Abstract
Data supporting the occurrence of adaptive trait transfer (i.e. the transfer of genes and thus the phenotype of an adaptive trait through viral recombination, lateral gene transfer or introgressive hybridization) are provided in this review. Specifically, we discuss examples of lateral gene transfer and introgressive hybridization that have resulted in the transfer or de novo origin of adaptations. The evolutionary clades in which this process has been identified include all types of organisms. However, we restrict our discussion to bacteria, fungi, plants and animals. Each of these examples reflects the same consequence, namely that the transfer of genetic material, through whatever mechanism, may result in adaptive evolution. In particular, each of the events discussed has been inferred to impact adaptations to novel environmental settings in the recipient lineage.
Keywords: introgressive hybridization, horizontal transfer, adaptation
1. Introduction
The role of introgressive hybridization or introgression (i.e. the transfer of genetic material between hybridizing taxa through backcrossing; Anderson & Hubricht 1938) in the evolutionary history of sexually reproducing organisms has gained much attention over the past decade and a half (e.g. Grant & Grant 1992; Arnold 1997, 2006; Mallet 2005). Of particular interest, and the subject of this review, are the potential effects from the introduction of ‘foreign’ genetic material into a novel (for the introduced material) genetic background. For example, genetic exchange events have been discussed as evolutionary catalysts for the origin of new species (see Arnold (1997, 2006) and Rieseberg (1997) for reviews) and in some cases entire clades (Seehausen 2004).
In this review, we will extend the discussion of genetic exchange to include not only those due to sexual reproduction and thus introgression but also those events that involve lateral gene transfer (i.e. the transfer of genetic material between individuals from two populations, or groups of populations, that are distinguishable on the basis of one or more heritable characters through the processes of transformation, transduction, conjugation or vector-mediated exchange; Arnold 2006). By discussing organisms that do not reproduce sexually, it is possible to illustrate the taxonomic breadth of similar effects from genetic transfer sensu lato. The observation of such widespread common effects reflects why the tree-of-life metaphor is now argued to be a less predictive hypothesis for biological evolution (e.g. Doolittle & Bapteste 2007). Indeed, we have argued for the adoption of a new metaphor, that of a web of life (Arnold & Larson 2004; Arnold 2006). Given the extent of genetic exchange that has occurred during the evolution of all clades of organisms, the web-of-life metaphor appears to be a more accurate descriptor of evolutionary pattern and process.
Though we will broaden the discussion to include organisms as diverse as bacteria and mammals, we will limit our review to examples consistent with a single evolutionary outcome—that of adaptive trait transfer (i.e. the transfer of genes and thus the phenotype of an adaptive trait through viral recombination, lateral gene transfer or introgressive hybridization; Arnold 2006). Our rationale for choosing to discuss only adaptive trait transfer reflects (i) our detection of this process in species under investigation by our group and (ii) the relatively recent increase in the number of examples that reflect this process. We do not intend to suggest that this is necessarily the most common outcome of natural hybridization, viral recombination or lateral gene transfer, though for certain clades it might well be. In addition, each of the examples given will benefit from further experimental manipulations or comparative analyses to provide a more rigorous test of the adaptive trait transfer hypothesis. Yet, the biological diversity reflected by these examples (Arnold 2006), and the inference of this process by such a wide array of researchers applying diverse approaches provides compelling evidence for adaptive trait transfer. Furthermore, the evolutionary effects that may be realized by the transfer of loci underlying adaptations are of profound importance. These include: (i) an increase in the ecological amplitude of the recipient lineage, (ii) the origin of novel adaptations in the recipient lineage, (iii) the evolution of hybrid taxa with a novel array of adaptations, and (iv) the adaptive radiation of entire taxonomic assemblages (see Anderson (1949), Arnold (1997, 2006) and Seehausen (2004) for reviews).
2. Bacterial species
(a) Lactobacillus salivarius
It is common to consider the evolutionary effects from lateral gene transfer among bacterial species from the viewpoint of species harmful to humans. This bias does not reflect a lack of transfer events involving bacterial species that provide benefits to their human (or other) hosts. It seems instead to reflect the understandable focus of research on control issues involving pathogens. However, in the present discussion, we will focus on two non-pathogenic bacterial examples, Lactobacillus salivarius (the topic of this section) and lactic acid bacterial species (discussed in §2b). These two examples illustrate the observation that beneficial bacterial taxa also possess genomic signatures indicative of gene transfer events during their evolution. Significantly, the species in these clades reflect the occurrence of adaptive trait transfers.
The genus Lactobacillus contains numerous species that provide their hosts ‘probiotic properties’ (i.e. ‘…upon ingestion, they confer a range of benefits on the host…’; Claesson et al. 2006). Like other members of this genus, L. salivarius is beneficial to its hosts. For example, this species has been shown to weaken the effects of both colitis and arthritis in mice (McCarthy et al. 2003; Sheil et al. 2004). Furthermore, it produces a bacteriocin (‘bacteriocins are loosely defined as biologically active protein moieties with a bacteriocidal mode of action’; Riley & Wertz 2002) that acts against harmful Gram-positive bacteria including some Staphylococcus species (Flynn et al. 2002; Claesson et al. 2006).
