Abstract
We report a fully quantitative spectroscopy imaging instrument for wide area detection of early cancer (dysplasia). This instrument provides quantitative maps of tissue biochemistry and morphology, making it a potentially powerful surveillance tool for objective early cancer detection. We describe the design, construction, calibration, and first clinical application of this new system. We demonstrate its accuracy using physical tissue models. We validate its diagnostic ability on a resected colon adenoma, and demonstrate feasibility of in vivo imaging in the oral cavity.
1. Introduction
Early detection is essential for managing cancer, since treatment is much more successful when lesions are diagnosed at an early, noninvasive stage. Current cancer diagnosis often employs visual inspection of a wide area of tissue followed by biopsy of suspicious sites. This practice is problematic for two reasons: (1) early cancers are not always detectable by visual inspection, so, unavoidably, unnecessary biopsies are taken for precautionary reasons and invisible lesions are missed; and (2) biopsy suffers from undersampling, and the results are subjective and the resulting pathology can be subject to low inter-observer agreement [1–4]. Furthermore, biopsy results often are not available immediately, resulting in delayed treatment and patient anxiety.
Much attention has been focused on spectroscopy, particularly reflectance and fluorescence spectroscopy, to overcome these problems. Reflectance and fluorescence are known to exhibit spectral features associated with the different morphology and biochemistry of normal and cancerous tissues. These techniques have the capability to detect invisible lesions, to provide quantitative diagnostic information for objective evaluation, and to be implemented for wide-area imaging, essential requirements for most applications. Wide-area imaging of spectroscopic information is important for complete acquisition of data over a large field of tissue, and to overcome the under-sampling problem associated with biopsy.
Studies of reflectance and fluorescence for tissue diagnosis using optical fiber contact probes for light delivery and collection have been performed by numerous groups in the cervix [5–8], oral cavity [9–14], esophagus [15, 16], colon [17], lung [18], and bladder [19], with various degrees of quantitative analysis. Our own work has focused on developing quantitative methods for tissue diagnosis. We use diffuse reflectance and fluorescence, in combination, to extract quantitative information about morphological and biochemical tissue constituents. We call the method quantitative spectroscopy (QS). Diffuse reflectance spectra from tissue are analyzed using a well-developed model to obtain information about hemoglobin concentration and saturation, light scattering parameters, and other tissue characteristics. This method is known as diffuse reflectance spectroscopy (DRS) [20]. Tissue fluorescence, collected from the same spot at the same time, is analyzed using the diffusely reflected light to remove spectral distortions, resulting in the “intrinsic fluorescence” that would be observed in the absence of scattering and absorption, from which contributions from tissue fluorophores can then be extracted. This method is known as intrinsic fluorescence spectroscopy (IFS) [21]. Histological parameters are then extracted by fitting the observed spectra to parameters such as tissue density, blood concentration and oxygenation, and concentrations of collagen and reduced nicotinamide adenine dinucleotide (NADH), determined from calibration of physical models of tissue with known features.
Contact probe techniques are promising, but, like biopsy, suffer from undersampling. To overcome this, various groups have been developing techniques to incorporate wide area imaging in fluorescence and reflectance tissue diagnosis, using a variety of approaches with different degrees of quantitative analysis [22–25]. Recently, our laboratory has extended our model-based, quantitative approach to wide field imaging. We call the new modality quantitative spectroscopic imaging (QSI). Data are collected by means of a non-contact “virtual” probe, imaged at the tissue surface. This virtual probe is then raster scanned to interrogate a wide tissue area (~4cm2), one spot (1mm2) at a time. The quantitative measurements of tissue properties enable the spectra for each pixel to be analyzed using our contact probe methodology. Hence, the QSI images are directly interpretable in terms of histological features, thus providing a robust diagnosis.
This paper describes the design, construction, calibration, and the first clinical application of our QSI system. Studies using physical tissue models (“phantoms”) demonstrate the accuracy of QSI. Ex vivo spectral images of a resected colon adenoma demonstrate its ability to diagnose and image malignant lesions. In addition, we present in vivo spectral images from a hyperkeratotic lesion on the ventral surface of the tongue to demonstrate clinical applicability.
The ultimate goal of the QSI system is imaging early cancer. The work described in this paper develops the quantitative mapping methodology required to accomplish this goal. To the best of our knowledge, this instrument is the first that provides quantitative maps of tissue biochemistry and morphology for wide area cancer surveillance.
2. Instrumentation
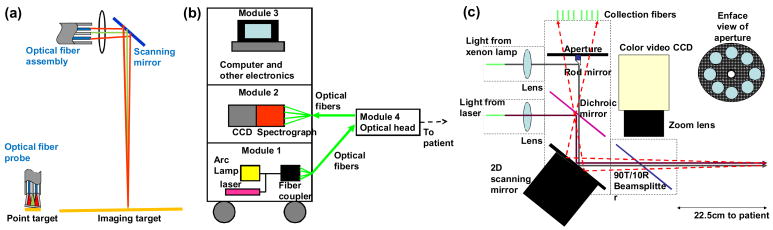
The QSI concept extends the quantitative methodology of our contact probe [26] to a wide area imaging instrument. In the contact probe geometry, the optical fiber probe consists of a single light delivery fiber surrounded by six collection fibers. All fibers are fused together at the tip to form an optical shield approximately 1mm long. This arrangement of fibers and quartz shield provides a reproducible geometry of overlapping excitation and collection cones, creating a fixed distance between fiber tips and tissue and a sampling spot on the tissue surface with a diameter of approximately 800 μm. In the imaging implementation, an optical fiber assembly is relayed by optics onto a small region of the tissue, providing a virtual noncontact probe (Fig. 1(a)). Wide area coverage is achieved by scanning the imaged spot over the tissue. Note that in this case the arrangement of the delivery and collection fibers can be precisely controlled, providing increased optical design flexibility.
Fig. 1.
(a). QSI instrumentation principle. Green beams are illumination and red beams are collection. Optics delivers a spot of light to the tissue surface. The spot is similar in diameter to that of an optical fiber contact probe. The area from which reflectance and fluorescence are collected is also similar. (b) Block diagram of the QSI instrument. (c) Schematic diagram of the optical head.
