that the animal chest space was enlarged as the tumor burden increased. We analyzed the time course data by grouping the KP and K mice together and compared chest space and T & V volume because both KP and K mice developed lung tumors in a similar fashion. As shown in Figure 4A, increases in T & V volume in both high- and low-dose groups was clearly associated with increases in chest space. To further confirm this phenomenon, several in-house experiments were pooled using 458 separate measurements. The data (Figure 4, B and C) demonstrated that increases in T & V volume were associated with increased chest spaces with Pearson's correlation R2 = 0.65 with 95% confidence and that this increased chest space volume was also associated with increased lung volume (R2 = 0.53). The solid lines in Figure 4, B and C, are 45-degree lines. Because either T & Vor lung volumes are smaller than total chest volumes, all data points must be below this upper-bound 45-degree line.
Erlotinib Suppressed Lung Tumor Growth in the Compound K-ras and p53 Lung Tumor Model
Previous reports demonstrated that the epidermal growth factor receptor inhibitor gefitinib suppressed tumor growth in a K-ras transgenic model [15] and that erlotinib had inhibitory effects on lung adenocarcinoma with both K-ras and p53 mutations in a chemically induced mouse tumor model [16]. To validate our imaging methodology, we tested the effect of erlotinib in the compound K-ras/p53 mouse model. Sixteen weeks after Adeno-Cre infection, mice were randomized into three groups (n = 12–15) and scanned with micro-CT before dosing to obtain baseline tumor burden. The mice were then orally treated once daily for 4 weeks with erlotinib at 75 and 150 mg/kg or vehicle and scanned with micro-CT. The total T & V volume significantly increased from 357 ± 28 to 451 ± 30 mm3 in vehicle-treated animals, whereas T & V volumes in erlotinib-treated animals slightly decreased from 448 ± 31 to 382 ± 33 mm3 for the 75-mg/kg group and from 431 ± 35 to 375 ± 26 mm3 for the 150-mg/kg group (Figure 5A). Tumor burden was calculated by subtracting the baseline vasculature volume of 128 mm3. We calculated percentage changes of tumor volumes (vasculature baseline subtracted) to normalize initial tumor burden among groups because it was noted that the predosing tumor burden in the vehicle group was significantly lower than the 75-mg/kg erlotinib group (Figure 5). Tumor burden was reduced to 81% and 94% of predosing levels for the 75- and 150-mg/kg groups, respectively, whereas tumor burden in the control group increased to 169% of the predosing level (Figure 5B). This result is similar to the previous report [15], further validating this imaging analysis methodology.
Discussion
The biopharmaceutical industry has been aggressively looking for more predictive animal models and advanced imaging techniques to increase the probability of success of advancing preclinical drug candidates to human clinical trials. For oncology, tumor xenograft models in immunodeficient mice remain the standard model to initially test and select compounds. However, these models poorly resemble human cancers and clinical trial results because the integrity of signaling pathways, mutation status, stromal cell interaction, etc., are altered by cell culture, implantation processes, and tumor-host interactions [19]. Genetically engineered mouse models (GEMMs) created by introducing oncogenes or altering tumor suppressor genes in the mouse genome can mimic human cancer development in several aspects. Genetically engineered mouse models have been proposed as additional models for preclinical assessment of drug candidates to complement the widely used xenograft models [20]. However, application of these GEMMs is hampered by relatively high cost, complex breeding process, mixed genetic backgrounds, extended tumor latency, and, in particular, difficulty of internal tumor detection and measurement.
