Abstract
Activating BRAF mutations have been reported in 40% of papillary thyroid carcinomas (PTCs). The involvement of BRAF pseudogene in thyroid tumorigenesis has not previously been studied. We investigated BRAF pseudogene expression in 68 thyroid tumors: 16 multinodular goiters, 43 classic PTCs, 6 follicular variants of PTC, and 3 anaplastic thyroid carcinomas. BRAF pseudogene function was studied by Western blots, soft agar assay, and tumorigenesis in nude mice. BRAF pseudogene expression was detected in 7 multinodular goiters, 18 classic PTC, and 1 follicular variants of PTC. There is an inverse correlation between BRAF pseudogene expression and BRAF mutation. The pseudogene transcripts were more frequently detected in tumors without BRAF mutation than those with BRAF mutation. Furthermore, BRAF pseudogene expression could activate the MAP kinase signaling pathway, transform NIH3T3 cells in vitro, and induce tumors in nude mice. These data suggest that BRAF pseudogene activation may play a role in thyroid tumor development.
Introduction
Papillary thyroid carcinomas (PTCs) are the most common type of differentiated thyroid carcinomas, accounting for some 85% thyroid malignancies [1]. The most common genetic events are involved in RAS-RAF-MEK-ERK-MAPK signaling pathway such as RAS point mutations, RET/PTC oncogenes, NTRK1 rearrangements (TRK), and BRAF mutations [2–8]. BRAF is a serine/threonine kinase that serves as an immediate downstream effector of RAS in the RAS-RAF-MEK-ERK-MAPK signaling cascade. Oncogenic mutations in BRAF are common in human cancers, nearly all of which are the T1799A transversion in exon 15, resulting in a V600E substitution in the protein [9,10]. This mutation is believed to produce a constitutively active kinase by disrupting hydrophobic interactions between residues in the activation loop and residues in the ATP binding site [11,12].
Two human BRAF loci (B-raf-1 and B-raf-2) have been identified [13]. B-raf-1 is located on chromosome 7q34 and encodes the functional 94-kDa BRAF protein. B-raf-2 is located on chromosome Xq13 and has been predicted to be an inactive processed pseudogene owing to the presence of point mutations, insertions, and deletions relative to B-raf-1, although its expression and function has not been investigated yet [13].
Pseudogenes have a similar DNA sequence to their functional genes; they are nonetheless unable to produce functional final protein products and have generally been considered as evolutionary “dead-end.” They may originate from reverse transcription (RT) of normal mRNA transcripts (processed pseudogenes) or from gene duplication (nonprocessed pseudogenes) [14]. One of the interesting findings from genome sequencing is the abundance of pseudogenes in mammalian genomes: roughly the same number of pseudogenes as protein-coding genes in all mammals sequenced so far [15]. Now there is a growing number of evidence suggesting that some of them may have active regulatory roles [16–18].
In the present study, we investigated BRAF pseudogene expression in 68 thyroid tumors from Saudi Arabia, its relationship with BRAF mutation, and its association with clinical and pathological features of PTC. The functional aspects of BRAF pseudogene expression were also studied.
Materials and Methods
Thyroid Tumor Specimens
All tumor tissues were obtained at surgery and stored at -70°C until processed. The clinical staging of thyroid cancer was based on the TNM classification [19]. Sixty-eight thyroid tumors were studied: 16 benign multinodular goiters, 43 classic PTCs, 6 follicular variants of PTC, and 3 anaplastic thyroid carcinomas (ATCs). The study was reviewed and approved by the institutional review board.
DNA Amplification and Sequence Analysis of Exon 15 of BRAF
Tumor DNA was extracted by standard proteinase-K treatment followed by phenol/chloroform extraction. Exon 15 was amplified by polymerase chain reaction (PCR) using the following two primers: 5′-TCATAATGCTTGCTCTGATAGGA-3′ and 5′-CTAGTAACTCAGCAGCATCTC-3′. The PCR conditions were 95°C for 10 minutes followed by 35 cycles of amplification (95°C for 1 minute, 54°C for 1 minute, and 72°C for 1 minute). The resulting PCR products were analyzed by gel electrophoresis. Each successfully amplified fragment was directly sequenced using the BigDye Terminator V3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA).
Reverse Transcription-PCR Analysis of BRAF Pseudogene Transcripts
Total RNA was extracted using the guanidinium thiocyanate-phenol-chloroform method. To amplify BRAF pseudogene transcripts, 2 µg of total RNA were first treated with DNase 1 (10 U at 37°C for 30 minutes, heat inactivation at 70°C for 5 minutes) to remove contaminating genomic DNA and then reverse-transcribed into cDNA using Oligo dT-based reverse transcription system (Proems, Madison, WI). Pseudogene transcripts were amplified by PCR using the following two specific primers: 5′-CTTGTATTACCTTCTCCATATA-3′ and 5′-CTAGACCAAAATCACCTATTTCC-3′. The sequences different from BRAF wild type gene were highlighted in bold. Polymerase chain reaction conditions were the same as described above. The resulting 185-bp PCR products were analyzed by gel electrophoresis and were directly sequenced to confirm specificity of pseudogene transcripts. To amplify BRAF transcripts covering exon 15, the forward primer (based on exon 14: 5′-GCATGGATTACTTACACGCCA-3′) and the reverse primer (based on exon 16: 5′-CAATTCATACAGAACAATCC-3′), were located in different exons to avoid amplification of contaminating genomic DNA. This primer pair can also amplify corresponding BRAF pseudogene transcripts.
