Abstract
Large white-headed gulls provide an interesting group of birds for studies of hybridization. The group is composed of 20 species of recent origin, often with weak reproductive barriers. Here we report the results from a study on the glaucous gull Larus hyperboreus, an Arctic species which has been breeding in Iceland for centuries, and the herring gull Larus argentatus which has a wide distribution in Europe but colonized Iceland in 1920s. Previous studies, based on morphological variation indicated hybridization between the two species in Iceland, have been questioned as it may just reflect variation within the species. Here we evaluate whether hybridization has occurred between the two species in Iceland by studying variation in microsatellites and mtDNA. The analysis is based on feathers taken from wings sampled in Iceland over a period of 40 years. The results are compared with samples obtained from East Greenland and published sequences of samples obtained throughout Europe. The genetic analysis reveals a distinctive grouping of the two species, although they present a shallow genealogy and an extensive sharing of the genetic variants between the two species. Several individuals show admixture for molecular markers, which may result from an incomplete lineage sorting although geographical patterns of both mtDNA haplotypes and microsatellites strongly indicate a recent hybridization in Iceland.
Keywords: gulls, introgression, Arctic, expansion, cytochrome b, phylogeography
1. Introduction
Species distributions and genetic population structure have been found to reflect climatic oscillations during the Quaternary glacial periods (Hewitt 2000; Newton 2003). Ancestral populations split up during glacial periods and evolved in allopatry in distinct refugia (Hewitt 1996). Following the retreat of the glaciers, populations of several species have expanded and formed secondary contact zones (Barton & Hewitt 1985; Hewitt 2004), often involving various forms of hybridization (Grant & Grant 1992).
Avian hybrids are widespread and more than 9% of all bird species are known to have hybridized in nature, although this is more common in some taxa than others (Grant & Grant 1992; Randler 2002). Hybridization among large white-headed gull species has been reported from many areas (McCarthy 2006), e.g. Larus smithsonianus×Larus hyperboreus in the Mackenzie Delta, Canada (Spear 1987), Larus glaucescens×Larus occidentalis on the Pacific coast of North America (Bell 1996), Larus argentatus×Larus fuscus in western France (Yesou 1991), L. argentatus×Larus cachinnans in Russia (Panov & Monzikov 1999) and Poland (Gay et al. 2007), and L. argentatus×L. hyperboreus in Iceland (Ingólfsson 1970). The widespread instances of incomplete reproductive isolation in this group provide an excellent system to study hybridization and introgression between species of recent origin. The large white-headed gull species originated ca 100–600 kyr ago, possibly in two main glacial refugia, the Atlantic and the Aralo-Caspian refugium (Crochet et al. 2003; Liebers et al. 2004).
The glaucous gull (L. hyperboreus) is an Arctic circumpolar species that has undoubtedly been a breeding bird in Iceland for a long time (Ingólfsson 1970). The breeding distribution of the European herring gull (L. argentatus) is confined to northern Europe, ranging from Kola Peninsula to France and recently to Iceland (Olsen & Larsson 2004). The distribution of L. argentatus increased markedly during the twentieth century, which led to its colonization in Iceland ca 1925, where it came into contact with L. hyperboreus (Ingólfsson 1970). In Iceland, both species breed in colonies, often large, the former usually on grassy slopes and cliffs by the sea (Guðmundsson 1955) and the latter more often on relatively level ground near the sea (Ingólfsson 1982, personal observations). Despite colonial breeding, dispersal is known to be substantial in large gulls (Cramps & Simmons 1983; Coulson 1991) and as is reflected by the recent colonization of Iceland.
Ingólfsson (1970, 1987) described hybridization among the two species based on morphological variation and observation of mated pairs. Apparently pure glaucous gulls predominated in western Iceland while apparently pure herring gulls and hybrids were common in southern and eastern Iceland. A colony that apparently consisted exclusively of glaucous gulls was known in southwestern Iceland in the early 1900, but by 1970 it consisted mostly of apparently pure L. argentatus and possible hybrids (Ingólfsson 1970). In another instance, a colony consisting mostly of apparently pure glaucous gulls in southeastern Iceland in 1963 had changed to a colony of mostly herring gull-like birds and putative hybrids by 1973 (Ingólfsson 1987). Studying the allozyme and morphological variation, Snell (1991a,b) argued against hybridization, claiming that the variation found in gulls in Iceland simply reflected the natural variation within the species (but see rebuttal by Ingólfsson 1993).
A previous study of the genetics of the herring gull failed to detect whether hybridization was taking place in Iceland (Snell 1991a). Studies by Bell (1996) and Crochet (2000) indicated that the situation in gulls was more complex than simple surveys of plumage phenotypes or allozyme variation had shown. Similarly, more recent genetic studies on herring gulls and related species have shown discrepancies between gene trees and the taxon phylogeny, resulting from incomplete lineage sorting and/or hybridization (Crochet et al. 2002, 2003; Liebers et al. 2004; Gay et al. 2007). Genetic work by Liebers et al. (2004) on L. hyperboreus from Novaja Semlija, Svalbard and Baffinland indicated a phylogeographical split between Europe and America, and a similar split was described between the European L. argentatus and the American L. smithsonianus.
In order to solve the dispute by Ingólfsson and Snell, on whether hybridization occurs among glaucous gulls (L. hyperboreus) and herring gulls (L. argentatus) in Iceland, a better understanding of various aspects of the biology of these two species is needed. Here we study variation at five microsatellite loci and a sequence variation in mtDNA (cytochrome b) in samples obtained in Iceland over a period of 40 years, and from Greenland. The mtDNA sequences are in addition compared with sequence data from samples obtained elsewhere in Europe and from Canada (Liebers et al. 2004). To look for signs of incomplete lineage sorting and hybridization, we examine firstly whether the mtDNA genealogy confines separate groups for the two species. Secondly, we look for signs of temporal and geographical patterns. As incomplete lineage sorting should be independent of geographical locations, a local sharing of haplotypes in a recent contact zone would point to hybridization.