We note that the characteristics of our non-contact probe (spot size ~ 1mm in diameter and NA ~ 0.02) differ somewhat from that of our contact probe studies (spot size ~ 0.8mm in diameter and NA ~ 0.22). As discussed below, the probe parameters are incorporated in our reflectance [20] and fluorescence [21] models, and differences in probe geometries can be taken into account, so the correct tissue parameters can be obtained from measurements with either probe. This robustness is an important virtue of our quantitative approach.
The QSI instrument has been developed for clinical settings, and must be portable. As shown in Fig. 1(b), portability is achieved by placing the heaviest components in a mobile cart and creating a separate lightweight, mobile optical head to be operated by the physician. The design is modular. The three larger modules, mounted in a cart, are connected to the optical head by means of optical fibers. Module 1, the light source module, consists of a 75W CW white light arc lamp (Simplicity series, Newport Corp.) and a nitrogen laser that delivers 337nm light pulses of duration <3.5 ns and energy 175 μJ at 20 Hz (NL100, Stanford Research Systems Inc.). Module 2 contains a spectrograph/CCD unit (Princeton Instruments Corp.). Module 3 is the computer, which controls data acquisition and analyzes the data. National Instruments Labview software and DAQ data acquisition hardware are used to control and coordinate the various components. Module 4, is mounted on an articulated arm for easy maneuvering. It is located on a smaller cart with wheels, so that the larger equipment cart can be located away from the patient, since space in hospital procedure rooms is limited.
Fig. 1(c) is a schematic diagram of the optical head. White light (with NA ~ 0.02) from the xenon arc lamp (CW) illuminates a 1mm diameter “diagnostic spot” on the tissue about 20cm away from the optical head. Diffusely reflected light from a 2mm diameter circle centered on the diagnostic spot is relayed back to the optical head and focused onto eight collection fibers coupled to a spectrograph and CCD. Each of the eight fibers collects light returning from the tissue in different directions, which helps avoid specular reflection. Next, 337nm light (with NA ~ 0.02) from the nitrogen laser illuminates the same diagnostic spot, and fluorescence is collected in the same manner as diffuse reflectance. These two measurements are equivalent to the reflectance and fluorescence measurements of a contact probe.
Wide area coverage is achieved by means of a 2D scanning mirror (OIM102, Optics in Motion LLC), which can tilt by up to ± 1.5° along two orthogonal axes, and thus raster scan the diagnostic spot across a 2.1cm × 2.1cm region of the tissue surface in a stepwise fashion. At each mirror position, a reflectance measurement is made, followed immediately by a fluorescence measurement. Each pair of measurements is associated with a tissue location which correlates with the mirror position. However, this correlation can be affected by patient movement during the procedure. Therefore, the patient’s movement needs to be tracked in order to take any shift in tissue position into account. An onboard color video camera (QICAM, QImaging Corp.) tracks the patient movement relative to the instrument by acquiring one photograph of the tissue every second during the procedure. Two white light LEDs (CCS Inc., not shown), which provide extra illumination for the video camera, are turned off during reflectance and fluorescence measurements. We have conducted experiments with phantoms to determine the impact of different working distances on the DRS and IFS measurements. From 21.5cm to 23.5cm, we found that the extracted parameters (A, B, etc.) varied by less than 10% of the values measured at 22.5cm (the optimal working distance). Since the color video camera and the white light LEDs allow us to place the instrument very close to the optimal working distance (well within the + 1cm range studied with phantoms) prior to acquiring data, the instrument always operates very close to the optimal working distance. The total time required for a 2.1cm × 2.1cm scan is approximately 90s at present, but this time can be considerably reduced by using a CCD camera (for spectra) with the on-board memory (to eliminate the data transfer time and greatly reduce the data writing time), and increasing the angle of optical collection (to decrease the CCD exposure time).
After the procedure, we apply the phase correlation technique to determine the lateral translation relative to the instrument of each photograph relative to the first photograph [27]. This technique involves computing the cross-correlation of two images and identifying the coordinates of the maximum value. The coordinates are the two coordinates of lateral translation. We found that phase correlation in the QSI instrument determines the two coordinates of translation within 0.2mm of their true values. Applying phase correlation to each photograph, we track the lateral shift in tissue position at the time of each photograph. This information is then used to correct the interrogated tissue location. We do not account for other types of motion such as rotation or tissue morphing because they are considerably less significant than translation in our studies.
3. System calibration
We used physical tissue models (“phantoms”) with known scattering, absorption and fluorescence parameters to calibrate the QSI system and establish its accuracy. These phantoms consist of mixtures of 10% intralipid (Fresenius Kabi AG), hemoglobin (Sigma Aldrich Co.), water, and furan (Lambda Physik) at various concentrations. Furan is a fluorescent dye with excitation and emission spectra similar to that of collagen, an endogenous tissue fluorophore. Spectralon (Labsphere SRS-20) was used as a reflectance standard. Measurements were made over the spectral range 387–707nm.
To test the accuracy of the QSI system’s reflectance measurement capability, we prepared nine combinations of intralipid diluted with water and hemoglobin. The three dilution ratios (with corresponding mass concentrations in parentheses) for intralipid were 1:9 (1%), 2:8 (2%), and 3:7 (3%). The three concentrations of hemoglobin were 0.5, 1.0, and 1.5mg/mL. We define the interrogated spot as a “pixel.” The reflectance spectra were collected from phantoms, over 441 pixels over a 2.1cm × 2.1cm region, were normalized by the corresponding reflectance spectra measured from the spectralon to remove spectral distortions and spatial inhomogeneity due to the instrument’s spectral and spatial responses. The solid lines of Fig. 2 show the resulting calibrated reflectance spectra taken from the same imaging position on different phantoms. DRS is used to extract diagnostic information: Each reflectance spectrum is fit (dashed lines) to the model described by Zonios et al. [20] to yield the reduced scattering μs′(λ) and absorption μa(λ) coefficients of the pixels, with μs′(λ) and μa(λ) of the form:
| (1) |
Fig. 2.