Quantitative Real-time RT-PCR Analysis of BRAF Transcripts
Quantitative RT-PCR analysis was performed as described previously [20]. Briefly, total RNA were reverse-transcribed into cDNA. LightCycler DNA Master SYBR Green kit was used for quantitative PCR analysis (Roche, Mannheim, Germany). The cDNA mix was diluted 10-fold, and 2 µl of the dilution was used for real-time PCR analysis. BRAF-specific PCR primers for 216-bp cDNA fragment were 5′-CAAACTTATAGATATTGCACA-3′ (sense) and 5′-TCTGGTGCCATCCACAAAATG-3′ (antisense). The sequence different from BRAF pseudogene was highlighted in bold, and the PCR primers span intron 14 so that contaminating genomic DNA will not be amplified. The mRNA level of housekeeping gene glyceraldehydes-3-phosphate dehydrogenase (GAPDH) was used as an internal control, and a 300-bp PCR product was amplified using the following two primers: 5′-ACAGTCAGCCGCATCTTCTT-3′ (sense) and 5′-TTGATTTTGGAGGGATCTCG-3′ (antisense). The PCR conditions were as follows: 95°C for 30 seconds followed by 35 cycles of amplification (95°C for 5 seconds, 48°C for 5 seconds, and 72°C for 10 seconds). The resulting concentrations of BRAF PCR products were normalized by comparison with GAPDH and were used to determine the BRAF mRNA level.
Cloning and Expression of BRAF-Wild Type, -V600E Mutant, and -Pseudogene
The full-length BRAF-wild type and -V600E mutant cDNA were amplified by RT-PCR from total RNA extracted from thyroid tissue using the following primers: 5′-GACAGCGGCCGCTCGGGCCC-3′ (sense), 5′-TCTCGGTTATAAGATGGCGGCG-3′ (sense, internal for nested PCR), and 5′-CCTGAACTCT CTCACTCATTTG-3 (antisense). The pseudogene cDNA was amplified by RT-PCR using primers corresponding to exon 2 (5′-GTATGGAATATCAAACAAATAA-3′) and 3′untranslated region of wild type BRAF: 5′-CCTGAACTCTCTCACTCATTTG-3 (internal for nested PCR, antisense) and 5-TCCTTTTGTTGCTACTCTCCTG-3′ (antisense). The resulting cDNA fragment was cloned into the expression vector pcDNA3.1, which is under the control of the CMV promoter (Invitrogen, Carlsbad, CA), and was verified by DNA sequencing with no mutation found. All three constructs (pBRAFWT, pBRAFV600E, and pBRAFPSEUDO) were transfected into CHO and NIH3T3 cells using Lipofectamine (Invitrogen). Cells were selected in the presence of 400 µg/ml G418 for 3 weeks (Sigma, St. Louis, MO), and stable clones were pooled for experiments. To detect protein expression of the pseudogene transcripts, the coding sequence was amplified by PCR from the pseudogene cDNA and cloned into a pcDNA3.1/V5-His expression vector in frame with the V5 epitope at the C-terminus (Invitrogen), using the following two primers: 5′-GGAACGGAACTGATTTTTCTGTT-3′ (sense) and 5′-GTGATCTTCATCTGCTGGT CGGAA-3′ (antisense). The resulting plasmid (pBRAFPSEUDO-V5) was stably transfected into CHO cells.
Western Blot Analysis
A total of 60 µg of protein from stably transfected CHO cells (pBRAFPSEUDO-V5, pBRAFWT, pBRAFV600E, and pBRAFPSEUDO) were loaded onto a 12% sodium dodecyl sulfate-polyacrylamide gel. Proteins were transferred to a polyvinylidene fluoride membrane and were subjected toWestern blot analysis with anti-V5 monoclonal antibody (1:5000 dilution; Invitrogen) or anti-total ERK 1/2 (1:2000 dilution) and anti-phospho-ERK 1/2 antibodies (1:1000; Santa Cruz Biotechnology Inc., Santa Cruz, CA).
Quantification of MAP Kinase Activation by BRAF Pseudogene in CHO Cells
pBRAFPSEUDO- and pBRAFPSEUDO-V5-transfected CHO cells were cultured in serum-free medium for 16 hours. The cells were then stimulated with 20% fetal bovine serum for 2 hours in the presence or absence of 20 µM MEK inhibitor U0126 or BRAF inhibitor SB590885 (EMD Chemicals, Inc., Gibbstown, NJ) for 2 hours. The amount of cellular phospho-ERK 1/2 was measured using a cell-based ELISA kit according to the manufacturer's instructions (SuperArray Bioscience Co., Frederick, MD).