2. Material and methods
(a) Samples
Samples were obtained from various breeding colonies in seven regions of Iceland (figure 1) and from Kulusuk in East Greenland (table 1). According to Ingólfsson (1970), western Iceland was dominated by L. hyperboreus-like birds, while eastern and southern Iceland was dominated by L. argentatus-like birds and apparent hybrids. In one colony in southeastern Iceland (location number 6 in table 1), both the types as well as their hybrids were common. Samples originated from three time periods. Samples from period 1 (collected by Ingólfsson 1964–1973) and period 2 (by Snell 1985–1986) were obtained from specimens at the collection of The Icelandic Institute of Natural History. Sampling from period 3 (2005–2006) was carried out in the field, by shooting or Cannon netting, near breeding colonies. For the purpose of the analysis, gulls were assigned to two groups on the basis of primary pattern (i.e. the coloration of wing primary feathers), those scoring 0.0–1.0 in hybrid index, HI (see Ingólfsson 1970) being termed L. hyperboreus, and those scoring between 1.1 and 5.0 being termed L. argentatus. The index reflects different degrees of black patterns, from no trace (HI=0) to black subterminal bands with sharp edges (HI=5). This grouping of genotypes based on a HI is supported by Bayesian clustering (BAPS; Marttinen et al. 2006). For comparison, mtDNA sequences from 137 individuals from Liebers et al. (2004) were included in this study.
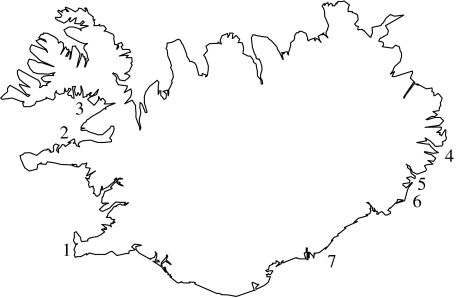
Figure 1.
Sampling locations of gulls in Iceland. Numbers refer to locations listed in table 1. Samples included in population comparisons per location are 1 (a3w), 2 (h1w, h2 and h3), 3 (h1w), 4 (a1, a2 and a3e) and 5 and 6 (a1 and h1e). The main nesting areas of L. hyperboreus are in northwestern Iceland (2 and 3), whereas L. argentatus is mainly nesting in eastern Iceland (4–6).
Table 1.
Numbers of individuals (n) sampled from each location in three time periods. (The numbers in sample names refer to the different periods: 1, 1964–1973; 2, 1985–1986 and 3, 2005–2006. The third letter in sample names refers to east (e) and west (w). Numbers (no) present locations in figure 1, the same number is given to locations in close vicinity to each other. Asterisks, samples were not included in population comparisons.)
| species | samples | no | location | latitude (N) | longitude (W) | n |
|---|---|---|---|---|---|---|
| L. arg. | a1 | 5 | Hrómundarey | 64°35 | 14°19 | 46 |
| — | — | 6 | Horn | 64°15 | 14°59 | 13 |
| — | — | 4 | Skrúður | 64°54 | 13°38 | 2 |
| — | 1 | Reykjanes* | 64°01 | 22°40 | 3 | |
| — | 7 | Hjörleifshöfði* | 63°25 | 18°45 | 1 | |
| — | a2 | 4 | Skrúður | 64°54 | 13°38 | 37 |
| — | a3e | 4 | Reyðarfjörður | 64°56 | 13°41 | 25 |
| — | a3w | 2 | Grundarfjörður | 64°55 | 23°15 | 3 |
| — | — | 1 | Reykjanes | 64°01 | 22°40 | 4 |
| L. hyper. | h1w | 2 | Bjarnarhafnarfjall | 64°59 | 23°01 | 45 |
| — | — | 2 | Búlandshöfði | 64°56 | 23°28 | 5 |
| — | — | 3 | Reykhólahreppur | 65°30 | 22°17 | 3 |
| — | h2 | 2 | Bjarnarhafnarfjall | 64°59 | 23°01 | 26 |
| — | 4 | Skrúður* | 64°54 | 13°38 | 1 | |
| — | h3 | 2 | Grundafjörður | 64°55 | 23°15 | 65 |
| — | 4 | Reyðarfjörður* | 64°56 | 13°41 | 1 | |
| — | h1e | 6 | Horn | 64°14 | 14°59 | 2 |
| — | — | 5 | Hrómundarey | 64°35 | 14°18 | 6 |
| — | hk | Kulusuk, Greenland | 65°34 | 37°10 | 19 |
(b) Molecular analysis
DNA was extracted from feathers in 300 μl of 6% Chelex 100 (Biorad) containing 3 μl proteinase K (0.5 mg ml−1), incubated at 65°C for 2 hours and at 95°C for 5 min. A 971 bp fragment of the mtDNA cytochrome b gene was amplified using primers L14967 and H15938, as in Crochet et al. (2003). For the museum specimens, two smaller overlapping segments of the cytochrome b gene were amplified separately, using primers L15440 (5′-GCCAAACCCTCGTAGAATGA-3′) with H15938, and H15619 (5′-GTAGGGGTGGAATGGGATTT-3′) with L15008 (Crochet et al. 2003). Polymerase chain reaction (PCR) amplifications were carried out in 10 μl volume containing 1×amplification buffer, 0.09 U Taq DNA polymerase, 1.5 mM MgCl2, 1 mM of each dNTP and 10 μM of each primer. Cycling conditions were 94°C for 40 s, 56°C for 30 s and 72°C for 60 s for 30 cycles for primers L14967-H15938. For primers used on museum specimens, the cycling conditions were 94°C for 60 s, 53°C for 30 s and 72°C for 60 s, for 38 cycles.
Prior to sequencing, excess primers and nucleotides were enzymatically removed from PCR amplification products using a mixture of exonuclease I and Antarctic phosphatase (New England BioLabs). Cycle sequencing was carried out using the BigDye Terminator v. 1.1 Cycle Sequencing Kit on ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
Five microsatellite loci (HG14, HG16, HG18, HG25 and HG27), developed for the North American herring gull L. smithsonianus, were amplified following the procedures in Crochet et al. (2003). PCR was performed in a 15 μl reaction containing the same concentrations as listed above; the products were sent for genotyping to GATCBiotech AG, Germany and scored with the Gene Marker v. 5.1 software package (SoftGenetics LLC 2004).