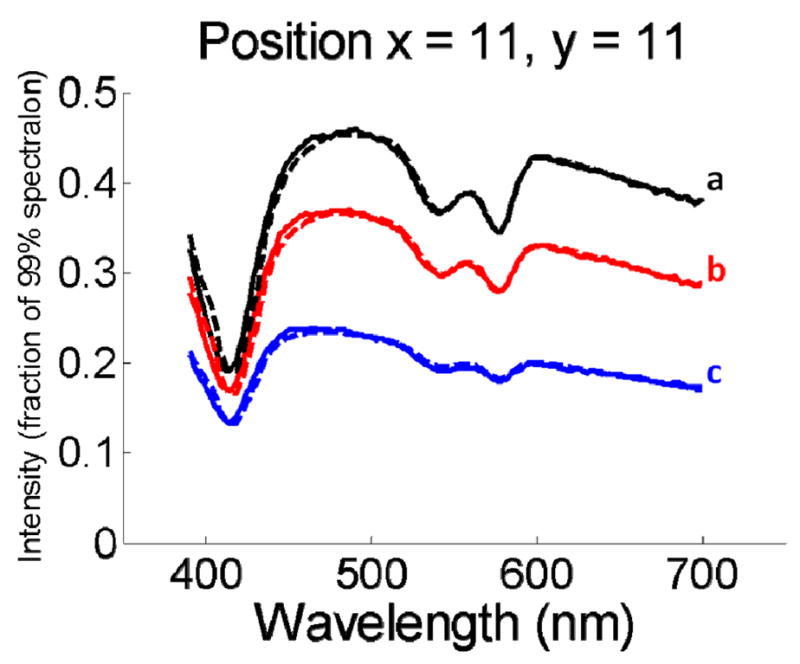
Calibrated reflectance spectra (solid lines) measured from the same imaging position on different tissue phantoms. The best fit spectra using the approach of Zonios et al. are also plotted (dashed lines). The characteristic absorption bands of hemoglobin at 420nm, 540nm, and 580nm are clearly visible.
For each pixel, four DRS parameters are extracted: A, the reduced scattering coefficient at the reference wavelength (i.e. A = μs′(λ0) with λ0 = 700nm); the exponent, B, related to the average scatter size [28]; and cHb and α, the concentration (mg/mL) and oxygen saturation of hemoglobin, respectively. εHb (λ) and εHbO2 (λ) are the extinction coefficients of 1mg/mL of deoxygenated and oxygenated hemoglobin, respectively [29]. As can be seen, the agreement between the measured and fitted spectra is excellent.
After extracting the four parameters, we obtained parameter maps across the 2.1cm × 2.1cm region with a spatial resolution of 1mm × 1mm. Table 1 summaries the extracted values for parameters A, B, and cHb. The mean value of A changes linearly with Intralipid concentration, as expected. The mean for cHb tracks the expected value with less than 10% difference. The mean values for A and B vary little with hemoglobin concentration, and the mean values for cHb vary little with intralipid concentration, indicating that QSI successfully decouples scattering from absorption. The standard deviations of the parameters across each phantom are less than 5% of the mean value, indicating a 5% variation in measured parameter values by QSI.
Table 1.
Reflectance parameters measured from tissue phantoms. The uncertainty of each parameter is the standard deviation of values measured from all 441 pixels on the phantom. Phantoms labeled (a), (b), and (c) are included in Fig. 2.
| Intralipid mass concentration | Hemoglobin concentration (mg/mL) | cHb(mg/mL) | A(λ0= 700nm) | B (λ in nm) |
|---|---|---|---|---|
| 1% (c) | 0.5 | 0.53±0.01 | 1.13±0.01 | 0.99±0.02 |
| 1% | 1.0 | 0.96±0.01 | 1.13±0.01 | 1.00±0.02 |
| 1% | 1.5 | 1.56±0.03 | 1.11±0.01 | 1.02±0.02 |
| 2% | 0.5 | 0.55±0.01 | 1.90±0.02 | 1.32±0.04 |
| 2% (b) | 1.0 | 1.04±0.01 | 1.86±0.02 | 1.36±0.04 |
| 2% | 1.5 | 1.49±0.01 | 1.86±0.02 | 1.40±0.04 |
| 3% | 0.5 | 0.53±0.01 | 2.67±0.04 | 1.38±0.04 |
| 3% | 1.0 | 1.05±0.01 | 2.63±0.04 | 1.45±0.04 |
| 3% (a) | 1.5 | 1.50±0.01 | 2.61±0.03 | 1.48±0.04 |
The above phantom experiments only establish that A measurements scale accurately, and do not address the absolute accuracy of A and B measurements. This is because the scattering properties of the batch of intralipid used were not known precisely. The optical properties of intralipid have been characterized [31], but from our experience, these properties vary considerably from batch to batch. To obtain the absolute accuracies of A and B measurements, we use QSI to measure a phantom consisting of 1μm diameter polystyrene spheres (Polysciences, Inc.) in water with number density 1.1 × 1010 spheres/mL. For this phantom, the reduced scattering coefficient can be computed with Mie theory [32]. To evaluate the absolute accuracy of QSI’s A and B measurements, we fit the μs′(λ) computed with Mie theory to the reduced scattering coefficient from and obtain AMie = 2.23mm−1 and BMie = 0.93. We then use QSI to conduct a reflectance measurement on the phantom, using the same procedures as with the nine intralipid phantoms above, and obtain mean values of A and B equal to 2.12mm−1 and 1.06, respectively. The excellent agreement of the parameters measured by DRS with the input parameters in both types of phantoms indicates that our system is properly calibrated.
To determine the accuracy of fluorescence measurements, we prepared six combinations of intralipid diluted with water, hemoglobin, and furan. Their concentrations are listed in. IFS is used to analyze the fluorescence for each pixel: Reflectance and bulk fluorescence spectra are measured from 441 spots on each phantom. The extracted reduced scattering and absorption coefficients are used to correct the bulk fluorescence spectra using the model described by Müller et al. [21] to extract the IFS spectra, which are the signals that would be measured in the absence of scattering and absorption.
We used our published fluorescence spectrum of furan in water excited by 337nm light as the basic spectrum (see Fig. 3(a) in [21]) for our fluorescence calibration. The instrument collection spectral response, which is extracted by taking the ratio between the basic spectrum and the measured spectrum of furan in water, is taken into account in all our fluorescence spectra shown in this paper. Fig. 3 shows several bulk and IFS spectra measured from on different phantoms (with hemoglobin and furan concentrations fixed but the intralipid concentration varied from 1% to 3%). The data indicates that bulk fluorescence spectra vary considerably with intralipid concentration while the IFS spectra do not, as expected. The intrinsic fluorescence spectra of Fig. 3 are the same as the fluorescence spectrum of furan in water excited by 337nm light, which is shown in green in Fig. 3. This excellent agreement indicates that IFS can be used to remove the distortion caused by tissue scattering and absorption and extract intrinsic fluorescence spectra.