Transformation Assays
A total of 2 x 104 stably transfected NIH3T3 cells (pBRAFWT, pBRAFV600E, and pBRAFPSEUDO) were plated into a six-well cell culture plate for an anchorage-independent growth by soft agar assay according to the manufacturer's instructions (Chemicon International, Temecula, CA). For in vivo transformation experiments, 1 x 106 of stably pBRAFPSEUDO-transfected NIH3T3 cells were injected subcutaneously into four female nude mice (The Jackson Laboratory, Bar Harbor, ME). Mice were observed twice weekly and killed when tumor reached 5 x 5 mm in size.
Results
BRAF Pseudogene Expression in Thyroid Tumors
Because BRAF pseudogene is a retrogene and has no intron, contaminating genomic DNA will be amplified by PCR from RNA preparations. To eliminate genomic DNA contamination, RNA was first digested with DNase 1 before being reverse-transcribed into cDNA. The pseudogene transcripts were detected in 7 (43.8%) of 16 multinodular goiters, 18 (41.9%) of 43 classic PTCs, and 1 (16.7%) of 6 follicular variants of PTC (FVPTCs; Figure 1 and Table 1). The transcripts were not detected in five normal thyroid tissues (data not shown). The transcripts were detected more frequently in tumors without BRAF mutation (20/27, 74.4%) than those with BRAF mutation (7/27, 25.9%; Table 1; P < .01, Fisher exact test). They were also found more frequently in benign tumors and in early stages of thyroid cancer (Figure 2A). By contrast, BRAF mutation is frequently found in patients with stages III and IV tumors (P < .01; Figure 2B). When using generic primers surrounding BRAF exon 15, wild type and mutated T1799A BRAF transcripts could be detected together with pseudogene transcripts by RT-PCR sequence analysis (Figure 3). The pseudogene transcripts harbored several sequence variations compared with wild type BRAF: T1794C (A1794A), C1807T (R603X), C1848T (S616S), A1849T (I617F), G1857T (W619C), C1894T (P632S), and G1915A (V639I) (Figure 3). If one overlooks the existence of pseudogene expression, these sequence variations could be mistakenly identified as BRAF mutations. However, these sequence variations were not present in the matched genomic DNA (Figure 3) because BRAF and its pseudogene are located on chromosomes Xq13 and 7q34, respectively. Sequence analysis showed that the pseudogene transcripts were more abundant than wild type or mutant BRAF transcripts in some tumor samples, as demonstrated by a single homozygous peak in the sequence chromatogram, whereas in other samples, heterozygous peaks could be detected, showing both functional BRAF and BRAF pseudogene transcripts (Figure 3).
Figure 1.
Detection of BRAF pseudogene expression by RT-PCR from thyroid tumor samples. A total of 2 µg of total RNA were first treated with DNase 1 to remove contaminating genomic DNA and then reverse-transcribed into cDNA. Pseudogene cDNA were amplified by PCR using specific primers. No amplification was seen when RNA was used as a template. Representative results from two cell lines and four samples were shown. Pseudogene transcripts were detected as a 185-bp cDNA fragment from SW579 cell line, one goiter and two PTC samples.
Table 1.
BRAF T1799A Mutation and Pseudogene Expression in Thyroid Carcinoma.
| No. | Diagnosis | Age (year) | Sex | Stage | BRAF T1799A Mutation | BRAF Pseudogene Expression |
| 1 | Goiter | 35 | F | No | No | |
| 2 | Goiter | 49 | M | No | No | |
| 3 | Goiter | 40 | F | No | Yes | |
| 4 | Goiter | 39 | F | No | Yes | |
| 5 | Goiter | 38 | F | No | Yes | |
| 6 | Goiter | 38 | F | No | Yes | |
| 7 | Goiter | 30 | F | No | No | |
| 8 | Goiter | 65 | F | No | No | |
| 9 | Goiter | 31 | F | No | No | |
| 10 | Goiter | 30 | F | No | Yes | |
| 11 | Goiter | 38 | F | No | Yes | |
| 12 | Goiter | 43 | M | No | Yes | |
| 13 | Goiter | 60 | F | No | No | |
| 14 | Goiter | 52 | M | No | No | |
| 15 | Goiter | 21 | F | No | No | |
| 16 | Goiter | 45 | F | No | No | |
| 17 | PTC | 28 | F | I | No | No |