Evidence for the adaptive transfer of genes into L. salivarius comes from the description of its 2.13 megabase (Mb) genome. In particular, Claesson et al. (2006) found that this genome consisted of four replicons: a 1.83 Mb circular chromosome, a 242 kb megaplasmid and two smaller plasmids (20.4 and 44 kb, respectively). The detection of the megaplasmid reflects the first characterization of this type of genomic component in intestinal lactobacilli, and Claesson et al. (2006) suggested that the megaplasmid itself resulted from a horizontal gene transfer event that conferred adaptations to the host L. salivarius. Several observations provide evidence for adaptive consequences resulting from the lateral transfer of components into the megaplasmid and from the incorporation of the megaplasmid replicon into L. salivarius. Characteristics of the megaplasmid, which are well-known signatures for horizontal transfers (see Ochman et al. (2000) for a review), include the presence of a high proportion of transposable elements and skewed G–C ratios in some genes (Claesson et al. 2006). However, most germane to the present discussion is the observation that the megaplasmid, ‘…although apparently dispensable for viability…confers on the strain a large number of contingency metabolic capabilities and traits directly related to GI tract survival or competitiveness’ (Claesson et al. 2006). This conclusion highlights the adaptive trait transfer event caused by the incorporation of this genomic element into L. salivarius. Intriguingly, one of the adaptive traits conferred by this transferred genome is the ability to produce a bacteriocin (Flynn et al. 2002; Claesson et al. 2006). Thus, the adaptive trait transfer that benefits L. salivarius has also resulted in one of the probiotic characteristics that benefits this bacterial species' hosts, including humans.
(b) Lactic acid bacteria
Makarova et al. (2006) define lactic acid bacteria (LAB) as ‘…microaerophilic, Gram-positive organisms that ferment hexose sugars to produce primarily lactic acid’. These authors also point out that the use of LAB by humans dates back to the dawn of agricultural systems and that at present they (i) provide the key bioconversions in the fermentation of dairy, meat and vegetable products and (ii) are crucial for the production of, for example, wine, coffee and silage (Makarova et al. 2006). These species are naturally associated with plant and milk environments as well as mucosal environments (e.g. small intestine) of animal hosts.
Like all other bacterial species (Ochman 2005), the predominant pattern of genome evolution in the LAB is the progressive loss of genes (Makarova et al. 2006). However, also like other bacterial lineages (Jordan et al. 2001), these species possess numerous horizontally acquired gene products (Makarova et al. 2006). Furthermore, analyses of the transferred elements indicate the presence of adaptive trait transfer. In particular, selective pressures generated by the nutrient-rich environments encountered by LABs appear to have favoured the acquisition of many peptidases. In addition, Makarova et al. (2006) provided an example involving the acquisition of the glycolytic enzyme, enolase. Though all bacterial species contain one copy of this gene, the lactobacilli possess two. The second copy of this gene was apparently acquired through horizontal gene transfer from another bacterial lineage (Makarova et al. 2006).
An additional signature of adaptive trait transfers in the lactobacilli involves the relative rate at which genes are either lost or gained (Makarova et al. 2006). The number of genes lost along lineages was strongly correlated with the length of the lineages. By contrast, the number of gene gain events was not correlated with the phylogenetic branch length. This observation led Makarova et al. (2006) to conclude that purifying selection was probably responsible for the genome reduction events, but that positive selection (i.e. for the acquisition of new adaptations) was a possible cause of the unequal rate of addition of new genes.
3. Fungal species
(a) Ophiostoma novo-ulmi
Unlike bacterial taxa, the process of introgressive hybridization, rather than horizontal gene exchange, is the basis of genetic transfer between fungal lineages. However, regardless of the mode of transfer, the acquisition of novel genes by the recipient fungal form has, in some cases, resulted in the transfer or de novo origin of adaptations (see Arnold (2006) for references). A clear example is provided by the plant pathogen Ophiostoma (Brasier 2001).
Brasier & Kirk (2001) recognized two subspecies of the causal agent of the Dutch elm disease pandemic, the ascomycete fungus Ophiostoma novo-ulmi. The fungal variants Ophiostoma novo-ulmi novo-ulmi and Ophiostoma novo-ulmi americana were originally designated as Eurasian and North American isolates, respectively. It was hypothesized that two separate hybrid subspeciation events had resulted in the two lineages (Brasier & Kirk 2001). Specifically, Brasier & Kirk (2001) suggested that subsequent to a Eurasian point of origin, O. novo-ulmi hybridized with Ophiostoma ulmi, resulting in the European subspecies and that the introduction of O. novo-ulmi into North America in the 1940s was followed by its hybridization with the North American O. ulmi, giving rise to O. n.-u. americana. The detection of diagnostic differences between the two subspecies that were shared with either the Eurasian or North American forms of O. ulmi provided support for this hypothesis (Brasier & Kirk 2001).
Subsequent to their formation, O. n.-u. novo-ulmi and O. n.-u. americana have been brought into contact in Europe. This contact has resulted in introgression between these two subspecies (Brasier 2001; Brasier & Kirk 2001). Though the recombinant forms demonstrated intermediate phenotypes relative to their parents, they were found to be just as pathogenic (Brasier 2001; Brasier & Kirk 2001). Brasier (2001) hypothesized that ‘From such a mélange of recombinants, natural selection may in the future favour…a new race or subspecies of the pathogen’. Brasier's (2001) hypothesis has indeed been supported by subsequent observations. In particular, the conclusions drawn by Paoletti et al. (2006) are of most significance for the present discussion. These authors argued that the introgression of the MAT-1 and vic loci reflected a ‘…rapid adaptation of invasive organisms to a new environment’ (Paoletti et al. 2006). Specifically, the transfer of these loci allowed the recipient lineages to reproduce sexually, thereby providing an escape mechanism from the viral infections that would otherwise be perpetuated through the clonal fungal lineages (Paoletti et al. 2006).