(c) Statistical analysis
Basic statistics of mtDNA diversity, including nucleotide and haplotype diversity for each sample, were calculated with Arlequin v. 3 (Excoffier et al. 2005). Deviations from equilibrium population dynamics, resulting from population expansion or admixture, were estimated by calculating pairwise differences among all sequences (Rogers & Harpending 1992). The introgression ratio, IG, was calculated according to Belahbib et al. (2001), with modifications. The original IG reflects the amount of locally shared haplotypes between two species and is expected to be one when there is no difference between the species and zero when they are totally different. Here we look at the IG ratio between a sample i of species x and all samples combined of species y, and vice versa. The standard errors and CIs of the IG values were estimated by non-parametric bootstrapping, with the program boot in the R package (available at http://www.r-project.org/). Bootstrap samples were obtained by resampling 1000 individuals within the subpopulations of each species. To detect signatures of population structure, hierarchical analyses of molecular variance (AMOVA) were calculated among species and populations (Excoffier et al. 1992), and for all pairwise comparisons between samples. The analyses were based on haplotype frequencies (Wright's FST) and also on variance in divergence between sequences (ΦST). The overall relationship among the haplotypes is presented with a median-joining network (Bandelt et al. 1999) constructed with Network v. 4.201 (www.fluxus-engineering.com). Equal weights were assumed for each variable position.
Genetic diversity parameters of the microsatellite variation, including number of alleles (na) and unbiased expected heterozygosity (He), were summarized with Genetix v. 4.05 (Belkhir et al. 2004). The effective number of alleles was calculated as ne=1/(1−He) (Weir 1990). Departures from Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium were tested with random permutations as implemented with Genetix. The hierarchical partition of variation among species and populations were conducted using Arlequin v. 3 (Excoffier et al. 2005). Pairwise FST values (Weir & Cockerham 1984) and the proportion of variation based on variance in allele size (RST) were calculated (Slatkin 1995). The pairwise differences were summarized with a multidimensional scale plot, also known as principal coordinate analysis (Venables & Ripley 1994). Bonferroni-adjusted probabilities were used (Sokal & Rohlf 1995) in cases of multiple and non-orthogonal tests.
The genetic structure, including both information from microsatellites and the mtDNA haplotypes together, was analysed by the Bayesian clustering method and the admixture analyses implemented in BAPS v. 4.14 (Marttinen et al. 2006). The analysis was based on samples that had been scored for three or more markers. In BAPS, the a priori information of the nine samples listed in table 2 was used for clustering. Evidence for admixture was considered significant for individuals with a Bayesian p<0.05, as recommended by Marttinen et al. (2006).
Table 2.
Mitochondrial DNA haplotypes, nucleotide diversity (π) and haplotype diversity (h) for each sample at a given time period in all locations. (Numbers of the haplotypes are given in brackets. IGs with standard errors in parenthesis are given for the samples from Iceland. Population names correspond to table 1. Asterisks, not calculated.)
| pop. | n | haplotypes | π*100 | h | IG |
|---|---|---|---|---|---|
| a1 | 28 | A12(13), A6(1) A25(1), A26(1), MA1(7), MA2(1), H2(4) | 0.194 | 0.718 | 0.849(0.086) |
| a2 | 26 | A12(7), A6(1), A14(1), A20(4), A23(3), MA1(7), MI3(3) | 0.446 | 0.830 | 0.464(0.120) |
| a3e | 24 | A12(9), A6(3), A20(2), A23(2), A24(1), MA1(7) | 0.373 | 0.793 | 0.558(0.120) |
| a3w | 7 | A12(2), A6(1), A21(1), H2(3) | 0.381 | 0.810 | 1.048(0.182) |
| h1w | 27 | A12(16), A20(2), H2(8), H8(1) | 0.188 | 0.576 | 0.798(0.061) |
| h2 | 21 | A12(6), H2(15) | 0.050 | 0.455 | 0.419(0.124) |
| h3 | 55 | A12(23), H1(1), H2(29), H6(1), H9(1) | 0.085 | 0.556 | 0.598(0.088) |
| h1e | 2 | A12, MA1 | 0.118 | 1.0 | * |
| Hk | 18 | A12(3), H1(7), H2(1), H7(1), G1(4), G3(2) | 0.285 | 0.798 | * |
3. Results
(a) Analysis of the mitochondrial cytochrome b
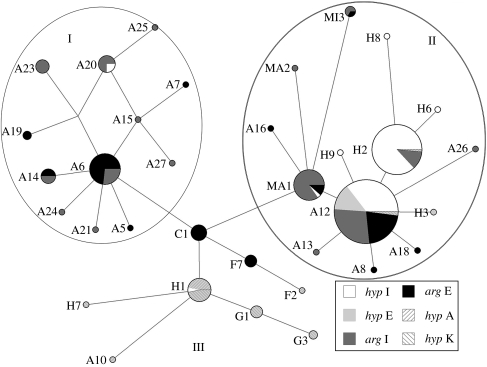
Sequences of a length of 850 bp of the cytochrome b were obtained from 214 gulls, with 41 polymorphic sites, defining 20 haplotypes (table 2). Sequences were deposited with GenBank (accession numbers: EU526315–EU526334). Haplotypes differ from each other by a maximum of 10 mutations (figure 2). Five haplotypes are identical to the haplotypes reported by Liebers et al. (2004). The haplotypes form three main clades: two European clades (I and II) and a North American clade (III; figure 2). Only one European L. hyperboreus, from west Iceland (h3), has an American haplotype (H1). Four L. hyperboreus from Kulusuk have European haplotypes, three A12 and one H2. Larus smithsonianus from N. America have haplotypes that differ by one substitution from H1, and Larus glaucoides (also N. American) has been found to share both H1 and G1.
Figure 2.
Network of the cytochrome b haplotypes obtained in this study and the study by Liebers et al. (2004). The size of the pies reflects the frequency of a particular haplotype. The length of the lines connecting the pies, measured from their centres, are in proportion to the number of base pair substitutions (1–3) separating the haplotypes. Clusters I and II are found within the European samples and cluster III represents the American clade. Species and geographical regions are noted with different shadings: hyp I, L. hyperboreus from Iceland (10 out of 114 specimens from Liebers et al. 2004); arg I, L. argentatus from Iceland (15 out of 100 specimens from Liebers et al. 2004); hyp K, from Kulusuk, Greenland; hyp E and arg E are specimens from other sites in Europe; and hyp A are from Canada (Liebers et al. 2004).