The K-ras and the K-ras/p53 compound conditional knock-in mouse models are two GEMMs that faithfully recapitulate characteristics of the human lung adenocarcinoma disease [6]. Micro-CT has been used to characterize lung tumor mouse models by qualitative assessment or quantitative measurement of individual tumors [21–23]. Most traditional micro-CT instruments take 10 to 20 minutes to scan the entire mouse body. This relatively low throughput limits micro-CT application to drug discovery because of the number of animals required in each study to reach statistical significance. For most GEMM studies, large cohorts of animals are needed owing to the complexity and variability of tumor latency, tumor penetrance, and growth kinetics. Therefore, development of a higher-throughput imaging protocol will greatly facilitate use of these models in drug efficacy studies. The high-throughput method for the quantification of lung tumor burden in this report used the GE eXplore Locus Ultra Pre-Clinical CT scanner, which allowed us to acquire micro-CT images of the whole body of two mice in only 16 seconds. This throughput translates into the realistic ability to perform imaging studies on more than 100 mice/day. The relatively fast acquisition times of this micro-CT scanner is offset by its relatively lower intrinsic resolution compared to other micro-CT instruments. Volumetric analysis of tumor burden is another limiting factor of throughput. Quantification of individual tumors by manually drawing regions of interest of individual tumors can be very time-consuming. For the K-ras/p53 model, in particular, mice normally develop multifocal small tumors that are visible but may not be quantifiable. The method described in this report is semiautomated by applying a threshold-based region grow algorithm and approximately 25 to 30 mice can be analyzed per day. This analytical approach measured not only the isolated tumors but also the small multifocal tumors that are not measurable by manually drawing regions of interest. Therefore, this method should represent more precisely the total tumor burden as confirmed by histologic data. The relatively poor correlation between histologic tumor areas and micro-CT tumor volumes among individual animals most likely arose from the limited histologic sampling. Indeed, a single slice of the left side of the lung does not capture and represent location, distribution, and irregular shapes of the tumors. The mean of tumor areas from a group of mice more accurately reflects tumor burden by averaging both underestimated and overestimated values (Figure 2C). Another advantage of this micro-CT analysis approach is that tumor development and response to drug treatment could be monitored longitudinally, whereas histologic examination is a terminal readout. As noted in the erlotinib experiment, with groups of 12 to 15 animals, the initial tumor burden before treatments varied markedly among three groups, although animals were randomly assigned. Noninvasive micro-CT imaging allows us to follow tumor growth more accurately by normalizing to predosing tumor burden. It makes it possible to prescreen and select animals with a certain range of tumor volumes as animal enrollment criteria to reduce group variability of tumor burden.
Using micro-CT, we detected increased lung opacity, which was determined to be macrophage infiltration with the histologic finding caused by viral infection 8 weeks after infection. Accumulation of macrophage cells in the lung may potentially interfere with tumor quantification. However, macrophage infiltration was mostly observed primarily in the early stages of infection when there was no significant tumor formation. This phenomenon was rarely observed at 14 weeks or later after viral infection when tumors become detectable. It will be necessary to prescreen animals and exclude animals that have acute inflammatory responses. Because micro-CT cannot distinguish infiltrated macrophages from tumor or vasculature, this method may falsely include local macrophage infiltration if animals do have lung infections. However, we did not observe significant macrophage infiltration in these two models after 14 weeks of viral infection either by imaging or by histologic analysis.
The method described here does not distinguish vascular tissue from tumor tissue. Non-contrast-enhanced micro-CT does not generate enough contrast between T & V volume to differentiate the tissues. Vasculature volume may increase as tumors grow and recruit additional vascular supply. However, the relative total tumor burden should be accurately approximated by measuring the total tumor and vessel volume. As noted, tumor burden measured by micro-CT was consistent with histologic assessment (Figure 2). We also found unexpectedly that tumor development was associated with the increase in the chest space volume. It is possible that tumor growth physically expanded the chest volume owing to size restraints. In addition, growth of tumor tissue may impair lung function that will reduce lung respiration. Another limitation of this method is that the micro-CT image acquisition was not respiratory-gated. Respiratory motion can introduce blurring of image edges [21], which would result in an over-estimation of the total tumor burden. Variability in the respiratory rate of individual mice may also affect tumor burden measurement.