4. Plant species
(a) Louisiana irises
Edgar Anderson (using data from Riley 1938) began his classic book, Introgressive hybridization, with a discussion of the morphological variation found in naturally occurring parental and hybrid populations of the plants known as Louisiana irises. He used these data to describe the process of introgressive hybridization. In a publication from a year earlier, Anderson (1948) also used these species to highlight the catalytic effect of human-mediated habitat disturbances on rates of hybridization and introgression. In both of these publications, Anderson stressed the exemplary nature of the Louisiana irises in defining the results of introgressive hybridization. In particular, Anderson (1949, p. 62) stressed repeatedly ‘A trickle of genes so slight as to be without any practical taxonomic result might still be many times more important than mutation…’
Recent analyses of both natural and experimental hybrid populations of Louisiana irises have supported the conclusions of Anderson (1948, 1949). For example, Cornman et al. (2004) determined both the spatial distribution of naturally occurring hybrid clones and the paternal contribution to the hybrid genotypes. Their detection of spatial genetic structuring and the recruitment of only a subset of the possible hybrid genotypes supported the hypothesis that certain hybrid genotypes possessed a higher fitness due to acquired adaptations (Cornman et al. 2004). These authors also hypothesized a role for differential selection that favoured or disfavoured different hybrids in ‘…the establishment of recombinant lineages that are more fit than the parental types in some habitats’ (Cornman et al. 2004).
Like the analysis of Cornman et al. (2004), linkage and quantitative trait locus (QTL) mapping experiments involving the Louisiana irises species, Iris fulva and Iris brevicaulis have supported the hypothesis of adaptive trait transfer and origin. First, a study by Bouck et al. (2005) detected genomic regions (in reciprocal backcross (BC1) individuals) characterized by significantly lower- or higher-than-expected levels of introgression. In the context of adaptive trait transfer, the detection of regions with significantly increased frequencies of introgression suggests that gene transfer led to positive selection reflected by the increased survivorship of these genotypes.
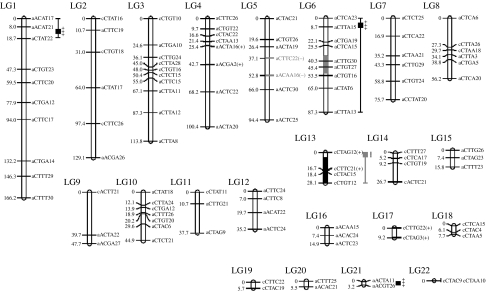
Consistent with the above inference, Martin et al. (2005) defined QTLs (figure 1) associated with the phenotype of long-term survivorship in the same greenhouse environment used by Bouck et al. (2005). Though the plants were watered regularly, the greenhouse environment reflected a water-limited setting for some of the Louisiana irises genotypes. Iris brevicaulis is often found in drier, greenhouse-like, natural environments. By contrast, natural populations of I. fulva most often occur in water-saturated soils (Viosca 1935; Cruzan & Arnold 1993; Johnston et al. 2001). One prediction from the habitat associations observed in the natural populations of I. brevicaulis and I. fulva is that alleles from the former should increase survivorship in the, relatively dry, greenhouse environment. The predicted patterns of mortality were indeed found between the two backcross (BC1) hybrid classes, with I. fulva backcrosses demonstrating twice the frequency of mortality as I. brevicaulis backcross plants. In addition, a QTL analysis detected four genomic regions in the I. fulva hybrids that were significantly associated with survivorship (figure 1; Martin et al. 2005). As expected, introgressed I. brevicaulis DNA increased survivorship at three of the four QTLs. However, the fourth QTL indicated that introgression of I. brevicaulis alleles was associated with decreased survivorship. For this latter locus, the presence of two copies of the I. fulva genomic region increased survivorship (Martin et al. 2005). Though not predicted, this result indicates the adaptive potential arising from the combination of genomic elements from different evolutionary lineages (Arnold 1997, 2006).
Figure 1.
Linkage map of dominant I. brevicaulis iris retroelements (IRRE) retrotransposon display markers (Kentner et al. 2003; Bouck et al. 2005) segregating in the F1 hybrid used to produce backcross hybrids towards I. fulva. Significant QTL for survival in greenhouse conditions are denoted (with 2-LOD CIs—LOD scores a distribution of distances from genetic markers estimated from recombination frequencies; CIs are the confidence intervals around these estimates) to the right of the marker names (Martin et al. 2005). Black or stippled bars (2-LOD CIs) denote QTLs for survival under natural conditions (Martin et al. 2006). Stippled bars represent regions where introgressed (hybrid/heterozygous) regions are favoured, while black bars represent regions where recurrent (parental/homozygous) regions are favoured.
Based on their data, Martin et al. (2005) constructed the following hypothesis: ‘The present findings have important implications for the evolutionary dynamics of naturally occurring hybrid zones. Regions of the genome that increase survivorship when in a heterozygous (i.e. hybrid) state should have an increased likelihood of passing across species boundaries, whereas those that decrease survivorship will be less likely to introgress’. A subsequent study (Martin et al. 2006), involving the transplantation of the same genotypes used in the analysis of survivorship under greenhouse conditions into natural settings, supported this hypothesis. This latter study also determined the genetic architecture, via QTL analyses, of survivorship. However, unlike the relatively dry greenhouse setting of the first two experiments, these same genotypes were exposed to a prolonged (lasting more than three months) flooding event. Under such a regime, it was predicted that alleles from the wet-adapted I. fulva species should positively affect survivorship. In general, the observed patterns of survivorship agreed with this prediction: (i) I. fulva individuals survived at significantly higher frequencies than I. brevicaulis plants, (ii) I. fulva backcross genotypes had a significantly higher frequency of survivorship than the reciprocal backcross genotypes towards I. brevicaulis, (iii) the frequency of survivorship of the I. brevicaulis backcross hybrids was increased by the presence of introgressed I. fulva alleles, and (iv) survivorship in the I. fulva backcross hybrids was affected by two epistatically interacting QTL of opposite effects (Martin et al. 2006). It should also be pointed out that the two QTLs that affected survivorship in the I. fulva BC1 hybrids were on two of the linkage groups that contained QTLs that impacted survivorship in the dry greenhouse environment. However, the effects of the QTLs under the two different environments were found to be in opposite directions. Specifically, the introgressed QTLs lowered survivorship in the dry habitat, while in the flooded environment the introgression of QTLs on these same linkage groups increased survivorship (Martin et al. 2006).