The two clusters I and II are connected through C1, described for L. cachinnans (Liebers et al. 2004). Cluster I is characterized by L. argentatus haplotypes and cluster II shows a mixture of haplotypes from both the species. Out of the 26 haplotypes in clusters I and II, 4 are shared between L. hyperboreus and L. argentatus, A20 in cluster I and MA1, A12, and Hy2 in cluster II. These four are found in both species in Iceland, but only A12 is found in other European L. hyperboreus. The wide occurrence of the A12 in both species in Europe (Liebers et al. 2004) is reflected in the IGs. IG values from samples from northern Europe are in the range of 0.53–0.75, being highest in the sample from northern Norway; in Germany, it is 0.33 and the IG values are equal to zero for the samples from France and The Netherlands, where A12 is absent. In Iceland, the IG values are high for samples a1 and a3w, but both samples had a geographical overlap with L. hyperboreus sampling sites. A high IG value is also observed for the h1w from the first time period, where two individuals share two haplotypes from cluster I (A20).
Cluster I is formed by 54 individuals, 80% from Iceland and western Europe (France, Netherlands and Faroe Islands), representing the Larus argentatus argenteus group (Barth 1968). Six individuals representing the L. a. argentatus group are from more southern range of the groups' distribution, Germany, Finland and Estonia. The proportion of cluster I among Icelandic L. argentatus increases significantly from time period 1 (7%) to periods 2 and 3 (42 and 32%; p<0.0039, Fisher's exact test). Cluster II holds 14 haplotypes among 249 individuals, 138 L. hyperboreus and 111 L. argentatus. Most individuals in this cluster originate from northerly areas, where the distribution of the two species is adjacent or overlapping. The majority are represented by haplotype A12 (59%) or the derivative H2 (29%). The H2 group is represented by almost exclusively Icelandic samples (99%), 60 L. hyperboreus from west Iceland and 8 L. argentatus only from Iceland (a1 and a3w).
Excluding h1e owing to the low sample size, the overall nucleotide and haplotype diversity was higher in L. argentatus groups compared with L. hyperboreus. Interestingly, L. hyperboreus from Kulusuk are more divergent than all hyperboreus groups from Iceland. Haplotype diversity is generally stable over time among the species. Nucleotide diversity changes through time; in the first time period, it is lowest in L. argentatus, whereas in L. hyperboreus, it is largest (table 2). The high diversity in L. hyperboreus during the first period results from the L. argentatus haplotype A20, present in two individuals.
(b) Microsatellite analysis
The effective numbers of alleles (ne) are similar for both species, ranging from 1.8 to 2.3 in L. argentatus and from 1.6 to 2.3 in hyperboreus. The total number of alleles per locus ranges from 3 to 11, with all populations combined. Most alleles are shared by both species, and only one allele (170 bp), at locus HG16 in L. hyperboreus, is in a frequency higher than 5% but absent in the other species. The average expected heterozygosities were also similar across samples, 0.38–0.57 in L. argentatus and L. hyperboreus. The most variable loci are HG25 (He=0.70) and HG18 (He=0.66), while the least variable locus is HG16 (He=0.31). The differences in allele composition between populations are mainly presented by frequency changes and not many low-frequency alleles were detected. In Iceland, a3w displays the largest variation (He=0.569 and ne=2.3). Populations from period 1, a1, h1e and h1w, have the lowest genetic variability (He=0.448, 0.377 and 0.510, ne=1.8, 1.6 and 2.0). FIS across loci is significant and positive in three populations after Bonferroni correction, in a1, h1w and h2. The largest deviation is seen for one locus (HG25), in a1, h1w and h3. Out of 100 tests of linkage disequilibrium, 7 give permutation values larger than or equal to the observed p values in the range of 0.8–3.1%. This weak signal of linkage disequilibrium can be explained by the random chance for unlinked markers.
(c) Population genetic differentiation
Hierarchical analysis of the genetic variance shows a significant differentiation among and within population. Differentiation among species is in all cases non-significant, representing an intraspecific variance, although it is close to significance when based on the mtDNA (table 3). The extent of differentiation among populations within species is larger when the variance in genetic differentiation among haplotypes is taken into account (increases from 3.4% to 7.4%). Similar results are obtained for microsatellites, the variance among populations within species increases to 40.21% in calculations based on RST.
Table 3.
Hierarchical analysis of molecular variance (AMOVA) of mtDNA and microsatellite variation among L. argentatus and L. hyperboreus in Iceland. (The p-values are based on 1000 permutations of the data, which resulted in equal or larger value than the observed statistic.)
| source of variation | % variation | fixation index | p | |
|---|---|---|---|---|
| mtDNA (FST) | among species | 13.9 | 0.139 | 0.088 |
| populations within species | 3.9 | 0.045 | 0.024 | |
| within populations | 82.3 | 0.177 | 0.000 | |
| mtDNA Φ(ST) | among species | 24.7 | 0.247 | 0.053 |
| populations within species | 6.7 | 0.089 | 0.004 | |
| within populations | 68.6 | 0.313 | 0.000 | |
| Microsat (FST) | among species | 0 | −0.009 | 0.523 |
| populations within species | 10.6 | 0.105 | 0.000 | |
| within populations | 25.5 | 0.282 | 0.000 |
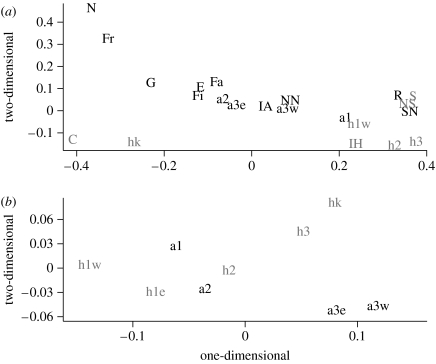
Pairwise comparisons among samples generally distinguish the L. argentatus and L. hyperboreus in Iceland, whether based on mtDNA haplotypes (figure 3a), the microsatellites (figure 3b) or both using the Bayesian clustering (figure 4). The population comparisons of the mtDNA haplotypes, including the samples from Liebers et al. (2004), locate the Icelandic samples between the L. argentatus and the L. hyperboreus and from rest of Europe. The figure shows also a close relationship between some geographically adjacent populations of the two species, e.g. in Iceland (a1 and h1w), Russia (R and NS) and Norway (SN and S), and the distinct grouping of the L. hyperboreus samples from Greenland and Canada.
Figure 3.