In summary, we have developed a high-throughput quantitative analytical method to measure total tumor burden in lung using micro-CT. This approach makes it possible to evaluate oncology therapeutic compounds in complex genetically engineered lung tumor mouse models.
Acknowledgments
The authors thank Nancy E. Kohl and Shailaja Kasibhatla for critical review of the manuscript and Nirah H. Shomer and Sada Breegi for providing animal support. The authors also thank Jacob Marcus for assisting histologic examination of tumor area of lung.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Meuwissen R, Linn SC, van der Valk M, Mooi WJ, Berns A. Mouse model for lung tumorigenesis through Cre/lox controlled sporadic activation of the K-Ras oncogene. Oncogene. 2001;20:6551–6558. doi: 10.1038/sj.onc.1204837. [DOI] [PubMed] [Google Scholar]
- 3.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 4.Chiba I, Takahashi T, Nau MM, D'Amico D, Curiel DT, Mitsudomi T, Buchhagen DL, Carbone D, Piantadosi S, Koga H, et al. Mutations in the p53 gene are frequent in primary, resected non-small cell lung cancer. Lung Cancer Study Group. Oncogene. 1990;5:1603–1610. [PubMed] [Google Scholar]
- 5.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 7.Hoess RH, Abremski K. Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc Natl Acad Sci USA. 1984;81:1026–1029. doi: 10.1073/pnas.81.4.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lakso M, Sauer B, Mosinger B, Jr, Lee EJ, Manning RW, Yu SH, Mulder KL, Westphal H. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci USA. 1992;89:6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Clerck NM, Meurrens K, Weiler H, Van Dyck D, Van Houtte G, Terpstra P, Postnov AA. High-resolution X-ray microtomography for the detection of lung tumors in living mice. Neoplasia. 2004;6:374–379. doi: 10.1593/neo.03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cody DD, Nelson CL, Bradley WM, Wislez M, Juroske D, Price RE, Zhou X, Bekele BN, Kurie JM. Murine lung tumor measurement using respiratory-gated micro-computed tomography. Invest Radiol. 2005;40:263–269. doi: 10.1097/01.rli.0000160070.67270.05. [DOI] [PubMed] [Google Scholar]
- 11.Wislez M, Spencer ML, Izzo JG, Juroske DM, Balhara K, Cody DD, Price RE, Hittelman WN, Wistuba II, Kurie JM. Inhibition of mammalian target of rapamycin reverses alveolar epithelial neoplasia induced by oncogenic K-ras. Cancer Res. 2005;65:3226–3235. doi: 10.1158/0008-5472.CAN-04-4420. [DOI] [PubMed] [Google Scholar]
- 12.Wislez M, Fujimoto N, Izzo JG, Hanna AE, Cody DD, Langley RR, Tang H, Burdick MD, Sato M, Minna JD, et al. High expression of ligands for chemokine receptor CXCR2 in alveolar epithelial neoplasia induced by oncogenic kras. Cancer Res. 2006;66:4198–4207. doi: 10.1158/0008-5472.CAN-05-3842. [DOI] [PubMed] [Google Scholar]
- 13.Grimm J, Kirsch DG, Windsor SD, Kim CF, Santiago PM, Ntziachristos V, Jacks T, Weissleder R. Use of gene expression profiling to direct in vivo molecular imaging of lung cancer. Proc Natl Acad Sci USA. 2005;102:14404–14409. doi: 10.1073/pnas.0503920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimoto N, Wislez M, Zhang J, Iwanaga K, Dackor J, Hanna AE, Kalyankrishna S, Cody DD, Price RE, Sato M, et al. High expression of ErbB family members and their ligands in lung adenocarcinomas that are sensitive to inhibition of epidermal growth factor receptor. Cancer Res. 2005;65:11478–11485. doi: 10.1158/0008-5472.CAN-05-1977. [DOI] [PubMed] [Google Scholar]
- 16.Zerbe LK, Dwyer-Nield LD, Fritz JM, Redente EF, Shroyer RJ, Conklin E, Kane S, Tucker C, Eckhardt SG, Gustafson DL, et al. Inhibition by erlotinib of primary lung adenocarcinoma at an early stage in male mice. Cancer Chemother Pharmacol. 2008;62:605–620. doi: 10.1007/s00280-007-0644-z. [DOI] [PubMed] [Google Scholar]
- 17.Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD, Bekele BN, Herbst RS, Wistuba II. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 18.Miller VA, Riely GJ, Zakowski MF, Li AR, Patel JD, Heelan RT, Kris MG, Sandler AB, Carbone DP, Tsao A, et al. Molecular characteristics of bronchioloalveolar carcinoma and adenocarcinoma, bronchioloalveolar carcinoma subtype, predict response to erlotinib. J Clin Oncol. 2008;26:1472–1478. doi: 10.1200/JCO.2007.13.0062. [DOI] [PubMed] [Google Scholar]
- 19.Johnson JI, Decker S, Zaharevitz D, Rubinstein LV, Venditti JM, Schepartz S, Kalyandrug S, Christian M, Arbuck S, Hollingshead M, et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer. 2001;84:1424–1431. doi: 10.1054/bjoc.2001.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olive KP, Tuveson DA. The use of targeted mouse models for preclinical testing of novel cancer therapeutics. Clin Cancer Res. 2006;12:5277–5287. doi: 10.1158/1078-0432.CCR-06-0436. [DOI] [PubMed] [Google Scholar]
- 21.Cavanaugh D, Johnson E, Price RE, Kurie J, Travis EL, Cody DD. In vivo respiratory-gated micro-CT imaging in small-animal oncology models. Mol Imaging. 2004;3:55–62. doi: 10.1162/15353500200403184. [DOI] [PubMed] [Google Scholar]
- 22.Li XF, Zanzonico P, Ling CC, O'Donoghue J. Visualization of experimental lung and bone metastases in live nude mice by X-ray micro-computed tomography. Technol Cancer Res Treat. 2006;5:147–155. [PubMed] [Google Scholar]
- 23.Greschus S, Savai R, Wolf JC, Rose F, Seeger W, Fitzgerald P, Traupe H. Non-invasive screening of lung nodules in mice comparing a novel volumetric computed tomography with a clinical multislice CT. Oncol Rep. 2007;17:707–712. [PubMed] [Google Scholar]