Abstract
Neutral genetic markers are useful for identifying species hybrids in natural populations, especially when used in conjunction with statistical methods like the one implemented in the software NewHybrids. Here, a short description of the extension of NewHybrids to dominant markers is given. Subsequently, an extensive series of simulations of amplified fragment length polymorphism (AFLP) data is performed to evaluate the prospects for hybrid identification with (possibly non-diagnostic) dominant markers. Distinguishing between F1's and F2's is shown to be difficult, possibly requiring upwards of 100 AFLP markers to be done accurately. Discriminating between pure-bred and non-pure (hybrid) individuals, however, is shown to be much easier, requiring perhaps as few as 10 dominant markers, even from relatively weakly diverged species.
Keywords: amplified fragment length polymorphism, mixture model, genetic admixture, receiver operating characteristic curve
1. Introduction
In the last decade, advances in biotechnology have brought a dramatic increase in the number of genetic markers available for the study of animal populations. This abundance of genetic information has allowed population geneticists and molecular ecologists to move beyond genetic inference at the population level (e.g. estimating the divergence between populations) and instead use genetic data to learn about particular individuals (Pearse & Crandall 2004) within animal assemblages (for example, identifying individuals in a population that appear to be migrants from another population or those which carry admixed ancestry). Statistical methods for such individual-based, multilocus genetic analysis have been available from early applications in Drosophila (Makela & Richardson 1977) and later for the estimation of mixture proportions in mixed stock fisheries (Fournier et al. 1984); however, since the late 1990s, there has been a proliferation of related methods. The new methods typically employ minor elaborations upon the standard mixed fishery model to allow the model to address a new characteristic feature of the data; for example, admixed individuals (Pritchard et al. 2000), species hybrids (Anderson & Thompson 2002), variable migration rates between demes (Wilson & Rannala 2003), etc.
By far the most commonly used method of these is the one implemented in the program Structure (Pritchard et al. 2000; Falush et al. 2003). The model underlying Structure was one of the first individual-based methods to allow individuals to have admixed ancestry, with various proportions of each sampled individual's genome originating from a different subpopulation. Structure accomplishes this by using a flexible model in which the origin of each gene copy within an individual is independently chosen from a vector of probabilities Q, which itself is drawn from a Dirichlet distribution. This model can be applied to a wide variety of circumstances, and it is particularly appropriate for identifying individuals with ancestry from two or more subpopulations or species, especially if the admixture has been ongoing for a long time. However, the fact that the origin of gene copies within an individual is conditionally independent given Q makes it impossible to distinguish between some classes of recent species hybrids. For example, because both F1 and F2 hybrids between two species, A and B, will have, on average, 50% of their genomes originating from each species, they are in the eyes of Structure essentially indistinguishable. For this reason, Anderson & Thompson (2002) introduced a model, implemented in the software NewHybrids, that computes the posterior probability that members in the sample belong to user-specified categories such as F1, F2 and backcross. NewHybrids takes explicit account of the fact that in some categories, the origin of the two gene copies at a locus is not independent. (For example, if the first gene copy at a locus in an F1 category is from species A, then the second gene copy must be from species B.)
Anderson & Thompson (2002) described the use of NewHybrids for inference from codominant genetic markers. Shortly after the article was published, however, the program was modified to allow the analysis with dominant markers such as amplified fragment length polymorphisms—AFLPs (Mueller & Wolfenbarger 1999)—as well. The user manual (Anderson 2003) describes how to format a dataset that includes dominant markers, but does not offer a clear explanation of how, mathematically, NewHybrids accommodates dominant data. The original article mentioned that the statistical framework would allow straightforward extension to dominant data, but did not provide many details. Now, having been available for some 5 years, NewHybrids has been applied to several AFLP datasets, including those from conifer species (Emelianov et al. 2004), Atlantic eels (Albert et al. 2006), cultivated and wild chicory (Kiær et al. 2007) and Swainson's thrush (Ruegg 2008). The purpose of this short paper is first to provide a formal, yet succinct, explanation of the extension of the NewHybrids model to dominant markers and second to summarize simulations that offer guidelines regarding the power available from AFLPs for identifying hybrids.
2. NewHybrids model
An extensive description of the model and Markov chain Monte Carlo (MCMC) methodology used in NewHybrids appears in Anderson & Thompson (2002). Here, a brief overview is given, enough to explain the variables in the model and their relationships to one another. NewHybrids is applicable to the situation where there are only two diploid species that seem to be hybridizing, and a sample of M individuals, possibly representing pure individuals as well as hybrid individuals, is taken and genotyped at L loci. The allelic types of the two gene copies at the ℓth locus in the ith member of the sample are denoted by and . We use Y to denote the data at all L loci from all M individuals. The loci are assumed to be independently segregating and to exhibit no Hardy–Weinberg disequilibrium or linkage disequilibrium when considered as part of a separate species A or species B ‘gene pool’. At the ℓth locus, we find Kℓ alleles in our sample and denote the (usually unknown) frequencies of these alleles in the pure species gene pools of A and B by and . The goal of inference is typically to use the genotypes of the individuals to determine which are pure representatives of the species and which have hybrid ancestry. Sometimes, pure representatives of each species may have been sampled separately from the mixture containing hybrids and can yield prior information about θA,ℓ and θB,ℓ; however, this is not absolutely necessary—with enough data, NewHybrids is able to infer the presence of two species and their hybrids, even in the absence of training data taken from the two species in isolation.