Fig. 3.
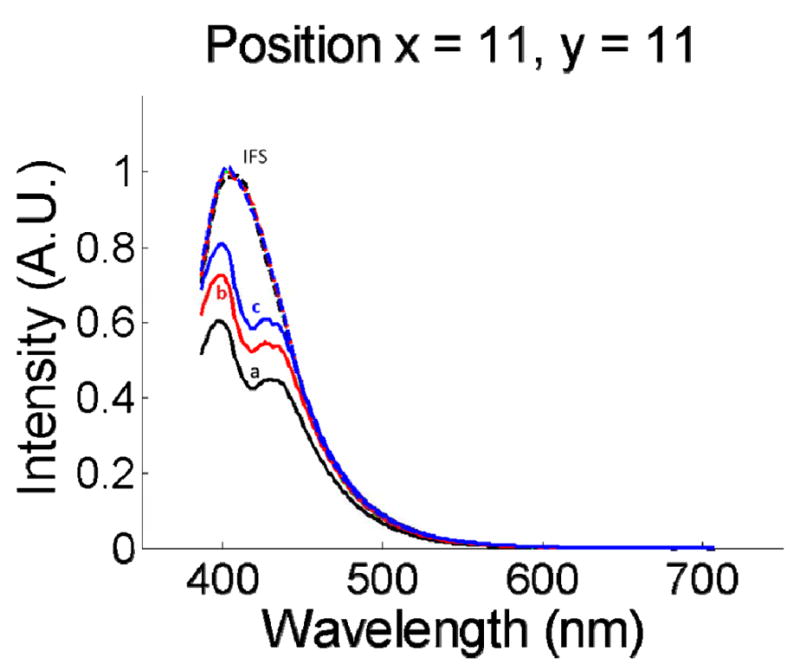
Bulk fluorescence spectra (a, b, c) measured from the same imaging position on different tissue phantoms (as indicated in Table 2). The corresponding intrinsic fluorescence spectra extracted using the method of Müller et al. are also plotted (IFS). The dashed green spectrum is the fluorescence measured from furan in water without intralipid or hemoglobin present. It is nearly identical to the red, black, and blue dashed lines. Note that the IFS spectra and the spectrum of furan in water all overlap, as they should.
Using one of the IFS spectra and the known concentration as a standard, we measured the furan concentrations in each pixel of each phantom by recording the amplitude difference between the spectrum from the pixel and the basis spectrum.
Table 2 shows the average furan concentrations measured from the 441 pixels on each phantom. The agreement between the prepared and measured furan concentrations is excellent. The variation in parameters measured across the homogeneous phantom is less than 5%. Therefore, 5% is a measure of the smallest difference between fluorescence properties measured from neighboring points of an inhomogeneous sample that can be resolved by the present QSI instrument.
Table 2.
Fluorescence parameters measured from tissue phantoms. The measured furan concentration for 1% intralipid has been normalized to 0.25 and all other measured concentrations are normalized by the same factor.
| Intralipid Mass concentration | Hemoglobin concentration (mg/mL) | Prepared furan concentration (μg/mL) | Measured furan concentration |
|---|---|---|---|
| 1% (a) | 0.5 | 0.25 | 0.25±0.004 |
| 2% (b) | 0.5 | 0.25 | 0.26±0.005 |
| 2% | 1.0 | 0.25 | 0.23±0.004 |
| 2% | 0.5 | 0.125 | 0.11±0.004 |
| 2% | 0.5 | 0.5 | 0.47±0.006 |
| 3% (c) | 0.5 | 0.25 | 0.26±0.005 |
4. Cancer diagnosis in ex vivo tissue
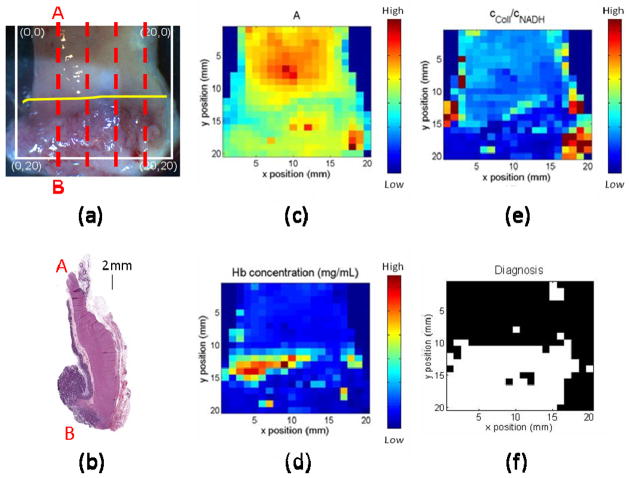
The ability to detect cancer using the QSI system was demonstrated using an ex vivo colon cancer specimen. Four histological sections were made at ~4 mm intervals through the tissue (indicated by dashed red lines in Fig. 4(a), all of which showed similar pathological features (Fig. 4(b) is one representative section). Given that the lesion was macroscopically contiguous, it was assumed that these 4 sections represent the entire lesion. Boundaries of the entire lesion were thus defined by interpolation of histopathological features between the 4 actual histological sections. The area below the yellow line was classified as the region of colon cancer. The area investigated by QSI (enclosed by the white square) was 2cm by 2cm with 400 pixels. From each pixel, we extracted four DRS parameters (A, B, cHb and α) and two fluorescence parameters (collagen concentration, cColl, and NADH concentration, cNADH). To measure the concentrations of two fluorophores at each point on the tissue requires a modification of the approach used for the tissue phantoms. As before, the IFS spectra were first extracted from the data. These spectra are a linear combination of collagen and NADH fluorescence. To extract the relative concentrations, the fluorescence basis spectra of collagen and NADH were determined by applying multivariate curve resolution [9], with the 400 intrinsic fluorescence spectra measured from the sample. Fluorescence from the two fluorophores can then be linearly combined to yield the net fluorescence. To obtain the best fit, the following equation is optimized over all wavelengths and at all points.
Fig. 4.