| 18 | PTC | 40 | M | I | No | No |
| 19 | PTC | 23 | F | I | No | No |
| 20 | PTC | 48 | F | I | No | No |
| 21 | PTC | 36 | F | I | No | No |
| 22 | PTC | 36 | F | I | No | Yes |
| 23 | PTC | 43 | F | I | No | Yes |
| 24 | PTC | 18 | M | I | No | Yes |
| 25 | Metastatic PTC | 33 | F | 1 | No | No |
| 26 | Metastatic PTC | 24 | F | I | No | No |
| 27 | Metastatic PTC | 35 | F | I | Yes | No |
| 28 | Metastatic PTC | 26 | F | I | No | Yes |
| 29 | Metastatic PTC | 36 | F | I | Yes | Yes |
| 30 | Metastatic PTC | 22 | F | II | No | No |
| 31 | Metastatic PTC | 41 | F | II | Yes | Yes |
| 32 | Metastatic PTC | 12 | F | I | No | Yes |
| 33 | Metastatic PTC | 40 | F | I | Yes | No |
| 34 | Metastatic PTC | 32 | F | I | No | No |
| 35 | Metastatic PTC | 24 | F | I | No | Yes |
| 36 | Metastatic PTC | 30 | F | I | Yes | No |
| 37 | Metastatic PTC | 25 | F | I | Yes | No |
| 38 | Metastatic PTC | 37 | F | I | No | Yes |
| 39 | Metastatic PTC | 26 | M | I | No | Yes |
| 40 | PTC | 82 | M | II | Yes | Yes |
| 41 | PTC | 57 | F | II | Yes | Yes |
| 42 | PTC | 52 | F | II | No | Yes |
| 43 | Metastatic PTC* | 27 | F | II | No | Yes |
| 44 | PTC | 55 | F | III | Yes | Yes |
| 45 | PTC | 46 | M | III | No | No |
| 46 | PTC | 60 | M | III | Yes | No |
| 47 | PTC | 58 | F | III | No | No |
| 48 | PTC | 73 | F | III | Yes | No |
| 49 | PTC | 72 | F | III | No | No |
| 50 | PTC | 59 | F | III | Yes | No |
| 51 | PTC | 50 | M | III | Yes | No |
| 52 | PTC | 72 | F | III | Yes | No |
| 53 | PTC | 66 | M | III | Yes | No |
| 54 | PTC | 71 | M | IV | No | Yes |
| 55 | PTC | 46 | F | IV | Yes | Yes |
| 56 | PTC | 70 | F | IV | Yes | No |
| 57 | PTC | 63 | M | IV | Yes | No |
| 58 | PTC | 49 | M | IV | Yes | No |
| 59 | PTC | 77 | M | IV | Yes | Yes |
| 60 | ATC | 81 | M | IV | Yes | No |
| 61 | ATC | 76 | F | IV | Yes | No |
| 62 | ATC | 43 | F | IV | Yes | No |
| 63 | FVPTC | 32 | F | I | No | No |
| 64 | FVPTC | 34 | F | I | No | No |
| 65 | Metastatic FVPTC | 21 | F | I | No | Yes |
| 66 | FVPTC | 43 | M | I | No | No |
| 67 | FVPTC | 68 | F | IV | No | No |
| 68 | FVPTC | 45 | F | II | No | No |
| ARO cell line | Yes | No | ||||
| NPA cell line | Yes | No | ||||
| SW579 cell line | No | Yes |
Metastatic PTC refers to stage I or II tumors with local lymph note metastasis.
PTC-1 mutation was detected.
Figure 2.
BRAF pseudogene expression in thyroid tumor specimens. (A) Inverse association of BRAF pseudogene expression with aggressiveness of PTC. The frequency of BRAF pseudogene expression was reduced in advanced stages of thyroid carcinoma. No BRAF pseudogene transcripts were detected in stage III PTC samples. (B) Association of BRAF mutation with aggressiveness of PTC. Higher frequency of V600E mutation was detected in advanced stages of thyroid carcinoma. Six FVPTC and three ATC samples were included in the analysis. No mutation was detected in the goiter.
Figure 3.
Detection of BRAF pseudogene transcripts by sequencing analysis of RT-PCR products. Three representative sequence chromatograms of matched DNA and RNA samples were shown. (A) Presence of both BRAF wild type and pseudogene transcripts in an RNA sample of multinodular goiter (no. 3 in Table 1). The pseudogene transcripts presented as heterozygous mutations at T1794C (A1794A) and C1807T (R603X), whereas matched DNA sample did not show the mutations. (B) Detection of only BRAF pseudogene transcripts in an RNA sample of multinodular goiter (no. 4 in Table 1). The pseudogene transcripts presented as homozygous mutations at T1794C (A1794A) and C1807T (R603X), whereas matched DNA sample did not show the mutations. (C) Presence of both BRAF mutant and pseudogene transcripts in an RNA sample of PTC (no. 44 in Table 1). The pseudogene transcripts presented as heterozygous mutations at T1794C (A1794A) and C1807T (R603X). Heterozygous T to A BRAF mutation was detected at nucleotide 1799 and was confirmed in the matched DNA sample.