Under the NewHybrids model, individuals belong to one of n different ‘hybrid categories’ that are characterized by the proportion of loci within an individual that are expected to carry 0, 1 or 2 gene copies derived from species A. For example, an F1 individual is expected to have 100% of its loci containing exactly one gene copy from species A, while the product of a mating between an and a B individual—a first-generation backcrossed individual that we will refer to as —is expected to have 50% of its loci containing zero gene copies from A and 50% of its loci containing one copy from species A and, of course, no loci with both gene copies originating from species A. The default configuration of NewHybrids considers the n=6 categories that represent all the possible products of two generations of random mating between two species, i.e. A, B, F1, F2, and . The sample of individuals is assumed to be drawn from a population that contains a mixture of individuals from the n categories in unknown proportions . To write down the probability model that relates the allele frequencies, θ, and the mixing proportions, π, to the observed genotypes in the sample, it is helpful to introduce some latent variables. These are pieces of information that we would like to know, but cannot observe directly. By including them in the model, we can make inference about them given the data that we actually can observe. First, Zi for every individual, i=1, …, M, in the sample tells us which hybrid category individual i belongs to and second, and for every individual i=1, …, M and every locus ℓ=1, …, L gives the species of origin of the first and second gene copies, respectively, at the ℓth locus in the ith individual.
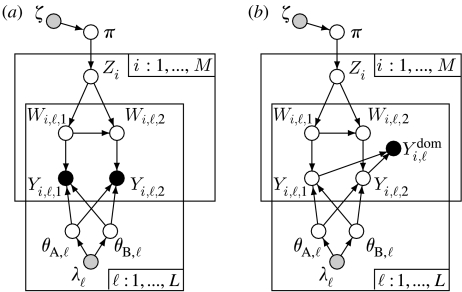
All of these variables are related together in the NewHybrids model as shown in the directed graph of figure 1a. This graph is a simple diagram in which variables in the model appear as nodes (circles) and the relationship between variables is depicted by arrows connecting the nodes. The directed graph expresses how the joint probability of all the variables in the model may be written as the product of simpler conditional densities, and it provides a visual representation that will help make it clear how the NewHybrids model is extended to accommodate dominant markers. See Jordan (2004) for a lucid introduction to graphical models in statistics. For the simplest interpretation of such a graph, you may regard it as a sort of flow chart that helps to keep track of the model that NewHybrids assumes for the data, as follows: first π gives the fraction of individuals in each hybrid category in the mixture from which the sample was taken. The value of π is unknown and not observed directly, so the node associated with π in the graph is unshaded. Moving down the graph, we see there is an arrow from π to Zi, which tells us that each Zi is a random draw from the mixing proportions π. This makes sense—the hybrid category of individuals in the sample depends on the frequency of that category in the population. Note that Zi is located upon a box (called a ‘plate’ in graphical model parlance) having ‘i=1, …, M’ given in the corner. This signifies that there are M such Zi variables, one for each of the M members of the sample, and they are conditionally independent given π. Continuing down in the graph, we see that arrows point from Zi to Wi,ℓ,1 and Wi,ℓ,2, and there is an arrow extending from Wi,ℓ,1 to Wi,ℓ,2. These relationships capture the fact that the origin of the two gene copies in individual i at locus ℓ are random variables that depend on the hybrid category to which individual i belongs. The arrow from Wi,ℓ,1 to Wi,ℓ,2 occurs because in NewHybrids (unlike in Structure), the origin of the two gene copies at a locus are not independent. For example, if individual i is an F1 (i.e. Zi=F1) and gene copy 1 at locus ℓ is from species A (Wi,ℓ,1=A), then we know that gene copy 2 must be from species B. Note that these nodes reside both in the plate with ‘i=1, …, M’ given in the corner as well as within the plate with ‘ℓ=1, …, L’ in the corner. This signifies that for each individual i, the ℓ pairs of variables, (Wi,ℓ,1,Wi,ℓ,2), one for each locus, are conditionally independent (across loci) given the value of Zi. Finally we arrive at the two nodes for Yi,ℓ,1 and Yi,ℓ,2. These variables, the allelic types at locus ℓ in individual i, are observed data when we have codominant genetic markers. Accordingly the nodes are shaded black. The arrangement of the nodes in this central portion of the graph indicates that the allelic types at a locus are random variables that depend immediately upon which gene pool the gene copy came from (the W's) and the frequencies of different alleles in that gene pool (the θ's). These relationships capture the assumption of the NewHybrids model that, given that a gene copy is from species A, say, the allelic type is drawn from the vector of allele frequencies in the species A gene pool, independently for each gene copy.
Figure 1.
Directed graphs describing the NewHybrids model. Black-shaded nodes represent observed data, grey-shaded nodes represent parameters of prior distributions and unshaded nodes represent unobserved variables. The boxes are ‘plates’ which denote multiple, conditionally independent replicates (indexed by the subscript ℓ over loci and i over individuals in the sample) of the enclosed nodes. (a) The model for codominant data. Note that the observed data are the allelic types Yi,ℓ,1 and Yi,ℓ,2 of the two gene copies at each locus in each individual (hence those nodes are shaded black). (b) The model for dominant data: each dominant locus is modelled as a diallelic locus with a recessive allele and a dominant one. These allele types are no longer observed, so they are latent variables. At each dominant locus, the observed datum is the presence or absence of a band, .
The foregoing described all the variables in the NewHybrids likelihood model. Conducting Bayesian inference also requires that prior distributions be placed upon π and the θA,ℓ's and θB,ℓ's. These priors are represented by ζ and the λℓ's, which are parameters of Dirichlet distributions. The values chosen for these prior-distribution parameters are assumptions of the model, thus their nodes in the graph are shaded grey. The goals of inference with NewHybrids can also be seen graphically in figure 1a—inference can be made for any unshaded node in the graph by computing the conditional distribution of that variable given the observed data. This conditional distribution, called the posterior distribution, cannot be computed exactly, in general, but it is not difficult to sample from it, and then use those samples to approximate the distribution. Thus, inferring the hybrid class of individual i can be done by summarizing the posterior distribution of Zi, estimating the allele frequencies at locus ℓ can be done by summarizing the posterior distributions of θA,ℓ and θB,ℓ, and estimating the mixing proportions of different hybrid classes in the sampled population can be done by summarizing the posterior distribution of π. Details of the MCMC used to sample from the posterior distribution may be found in Anderson & Thompson (2002).