(a). White light image of ex vivo colon specimen. (b): Ex vivo colon pathology. The specimen is sectioned from the region indicated by the left red line in (a). Parameter maps of A, cHb, and cColl/cNADH are shown in (c) – (e). Maps can be compared to the picture in (a) by matching the coordinates with units in millimeters. (f): Diagnosis map with black normal and white cancerous tissue. The diagnosis using QSI is constant to the diagnosis using histopathology which indicates area below yellow line in (a) is cancerous.
| (2) |
Here, Idata(x,y) is the experimentally measured intrinsic fluorescence spectrum from point (x, y); Ibasis are the basis spectra of collagen and NADH. Ibasis is normalized such that its peak emission is 1. c(x,y) are the concentrations of collagen and NADH at point (x, y).
Bayes’ theorem [30] was used to develop a classification algorithm to distinguish between normal and cancer groups. The training set consisted of 16 normal and 16 cancer data points chosen from two equal sized regions of the sample located away from the boundary between normal and cancer (Fig. 4(a)). A two-tailed Student’s t-test was used to compare means of extracted spectroscopy parameters between normal and cancer groups. A p-value < 0.05 was considered significant. We found that the three most significant parameters to differentiate between the normal and cancer groups are A, cHb, and the cColl/cNADH ratio. The maps for these parameters are shown in Fig. 4(c), 4(d), and 4(e). These parameters represent three different types of tissue information: structure, hemoglobin absorption, and biochemistry, respectively. Multivariate Gaussian probability distributions were used to model the data. We applied Bayes’ rule prospectively to calculate the posterior probability of cancer (“pc”) for each of the imaged pixels. The prior probablity was estimated from the pathology image to be 0.5. Fig. 4(f) represents the resulting diagnostic map, constructed by using the threshold for assigning a pixel to the cancer group of pc > 0.5. The QSI classification is consistent with the histological classification of Fig. 4(a), and it demonstrates the ability of our QSI system to distinguish between cancer and normal tissue in this specimen.
5. In vivo feasibility
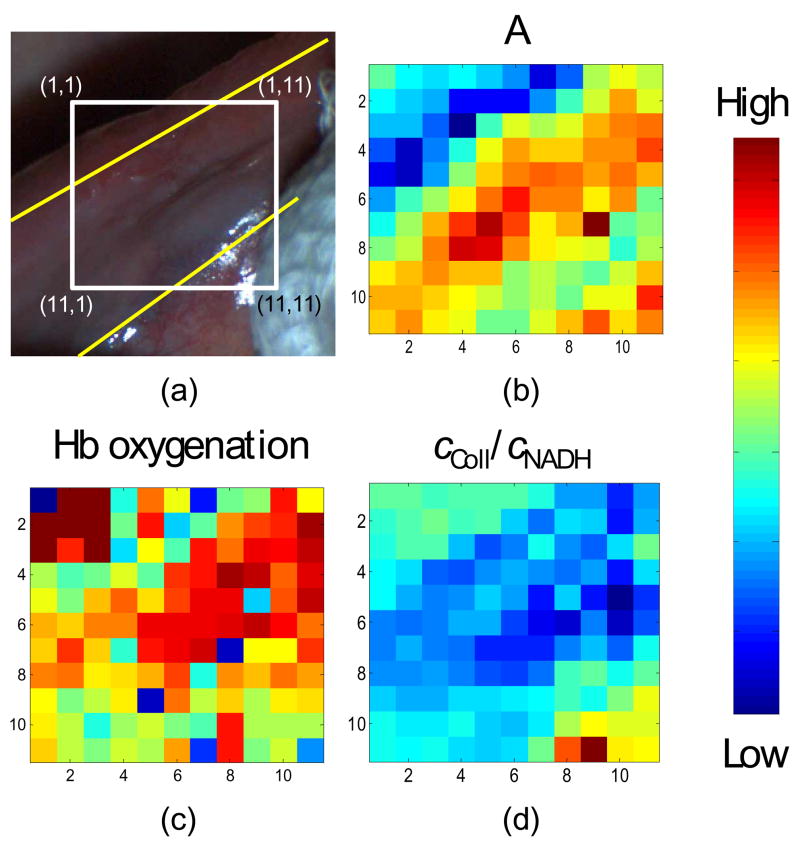
The QSI system was then used to examine a suspicious tissue site, as determined by the physician using conventional white light examination, on the ventral tongue of a patient. This was the first in vivo measurement conducted with the QSI system, and more will be conducted in the near future. The purpose of this study is to demonstrate the in vivo applicability of the QSI instrument instead of making correlations. Fig. 5(a) is the white light image of the ventral tongue. The yellow lines indicate the boundaries between normal areas and a region of hyperkeratosis identified by the oral surgeon. Subsequent histopathology analysis of biopsied tissue determined the site to be a benign area of hyperplasia. The gauze (shown on the lower right corner) was used by the oral surgeon to hold the tongue in order to reveal the hyperkeratosis for interrogation. The tissue within the white box was interrogated with QSI system. Fig. 5(b)–5(d) show reflectance and fluorescence parameters measured from 121 points on the site over a 1.1cm × 1.1cm area in approximately 15s. Fig. 5(b) indicates an increase in scattering in the hyperkeratotic region as compared to the surrounding normal mucosa. Fig. 5(d) shows reductions in cColl/cNADH in the hyperkeratotic region. This is consistent with the understanding that keratin reduces the amount of light reaching the stroma, thus reducing collagen fluorescence. This study represents our first in vivo measurement. Detailed comparisons between spectroscopy and histology in multiple patients in various tissue types will be used to provide accurate diagnostic image information, as well as insights into the correlation between QSI parameters and tissue state.
Fig. 5.
(a). White light image of ventral tongue site with the boundaries, between hyperkeratosis and normal areas, delineated by the dashed yellow lines. The coordinates are in millimeters. Data in (b) – (d) can be compared to the picture in (a) by matching the coordinates. (b) A parameter. (c) α parameter. (d) cColl/cNADH parameter.
6. Discussion
Current cancer diagnosis often employs visual inspection of a wide area of tissue (sometimes assisted by endoscopy), followed by biopsy of suspicious sites. As mentioned in the introduction, this leads to unnecessary biopsies and delays and inaccuracies in pathology. Quantitative spectroscopy (QS), which combines DRS and IFS, for early cancer detection, seeks to address those shortcomings by extracting parameters that characterize a tissue sample without tissue removal. The extracted parameters can then be combined to form a diagnostic algorithm to quantify the probability of the interrogated tissue being dysplastic. The analysis can be performed by computer in real time. Although QS as a contact probe technique is effective in various organs, it is essential to extend it to the imaging mode so that wide areas of tissues can be studied.