Multidimensional scaling plot based on pairwise FSTs. (a) Comparisons based on mtDNA data from this study (denoted with small letters, see table 1) and from Liebers et al. (2004) in capital letters. Larus argentatus (black): N, Netherlands; Fr, France; G, Germany; Fa, Faroe Islands; E, Estonia; Fi, Finland; SN, southern Norway; NN, northern Norway; R, Russia; IA, Iceland. Larus hyperboreus (grey): S, Svalbard; NS, Novaja Semlija; IH, Iceland; C, Canada Baffinland. (b) Comparisons based on the microsatellite data.
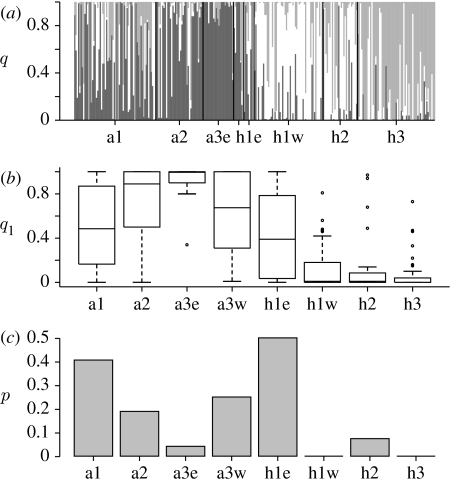
Figure 4.
Admixture analysis by BAPS (Marttinen et al. 2006) for samples from Iceland. (a) Larus shadings: dark grey, white and grey vertical lines represent the admixture coefficients for each individual, i.e. the proportion of the genome of each individual (qi) which is traced to clusters 1–3, respectively. (b) Boxplot of the admixture coefficient q1 for each sample. (c) Proportion (p) of individuals per sample, which were assigned to another cluster (Bayesian p-value<0.05).
The composition of mtDNA frequencies within species in the Icelandic samples are not significantly different from each other. The L. hyperboreus samples differ from all but one L. argentatus population, a3w, and h1w is not significantly different from a1. Interestingly, h1w is significantly different from h2 and h3 (p<0.05). Pairwise differences based on the ΦST method are larger than the conventional FST, and this is observed both in comparisons of conspecifics at different periods (h3 and h2 versus h1w and h1e; a3e and a2 versus a1) and also between different species at the same locations (a3w versus h3 and h2).
Pairwise estimates of genetic differentiation between all nine groups, using F-statistics, are more often significant for the microsatellite data (32 out of 36) than for the cytochrome b (18 out of 36). Non-significant pairwise comparisons based on microsatellites are between a1, a2 and h1e, and between h1w and h1e (table 1).
Based on both the mtDNA and the microsatellite data, the Bayesian clustering method (BAPS) gives an optimal partition of four clusters with a probability of 99.9%. The log likelihoods for different numbers of clusters k from 2 to 4 were −3673.6, −3656.5 and −3655.3, respectively. Larger numbers of initial groups (5–8) resulted in four clusters. All L. argentatus groups cluster together in cluster 1, plus the h1e group. All L. hyperboreus from the west of Iceland are in clusters 2 (h1w, h2) and 3 (h3), and they are equally distant from cluster 1. L. hyperboreus from Kulusuk (cluster 4) clusters further apart from the previously mentioned clusters. The same partitioning was obtained among the Icelandic samples when the Kulusuk sample was omitted from the analysis (figure 4), showing a clear evidence of admixture. A considerable proportion of the individuals have admixture coefficients (q1) which are intermediate (figure 4a,b). This is especially evident for a1, a3w and h1e. The frequency of individuals per sample, which have a higher proportion of their genome originating from the other species, is largest at the first time period in eastern Iceland where it decreases with time for the L. argentatus samples, and an asymmetry is observed in the introgression (figure 4c). In addition, the two L. hyperboreus individuals sampled in eastern Iceland during the second and third periods (see table 1) have the largest genetic similarities to L. argentatus, the second non-significant (q1=0.94 and 0.73). Three out of four L. argentatus sampled in western Iceland and Hjörleifshöfði during period 1 have a significantly higher proportion of the L. hyperboreus genome. One-quarter of the L. argentatus individuals (36 out of 143) have a significantly larger proportion of the genetic markers that are characteristic for the L. hyperboreus samples. However, only 4% (6 out of 150) of L. hyperboreus individuals have a significantly larger proportion of the genetic markers that are characteristic for the L. argentatus samples. Four of these are L. hyperboreus from h1e. The association is significant (p=2.2×10−8, Fisher's exact test). When considering the samples from Greenland, 2 out of 19 are assigned to cluster 3 (H3).
4. Discussion
(a) Phylogeography
A previous study of mtDNA phylogeography by Liebers et al. (2004) shows that L. argentatus and L. hyperboreus do not correspond strictly to distinct monophyletic mtDNA haplogroups within the phylogeny of large white-headed gulls. Whereas L. hyperboreus appear biphyletically in an American and an European clade, L. argentatus presents a large ramificated tree consisting of two distinct clusters (I and II) within the European clade, separated by haplotypes that are shared by other gull species, namely L. cachinnans, L. fuscus and L. marinus (Liebers et al. 2004). European L. hyperboreus is mainly found in one of these clusters (II), sharing haplotypes with L. argentatus. The observed polyphyly of the two species could be due to the incomplete sorting of mtDNA lineages from a polymorphic ancestral gene pool but, in some cases, is more likely as a result of hybridization, as discussed below.
The mtDNA of L. hyperboreus is characterized by low diversity, as it has been observed for several Arctic species (Hewitt 2004). The low diversity and shape of the phylogenetic network may reflect fluctuations in population size which may have been more severe for the mtDNA than the nuclear loci due to smaller effective population size. The Icelandic L. hyperboreus population belongs to the European L. hyperboreus clade, but is clearly distinct from the populations sampled elsewhere. Only 1 L. hyperboreus out of 117 sampled in Iceland and 20 from Svalbard and Novaja Zemlija shared an American haplotype (H1) with L. hyperboreus from Kulusuk, Greenland and Baffinland, northeastern Canada. This points to little contact between the Icelandic and the East Greenland population, reflecting the division between the Palaearctic and the Nearctic L. hyperboreus. This division is also supported by the microsatellite data, although the most recent samples are most similar, possibly reflecting sex-biased migration.