3. Extension to dominant markers
A dominant marker such as an AFLP is resolved by the presence or absence of a band of a certain length on a gel. A standard model for such presence/absence phenotypes assumes that each band is uniquely associated with a locus that has two alleles: the recessive r allele and the dominant d allele (Weir 1996). If an individual carries at least one copy of the d allele, then it will produce a band, and otherwise it will not. Hence the rr homozygote does not produce a band while heterozygous individuals and the dd homozygotes do produce a band. The model described above is adopted to allow NewHybrids to do inference from dominant markers. It makes certain assumptions that may not be met at all times. Most importantly, bands of a certain length may be homoplasic; that is, different sections of the genome might comigrate. In this case, there may be two or more loci responsible for what appears to be a single band. To reduce homoplasy, it seems prudent to avoid scoring the shortest bands from an AFLP gel (Vekemans et al. 2002)
Because an individual that is heterozygous or homozygous for the d allele at a locus will produce the same phenotype, it is not possible to directly observe the underlying genotype, so the allelic types carried by the individual must be treated as latent variables. This is captured in figure 1b that shows the graphical model for NewHybrids with dominant data. The observed variable at locus ℓ in individual i is now , which has two possible states—1 for ‘band present’ and 0 for ‘band absent’—that depend on Yi,ℓ,1 and Yi,ℓ,1 in a deterministic fashion following the standard model for dominant markers:
Since there are only two alleles, r and d, at each locus, we can designate the allele frequencies in the species A gene pool as , and analogously for species B. Using this, we can easily compute the conditional probability of Yi,ℓ,1 and Yi,ℓ,2 given , Wi,ℓ,1, Wi,ℓ,2, θA,ℓ and θB,ℓ. For instance, the probability that Yi,ℓ,1=r and Yi,ℓ,2=r is 1 if , and 0 if . Additionally, if , then the probability that the underlying genotype is homozygous (dd) or is one of the heterozygotes (rd or dr) is simply proportional to the expected frequencies of a dd homozygote or the heterozygotes given the species of origin of each gene copy and the frequency of the r and d alleles in those species. Thus we have
for all , when . And when :
for all and for such that j and k are not both equal to r. Of course, , always.
These relationships allow us to use MCMC to sample over the latent states of Yi,ℓ,1 and Yi,ℓ,2 with ease. Analogous to the way that updates for Zi with codominant loci are done by integrating out all possible states of Wi,ℓ,1 and Wi,ℓ,2, updates for Zi with dominant markers involve integrating out all possible states of Wi,ℓ,1, Wi,ℓ,2 and Yi,ℓ,1, Yi,ℓ,2. Since there are only two possible alleles assumed to be underlying every dominant marker, this involves a small tractable sum, i.e.
| (3.1) |
After the quantities in (3.1) are normalized to sum to 1 over all the hybrid categories, z, a new value of Zi can easily be drawn from the distribution. Once that is done, new values of Wi,ℓ,1 and Wi,ℓ,2, and then of Yi,ℓ,1 and Yi,ℓ,2 may be sampled from their full conditional distributions using probabilities that were computed and stored during the execution of the sums in (3.1). MCMC updates for the allele frequencies and for π proceed as described in Anderson & Thompson (2002). Inference for any latent variable in the model, including the allelic types, proceeds as before by summarizing its posterior distribution.
4. Simulation methods
To test NewHybrids' inference with dominant markers, we prepared and analysed a large number of simulated datasets under different conditions. Two species (which we will call A and B) were simulated from an allopatric population divergence model with no migration, using the coalescent framework implemented in Makesamples (Hudson 2002). The species were assumed to exist in populations of effective size N, and samples of 450 individuals from each species were simulated. Two different scenarios were investigated: a low-divergence (LD) scenario in which the species split 0.6N generations in the past and a high-divergence (HD) scenario with the species split occurring at 1.2N generations in the past. For each simulation, either H=100 or 1000 separate coalescent trees were simulated, each one corresponding to an independently segregating, non-recombining genomic region, and 100 mutations at unique nucleotide positions were simulated in the region. Exactly one of these mutations from each genomic segment was chosen to be a mutation underlying a dominant marker using the following scheme: first, each mutation was independently decided to have produced either a dominant band-producing allele (with probability 1/2) or a recessive allele (with probability 1/2), with the wild-type being the opposite allele; then, eight individuals from each species (16 individuals in total) were randomly selected and the mutations within them investigated in order along the genomic sequence until the first one was encountered at which at least 3 of the 16 individuals produced bands and at least 3 produced no bands. The first mutation fitting this criterion was declared the locus underlying a dominant marker associated with a band of unique length, and the other mutations in the genomic region were discarded. This emulates the use of a small ascertainment panel of individuals to discover polymorphic bands. Note that this method assumes no homoplasy between bands and also assumes that the markers are independently segregating and are not in linkage disequilibrium.