More recent analyses of natural and experimental Louisiana iris hybrid populations provide support for Anderson's (1949) conclusion that introgression has affected the evolutionary trajectory of this species complex. In general, these findings ‘…demonstrate the potential for adaptive trait introgression between these…species and may help to explain patterns of genetic variation observed in naturally occurring hybrid zones’ (Martin et al. 2006).
5. Animal species
(a) Heliconius
Species belonging to the butterfly genus Heliconius provide striking examples of Müllerian mimicry (mimicry between taxa that are unpalatable to predators; see Jiggins et al. (2001) for references). In addition to acting as a warning colour-mediated defence mechanism, the variation in wing markings also affects mate choice thus leading to elevated levels of assortative reproduction (e.g. Jiggins et al. 1997, 2001). Notwithstanding the limitations that divergent wing patterns place on interspecific hybridization, there are now numerous datasets that confirm the role of introgressive hybridization in the evolution of Heliconius and other butterfly genera that display Müllerian mimicry (e.g. Jiggins et al. 2006; Kronforst et al. 2006; Mavárez et al. 2006). For example, Jiggins et al. (2006) used a species-level phylogenetic approach for the genus Ithomia to test for cladogenesis due to colour pattern change. In general, their results supported this hypothesis. However, they also found signatures of divergence accompanied by gene flow. In particular, the phylogenetically most closely related species were determined to be those that were either sympatric or parapatric. One explanation for this phylogenetic signal was ‘…divergence driven by ecological change with ongoing gene flow…’ (Jiggins et al. 2006). Like Ithomia, Heliconius provides excellent examples of divergence accompanied by introgression. Furthermore, because hybrid derivatives demonstrate novel wing patterning, and thus novel adaptations to their ecological settings (i.e. predators), their evolution reflects the process of introgressive hybridization-driven adaptive evolution.
Kronforst et al. (2006) used 657 AFLP markers (a method for generating anonymous genetic markers via polymerase chain reaction) and the sequence variation at 1 mitochondrial and 14 nuclear loci to test for introgression among Heliconius cydno, Heliconius pachinus and Heliconius melpomene. The patterns of variation revealed by this analysis indicated the occurrence of contemporary and historical introgressive hybridization (Kronforst et al. 2006). Significantly, Kronforst et al. (2006) also suggested that H. pachinus' demonstration of a wing pattern with components typical of the other two species studied might reflect a hybrid origin for H. pachinus. This conclusion is, however, not restricted to H. pachinus. Mavárez et al. (2006) also documented the evolution of a novel wing pattern adaptation, and thus the origin of the hybrid species, Heliconius heurippa.
The detection of novel hybrid wing patterning and molecular genetic variation indicating both ancient and recent introgression lends ‘…support to the hypothesis that phenotypic diversification in the genus Heliconius has been fuelled by introgressive hybridization…’ (Kronforst et al. 2006). This conclusion is likely to be true as well for other clades marked by Müllerian mimicry (e.g. Jiggins et al. 2006).
(b) Manacus
Birds provide some of the clearest, and best documented, zoological examples of the evolutionary role of introgressive hybridization (see Grant & Grant (1992) and Arnold (1997, 2006) for references). Indeed, the most well-characterized avian example, and one that includes evidence for adaptive trait transfer, the Darwin's finches, is discussed elsewhere in this volume (Grant & Grant 2008). However, the genus Manacus also provides an illustration of not only the effect of introgressive hybridization but also the process of adaptive trait transfer.
Manacus candei (white-collared manakin) and Manacus vitellinus (golden-collared manakin) form a hybrid zone, the centre of which is proximal to Rio Robalo in the Panamanian province of Bocas del Toro (Parsons et al. 1993; Stein & Uy 2006). Within this hybrid zone, morphological and genetic characters diagnostic for the two species demonstrate significantly different patterns of changeover. In particular, the patterns are typified by non-coincident, clinal changeover for nuclear and mitochondrial molecular markers relative to the male plumage characteristics (Parsons et al. 1993; Brumfield et al. 2001). Importantly, the male plumage traits of the golden-collared species were found to have introgressed into populations of the white-collared phenotype approximately 50 km further than other morphological traits and the DNA markers (Brumfield et al. 2001).
Parsons et al. (1993) and Brumfield et al. (2001) hypothesized that introgression of the male plumage characteristics from the golden- into the white-collared species reflected a selective advantage for males carrying these traits. Furthermore, Parsons et al. (1993) argued that this advantage was most likely due to highly skewed male mating success at the lek sites of Manacus. Support for the hypothesis of sexual selection-driven introgression of the golden-collared traits has come from two studies of behaviour and/or mating success for the two species and their hybrids (McDonald et al. 2001; Stein & Uy 2006). The study conducted by McDonald et al. (2001) involved an analysis of male behaviour demonstrated by three classes of Manacus: (i) white collared, (ii) golden collared, and (iii) ‘lemon collared’. These classes contained M. candei, M. vitellinus and natural hybrid genotypes, respectively. McDonald et al. (2001) hypothesized that higher levels of aggressiveness by the golden- and lemon-collared males, relative to the white-collared form, might be an important contributor to sexual selection-derived introgression. In support of this hypothesis, the golden and lemon males demonstrated a significantly higher attack frequency on taxidermy-mounted specimens. In addition, males belonging to the hybrid (i.e. lemon) class produced more vocalizations than either of the parental classes (McDonald et al. 2001).