Overall, L. argentatus harbour larger number of haplotypes and higher diversity in mtDNA in Iceland compared with L. hyperboreus, in agreement with the previous genetic studies of L. argentatus in Europe (Crochet et al. 2002, 2003; Liebers et al. 2004). The Icelandic population differs from the rest of Europe, showing greatest similarity to the samples from northern Europe. Interestingly, comparisons with other samples from Europe, obtained by Liebers et al. (2004), show that the earliest L. argentatus samples in Iceland were actually more similar to populations of L. hyperboreus from northern regions (Norway, Russia) than to other L. argentatus populations. Samples of L. argentatus from time periods 2 and 3 are more similar to populations with more southern and western distribution, sharing haplotypes with individuals from, for example, France, Faroe Islands and Germany. A closer look at the composition of the haplotypes in Icelandic L. argentatus shows that the proportion of the clusters I and II changes over time. Cluster II is dominant during the first time period (92%), whereas haplotypes in cluster I increase in frequency with time, up to 40% during 1985–1986. This change is also reflected in the nucleotide and haplotype diversity. This could be explained by a high frequency of hybrid individuals or indication of introgression during the first period among L. argentatus, or alternatively by later waves of immigration of L. argentatus to Iceland. This may also explain a bimodal mismatch distribution observed for L. argentatus, which results from comparisons between sequences in clusters I and II (data not shown). Bimodality is generally interpreted as a sign of stable evolutionary population size; here it may have resulted from an admixture of L. argentatus populations, or alternatively hybridization over a long period, where cluster II has spread from L. hyperboreus to L. argentatus by introgression.
(b) Hybridization
Genetic variation in large white-headed gulls appears to be shaped by both the retention of ancestral polymorphism and introgression. Liebers et al. (2004) suggested that the haplotypes, here presented in cluster II, were ancestral in L. argentatus and that cluster I had resulted from hybridization with descendants from the Aralo-Caspian refugium. The argument was that cluster II harboured more variation in L. argentatus and that it had been supported by nuclear AFLP (de Knijff et al. 2001). This is also supported by the recent study by Gay et al. (2007) on introgression between L. cachinnans (a descendant from the Aralo-Caspian refugium) and L. argentatus.
A characteristic result of the study on the mtDNA in European L. hyperboreus is a lack of haplotypes that are not shared by L. argentatus. Most of the L. hyperboreus haplotypes are found in cluster II and only three of them have not been found in L. argentatus. These three are singletons. The most common haplotype (A12) is shared by L. argentatus in northern and northwestern Europe, and is the only haplotype shared between L. hyperboreus and L. argentatus outside Iceland. The widespread occurrence of A12 results in high IG values and reflects possibly past introgression or incomplete lineage sorting. Sharing of other haplotypes among the species is, however, only found in Iceland, including the common haplotype H2. As L. argentatus is a recent settler in Iceland, this suggests recent introgression rather than shared ancestry. Three instances are observed where widely distributed L. argentatus haplotypes (MA1 and A20) are found only in L. hyperboreus in Iceland, all in samples from the first time period (1964–1973), which had high IG values. One of these haplotypes is from cluster I (A20), and it caused the high nucleotide diversity seen in L. hyperboreus during the first time period. If shared ancestry would be the case, these haplotypes would most likely have been found in other L. hyperboreus elsewhere. Less sharing of the haplotypes in cluster I, among the two species, may indicate that the introgression of mtDNA from L. hyperboreus to L. argentatus has been more common than that from L. argentatus to L. hyperboreus, or that the L. argentatus with haplotypes found in cluster I have arrived on Iceland more recently. As concluded by Liebers et al. (2004), a study including more nuclear markers will be needed to solve the question of which direction the cytonuclear replacement occurred. The genetic difference between the European and the American L. hyperboreus remains to be explained; it might reflect a parallel morphological evolution or ancient introgression from L. argentatus.
The lack of significant differences between the species in the hierarchical analysis points to a high intraspecific variability, agreeing with gene flow between the species and the retention of ancestral polymorphism. This also suggests that a larger number of nuclear markers may be needed to distinguish between the two species. It is also possible that there are only few genes that are responsible for the species differences. The similarity of sympatric groups of the two species was, however, clearly seen in all pairwise comparisons based on microsatellites, implying gene flow. Bayesian clustering analysis (BAPS) based on mtDNA and microsatellite results gave similar findings as the FST's analysis, where the L. hyperboreus group (h1e) sharing the L. argentatus types, clustered among the L. argentatus. Supporting these signs of hybridization, admixture analyses revealed a number of L. argentatus individuals that were more likely to be characterized with L. hyperboreus genotypes. The admixture analysis indicated that 27% of individuals showing L. argentatus plumage pattern should be classified as L. hyperboreus based on genetic markers and only 4% of the L. hyperboreus as L. argentatus, indicating that introgression occurs more frequently from L. hyperboreus to L. argentatus. In agreement with these findings is the fact that the L. hyperboreus that were breeding in a colony in southeast Iceland, which also harboured a large number of L. argentatus, were clustered with all L. argentatus groups. Similarly, most of the L. argentatus that were assigned to L. hyperboreus clusters were sampled during the first time period in southeast Iceland. The observed directional introgression may possibly reflect the different numbers of L. argentatus and L. hyperboreus studied from this area, where hybridization appears to have been common. It seems that the hybridization during this early period has not had a lasting impact on the genetic composition in eastern Iceland. Today, the main hybridization appears in western Iceland, where L. argentatus, although in low numbers, are highly admixed. A monitoring of the populations in western Iceland may provide further insights into the dynamics of this secondary contact zone.
One caveat about the results presented in this study is that the species classification is originally based on morphology. Although the classification is well supported by the Bayesian analysis, some individuals may be wrongly identified due to introgression of genes behind the morphological trait. A further analysis of these individuals may show that the extent of hybridization can be even more common than that revealed in this study. Such admixed individuals may cause admixture in the pool of the reference group. To solve this problem, it would be good to include just ‘pure’ populations for reference for the putative hybrid individuals. However, such populations may be hard to find, considering the population structure, recent origin of the species, incomplete lineage sorting and even widespread hybridization.