Theme of this paper is extension of QS to the imaging mode. Our QS methodology is a model-based approach that extracts tissue morphological and biochemical information from tissue reflectance (DRS) and fluorescence (IFS) spectra. The method is based on the correlation of tissue parameters with disease state. Many groups employ spectral changes to develop algorithms for diagnosing cancer without analyzing the underlying tissue parameters, can be used to understand tissue composition and chemical makeup. In contrast, QSI provides a method of analysis based on understanding the biochemical and morphological structure of the tissue. This information can provide more accurate and robust diagnosis.
Our spectral imaging approach of raster scanning small spots is distinct of those of other groups, most of which use full-field illumination [22–25]. Although full-field illumination can be used for wide area detection using a CCD camera, it cannot be used to provide the desired quantitative information. Cross talk between spatial locations can occur, so the information extracted from one location can be influenced by neighboring locations. UV excitation power densities are small since the light is distributed over the entire area. This imaging significantly limits the speed at which fluorescence measurements can be performed. Moreover, there is a fundamental reason why we employ raster scanning rather than full-field illumination. To extract tissue parameters, A, B, cHb and α in Eq. (1), we need to measure the reduced scattering μs′(λ) and absorption μa(λ) coefficients independently. However, with full field illumination, only the ratio of μs′(λ) and μa(λ) can be measured. Use of a probe with small delivery and collection spot size provides a scale parameter rc′, for the measurement, which provides information about two dimensionless parameters μs′(λ)*rc′ and μa(λ)*rc′ measured at each pixel [20].
Our approach accomplishes wide field coverage by employing a virtual probe to sample a small area (defined as one pixel) of tissue at any one time. As discussed above, this probe has effectively all of the features of a contact probe, without the need to make contact with the tissue. We then employ raster scanning to cover a large area of tissue, pixel by pixel. This would not be possible with a contact probe. Because we retain the contact-probe feature of fixed delivery-collection geometry, this method allows us to directly transfer the data analysis procedures and results obtained from our contact-probe studies to our imaging studies. To the best of our knowledge, this is the first spectral imaging system capable of extracting tissue biochemical and morphological information quantitatively.
We have successfully demonstrated accurate extraction of tissue parameters using physical tissue models (“phantoms”). The ability of our QSI imaging system to identify cancerous lesions was demonstrated on excised tissue, and in vivo use of this imaging system was demonstrated in the oral cavity.
Several spectral imaging systems have been reported in the literature, with promising results. Benavides, et al. [22] have constructed a multispectral digital colposcope. Multispectral autofluorescence and reflectance images of the cervix are acquired using an inexpensive color CCD camera for in vivo detection of cervical cancer. Spectral sensitivity is provided by the three color channels of the CCD. Vo-Dinh, et al. [23] have used an acoustooptic tunable filter (AOTF) to select the wavelength of the tissue fluorescence image acquired by a CCD. This AOTF-based spectral imaging system can record spectral images at a series of wavelengths of interest to provide spectral contracts for diagnosis. Using a similar approach, Gebhart et al. [25] have developed a spectral imaging system using a liquid-crystal tunable filter (LCTF) to select wavelength of the tissue images acquired by a CCD. Both reflectance and fluorescence spectral contrasts were shown between cortex and white matter on the in vitro mouse brain and between tumor and normal cortex on the in vivo human brain. All three systems mentioned above used full-field illumination and collection with different types of wavelength selection mechanisms. The spectral imaging system reported by Schomacker et al. [24] studies cervical tissue, and uses different illumination/collection geometries for measuring diffuse reflectance and fluorescence spectra consecutively from a 1 mm interrogation region. For reflectance, full-field illumination was used, whereas fluorescence was implemented by delivering a 1mm diameter spot of UV excitation and collecting fluorescence from the same location. Full cervical scans employed 499 interrogation locations. Spectral differences were observed between normal squamous epithelium and highgrade cervical intraepithelial neoplasia.
All of these spectral imaging systems reported observation of spectral contrast between benign and malignant tissues in reflectance and/or fluorescence. Some also demonstrated the effectiveness of using this contrast in tissue diagnosis. This is encouraging, and provides motivation for further advances using quantitative spectral imaging modalities such as QSI for detecting early cancer noninvasively and objectively. As discussed above, QSI extracts quantitative information about morphological and biochemical tissue constituents that give rise to spectral contrast. This provides two important benefits: (1) Because the extracted parameters are tissue properties, the results should be more robust and instrument independent; (2) the extracted parameters have physical meaning which can be correlated with morphological and biochemical information. Since the information extracted is intrinsic to the tissue, the diagnostic algorithm developed using tissue parameters should be more robust and can be directly transferred from contact probe application to imaging mode. This is true even though the illumination and collection geometries may differ, as such difference can be taken into account in the modeling. Therefore, for the QSI study presented in this paper, we used the same DRS and IFS methodologies as the ones used in our probe-based studies. No changes are needed for tissue parameter extraction except for instrument-dependent constants such as effective probe radius rc′ [20] and the wavelength range used in the models.
QSI uses tissue parameters for contrast to give diagnoses. Knowledge about morphology and biochemistry of cancer as it evolves can be used to develop QSI algorithms and authenticate their validity. As an example, collagen in the tissue matrix degrades in cancerous tissue [33,34]. Therefore, the scattering intensity for cancerous tissue should decrease, which is consistent with our DRS images (Fig. 4(c)). It is also known that higher NADH concentration accompanies cancer progression (since NADH is a measure of intracellular oxidation-reduction states in vivo [35]), due to greater metabolic activity of cancerous cells. Therefore, the concentration ratio between collagen and NADH should be lower for cancerous tissue, which is again confirmed by the IFS images in the ex vivo study (Fig. 4(e)). Moreover, combining DRS and IFS provides more complete tissue morphological and biochemical information, which can be advantageous in cancer detection.