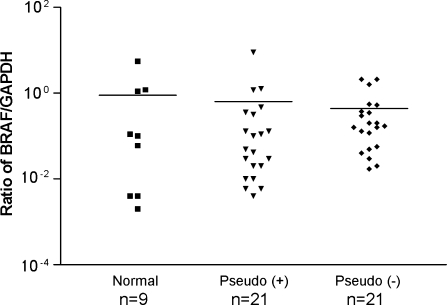
To determine whether there is a correlation between BRAF gene expression and BRAF pseudogene expression, we performed quantitative PCR comparing BRAF gene expression among normal, pseudogene-positive, and pseudogene-negative tumor samples. As shown in Figure 4, no correlation of BRAF expression was found between pseudogene-positive and pseudogene-negative tumors. In pseudogene-positive samples, the mean of BRAF/GAPDH ratio is 0.63 ± 0.43 compared with 0.44 ± 0.14 in pseudogene-negative tumors (P > .05, Student's t test). These data suggest that BRAF pseudogene expression may not regulate BRAF expression.
Figure 4.
Real-time RT-PCR analysis of BRAF mRNA among normal, BRAF pseudogene-positive, and pseudogene-negative thyroid tumor samples. BRAF transcripts were quantified and normalized by comparison with GAPDH. Data are expressed as ratio of BRAF/GAPDH mRNA among different groups. Because GAPDH mRNA is more abundant than BRAF, log scale is used.
Activation of MAP Kinase by BRAF Pseudogene Expression
Analysis of the pseudogene cDNA sequence revealed that it has a long open reading frame of 244 amino acids and corresponds to the wild type CR1 domain (Figure 5), which has been shown to mediate autoinhibition of the C-terminal protein kinase domain (CR3) [21]. Amino acid sequence alignment between wild type BRAF and its pseudogene showed 85.7% homology, suggesting that they may interact with each other resulting in modulation of the MAP kinase signaling pathway. To further investigate the functional role of BRAF pseudogene expression, we cloned and expressed the pseudogene in both CHO and NIH3T3 cells. As shown in Figure 6A, BRAF pseudogene expression could activate MAP kinase in CHO cells. Similar results were also found in NIH3T3 cells. We next quantified MAP kinase activation by measuring phospho-ERK in BRAF pseudogene (full-length) and N-terminal part of BRAF pseudogene-transfected CHO cells. As shown in Figure 6B, both BRAF pseudogene and N-terminal part of BRAF pseudogene could induce a higher level of MAP kinase activation in the absence of serum stimulation. Both MEK inhibitor U0126 and BRAF inhibitor SB590885 could reduce the kinase activation by more than 70%. These data suggest that BRAF pseudogene could lead to constitutive MAP kinase activation through interaction with wild type BRAF and MEK, likely through the N-terminal part of BRAF pseudogene. For confirmation that pseudogene transcripts could be translated into protein, we cloned the potential pseudogene coding sequence (244 amino acids N-terminus) into a pcDNA3.1/V5 expression vector in frame with the V5 epitope tag (pBRAFPSEUDO-V5). Using V5 monoclonal antibody, we were able to detect a predicted 32-kDa protein in CHO cells stably transfected with the vector, whereas no protein was detected in CHO cells transfected with vector alone (Figure 6C).
Figure 5.
(A) Schematic representation of human BRAF gene and its pseudogene in scale. Each BRAF exon is indicated by its number inside the boxes. The three conserved regions of the protein with the ARAF and RAF-1 genes are indicated by CR1, CR2, and CR3, respectively. Ras binding domain (RBD), cysteine-rich domain (CRD), and kinase domain (KD) are indicated by three bars. G-loop is a conserved glycine motif in exon 11, and AS is the activation segment in exon 15. Black arrows indicate the major phosphorylation sites of the protein. The longest open reading frame of BRAF pseudogene is indicated by a black box and corresponds to wild type CR1 region, which plays a regulative role in kinase activity. (B) Amino acid sequence alignment between wild type BRAF and its pseudogene. Sequence difference is highlighted in bold. There are a total of 244 amino acids in the pseudogene coding region, and only 35 amino acid difference compared with wild type (85.68% homology).
Figure 6.
(A) Activation of MAP kinase by BRAF pseudogene. Chinese hamster ovary cells were stably transfected by pBRAFWT, pBRAFV600E, or pBRAFPSEUDO, respectively. A total of 60 µg of proteins from each cell lysate were used for Western blot analysis. Two blots with identical amount of protein loading were probed with antibodies to phospho-ERK 1/2 (pERK, indicating MAP kinase activation) and total ERK 1/2 (for monitoring protein loading), respectively. (B) Quantification of MAP kinase activation by BRAF pseudogene and N-terminal part of BRAF pseudogene (pBRAFPSEUDO-V5) in CHO cells. pBRAFPSEUDO- and pBRAFPSEUDO-V5-transfected CHO cells were cultured in serum-free medium for 16 hours. The cells were then stimulated with 20% fetal bovine serum for 2 hours in the presence or absence of 20 µM MEK inhibitor U0126 or BRAF inhibitor SB590885. The amount of phospho-ERK in the cells was measured using a cell-based ELISA kit at OD 450 nm and was normalized by dividing the OD 595 nm readings of relative cell number. Data are expressed as means ± SEM of two separate experiments. (C) Protein expression of BRAF pseudogene. The putative BRAF pseudogene coding region of 244 amino acids was cloned into a pcDNA3.1/V5-His vector (pBRAFPSEUDO-V5) and was expressed as a fusion protein with a V5 epitope tag at the C-terminus in CHO cells. The expected 32-kDa fusion protein was detected by Western blot using V5 monoclonal antibody (lane 1). No protein was detected when CHO cells were transfected with a pcDNA3.1/V5-His vector alone (lane 2).