Like McDonald et al. (2001), Stein & Uy (2006) investigated the possible role of male–male interactions as a catalyst for the adaptive trait introgression of male plumage characteristics from M. vitellinus into M. candei. Stein & Uy (2006) also tested for the role played by female mate choice. In contrast to McDonald et al. (2001), Stein & Uy (2006) found that the two classes demonstrated equivalent aggressiveness and ‘…held comparable positions within leks’ (Stein & Uy 2006). Furthermore, the golden and white males had equivalent courtship visits from females. Yet, the golden class had significantly more matings. Overall, Stein & Uy's (2006) observations supported a role for sexual selection-driven adaptive trait transfer between the two manakin species. However, their data led them to conclude that male–male interactions were not the major process determining the introgression of male plumage traits. Instead, they concluded rather that sexual selection provided by female mate choice was the primary causal factor. Relevant to the present context, their findings also provided ‘…evidence for hybrid zones as an important source for attractive and adaptive display traits’ (Stein & Uy 2006).
(c) Ficedula
The collared and pied flycatchers (Ficedula albicollis and Ficedula hypoleuca, respectively) reflect our second exemplar of the potential for adaptive trait transfer between avian taxa. Though these species have primarily been used as a model system for studying reproductive isolation and biological speciation (e.g. Sætre et al. 2001), natural hybrid zones characterized by introgression have been detected (Tegelström & Gelter 1990; Sætre et al. 2001, 2003). Furthermore, introgression between these species has resulted in mosaic hybrid genomes characterized by introgressed and non-introgressed regions. For example, Sætre et al. (2003) observed restricted transfer of sex chromosome loci, but ‘…rather extensive introgression and recombination of autosomal genes’.
As with the other examples described in this review, the elevated frequency of transfer of some loci suggests the action of positive selection. Support for the occurrence of adaptive introgression between F. albicollis and F. hypoleuca was discovered in an analysis of the frequency of introgression among nine autosomal and four sex chromosome loci (Borge et al. 2005). In particular, the F. hypoleuca ‘Alasy’ locus allele was found in very high frequencies in F. albicollis. Borge et al. (2005) hypothesized that this allele was linked to a locus under positive selection when in the hybrid background. Given this, the higher-than-expected frequency of introgression of the pied into the collared flycatcher background would indicate that ‘…the collared flycatcher has inherited an adaptive allelic state from the pied flycatcher through introgressive hybridization…’ (Borge et al. 2005).
(d) Homo
It would seem that one of the more contentious issues concerning human origins concerns whether or not introgressive hybridization occurred between anatomically modern (i.e. Homo sapiens) and archaic (e.g. Homo neanderthalensis and Homo erectus) lineages (Stringer & Andrews 1988; Arnold & Meyer 2006; Evans et al. 2006). However, the view that such genetic exchange did not occur seems unsupported given (i) the widespread introgression between contemporary primate taxa (see Jolly (2001) and Arnold & Meyer (2006) for reviews), (ii) the fossil evidence for the spatial and temporal overlap between H. sapiens and archaic Homo taxa (Stringer & Andrews 1988; Finlayson 2005), and (iii) the genetic variation indicating reticulate evolution within the clade containing H. sapiens (e.g. Navarro & Barton 2003; Garrigan et al. 2005; Osada & Wu 2005).
In addition to the large body of data supporting the hypothesis that the divergence of Homo lineages was accompanied by introgressive hybridization, there is also evidence that some of the genetic exchange events resulted in the transfer/origin of adaptations. One such example comes from the analysis of allelic variation present at the microcephalin locus (Evans et al. 2006). Patterns of genetic variation at this locus suggested the introgression of anciently derived alleles from an archaic Homo taxon (possibly H. neanderthalensis) into H. sapiens ca 37 000 yr BP (figure 2; Evans et al. 2006). The significance of this finding was twofold. First, it is additional evidence of the genetic interaction between archaic and modern forms of Homo. Second, it provided evidence of adaptive trait transfer (Evans et al. 2006).
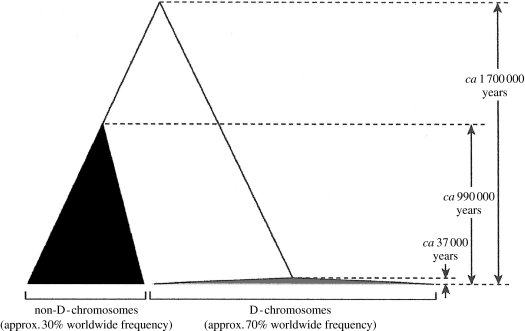
Figure 2.
Genealogical representation of coalescent times for D- and non-D-haplogroup alleles of the microcephalin gene (two different groups of closely related haplotypes at this locus; Evans et al. 2006). The extremely recent coalescence time for the D-haplogroup alleles, and their ancient divergence from the non-D-haplogroup alleles, supports the hypothesis of introgression of the D-alleles from an archaic Homo lineage into H. sapiens ca 37 000 yr BP (Evans et al. 2006).
The microcephalin gene is known to contribute to the regulation of brain size in humans (Jackson et al. 2002). In addition, the inference of positive selection acting on this locus, both in the ancestral lineage of humans (Evans et al. 2004; Wang & Su 2004) and in the Homo lineage as well (Evans et al. 2005), suggests its adaptive significance. The pattern indicative of strong positive selection in the Homo clade is illustrated in figure 2. In this regard, Evans et al. (2006) detected a recent (ca 37 000 yr BP) coalescence age for the D-haplotype alleles (a group of closely related haplotypes at the microcephalin locus; Evans et al. 2006). Yet, these alleles occur at an extremely high frequency and are distributed worldwide (figure 2; Evans et al. 2006). The combination of their recent coalescence and high worldwide frequency lead to the conclusion that the D-haplotype alleles have been increased in frequency by positive selection. Evans et al. (2006) also concluded that the introduction of these haplotypes into the H. sapiens population was via introgressive hybridization with an archaic lineage that had been separated from the lineage leading to modern humans for ca 1.1 Myr (figure 2).