Simulations were performed with two different values of H, the number of ascertained, polymorphic, dominant markers. The ‘many-markers’ condition included H=1000 polymorphic markers, corresponding to a survey of AFLP variation using many different adapter pairs (see Mueller & Wolfenbarger 1999). The ‘few-markers’ scenario included H=100 polymorphic markers. The 450 individuals from each species were used to create samples for analysis with NewHybrids. First, 125 individuals from A and 125 individuals from B were included in the sample as known representatives of their species that were sampled separately from the remaining mixture of individuals, using NewHybrids' individual-specific z and s options. Then, included in the mixture were 40 A's, 30 B's, 15 F1's, 10 's, 3 's and 2 F2's. F1's were created by randomly mating 15 A–B pairs, each one creating a single offspring. F2's were created by randomly mating F1's, etc. Parents of hybrids in the dataset were unique—i.e. no two hybrids in the dataset shared parents or grandparents, and none of their parents or grandparents were also included as pure individuals in the dataset. The individuals in the sample were all of the same cohort, no individuals were parents or offspring of any others.
Each dataset simulated as above was analysed using the L ‘most informative’ loci with , (L=400 was also used in the H=1000 condition). This feature of the simulations was intended to mimic the ‘high grading’ of bands showing large differences between species. This approach might be taken if very many polymorphic bands were discovered, and it was not feasible or reasonable to score all of those bands on all the individuals in the dataset. Such an approach was used in Ruegg (2008).
Loci were ranked by informativeness using the Kullback–Leibler divergence applied to the frequency of band presence and absence among the 125 known representatives of each species. That is, if RA is the relative frequency of the recessive phenotype and DA the relative frequency of the band-producing phenotype among the 125 known representatives of species A, and RB and DB are the same for species B, then the Kullback–Leibler divergence of the distribution of phenotypes in species A relative to species B is
Loci were ranked according to the larger, at each locus, of and .
NewHybrids was run using Jeffrey's priors on the mixing proportions and the allele frequencies. During exploratory NewHybrids runs, visual inspection of the MCMC showed that convergence to a region of high posterior probability was almost immediate and the chain was mixing well, so 200 sweeps of burn-in were used, along with 800 sweeps of data collection. This is a much smaller number of sweeps than would typically be used for a single real dataset, but doing such short runs allows many replicate datasets to be analysed. For each set of conditions, 50 replicate datasets were simulated and analysed. The conditions included all factorial combinations of the two divergence scenarios, the two values of H and the five (for H=100) or six (for H=1000) values of L. Accordingly, 1100 datasets, in total, were simulated and analysed.
5. Simulation results
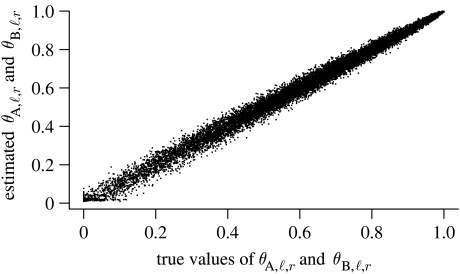
For our first check of the simulation results, we verify that the estimated frequency of the r allele at each locus is close to the true frequency of the r allele among the 450 simulated individuals in each population. Figure 2 shows that NewHybrids is capable of estimating the allele frequencies at dominant loci well. As one would expect, the frequency of allele r is estimated best when it is close to 1.0, because at those loci, most of the individuals are homozygous for the r allele and the allelic state of such individuals, at both gene copies, may be inferred without ambiguity. When a higher fraction of the sample is composed of band-producing individuals, there is more uncertainty in the estimation of θA,ℓ,r and θB,ℓ, but NewHybrids still performs quite well. Note that since the points in figure 2 are smoothly dispersed around the y=x line with no obvious outlying clusters, it suggests that 200 sweeps of burn-in were sufficient for the MCMC to have converged to the correct part of the parameter space.
Figure 2.
Posterior mean estimates (y-axis) versus true values (x-axis) of the frequency of the recessive r allele from either species gene pool (A or B) across all simulated datasets (both LD and HD conditions) with H=100 and L=100.There are 20 000 points in total, each one corresponding to an allele frequency at a single locus in species A or B.
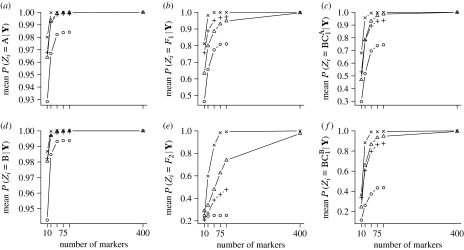
We next summarize how well the hybrid category of different individuals may be inferred. We do this in several ways. First, for each simulation condition (combination of divergence, H and L), we computed the average across the individuals in each hybrid category, and across all 50 simulated datasets, of the mean posterior probability of belonging to the correct hybrid category. In other words, over all individuals in the simulations from hybrid class HC, the average value of was recorded. If enough data were available so that individuals could be assigned to their correct hybrid category with no uncertainty, then each one would have a posterior probability of 1.0 of belonging to their true hybrid category, and the average value of the posterior probability over all members of that hybrid category would be 1.0 as well. Values less than 1.0 indicate uncertainty. Figure 3 shows that with the most informative L=400 loci from H=1000 ascertained markers, there is almost no uncertainty about hybrid category assignments reflected in the posterior probabilities for the six hybrid categories investigated. However, with fewer markers, there is some uncertainty. As expected, performance is better with higher divergence and also with a larger number, H, of ascertained polymorphic markers. Particularly striking are the posterior probabilities for individuals in the F2 category in the LD scenario with H=100. Even with 100 markers, the average posterior probability with which an F2 individual belongs in the F2 category is less than 0.25. Interestingly, the influence of divergence (LD and HD) and number of markers ascertained (H) varies for different hybrid categories. For all categories, the least accurate scenario is LD with H=100 and the most accurate scenario is HD with H=1000. However, the accuracy of the two remaining scenarios is reversed between F1's and F2's. F2's are better discriminated in the LD scenario with H=1000 than in the HD scenario with H=100, while F1's are better discriminated in the HD scenario with H=100 than in the LD scenario with H=1000.