7. Conclusion
This research has demonstrated the successful extension of quantitative spectroscopy from a contact probe modality to a wide-area imaging technique. Its ability to provide spectral contrast based on tissue parameters has been demonstrated in the example of ex vivo colonic tissue. In addition, QSI’s ability to measure spectra in vivo has been established. The current QSI instrument is designed to image openly accessible sites such as the cervix, oral cavity and skin. QSI can also be applied in an endoscope-type delivery system to image the hollow organs of the body. This offers the potential of using the quantitative diagnostic ability of spectroscopy to diagnose cancer, atherosclerosis, and other disease states throughout the body. In addition, QSI can readily be used to conduct margin detection. This will potentially reduce the amount of time the patient and medical staff have to wait for results from pathology and the number of return visits required.
Two future directions are being pursued in our research. We are conducting a clinical in vivo feasibility study in imaging of cervical dysplasia using our current QSI instrument. We are also developing a next generation QSI instrument in an endoscopic configuration.
Acknowledgments
We gratefully acknowledge CCS America, Inc. (http://www.ccsamerica.com/) for their generous support in providing LED illumination system used in the QSI instrument. This research was conducted at the Massachusetts Institute of Technology Laser Biomedical Research Center and supported by the National Institute of Health (grants P41-RR02594 AND R01-CA97966).
Footnotes
OCIS codes: (300.0300) Spectroscopy; (170.0110) Imaging system; (170.3890) Medical optics instrumentation; (170.4580) Optical diagnostics for medicine; (170.1610) Clinical applications.
References and links
- 1.Yoon H, Martin A, Benamouzig R, Longchampt E, Deyra J, Chaussade S, Grp A. Inter-observer agreement on histological diagnosis of colorectal polyps: the APACC study. Gastroenterol Clin Et Biol. 2002;26:220–224. [PubMed] [Google Scholar]
- 2.McCluggage WG, Walsh MY, Thornton CM, Hamilton PW, Date A, Caughley LM, Bharucha H. Inter- and intra-observer variation in the histopathological reporting of cervical squamous intraepithelial lesions using a modified Bethesda grading system. Br J Obstet Gynaecol. 1998;105:206–210. doi: 10.1111/j.1471-0528.1998.tb10054.x. [DOI] [PubMed] [Google Scholar]
- 3.Ismail SM, Colclough AB, Dinnen JS, Eakins D, Evans DMD, Gradwell E, Osullivan JP, Summerell JM, Newcombe R. Intrapathologist and Interpathologist Variation in Reporting Cervical Intra- Epithelial Neoplasia and Factors Associated with Disagreement. J Pathol. 1990;160:A176–A176. doi: 10.1111/j.1365-2559.1990.tb01141.x. [DOI] [PubMed] [Google Scholar]
- 4.Reid BJ, Haggitt RC, Rubin CE, Roth G, Surawicz CM, Vanbelle G, Lewin K, Weinstein WM, Antonioli DA, Goldman H, Macdonald W, Owen D. Observer Variation in the Diagnosis of Dysplasia in Barretts Esophagus. Hum Pathol. 1988;19:166–178. doi: 10.1016/s0046-8177(88)80344-7. [DOI] [PubMed] [Google Scholar]
- 5.Georgakoudi I, Sheets EE, Muller MG, Backman V, Crum CP, Badizadegan K, Dasari RR, Feld MS. Trimodal spectroscopy for the detection and characterization of cervical precancers in vivo. Am J Obstet Gynecol. 2002;186:374–382. doi: 10.1067/mob.2002.121075. [DOI] [PubMed] [Google Scholar]
- 6.Freeberg JA, Serachitopol DM, McKinnon N, Price R, Atkinson EN, Cox DD, MacAulay C, Richards-Kortum R, Follen M, Pikkula B. Fluorescence and reflectance device variability throughout the progression of a phase II clinical trial to detect and screen for cervical neoplasia using a fiber optic probe. J Biomed Opt. 2007;12:034015. doi: 10.1117/1.2750332. [DOI] [PubMed] [Google Scholar]
- 7.Chang SK, Mirabal YN, Atkinson EN, Cox D, Malpica A, Follen M, Richards-Kortum R. Combined reflectance and fluorescence spectroscopy for in vivo detection of cervical pre-cancer. J Biomed Opt. 2005;10:024031. doi: 10.1117/1.1899686. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell MF, Cantor SB, Ramanujam N, Tortolero-Luna G, Richards-Kortum R. Fluorescence spectroscopy for diagnosis of squamous intraepithelial lesions of the cervix. Obstet Gynecol. 1999;93:462–470. doi: 10.1016/s0029-7844(98)00385-8. [DOI] [PubMed] [Google Scholar]
- 9.Muller MG, Valdez TA, Georgakoudi I, Backman V, Fuentes C, Kabani S, Laver N, Wang ZM, Boone CW, Dasari RR, Shapshay SM, Feld MS. Spectroscopic detection and evaluation of morphologic and biochemical changes in early human oral carcinoma. Cancer. 2003;97:1681–1692. doi: 10.1002/cncr.11255. [DOI] [PubMed] [Google Scholar]
- 10.de Veld DCG, Skurichina M, Wities MJH, Duin RPW, Sterenborg HJCM, Roodenburg JLN. Autofluorescence and diffuse reflectance spectroscopy for oral oncology. Lasers Surg Med. 2005;36:356–364. doi: 10.1002/lsm.20122. [DOI] [PubMed] [Google Scholar]
- 11.Gillenwater A, Jacob R, Ganeshappa R, Kemp B, El-Naggar AK, Palmer JL, Clayman G, Mitchell MF, Richards-Kortum R. Noninvasive diagnosis of oral neoplasia based on fluorescence spectroscopy and native tissue autofluorescence. Arch Otolaryngol Head Neck Surg. 1998;124:1251–1258. doi: 10.1001/archotol.124.11.1251. [DOI] [PubMed] [Google Scholar]