Pairwise comparisons based on microsatellites were in general similar to comparisons based on mtDNA, although comparisons between populations within species based on the microsatellites are more often significant. The difference may result from a larger statistical power obtained when studying the microsatellites, as they are based on more markers and on 93 more individuals than the mtDNA analyses. Interestingly, the extent of the differentiation varies in some cases based on the markers studied, for example, the a2 and h3k samples are more differentiated from the L. hyperboreus in Iceland for the mtDNA than the microsatellites (figure 3). Similarly, a larger genetic differentiation for the mtDNA was observed in the study by Gay et al. (2007) on L. argentatus and L. cachinnans, where a complete lineage sorting was observed for the mtDNA, despite shared polymorphism in microsatellites. Such differences may result from stronger drift on mtDNA due to lower effective population size, behavioural effects and/or to different post-zygotic isolation in male and female hybrids (e.g. Orr 1997; Crochet et al. 2003; Gay et al. 2007). The observation of larger differences for the mtDNA is in accordance to the prediction of Haldane's rule (Haldane 1922); the genetic markers transmitted by the heterogametic sex should show more differentiation. A previous study of the L. hyperboreus and L. argentatus in Iceland (Ingólfsson 1970) did not detect any clear evidence of assortative mating.
The result of this study concurs with numerous reports of introgression in areas of sympatry among various species of plants (e.g. Palmé et al. 2004) and animals of recent origin (e.g. overview on studies on birds in McCarthy 2006). The hierofalcons provide one recent example where variation reflects both hybridization and incomplete lineage sorting (Nittinger et al. 2007). In such studies of closely related species, it has become evident that both phylogeographical patterns as well as genetic information from the different species are needed in order to disentangle the impact of incomplete sorting of lineages, and current and past hybridizations among taxa. A more common haplotype sharing in areas of sympatry points to hybridization. If a biphyly is due to shared ancestry, there should be no correspondence to geographical distribution. A biphyly has been detected in a previous study of gulls by Liebers et al. (2004) where introgression was implied along with shared ancestry. As suggested by Crochet et al. (2002, 2003), sharing of the most divergent mitochondrial lineages clearly results from introgression. Where more than one haplotype is found in a species and the less frequent haplotype is identical to the common haplotype in another species, the geographical distribution of haplotypes is indicative of interspecific horizontal transfer. Such events of a specific haplotype sharing between a recent colonizer (here L. argentatus) and a settled species (here L. hyperboreus) was seen in the North American L. marinus (a recent colonizer), which acquired the L. smithsonianus (the settled one) haplotypes, presumably through hybridization (Crochet et al. 2003).
5. Concluding remarks
Earlier claims of hybridization between L. hyperboreus and L. argentatus in Iceland (Ingólfsson 1970), based on morphology, were questioned by Snell (1991a,b) who argued that the observed intrapopulation variability in L. argentatus resulted from a founder effect. Snell suggested that the claimed hybrids in Iceland were light-winged argentatus originating from Scandinavia and that no hybridization between the two species in Iceland occurs. Along with all the previously mentioned cases of hybridization in gulls, genetic research strongly suggests that gene flow among large white-headed gulls is in fact extensive in many parts of the world (Crochet et al. 2002, 2003; Liebers et al. 2004; Gay et al. 2007). Thus, the haplotype and allele sharing in Icelandic L. hyperboreus and L. argentatus observed in this study should not be surprising. The genetic sharing in Iceland follows a geographical pattern and is most obvious in an area where both species were common, strongly indicating that hybridization occurred after the arrival of L. argentatus on Iceland.
Acknowledgments
We thank Guðmundur A. Guðmundsson, Gunnar Þór Hallgrímsson, Jóhann Brandsson and Páll Leifsson who provided samples or assisted in the field, and Guðmundur Guðmundsson at The Icelandic Institute of Natural History for assistance with museum samples. We also want to thank the editors and three anonymous reviewers for their useful comments. The study was supported by the Icelandic Research Council and the University of Iceland Science Fund.
Footnotes
One contribution of 16 to a Theme Issue ‘Hybridization in animals: extent, processes and evolutionary impact’.
References
- Bandelt H.J, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barth E.K. The circumpolar systematics of Larus argentatus and Larus fuscus with special reference to the Norwegian populations. Nytt Mag. Zool. 1968;15:1–50. [Google Scholar]
- Barton N.H, Hewitt G.M. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 1985;16:113–148. doi:10.1146/annurev.es.16.110185.000553 [Google Scholar]
- Belahbib N, Pemonge M.-H, Ouassou A, Sbay H, Kremer A, Petit R.J. Frequent cytoplasmic exchange between oak species that are not closely related: Quercus suber and Q. ilex in Morocco. Mol. Ecol. 2001;10:2003–2012. doi: 10.1046/j.0962-1083.2001.01330.x. doi:10.1046/j.0962-1083.2001.01330.x [DOI] [PubMed] [Google Scholar]
- Belkhir, K., Borsa, P., Chikhi, L., Raufaste, N. & Bonhomme, F. 2004 Genetix version 4.05, Logiciel sous Windows pour la génétique des populations. Laboratoire génome, populations, interactions. CNRS UMR 5000, Université de Montpellier II, Montpellier, France.