Figure 3.
Mean posterior probability of hybrid category membership. (a–f) The mean over all simulations with different simulation scenarios denoted by the plot symbols as follows: circles, LD and H=100; triangles, LD and H=1000; pluses, HD and H=100; crosses, HD and H=1000. The x-axis of (a–f) shows L, the number of markers used. (a–f) Correspond to different hybrid categories as indicated on each plot. Note that the y-axis scale differs on each plot: (a) A, (b) F1, (c) , (d) B, (e) F2, (f) .
While mean posterior probabilities provide one summary of the data, it should be recognized that posterior probabilities of 1.0 are not necessarily required to accurately discriminate between hybrid categories—there may still be a clear separation of the distribution of posterior probability values for individuals in different categories. Since we know the true hybrid categories of the individuals from the simulations, we can investigate this by quantifying allocation rates between the different hybrid categories. To allocate individuals to hybrid categories, we used a maximum a posteriori rule: each individual was allocated to the hybrid category for which it had the highest posterior probability.
The resulting allocation rates are summarized for L=50 and 100 across all simulation conditions in table 1. The results show that distinguishing between the hybrid categories F1, F2, and can require a large number of very divergent markers. For example, even with L=100 loci used from H=100 markers ascertained, under the LD scenario, 59% of the F2's get allocated to the F1 category. In fact, accurate discrimination between F1's and F2's occurs only under the HD scenario with H=1000 markers. These findings are concordant with Anderson & Thompson's (2002) original conclusion with codominant markers that it is difficult to distinguish the different truly hybrid categories.
Table 1.
Allocation rates to various categories using maximum a posteriori assignment. (The rows correspond to the true hybrid category of individuals and the columns correspond to the inferred (maximum posterior probability) hybrid category. For example, the first row shows that under the LD scenario with L=50 and H=1000, 100% of the individuals in the simulation had the highest posterior probability of belonging to the category. Non-zero values are given in italic.)
| div. | H | G | L=50 | L=100 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||
| A | B | F1 | F2 | A | B | F1 | F2 | |||||||
| LD | 100 | A | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| LD | 100 | B | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| LD | 100 | F1 | 0.00 | 0.00 | 0.90 | 0.02 | 0.05 | 0.02 | 0.00 | 0.00 | 0.93 | 0.01 | 0.05 | 0.01 |
| LD | 100 | F2 | 0.02 | 0.00 | 0.56 | 0.23 | 0.13 | 0.06 | 0.00 | 0.00 | 0.59 | 0.24 | 0.13 | 0.04 |
| LD | 100 | 0.09 | 0.00 | 0.11 | 0.01 | 0.78 | 0.00 | 0.08 | 0.00 | 0.09 | 0.01 | 0.82 | 0.00 | |
| LD | 100 | 0.00 | 0.21 | 0.35 | 0.07 | 0.00 | 0.37 | 0.00 | 0.19 | 0.31 | 0.06 | 0.00 | 0.45 | |
| LD | 1000 | A | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| LD | 1000 | B | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| LD | 1000 | F1 | 0.00 | 0.00 | 0.94 | 0.03 | 0.02 | 0.01 | 0.00 | 0.00 | 0.98 | 0.01 | 0.00 | 0.00 |
| LD | 1000 | F2 | 0.00 | 0.00 | 0.36 | 0.48 | 0.11 | 0.05 | 0.00 | 0.00 | 0.19 | 0.77 | 0.03 | 0.01 |
| LD | 1000 | 0.02 | 0.00 | 0.02 | 0.01 | 0.96 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.99 | 0.00 | |
| LD | 1000 | 0.00 | 0.01 | 0.06 | 0.05 | 0.00 | 0.88 | 0.00 | 0.00 | 0.01 | 0.03 | 0.00 | 0.95 | |
| HD | 100 | A | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| HD | 100 | B | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| HD | 100 | F1 | 0.00 | 0.00 | 0.99 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.99 | 0.00 | 0.01 | 0.00 |
| HD | 100 | F2 | 0.00 | 0.00 | 0.34 | 0.39 | 0.16 | 0.11 | 0.00 | 0.00 | 0.30 | 0.46 | 0.12 | 0.12 |
| HD | 100 | 0.03 | 0.00 | 0.04 | 0.00 | 0.93 | 0.00 | 0.01 | 0.00 | 0.04 | 0.00 | 0.95 | 0.00 | |
| HD | 100 | 0.00 | 0.02 | 0.09 | 0.03 | 0.00 | 0.85 | 0.00 | 0.01 | 0.08 | 0.02 | 0.00 | 0.89 | |
| HD | 1000 | A | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| HD | 1000 | B | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| HD | 1000 | F1 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| HD | 1000 | F2 | 0.00 | 0.00 | 0.05 | 0.90 | 0.04 | 0.01 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| HD | 1000 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | |
| HD | 1000 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.99 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.99 | |
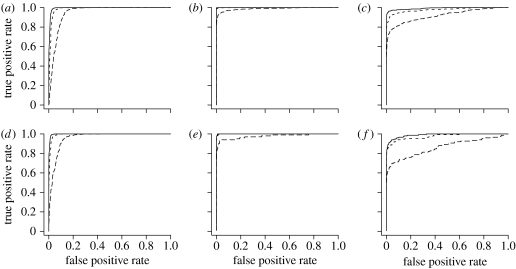
Finally, we use the simulation output to quantify how well pure individuals (categories A and B) can be distinguished from non-pure ones (F1, F2, and ). The power available for doing this can be summarized graphically using the receiver operating characteristic (ROC) curves (Metz 1978) of figure 4. ROC curves measure the power of a statistical classification rule to correctly allocate an observation into one of the two different categories. Our classification rule is based upon the posterior probability that an individual belongs to a pure species category, . If this quantity is below a certain threshold C, we allocate the individual to the non-pure class, and if it is above C then we allocate it to the pure class. The value of C determines the false positive rate (fraction of individuals assigned to the wrong class) and the true positive rate (fraction of individuals assigned to the correct class). The ROC curve plots the pairs of false positive rate and false negative rate as the threshold C varies between 0 and 1. In figure 4, the ROC curves make it clear that distinguishing pure from non-pure is a much easier problem than distinguishing between different non-pure hybrid categories. For example, even with only L=10 dominant markers, chosen from H=100 in the LD scenario, it is possible to classify individuals on the basis of P(Pure) so that 90% of the F1's, 81% of the F2's, 62% of the 's and 59% of the 's are correctly classified as non-pure, while fewer than 1% of the pure individuals would be incorrectly classified as non-pure. In our example, the and categories are the most difficult to distinguish from the pure species. Later-generation backcrosses (i.e. or ) would be even more difficult to distinguish from pure individuals. Boecklen & Howard (1997) provided calculations for determining how many diagnostic dominant markers would be required for discriminating such backcross categories. It should be kept in mind that it would require far more non-diagnostic markers.