Like each of the examples in §5a–c, the apparent introgression of the microcephalin allele from an archaic form into an anatomically modern Homo species had the potential to affect adaptive evolution. Indeed, Evans et al. (2006) suggested that ‘It is perhaps not surprising then that modern humans…could in theory benefit from adopting some adaptive alleles from the populations they replaced’. Like bacteria, fungi, plants and other animals, the introgression of this key determinant of human brain size supports the hypothesis that adaptive trait transfer has profoundly impacted the evolution of H. sapiens as well.
6. Conclusions
Genetic exchange, involving introgression or lateral exchange, has been a frequent and evolutionary significant factor for all organismic clades (Arnold 1997, 2006). Each genetic transfer event has had the potential to affect the fitness of the recipient individual. When the introduction of the foreign DNA has resulted in an increase in fitness in certain environmental settings, the transfer or de novo origin of an adaptive trait can be inferred. QTL mapping experiments, genome-sequencing projects, experimental determinations of fitness and gene functional assays have identified numerous examples in which horizontal gene transfer or introgressive hybridization has resulted in such adaptive exchanges/origins. Though each of the findings discussed in this review are consistent with the hypothesis of adaptive trait transfer, they will be strengthened by additional experimental and comparative studies. As additional datasets are produced, we expect that the adaptive consequences inferred for the above examples will be identified as a common outcome of genetic exchange.
Acknowledgments
During the writing of this review, M.L.A. and Y.S. were supported by NSF grant DEB-0345123. The authors wish to express their deepest gratitude to N. Brede, K. Schwenk and B. Streit who were both the organizers of the symposium that was the stimulus for these proceedings and the editors of this volume.
Footnotes
One contribution of 16 to a Theme Issue ‘Hybridization in animals: extent, processes and evolutionary impact’.
References
- Anderson E. Hybridization of the habitat. Evolution. 1948;2:1–9. doi:10.2307/2405610 [Google Scholar]
- Anderson E. Wiley; New York, NY: 1949. Introgressive hybridization. [Google Scholar]
- Anderson E, Hubricht L. Hybridization in Tradescantia. III. The evidence for introgressive hybridization. Am. J. Bot. 1938;25:396–402. doi:10.2307/2436413 [Google Scholar]
- Arnold M.L. Oxford University Press; Oxford, UK: 1997. Natural hybridization and evolution. [Google Scholar]
- Arnold M.L. Oxford University Press; Oxford, UK: 2006. Evolution through genetic exchange. [Google Scholar]
- Arnold M.L, Larson E.J. Evolution's new look. Wils. Q. (Autumn) 2004:60–72. [Google Scholar]
- Arnold M.L, Meyer A. Natural hybridization in primates: one evolutionary mechanism. Zoology. 2006;109:261–276. doi: 10.1016/j.zool.2006.03.006. doi:10.1016/j.zool.2006.03.006 [DOI] [PubMed] [Google Scholar]
- Borge T, Lindroos K, Nádvorník P, Syvänen A.-C, Sætre G.-P. Amount of introgression in flycatcher hybrid zones reflects regional differences in pre and post-zygotic barriers to gene exchange. J. Evol. Biol. 2005;18:1416–1424. doi: 10.1111/j.1420-9101.2005.00964.x. [DOI] [PubMed] [Google Scholar]
- Bouck A.C, Peeler R, Arnold M.L, Wessler S.R. Genetic mapping of species boundaries in Louisiana irises using IRRE retrotransposon display markers. Genetics. 2005;171:1289–1303. doi: 10.1534/genetics.105.044552. doi:10.1534/genetics.105.044552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasier C.M. Rapid evolution of introduced plant pathogens via interspecific hybridization. BioScience. 2001;51:123–133. doi:10.1641/0006-3568(2001)051[0123:REOIPP]2.0.CO;2 [Google Scholar]
- Brasier C.M, Kirk S.A. Designation of the EAN and NAN races of Ophiostoma novo-ulmi as subspecies. Mycol. Res. 2001;105:547–554. doi:10.1017/S0953756201004087 [Google Scholar]
- Brumfield R.T, Jernigan R.W, McDonald D.B, Braun M.J. Evolutionary implications of divergent clines in an avian (Manacus: Aves) hybrid zone. Evolution. 2001;55:2070–2087. doi: 10.1111/j.0014-3820.2001.tb01322.x. doi:10.1111/j.0014-3820.2001.tb01322.x [DOI] [PubMed] [Google Scholar]
- Claesson M.J, et al. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl Acad. Sci. USA. 2006;103:6718–6723. doi: 10.1073/pnas.0511060103. doi:10.1073/pnas.0511060103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman R.S, Burke J.M, Wesselingh R.A, Arnold M.L. Contrasting genetic structure of adults and progeny in a Louisiana iris hybrid population. Evolution. 2004;58:2669–2681. doi: 10.1111/j.0014-3820.2004.tb01620.x. doi:10.1554/04-321 [DOI] [PubMed] [Google Scholar]
- Cruzan M.B, Arnold M.L. Ecological and genetic associations in an Iris hybrid zone. Evolution. 1993;47:1432–1445. doi: 10.1111/j.1558-5646.1993.tb02165.x. doi:10.2307/2410158 [DOI] [PubMed] [Google Scholar]