The transformation potential of BRAF pseudogene was assessed in vitro by a soft agar assay in pBRAFPSEUDO-transfected NIH3T3 and CHO cells and in vivo by subcutaneous injection of stably pBRAFPSEUDO-transfected NIH3T3 cells into four nude mice. As shown in Figure 7, A and B, the soft agar assay resulted in six-fold increases in the number of colonies from pBRAFPSEUDO-transfected NIH3T3 cells and a four-fold increase from pBRAFPSEUDO-transfected CHO cells, respectively. All four nude mice developed tumors within 2 weeks after injection of 1 x 106 pBRAFPSEUDO-transfected NIH3T3 cells.
Figure 7.
(A) Anchorage-independent growth of NIH3T3 and CHO cells stably transfected by pBRAFWT, pBRAFV600E, or pBRAFPSEUDO, respectively. A total of 2 x 104 cells in 0.4% soft agar were cultured in duplicate in a six-well cell culture plate for 28 days. The colonies were stained and counted. The results are the average number of colonies per well from four wells. (B) Representative results of colony formation of NIH3T3 cells or CHO cells stably transfected by pBRAFWT, pBRAFV600E, or pBRAFPSEUDO, respectively. Original magnification, x40.
Discussion
In the present study, we reported BRAF pseudogene expression in thyroid tumors, which could transactivate MAP kinase and induce tumors in nude mice. Furthermore, reciprocal BRAF mutation and its pseudogene expression were also demonstrated.
BRAF pseudogene is a processed pseudogene that arises by retrotransposition. In the process of retrotransposition, a portion of the mRNA transcript of a gene is spontaneously reverse-transcribed back into DNA and inserted into chromosomal DNA. Because it is derived from a mature mRNA product, a processed pseudogene lacks introns and the upstream promoter of a normal gene and is thus considered “dead on arrival.” Pseudogenes also have many genetic alterations such as point mutations, deletions, and insertions compared to their normal genes [22]. However, many exceptions exist where a pseudogene can be transcribed and even translated into an active protein [22–28]. Indeed, BRAF pseudogene produces mRNA transcripts that have many stop codons and cannot be translated into a functional, full-length protein. The longest open reading frame can only produce a polypeptide of 244 amino acids. This polypeptide has no kinase domain and, therefore, cannot be functional by itself. However, expression of BRAF pseudogene in some thyroid tumors suggests that it may play a role in thyroid tumor development. The functional studies confirmed that the BRAF pseudogene plays an oncogenic role in thyroid tumorigenesis by transactivating the MAP kinase pathway, likely through wild type BRAF, because BRAF inhibitor abolishes its effect. Given the high sequence homology between the CR1 domain of wild type BRAF and its pseudogene, it is likely that the BRAF pseudogene peptide interacts with the CR1 domain of wild type BRAF and interferes with its normal physiologic role of negatively autoregulating its C-terminal protein kinase domain. Hitotsune et al. [29,30] demonstrated that an expressed pseudogene Mkrn1-p1 could regulate the expression of its homologous functional coding gene by modulating messenger RNA stability. However, their results have been challenged recently by Gray et al. [31], who showed that the Mkrn1-p1 pseudogene is neither expressed nor imprinted, nor does it transregulate its source gene. Apparently, the contradictory reports remain to be settled.
There is an inverse correlation between BRAF pseudogene activation and BRAF mutation in thyroid tumors. BRAF pseudogene transcripts are more frequently detected in benign goiters and early stages of thyroid cancer. It becomes silenced in late stages of cancer, especially when BRAF mutation is present, suggesting that only one of the events is enough to drive oncogenic transformation of normal thyroid epithelial cells. A previous study by Zhang et al. [32] showed that PTEN pseudogene expression might play a pathological role in glioblastomas and was complementary to PTEN mutation. In 14 of 18 glioblastomas, either PTEN mutation or PTEN pseudogene expression was found, and only one case showed both PTEN mutation and pseudogene expression. Their study reported similar observations to the current study, that is, BRAF or PTEN pseudogene expression could act as an alternative to BRAF or PTEN mutation [32].
The high frequency of BRAF pseudogene expression in benign goiters suggests that it may play a role in initiating goiter formation or that a subset of goiters may contain precursor lesions such as BRAF pseudogene expression, which may progress to malignancy in the context of additional genetic or environmental factors. Recent studies suggest that there may be abnormal BRAF in precursor lesions and BRAFE600 contributes to the formation of some benign lesions [33]. Our data suggest that the RAS-RAF-MEK-ERK-MAP kinase-signaling cascade may be involved in some benign thyroid goiters (43.8% in our study). Because the female-to-male ratio in thyroid epithelial cell tumors is 3:1, one may wonder whether BRAF pseudogene location on chromosome Xq13 contributes to this difference, although in female mammals, most genes on one X chromosome are silenced because of X-chromosome inactivation to achieve dosage compensation between males and females. However, recent studies have shown that some genes can escape X-inactivation and are expressed from both the active and inactive X chromosomes [34–36]. BRAF pseudogene may escape X-inactivation and remains active in some thyroid tumors. Further studies are needed to confirm this speculation. Finally, BRAF pseudogene expression in a subset of goiters may also have significant clinical implications. These patients may need to be followed up more closely for early sign of cancer development.