Figure 4.
ROC curves based on P(Pure) (see text) computed from simulation output for the LD scenario with H=100 and L equal to 10 (long-dashed line), 25 (short dashed line) and 50 (solid line). A separate ROC curve has been plotted for each true hybrid category, i.e. (a) shows the rate at which pure A individuals are allocated to the pure category as a function of the rate at which individuals from the non-pure categories are incorrectly assigned to the pure category; (b) shows the rate that F1's are allocated to the non-pure category as a function of the rate at which pure A and B individuals are incorrectly assigned to the non-pure category, etc. (a) A, (b) F1, (c) , (d) B, (e) F2, (f) .
6. Discussion
This paper provides the first formal description of the model implemented in NewHybrids for dominant-marker data. The extension of the original model is shown in the graphical model of figure 1 to be quite simple, structurally. The model underlying the phenotype expression is the standard one, which is also adopted in the recent extension of Structure to dominant data (Falush et al. 2007). However, the Structure model can also be applied to loci with multiple alleles, only one of them being recessive. This allows it to model null alleles in, for example, microsatellite markers. The NewHybrids model includes only two alleles (r and d), and is thus specialized for strictly dominant markers, which carries certain computational advantages, like allowing blocked Gibbs updates for the Zi's. There is no restriction in NewHybrids, however, on mixing different marker types in a dataset. It is perfectly acceptable, for example, to type individuals at both microsatellites and AFLP markers and include all the markers in the dataset. The NewHybrids model allows both data types to be analysed simultaneously.
In the course of analysing various real and simulated AFLP datasets, I have seen that NewHybrids performs best with dominant data when some known representatives of the pure species A and B are included (with NewHybrids' z option) in the dataset. Without these learning samples, the MCMC sampler may require long burn-in times. This can often be observed in NewHybrids runs initialized with random starting values: the chain may remain for many sweeps in a configuration in which all individuals are inferred to be F1's or F2's and the estimated allele frequencies bear no relation to the true allele frequencies in the species. For example, in the simulations performed in this paper, having large learning samples meant that the MCMC sampler converged to the proper part of the parameter space within the first several sweeps. By contrast, when I repeated the simulations without notifying NewHybrids that 125 A individuals and 125 B individuals were known representatives of their species, it took much longer for the Markov chain to converge to the correct region. In general, it took longer for datasets with more loci to converge. For example, at the high divergence level with H=100, NewHybrids had converged within 1000 sweeps on all 50 replicate datasets with L=10. With L=50, NewHybrids had failed to converge within 1000 sweeps in 8 of 50 datasets; for L=100, 22 of 50 had not yet converged. Results were similar for the LD scenario. This indicates that care should be taken when analysing datasets with dominant markers (especially with many dominant markers). Every effort should be made to obtain learning samples. If they are not available, then multiple NewHybrids runs should be performed with very long burn-in periods (of the order of 75 000 sweeps), and the individual runs should be compared to each other to ensure concordance.
As noted by Anderson & Thompson (2002), distinguishing between the non-pure hybrid categories with genetic data is difficult and requires many markers. The same is true with dominant markers. The simulations performed here show that, depending on the degree of divergence of the species, even 100 AFLPs may not allow clear separation of F1's and F2's. On the other hand, distinguishing between pure and non-pure individuals can be done with far fewer markers. Even under a ‘LD’ scenario, simulations showed that as few as the 10 most informative markers from 100 ascertained polymorphic bands provide ample power to discriminate F1's and F2's from pure individuals.
In the simulations, we mimicked the process of ascertainment of AFLPs using a small set of pure individuals from both the species. We further investigated the effect of high grading a small number of the most informative markers. Under the divergence in allopatry model used in the simulations, this is an acceptable course of action, as borne out by the simulations: figure 3 shows that some power is lost, but, if a large number of bands (such as 1000) are available, the 50–100 most informative bands may capture most of the resolution in the data. If the species diverged recently in sympatry, then there is some possibility that the markers that are most divergent in the two species may be involved in their reproductive isolation. This would violate the NewHybrids assumption of neutrality of the markers.