- Doolittle W.F, Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc. Natl Acad. Sci. USA. 2007;104:2043–2049. doi: 10.1073/pnas.0610699104. doi:10.1073/pnas.0610699104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.D, Anderson J.R, Vallender E.J, Choi S.S, Lahn B.T. Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Hum. Mol. Genet. 2004;13:1139–1145. doi: 10.1093/hmg/ddh126. doi:10.1093/hmg/ddh126 [DOI] [PubMed] [Google Scholar]
- Evans P.D, Gilbert S.L, Mekel-Bobrov N, Vallender E.J, Anderson J.R, Vaez-Azizi L.M, Tishkoff S.A, Hudson R.R, Lahn B.T. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science. 2005;309:1717–1720. doi: 10.1126/science.1113722. doi:10.1126/science.1113722 [DOI] [PubMed] [Google Scholar]
- Evans P.D, Mekel-Bobrov N, Vallender E.J, Hudson R.R, Lahn B.T. Evidence that the adaptive allele of the brain size gene microcephalin introgressed into Homo sapiens from an archaic Homo lineage. Proc. Natl Acad. Sci. USA. 2006;103:18 178–18 183. doi: 10.1073/pnas.0606966103. doi:10.1073/pnas.0606966103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson C. Biogeography and evolution of the genus Homo. Trends Ecol. Evol. 2005;20:457–463. doi: 10.1016/j.tree.2005.05.019. doi:10.1016/j.tree.2005.05.019 [DOI] [PubMed] [Google Scholar]
- Flynn S, van Sinderen D, Thornton G.M, Holo H, Nes I.F, Collins J.K. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology. 2002;148:973–984. doi: 10.1099/00221287-148-4-973. [DOI] [PubMed] [Google Scholar]
- Garrigan D, Mobasher Z, Severson T, Wilder J.A, Hammer M.F. Evidence for archaic Asian ancestry on the human X chromosome. Mol. Biol. Evol. 2005;22:189–192. doi: 10.1093/molbev/msi013. doi:10.1093/molbev/msi013 [DOI] [PubMed] [Google Scholar]
- Grant P.R, Grant B.R. Hybridization of bird species. Science. 1992;256:193–197. doi: 10.1126/science.256.5054.193. doi:10.1126/science.256.5054.193 [DOI] [PubMed] [Google Scholar]
- Grant B.R, Grant P.R. Fission and fusion of Darwin's finches populations. Phil. Trans. R. Soc. B. 2008;363:2821–2829. doi: 10.1098/rstb.2008.0051. doi:10.1098/rstb.2008.0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.P, et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am. J. Hum. Genet. 2002;71:136–142. doi: 10.1086/341283. doi:10.1086/341283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins C.D, McMillan W.O, King P, Mallet J. The maintenance of species differences across a Heliconius hybrid zone. Heredity. 1997;79:495–505. [Google Scholar]
- Jiggins C.D, Naisbit R.E, Coe R.L, Mallet J. Reproductive isolation caused by colour pattern mimicry. Nature. 2001;411:302–305. doi: 10.1038/35077075. doi:10.1038/35077075 [DOI] [PubMed] [Google Scholar]
- Jiggins C.D, Mallarino R, Willmott K.R, Bermingham E. The phylogenetic pattern of speciation and wing pattern change in neotropical Ithomia butterflies (Lepidoptera: Nymphalidae) Evolution. 2006;60:1454–1466. doi: 10.1554/05-483.1. doi:10.1554/05-483.1 [DOI] [PubMed] [Google Scholar]
- Johnston J.A, Wesselingh R.A, Bouck A.C, Donovan L.A, Arnold M.L. Intimately linked or hardly speaking? The relationship between genotypic variation and environmental gradients in a Louisiana iris hybrid population. Mol. Ecol. 2001;10:673–681. doi: 10.1046/j.1365-294x.2001.01217.x. doi:10.1046/j.1365-294x.2001.01217.x [DOI] [PubMed] [Google Scholar]
- Jolly C.J. A proper study for mankind: analogies from the papionin monkeys and their implications for human evolution. Yearb. Phys. Anthropol. 2001;44:177–204. doi: 10.1002/ajpa.10021. doi:10.1002/ajpa.10021 [DOI] [PubMed] [Google Scholar]
- Jordan I.K, Makarova K.S, Spouge J.L, Wolf Y.I, Koonin E.V. Lineage-specific gene expansions in bacterial and archaeal genomes. Genome Res. 2001;11:555–565. doi: 10.1101/gr.166001. doi:10.1101/gr.GR-1660R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentner E.K, Arnold M.L, Wessler S.R. Characterization of high copy number retrotransposons from the large genomes of the Louisiana iris species and their use as molecular markers. Genetics. 2003;164:685–697. doi: 10.1093/genetics/164.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst M.R, Young L.G, Blume L.M, Gilbert L.E. Multilocus analyses of admixture and introgression among hybridizing Heliconius butterflies. Evolution. 2006;60:1254–1268. [PubMed] [Google Scholar]
- Makarova K, et al. Comparative genomics of the lactic acid bacteria. Proc. Natl Acad. Sci. USA. 2006;103:15 611–15 616. doi: 10.1073/pnas.0607117103. doi:10.1073/pnas.0607117103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. doi:10.1016/j.tree.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Martin N.H, Bouck A.C, Arnold M.L. Loci affecting long-term hybrid survivability in Louisiana irises: implications for reproductive isolation and introgression. Evolution. 2005;59:2116–2124. [PubMed] [Google Scholar]