Acknowledgments
The authors thank Raafat M. El-Sayed from the Animal Facility for his excellent support.
Footnotes
The research was funded by King Faisal Specialist Hospital and Research Centre (grant no. 2070064) and King Abdulaziz City for Science and Technology (grant no. ARP-24-11).
Disclosure of conflicts of interest
All authors have nothing to declare.
References
- 1.Hundahl SA, Fleming ID, Fremgen AM, Menck HR. A National Cancer Data Base report on 53,856 cases of thyroid carcinoma treated in the U.S., 1985–1995. Cancer. 1998;83:2638–2648. doi: 10.1002/(sici)1097-0142(19981215)83:12<2638::aid-cncr31>3.0.co;2-1. [see comments] [DOI] [PubMed] [Google Scholar]
- 2.Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63:1454–1457. [PubMed] [Google Scholar]
- 3.Cohen Y, Xing M, Mambo E, Guo Z, Wu G, Trink B, Beller U, Westra WH, Ladenson PW, Sidransky D. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95:625–627. doi: 10.1093/jnci/95.8.625. [DOI] [PubMed] [Google Scholar]
- 4.Soares P, Trovisco V, Rocha AS, Lima J, Castro P, Preto A, Maximo V, Botelho T, Seruca R, Sobrinho-Simoes M. BRAF mutations and RET/PTC rearrangements are alternative events in the etiopathogenesis of PTC. Oncogene. 2003;22:4578–4580. doi: 10.1038/sj.onc.1206706. [DOI] [PubMed] [Google Scholar]
- 5.Namba H, Nakashima M, Hayashi T, Hayashida N, Maeda S, Rogounovitch TI, Ohtsuru A, Saenko VA, Kanematsu T, Yamashita S. Clinical implication of hot spot BRAF mutation, V599E, in papillary thyroid cancers. J Clin Endocrinol Metab. 2003;88:4393–4397. doi: 10.1210/jc.2003-030305. [DOI] [PubMed] [Google Scholar]
- 6.Xu X, Quiros RM, Gattuso P, Ain KB, Prinz RA. High prevalence of BRAF gene mutation in papillary thyroid carcinomas and thyroid tumor cell lines. Cancer Res. 2003;63:4561–4567. [PubMed] [Google Scholar]
- 7.Xing M. BRAF mutation in thyroid cancer. Endocr Relat Cancer. 2005;12:245–262. doi: 10.1677/erc.1.0978. [DOI] [PubMed] [Google Scholar]
- 8.Fagin JA. How thyroid tumors start and why it matters: kinase mutants as targets for solid cancer pharmacotherapy. J Endocrinol. 2004;183:249–256. doi: 10.1677/joe.1.05895. [DOI] [PubMed] [Google Scholar]
- 9.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6:313–319. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 10.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. (Epub 2002 Jun 2009) [DOI] [PubMed] [Google Scholar]
- 11.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 12.Dhillon AS, Kolch W. Oncogenic B-Raf mutations: crystal clear at last. Cancer Cell. 2004;5:303–304. doi: 10.1016/s1535-6108(04)00087-x. [DOI] [PubMed] [Google Scholar]
- 13.Eychene A, Barnier JV, Apiou F, Dutrillaux B, Calothy G. Chromosomal assignment of two human B-raf(Rmil) proto-oncogene loci: B-raf-1 encoding the p94Braf/Rmil and B-raf-2, a processed pseudogene. Oncogene. 1992;7:1657–1660. [PubMed] [Google Scholar]
- 14.Mighell AJ, Smith NR, Robinson PA, Markham AF. Vertebrate pseudogenes. FEBS Lett. 2000;468:109–114. doi: 10.1016/s0014-5793(00)01199-6. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Carriero N, Gerstein M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 2004;20:62–67. doi: 10.1016/j.tig.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasidharan R, Gerstein M. Genomics: protein fossils live on as RNA. Nature. 2008;453:729–731. doi: 10.1038/453729a. [DOI] [PubMed] [Google Scholar]
- 18.Piehler AP, Hellum M, Wenzel JJ, Kaminski E, Haug KB, Kierulf P, Kaminski WE. The human ABC transporter pseudogene family: evidence for transcription and gene-pseudogene interference. BMC Genomics. 2008;9:165. doi: 10.1186/1471-2164-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greene FL, Page DL, Fleming ID, Fritz AG, Balch CM, Haller DG, Morrow M. American Joint Committee on Cancer: AJCC Cancer Staging Manual. 6th ed. New York, NY: Springer; 2002. Thyroid; pp. 77–87. [Google Scholar]