In the present paper, we have characterized the statistical power to discriminate between different categories using the ROC curve (figure 4). In our simulations, since we know the true category that each individual belongs to, it is possible to compute the ROC curve for a range of thresholds, C. This will not typically be the case in a real-life situation with a real dataset. In fact, this is a ubiquitous problem when analysing data from closely related species or subpopulations with Bayesian clustering methods—the methods yield posterior probabilities that may be difficult to interpret. For example, NewHybrids might tell you that individual i has posterior probability of 85% of belonging to the F2 category. This quantity is not, however, an estimate of the actual probability that you would be correct if you were to declare the individual an F2. This latter probability cannot be directly obtained from the typical output of a program such as Structure or NewHybrids, so various ad hoc approaches involving comparison to simulated data have been used for assessing the accuracy of allocation based on the posterior probability, or for testing hypotheses such as the hypothesis of hybridization versus non-introgressive mixing of pure individuals (Nielsen et al. 2003).
7. Future prospects for the model-based analysis of species hybrids
I would like to conclude with some perspectives on the current limitations of NewHybrids and the interesting opportunities and challenges available to those interested in extending the capabilities of programs such as NewHybrids and Structure for the analysis of animal hybridization.
It must be noted that the model underlying NewHybrids relies on the assumption that the genetic markers in the dataset are independently segregating. This assumption is likely to be violated when many markers are used; with 400 AFLP markers, for example, many loci will occur together on common linkage groups. If markers are physically linked, then their segregation is not independent and, because NewHybrids treats their segregations as independent, this will cause NewHybrids to underestimate the uncertainty in its estimates of the Zi variables. There is currently no general method implemented in NewHybrids for dealing with this. One way to mitigate the problem would be to use a small number of the most informative loci (i.e. those with the highest degree of interspecies differentiation); however, this necessarily discards some information. If there is a physical or genetic map for the markers, then the linkage between markers could be modelled. Version 2 of the program Structure (Falush et al. 2003) provides a way to model the dependence between markers using a hidden Markov chain model. No such method is currently available with NewHybrids.
As noted in §6, the interpretation of posterior probabilities from any model-based genetic clustering method is not straightforward, especially if the data include individuals from closely related subpopulations or species, and if there are various plausible biological models (e.g. admixture versus mixture) for the data. Model assessment and model checking via ‘posterior predictive checking’ are now all but expected as elements of a complete Bayesian analysis of any statistical problem (see Gelman et al. 2004, ch. 6). However, none of the Bayesian clustering methods for multilocus genetic data (such as NewHybrids or Structure) provide such model checking as an option. Instead of leaving it to the software users to design simulations to assess and interpret the output of Structure and NewHybrids, there seems to be an opportunity to expand the programs themselves to allow for simulation-based model assessment. It may be possible to use simulated values from the posterior predictive distribution to provide an estimate of ROC curves and/or related quantities, though this is a difficult problem, especially in the absence of training samples, because a gold standard is not available for computing the ROC curve (Zhou et al. 2005). This is currently an area of development in NewHybrids.
Regions where two species meet and form stable ‘hybrid zones’ have received considerable attention from zoologists as ‘windows on the evolutionary process’ (Harrison 1990) and ‘natural laboratories’ (Hewitt 1988) for the study of evolution. One of the primary means of mathematically analysing such hybrid zones has been to investigate clines of allele frequencies along transects through them. A great deal of theory has been developed regarding the rate at which alleles from one species decline in frequency and give way to the alleles of another species as one travels through a hybrid zone (Barton 1979; Barton & Gale 1993). Much of this theory, however, has been developed for alleles that are alternately fixed in the different species, which complicates the estimation of clines with non-fixed allelic differences. Hierarchical Bayesian models, such as those in NewHybrids and Structure, that use variables to denote the species of origin of a particular gene copy (Wi,ℓ,1 and Wi,ℓ,2 in figure 1) would be well suited to addressing the estimation of clines with non-fixed allelic differences: instead of estimating clines by focusing on particular allelic states, the clines can be estimated using the origins (species A or B) of different gene copies. Since it is possible to sample from the full posterior distribution of these gene origins (the Wi,ℓ,1's and Wi,ℓ,2's), it would be possible to propagate that uncertainty into the estimate of the genetic cline between species.
Finally, both NewHybrids and Structure make the assumption that the sampled genes behave neutrally and are not influenced by selection. This is an undesirable assumption when using large datasets with many markers and coverage throughout the genome. The non-neutrality of genetic transmission to hybrids was documented in Drosophila by Dobzhansky (1936) and recently has been observed in other animal and plant species (e.g. Jiang et al. 2000; Martinsen et al. 2001; Teeter et al. 2008). It would be interesting (albeit challenging) to try to modify a program such as NewHybrids or Structure to allow for the fact that genes introgress selectively between species. Accounting for such selective effects should allow more accurate inference of hybrid category or degree or admixture, and also would provide for a model-based assessment of the degree of non-neutrality of introgression among loci, which could potentially illuminate important evolutionary processes.
Acknowledgements
I would like to thank the users of NewHybrids that I have had the pleasure of working with. Their suggestions and insights have led to many enhancements in the program. Special thanks to Kristen Ruegg for extensive feedback on NewHybrids' facilities for AFLP markers. I am particularly thankful to Klaus Schwenk, Bruno Streit and Nora Brede for organizing the very interesting symposium upon which this issue of Philosophical Transactions B is based.
Footnotes
One contribution of 16 to a Theme Issue ‘Hybridization in animals: extent, processes and evolutionary impact’.
References
- Albert V, Jónsson B, Bernatchez L. Natural hybrids in Atlantic eels (Anguilla anguilla, A. rostrata): evidence for successful reproduction and flucuating abundance in space and time. Mol. Ecol. 2006;15:1903–1916. doi: 10.1111/j.1365-294X.2006.02917.x. [DOI] [PubMed] [Google Scholar]
- Anderson, E. C. 2003 User's guide to the program NewHybrids, version 1.1 beta. Technical report, 7 April, 2003.