- 12.Heintzelman DL, Utzinger U, Fuchs H, Zuluaga A, Gossage K, Gillenwater AM, Jacob R, Kemp B, Richards-Kortum RR. Optimal excitation wavelengths for in vivo detection of oral neoplasia using fluorescence spectroscopy. Photochem Photobiol. 2000;72:103–113. doi: 10.1562/0031-8655(2000)072<0103:OEWFIV>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 13.Majumder SK, Mohanty SK, Ghosh N, Gupta PK, Jain DK, Khan F. A pilot study on the use of autofluorescence spectroscopy for diagnosis of the cancer of human oral cavity. Curr Sci. 2000;79:1089–1094. [Google Scholar]
- 14.Amelink A, Kaspers OP, Sterenborg HJCM, van der Wal JE, Roodenburg JLN, Witjes MJH. Non-invasive measurement of the morphology and physiology of oral mucosa by use of optical spectroscopy. Oral Oncol. 2008;44:65–71. doi: 10.1016/j.oraloncology.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Georgakoudi I, Jacobson BC, Van Dam J, Backman V, Wallace MB, Muller MG, Zhang Q, Badizadegan K, Sun D, Thomas GA, Perelman LT, Feld MS. Fluorescence, reflectance, and lightscattering spectroscopy for evaluating dysplasia in patients with Barrett’s esophagus. Gastroenterology. 2001;120:1620–1629. doi: 10.1053/gast.2001.24842. [DOI] [PubMed] [Google Scholar]
- 16.Vo-Dinh T, Panjehpour M, Overholt BF. Laser-induced fluorescence for esophageal cancer and dysplasia diagnosis. Adv Opt Biopsy Opt Mammography. 1998;838:116–122. doi: 10.1111/j.1749-6632.1998.tb08192.x. [DOI] [PubMed] [Google Scholar]
- 17.Fiarman GS, Nathanson MH, West AB, Deckelbaum LI, Kelly L, Kapadia CR. Differences in Laser-Induced Autofluorescence between Adenomatous and Hyperplastic Polyps and Normal Colonic Mucosa by Confocal Microscopy. Dig Dis Sci. 1995;40:1261–1268. doi: 10.1007/BF02065535. [DOI] [PubMed] [Google Scholar]
- 18.Bard MPL, Amelink A, Skurichina M, Hegt VN, Duin RPW, Sterenborg HJCM, Hoogsteden HC, Aerts JGJV. Optical spectroscopy for the classification of malignant lesions of the bronchial tree. Chest. 2006;129:995–1001. doi: 10.1378/chest.129.4.995. [DOI] [PubMed] [Google Scholar]
- 19.Mourant JR, Bigio IJ, Boyer J, Conn RL, Johnson T, Shimada T. Spectroscopic diagnosis of bladder cancer with elastic light scattering. Lasers Surg Med. 1995;17:350–357. doi: 10.1002/lsm.1900170403. [DOI] [PubMed] [Google Scholar]
- 20.Zonios G, Perelman LT, Backman VM, Manoharan R, Fitzmaurice M, Van Dam J, Feld MS. Diffuse reflectance spectroscopy of human adenomatous colon polyps in vivo. Appl Opt. 1999;38:6628–6637. doi: 10.1364/ao.38.006628. [DOI] [PubMed] [Google Scholar]
- 21.Muller MG, Georgakoudi I, Zhang QG, Wu J, Feld MS. Intrinsic fluorescence spectroscopy in turbid media: disentangling effects of scattering and absorption. Appl Opt. 2001;40:4633–4646. doi: 10.1364/ao.40.004633. [DOI] [PubMed] [Google Scholar]
- 22.Benavides JM, Chang S, Park SY, Richards-Kortum R, Mackinnon N, MacAulay C, Milbourne A, Malpica A, Follen M. Multispectral digital colposcopy for in vivo detection of cervical cancer. Opt Express. 2003;11:1223–1236. doi: 10.1364/oe.11.001223. [DOI] [PubMed] [Google Scholar]
- 23.Vo-Dinh T, Stokes DL, Wabuyele MB, Martin ME, Song JM, Jagannathan R, Michaud E, Lee RJ, Pan XG. A hyperspectral imaging system for in vivo optical diagnosis. IEEE Eng Med Biol Mag. 2004;23:40–49. doi: 10.1109/memb.2004.1360407. [DOI] [PubMed] [Google Scholar]
- 24.Schomacker KT, Meese TM, Jiang CS, Abele CC, Dickson K, Sum ST, Flewelling RF. Novel optical detection system for in vivo identification and localization of cervical intraepithelial neoplasia. J Biomed Opt. 2006;11:034009. doi: 10.1117/1.2208987. [DOI] [PubMed] [Google Scholar]
- 25.Gebhart SC, Thompson RC, Mahadevan-Jansen A. Liquid-crystal tunable filter spectral imaging for brain tumor demarcation. Appl Opt. 2007;46:1896–1910. doi: 10.1364/ao.46.001896. [DOI] [PubMed] [Google Scholar]
- 26.Tunnell JW, Desjardins AE, Galindo L, Georgakoudi I, McGee SA, Mirkovic J, Mueller MG, Nazemi J, Nguyen FT, Wax A, Zhang QG, Dasari RR, Feld MS. Instrumentation for multimodal spectroscopic diagnosis of epithelial dysplasia. Technol Cancer Res Treat. 2003;2:505–514. doi: 10.1177/153303460300200603. [DOI] [PubMed] [Google Scholar]
- 27.Kuglin CD, Hines DC. IEEE Int Conf on Cybernetics and Society. 1975. The phase correlation image alignment method. [Google Scholar]
- 28.Mourant JR, Fuselier T, Boyer J, Johnson TM, Bigio IJ. Predictions and measurements of scattering and absorption over broad wavelength ranges in tissue phantoms. Appl Opt. 1997;36:949–957. doi: 10.1364/ao.36.000949. [DOI] [PubMed] [Google Scholar]
- 29.Assendelft OWV. Spectrophotometry of Haemoglobin Derivatives. Royal Vangorcum; Assen: 1970. [Google Scholar]
- 30.Rosner B. Fundamentals of biostatistics. 6. Thomson-Brooks/Cole; Belmont, CA: 2006. p. 868. [Google Scholar]
- 31.Vanstaveren HJ, Moes CJM, Vanmarle J, Prahl SA, Vangemert MJC. Light-Scattering in Intralipid-10-Percent in the Wavelength Range of 400–1100 Nm. Appl Opt. 1991;30:4507–4514. doi: 10.1364/AO.30.004507. [DOI] [PubMed] [Google Scholar]
- 32.Wang LV, Wu H-i. Biomedical Optics: Principles and Imaging. Wiley; 2007. [Google Scholar]
- 33.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 34.Nagase H, Woessner JF. Matrix metalloproteinases. J Biol Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 35.Chance B, Cohen P, Jobsis F, Schoener B. Intracellular oxidation-reduction states in vivo. Science. 1962;137:499–508. doi: 10.1126/science.137.3529.499. [DOI] [PubMed] [Google Scholar]