- 20.Shi Y, Zou M, Baitei EY, Alzahrani AS, Parhar RS, Al-Makhalafi Z, Al-Mohanna FA. Cannabinoid 2 receptor induction by IL-12 and its potential as a therapeutic target for the treatment of anaplastic thyroid carcinoma. Cancer Gene Ther. 2008;15:101–107. doi: 10.1038/sj.cgt.7701101. [DOI] [PubMed] [Google Scholar]
- 21.Chong H, Guan KL. Regulation of Raf through phosphorylation and N terminus-C terminus interaction. J Biol Chem. 2003;278:36269–36276. doi: 10.1074/jbc.M212803200. [DOI] [PubMed] [Google Scholar]
- 22.Balakirev ES, Ayala FJ. Pseudogenes: are they “junk” or functional DNA? Annu Rev Genet. 2003;37:123–151. doi: 10.1146/annurev.genet.37.040103.103949. [DOI] [PubMed] [Google Scholar]
- 23.Renaudie F, Yachou AK, Grandchamp B, Jones R, Beaumont C. A second ferritin L subunit is encoded by an intronless gene in the mouse. Mamm Genome. 1992;2:143–149. doi: 10.1007/BF00302872. [DOI] [PubMed] [Google Scholar]
- 24.Sorge J, Gross E, West C, Beutler E. High level transcription of the glucocerebrosidase pseudogene in normal subjects and patients with Gaucher disease. J Clin Invest. 1990;86:1137–1141. doi: 10.1172/JCI114818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kandouz M, Bier A, Carystinos GD, Alaoui-Jamali MA, Batist G. Connexin43 pseudogene is expressed in tumor cells and inhibits growth. Oncogene. 2004;23:4763–4770. doi: 10.1038/sj.onc.1207506. [DOI] [PubMed] [Google Scholar]
- 26.Chakrabarti R, McCracken JB, Jr, Chakrabarti D, Souba WW. Detection of a functional promoter/enhancer in an intron-less human gene encoding a glutamine synthetase-like enzyme. Gene. 1995;153:163–199. doi: 10.1016/0378-1119(94)00751-d. [DOI] [PubMed] [Google Scholar]
- 27.Sun D, Elsea SH, Patel PI, Funk CD. Cloning of a human “epidermal-type” 12-lipoxygenase-related gene and chromosomal localization to 17p13. Cytogenet Cell Genet. 1998;81:79–82. doi: 10.1159/000014993. [DOI] [PubMed] [Google Scholar]
- 28.Thiele H, Berger M, Skalweit A, Thiele BJ. Expression of the gene and processed pseudogenes encoding the human and rabbit translationally controlled tumour protein (TCTP) Eur J Biochem. 2000;267:5473–5481. doi: 10.1046/j.1432-1327.2000.01609.x. [DOI] [PubMed] [Google Scholar]
- 29.Hirotsune S, Yoshida N, Chen A, Garrett L, Sugiyama F, Takahashi S, Yagami K, Wynshaw-Boris A, Yoshiki A. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature. 2003;423:91–96. doi: 10.1038/nature01535. [DOI] [PubMed] [Google Scholar]
- 30.Yano Y, Saito R, Yoshida N, Yoshiki A, Wynshaw-Boris A, Tomita M, Hirotsune S. A new role for expressed pseudogenes as ncRNA: regulation of mRNA stability of its homologous coding gene. J Mol Med. 2004;82:414–422. doi: 10.1007/s00109-004-0550-3. [DOI] [PubMed] [Google Scholar]
- 31.Gray TA, Wilson A, Fortin PJ, Nicholls RD. The putatively functional Mkrn1-p1 pseudogene is neither expressed nor imprinted, nor does it regulate its source gene in trans. Proc Natl Acad Sci USA. 2006;103:12039–12044. doi: 10.1073/pnas.0602216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang CL, Tada M, Kobayashi H, Nozaki M, Moriuchi T, Abe H. Detection of PTEN nonsense mutation and psiPTEN expression in central nervous system high-grade astrocytic tumors by a yeast-based stop codon assay. Oncogene. 2000;19:4346–4353. doi: 10.1038/sj.onc.1203795. [DOI] [PubMed] [Google Scholar]
- 33.Michaloglou C, Vredeveld LC, Mooi WJ, Peeper DS. BRAF(E600) in benign and malignant human tumours. Oncogene. 2008;27:877–895. doi: 10.1038/sj.onc.1210704. [DOI] [PubMed] [Google Scholar]
- 34.Brown CJ, Greally JM. A stain upon the silence: genes escaping X inactivation. Trends Genet. 2003;19:432–438. doi: 10.1016/S0168-9525(03)00177-X. [DOI] [PubMed] [Google Scholar]
- 35.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 36.Disteche CM, Filippova GN, Tsuchiya KD. Escape from X inactivation. Cytogenet Genome Res. 2002;99:36–43. doi: 10.1159/000071572. [DOI] [PubMed] [Google Scholar]