- Anderson E.C, Thompson E.A. A model-based method for identifying species hybrids using multilocus genetic data. Genetics. 2002;160:1217–1229. doi: 10.1093/genetics/160.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N.H. The dynamics of hybrid zones. Heredity. 1979;43:341–359. doi: 10.1038/hdy.1979.87. [DOI] [Google Scholar]
- Barton N.H, Gale K.S. Genetic analysis of hybrid zones. In: Harrison R.G, editor. Hybrid zones and the evolutionary process. Oxford University Press; Oxford, UK: 1993. pp. 13–45. [Google Scholar]
- Boecklen W.J, Howard D.J. Genetic analysis of hybrid zones: numbers of markers and power of resolution. Ecology. 1997;78:2611–2616. doi: 10.2307/2265918. [DOI] [Google Scholar]
- Dobzhansky T. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics. 1936;21:113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelianov I, Marec F, Mallet J. Genomic evidence for divergence with gene flow in host races of the larch budmoth. Proc. R. Soc. B. 2004;271:97–105. doi: 10.1098/rspb.2003.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard J.K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard J.K. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier D.A, Beacham T.D, Riddell B.E, Busack C.A. Estimating stock composition in mixed stock fisheries using morphometric, meristic, and electrophoretic characteristics. Can. J. Fish. Aquat. Sci. 1984;41:400–408. [Google Scholar]
- Gelman A, Carlin J.B, Stern H.S, Rubin D.B. 2nd edn. Chapman and Hall; New York, NY: 2004. Bayesian data analysis. [Google Scholar]
- Harrison R.G. Hybrid zones: windows on the evolutionary process. Oxf. Surv. Evol. Biol. 1990;7:69–128. [Google Scholar]
- Hewitt G.M. Hybrid zones—natural laboratories for evolutionary studies. Trends Ecol. Evol. 1988;3:158–167. doi: 10.1016/0169-5347(88)90033-X. [DOI] [PubMed] [Google Scholar]
- Hudson R.R. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics (Oxf.) 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Jiang C.-X, Chee P.W, Draye X, Morrell P.L, Smith C.W, Paterson A.H. Multilocus interactions restrict gene introgression in interspecific populations of polyploid Gossypium (cotton) Evolution. 2000;54:798–814. doi: 10.1111/j.0014-3820.2000.tb00081.x. [DOI] [PubMed] [Google Scholar]
- Jordan M.I. Graphical models. Stat. Sci. 2004;19:140–155. doi: 10.1214/088342304000000026. [DOI] [Google Scholar]
- Kiær L.P, Philipp M, Jørgensen R.B, Hauser T.P. Genealogy, morphology and fitness of spontaneous hybrids between wild and cultivated chicory (Cichorium intybus) Heredity. 2007;99:112–120. doi: 10.1038/sj.hdy.6800973. [DOI] [PubMed] [Google Scholar]
- Makela M.E, Richardson R.H. The detection of sympatric sibling species using genetic correlation analysis. I. Two loci, two gamodemes. Genetics. 1977;86:665–678. doi: 10.1093/genetics/86.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinsen G.D, Whitham T.G, Turek R.J, Keim P. Hybrid populations selectively filter gene introgression between species. Evolution. 2001;55:1325–1335. doi: 10.1554/0014-3820(2001)055[1325:HPSFGI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Metz C. Basic principles of ROC analysis. Semin. Nucl. Med. 1978;8:283–298. doi: 10.1016/S0001-2998(78)80014-2. [DOI] [PubMed] [Google Scholar]
- Mueller U.G, Wolfenbarger L.L. AFLP genotyping and fingerprinting. Trends Ecol. Evol. 1999;14:389–394. doi: 10.1016/S0169-5347(99)01659-6. [DOI] [PubMed] [Google Scholar]
- Nielsen E.E, Hansen M.M, Ruzzante D.E, Meldrup D, Gronkjaer P. Evidence of a hybrid-zone in Atlantic cod (Gadus morhua) in the Baltic and the Danish Belt Sea revealed by individual admixture analysis. Mol. Ecol. 2003;12:1497–1508. doi: 10.1046/j.1365-294X.2003.01819.x. [DOI] [PubMed] [Google Scholar]
- Pearse D.E, Crandall K.A. Beyond FST: analysis of population genetic data for conservation. Conserv. Genet. 2004;5:585–602. doi: 10.1007/s10592-003-1863-4. [DOI] [Google Scholar]
- Pritchard J.K, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegg K.C. Genetic, morphological, and ecological characterization of a hybrid zone that spans a migratory divide. Evolution. 2008;62:452–456. doi: 10.1111/j.1558-5646.2007.00263.x. [DOI] [PubMed] [Google Scholar]
- Teeter K.C, et al. Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Res. 2008;18:67–76. doi: 10.1101/gr.6757907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekemans X, Beauwens T, Lemaire M, Roldán-Ruiz I. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and a relationship between degree of homoplasy and fragment size. Mol. Ecol. 2002;11:139–151. doi: 10.1046/j.0962-1083.2001.01415.x. [DOI] [PubMed] [Google Scholar]
- Weir B.S. Sinauer Associates; Sunderland, MA: 1996. Genetic data analysis II. [Google Scholar]
- Wilson G.A, Rannala B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics. 2003;163:1177–1191. doi: 10.1093/genetics/163.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.-H, Castelluccio P, Zhou C. Nonparametric estimation of ROC curves in the absence of a gold standard. Biometrics. 2005;61:600–609. doi: 10.1111/j.1541-0420.2005.00324.x. [DOI] [PubMed] [Google Scholar]