- Bell D.A. Genetic differentiation, geographic variation and hybridization in gulls of the Larus glaucescens–occidentalis complex. Condor. 1996;98:527–546. doi:10.2307/1369566 [Google Scholar]
- Coulson J.C. The population dynamics of culling herring gulls and lesser black-backed gulls. In: Perrins C.M, Lebreton J.D, Hirons G.J.M, editors. Bird population studies, relevance to conservation and management. Oxford University Press; Oxford, UK: 1991. pp. 479–497. [Google Scholar]
- Cramps S, Simmons K.E.L. Oxford University Press; Oxford, UK: 1983. Handbook of the birds of Europe, the Middle East and North Africa. The birds of the Western Palearctic. [Google Scholar]
- Crochet P.A. Genetic structure of avian populations—allozymes revisited. Mol. Ecol. 2000;9:1463–1469. doi: 10.1046/j.1365-294x.2000.01026.x. doi:10.1046/j.1365-294x.2000.01026.x [DOI] [PubMed] [Google Scholar]
- Crochet P.A, Lebreton J.D, Bonhomme F. Systematics of large white-headed gulls: patterns of mitochondrial DNA variation in western European taxa. Auk. 2002;119:603–620. doi:10.1642/0004-8038(2002)119[0603:SOLWHG]2.0.CO;2 [Google Scholar]
- Crochet P.A, Chen J.Z, Pons J.M, Lebreton J.D, Hebert P.D, Bonhomme F. Genetic differentiation at nuclear and mitochondrial loci among large white-headed gulls: sex-biased interspecific gene flow? Evolution. 2003;57:2865–2878. doi: 10.1111/j.0014-3820.2003.tb01527.x. doi:10.1111/j.0014-3820.2003.tb01527.x [DOI] [PubMed] [Google Scholar]
- de Knijff P, Denkers F, van Swelm N.D, Kuiper M. Genetic affinities within the Larus argentatus assemblage revealed by AFLP genotyping. J. Mol. Evol. 2001;52:85–93. doi: 10.1007/s002390010137. doi:10.1007/s002390010137 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse P.E, Quattro J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial-DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver 3.0: an integrated software package for population genetic data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Gay L, Neubauer G, Zagalska-Neubauer M, Debain C, Pons J.-M, David P, Crochet P.-A. Molecular and morphological patterns of introgression between two large white-headed gull species in a zone of recent secondary contact. Mol. Ecol. 2007;16:215–227. doi: 10.1111/j.1365-294X.2007.03363.x. doi:10.1111/j.1365-294X.2007.03363.x [DOI] [PubMed] [Google Scholar]
- Grant P.R, Grant B.R. Hybridization of bird species. Science. 1992;256:193–197. doi: 10.1126/science.256.5054.193. doi:10.1126/science.256.5054.193 [DOI] [PubMed] [Google Scholar]
- Guðmundsson F. Íslenskir fuglar XI. Hvítmáfur (Larus hyperboreus) Náttúrufræðingurinn. 1955;25:24–35. [Google Scholar]
- Haldane J.B.S. Sex-ratio and unisexual sterility in hybrid animals. J. Genet. 1922;12:101–109. [Google Scholar]
- Hewitt G.M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996;58:247–276. [Google Scholar]
- Hewitt G.M. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. doi:10.1038/35016000 [DOI] [PubMed] [Google Scholar]
- Hewitt G.M. Genetic consequences of climatic oscillations in the Quaternary. Phil. Trans. R. Soc. B. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. doi:10.1098/rstb.2003.1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingólfsson A. Hybridization of glaucous gulls Larus hyperboreus and herring gulls Larus argentatus in Iceland. Ibis. 1970;112:340–362. doi: 10.1098/rstb.2008.0042. doi:10.1111/j.1474-919X.1970.tb00112.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingólfsson, A. 1982 Máfar, kjóar og skúmar. In Fuglar (ed. A. Garðarsson). Reykjavík, Iceland: Landvernd.
- Ingólfsson A. Hybridization of glaucous and herring gulls in Iceland. Stud. Avian Biol. 1987;10:131–140. [Google Scholar]
- Ingólfsson A. The variably plumaged gulls of Iceland. Auk. 1993;110:409–410. [Google Scholar]
- Liebers D, de Knijff P, Helbig A.J. The herring gull complex is not a ring species. Proc. R. Soc. B. 2004;271:893–901. doi: 10.1098/rspb.2004.2679. doi:10.1098/rspb.2004.2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttinen P, Corander J, Törönen P, Holm L. Bayesian search of functionally divergent protein subgroups and their function specific residues. Bioinformatics. 2006;22:2466. doi: 10.1093/bioinformatics/btl411. doi:10.1093/bioinformatics/btl411 [DOI] [PubMed] [Google Scholar]
- McCarthy E.M. Oxford University Press; Oxford, UK: 2006. Handbook of avian hybrids of the world. [Google Scholar]
- Newton I. Academic Press; San Diego, CA: 2003. The speciation and biogeography of birds. [Google Scholar]
- Nittinger F, Gamauf A, Pinsker W, Wink M, Haring E. Phylogeography and population structure of the saker falcon (Falco cherrug) and the influence of hybridization: mitochondrial and microsatellite data. Mol. Ecol. 2007;16:1497–1517. doi: 10.1111/j.1365-294X.2007.03245.x. doi:10.1111/j.1365-294X.2007.03245.x [DOI] [PubMed] [Google Scholar]
- Olsen K.M, Larsson H. A & C Black Publishers; London, UK: 2004. Gulls of Europe, Asia and North America. [Google Scholar]
- Orr H.A. Haldane's rule. Annu. Rev. Ecol. Syst. 1997;28:195–218. doi:10.1146/annurev.ecolsys.28.1.195 [Google Scholar]
- Palmé A.E, Su Q, Pálsson S, Lascoux M. Extensive sharing of chloroplast haplotypes among European birches: Betula pendula, B. pubescens and B. nana. Mol. Ecol. 2004;13:167–178. doi: 10.1046/j.1365-294x.2003.02034.x. doi:10.1046/j.1365-294X.2003.02034.x [DOI] [PubMed] [Google Scholar]
- Panov E.N, Monzikov D.G. Intergradation between the herring gull Larus argentatus and the southern herring gull Larus cachinnans in European Russia. Zool. Zh. 1999;78:334–348. [Google Scholar]
- Randler C. Avian hybridization, mixed pairing and female choice. Anim. Behav. 2002;63:103–119. doi:10.1006/anbe.2001.1884 [Google Scholar]
- Rogers A.R, Harpending H. Population-growth makes waves in the distribution of pairwise genetic-differences. Mol. Biol. Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell R.R. Interspecific allozyme differentiation among North-Atlantic white-headed Larid gulls. Auk. 1991a;108:319–328. [Google Scholar]
- Snell R.R. Variably plumaged Icelandic herring-gulls reflect founders not hybrids. Auk. 1991b;108:329–341. [Google Scholar]
- Sokal R.R, Rohlf F.J. W. H. Freeman and Company; New York, NY: 1995. Biometry. [Google Scholar]
- Spear L.B. Hybridization of glaucous and herring-gulls at the Mackenzie Delta, Canada. Auk. 1987;104:123–125. [Google Scholar]
- Venables W, Ripley B. Springer; New York, NY: 1994. Modern applied statisticswith S-plus. [Google Scholar]
- Weir B.S. Sinauer Associates; Sunderland, MA: 1990. Genetic data analysis. [Google Scholar]
- Weir B.S, Cockerham C.C. Estimating F-statistics for the analysis of population-structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. doi:10.2307/2408641 [DOI] [PubMed] [Google Scholar]
- Yesou P. The sympatric breeding of Larus fuscus, L. cachinnans and L. argentatus in Western France. Ibis. 1991;133:256–263. doi:10.1111/j.1474-919X.1991.tb04567.x [Google Scholar]