- Martin N.H, Bouck A.C, Arnold M.L. Detecting adaptive trait introgression between Iris fulva and I. brevicaulis in highly selective field conditions. Genetics. 2006;172:2481–2489. doi: 10.1534/genetics.105.053538. doi:10.1534/genetics.105.053538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavárez J, Salazar C.A, Bermingham E, Salcedo C, Jiggins C.D, Linares M. Speciation by hybridization in Heliconius butterflies. Nature. 2006;441:868–871. doi: 10.1038/nature04738. doi:10.1038/nature04738 [DOI] [PubMed] [Google Scholar]
- McCarthy J, et al. Double blind, placebo controlled trial of two probiotic strains in interleukin 10 knockout mice and mechanistic link with cytokine balance. Gut. 2003;52:975–980. doi: 10.1136/gut.52.7.975. doi:10.1136/gut.52.7.975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D.B, Clay R.P, Brumfield R.T, Braun M.J. Sexual selection on plumage and behavior in an avian hybrid zone: experimental tests of male–male interactions. Evolution. 2001;55:1443–1451. doi: 10.1111/j.0014-3820.2001.tb00664.x. doi:10.1111/j.0014-3820.2001.tb00664.x [DOI] [PubMed] [Google Scholar]
- Navarro A, Barton N.H. Chromosomal speciation and molecular divergence—accelerated evolution in rearranged chromosomes. Science. 2003;300:321–324. doi: 10.1126/science.1080600. doi:10.1126/science.1080600 [DOI] [PubMed] [Google Scholar]
- Ochman H. Genomes on the shrink. Proc. Natl Acad. Sci. USA. 2005;102:11 959–11 960. doi: 10.1073/pnas.0505863102. doi:10.1073/pnas.0505863102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence J.G, Groisman E.A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. doi:10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- Osada N, Wu C.-I. Inferring the mode of speciation from genomic data: a study of the great apes. Genetics. 2005;169:259–264. doi: 10.1534/genetics.104.029231. doi:10.1534/genetics.104.029231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M, Buck K.W, Brasier C.M. Selective acquisition of novel mating type and vegetative incompatibility genes via interspecies gene transfer in the globally invading eukaryote Ophiostoma novo-ulmi. Mol. Ecol. 2006;15:249–262. doi: 10.1111/j.1365-294X.2005.02728.x. doi:10.1111/j.1365-294X.2005.02728.x [DOI] [PubMed] [Google Scholar]
- Parsons T.J, Olson S.L, Braun M.J. Unidirectional spread of secondary sexual plumage traits across an avian hybrid zone. Science. 1993;260:1643–1646. doi: 10.1126/science.260.5114.1643. doi:10.1126/science.260.5114.1643 [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H. Hybrid origins of plant species. Annu. Rev. Ecol. Syst. 1997;28:359–389. doi:10.1146/annurev.ecolsys.28.1.359 [Google Scholar]
- Riley H.P. A character analysis of colonies of Iris fulva, Iris hexagona var. giganticaerulea and natural hybrids. Am. J. Bot. 1938;25:727–738. doi:10.2307/2436599 [Google Scholar]
- Riley M.A, Wertz J.E. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie. 2002;84:357–364. doi: 10.1016/s0300-9084(02)01421-9. doi:10.1016/S0300-9084(02)01421-9 [DOI] [PubMed] [Google Scholar]
- Sætre G.-P, Borge T, Lindell J, Moum T, Primmer C.R, Sheldon B.C, Johnsen A, Ellegren H. Speciation, introgressive hybridization and nonlinear rate of molecular evolution in flycatchers. Mol. Ecol. 2001;10:737–749. doi: 10.1046/j.1365-294x.2001.01208.x. doi:10.1046/j.1365-294x.2001.01208.x [DOI] [PubMed] [Google Scholar]
- Sætre G.-P, Borge T, Lindroos K, Haavie J, Sheldon B.C, Primmer C, Syvänen A.-C. Sex chromosome evolution and speciation in Ficedula flycatchers. Proc. R. Soc. B. 2003;270:53–59. doi: 10.1098/rspb.2002.2204. doi:10.1098/rspb.2002.2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. doi:10.1016/j.tree.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Sheil B, McCarthy J, O'Mahony L, Bennett M.W, Ryan P, Fitzgibbon J.J, Kiely B, Collins J.K, Shanahan F. Is the mucosal route of administration essential for probiotic function? Subcutaneous administration is associated with attenuation of murine colitis and arthritis. Gut. 2004;53:694–700. doi: 10.1136/gut.2003.027789. doi:10.1136/gut.2003.027789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A.C, Uy J.A.C. Unidirectional introgression of a sexually selected trait across an avian hybrid zone: a role for female choice? Evolution. 2006;60:1476–1485. doi:10.1554/05-575.1 [PubMed] [Google Scholar]
- Stringer C.B, Andrews P. Genetic and fossil evidence for the origin of modern humans. Science. 1988;239:1263–1268. doi: 10.1126/science.3125610. doi:10.1126/science.3125610 [DOI] [PubMed] [Google Scholar]
- Tegelström H, Gelter H.P. Haldane's rule and sex biased gene flow between two hybridizing flycatcher species (Ficedula albicollis and F. hypoleuca, Aves: Muscicapidae) Evolution. 1990;44:2012–2021. doi: 10.1111/j.1558-5646.1990.tb04307.x. doi:10.2307/2409611 [DOI] [PubMed] [Google Scholar]
- Viosca P., Jr The irises of southeastern Louisiana—a taxonomic and ecological interpretation. Bull. Am. Iris Soc. 1935;57:3–56. [Google Scholar]
- Wang Y.-Q, Su B. Molecular evolution of microcephalin, a gene determining human brain size. Hum. Mol. Genet. 2004;13:1131–1137. doi: 10.1093/hmg/ddh127. doi:10.1093/hmg/ddh127 [DOI] [PubMed] [Google Scholar]