Abstract
Interspecific hybridization may result in asexual hybrid lineages that reproduce via parthenogenesis. Contrary to true parthenogens, sperm-dependent asexuals (gynogens and hybridogens) are restricted to the range of bisexual species, generally the parental taxa, by their need for a sperm donor. It has been documented that asexual lineages may rarely use sperm from a non-parental species or even switch a host. The available literature reports do not allow distinguishing, between whether such host switches arise by the expansion of asexuals out of their parental's range (and into that of another's) or by the local extinction of a parental population followed by a host switch. The present study combines new and previously collected data on the distribution and history of gynogenetic spined loaches (Cobitis) of hybrid origin. We identified at least three clonal lineages that have independently switched their sperm dependency to different non-parental Cobitis species, and in cases incorporated their genomes. Our current knowledge of European Cobitis species and their hybrids suggests that this pattern most probably results from the expansion of gynogenetic lineages into new areas. Such expansion was independent of the original parental species. This suggests that sperm dependence is not as restrictive to geographical expansion when compared with true parthenogenesis as previously thought.
Keywords: hybridization, gynogenesis, polyploidy, asexual population, sperm dependence, biogeography
1. Introduction
Interspecific hybridization has long been considered a significant generator of diversity in plant evolution, but its role in the animal kingdom has only recently been appreciated (reviewed in Arnold 1997; Seehausen 2004). Normally, natural mating between two distinct animal species yields either fertile hybrids with normal meiosis or F1s that are not viable or sterile. However, in some instances, the crossing of two distinct species can result in asexual (unisexual) hybrids due to disrupted meiosis (reviewed in Dawley 1989; Vrijenhoek 1989). In asexual animals, two basic reproductive modes have been recognized that differ in whether or not they require sperm: (i) sperm-independent parthenogenesis, where sperm is not required for the development of clonal progeny and (ii) sperm-dependent parthenogenesis where sperm is required, either to (i) trigger cleavage in the absence of syngamy resulting in fully clonal progeny (gynogenesis) or (ii) fuse with the clonal maternal genome (to restore hybridity) without contributing genetically to the next generation (hybridogenesis).
Although hybrid asexuality is assumed to pose serious constraints on the evolutionary longevity of clones (for reviews see, Maynard Smith 1978; Vrijenhoek 1998; Simon et al. 2003; Schlupp 2005), parthenogens may profit from some short-term advantages over sexual forms (Vandel 1928; Baker 1955; Kirkendall 1990). However, sperm-dependent parthenogenetic hybrids suffer one important constraint—they are reproductively dependent on sexual individuals and thus may neither be able to out-compete their sexual counterparts nor escape from their competition (Beukeboom & Vrijenhoek 1998). So far, the evolution of sperm-dependent parthenogenesis has mostly been studied from the point of view of the fine balance between ecological and reproductive parameters that allow for the coexistence of gynogens or hybridogens and their sexual progenitors (Baker 1965; Vrijenhoek 1979, 1989; Semlitsch et al. 1997; Doncaster et al. 2000; Alves et al. 2001; Engeler & Reyer 2001; Pound et al. 2002; Schley et al. 2004).
Modern molecular genetic tools have allowed the identification of many hybrid forms. This has provided an opportunity to assess critically whether sperm-dependent parthenogens need to coexist with their sexual parental species or whether they may switch to a new sperm-host species. To our knowledge, host switches have been documented in the asexual lineages of fishes (Cobitis: Janko et al. 2005a; Poecilia: Niemeitz et al. 2002, Schlupp et al. 2002; Poeciliopsis: Mateos & Vrijenhoek 2002, 2005; Squalius: Alves et al. 1997, Cunha et al. 2004, Sousa-Santos et al. 2006) and amphibians (Ambystoma: Hedges et al. 1992, Spolsky et al. 1992; Rana: Arano et al. 1995). However, it is still unclear how such host switches evolve in general.
Two alternative hypotheses may explain the evolution of host switches: (i) all-female asexuals may either escape from their parents by invading new areas and ‘infecting’ local populations of a new host species or (ii) they may locally out-compete their parents and switch to a new suitable sympatric host. Both hypotheses assume different population processes and would probably differentially affect the evolution of the (species–hybrid) complex as a whole. While the first scenario would blur the distinction between sperm-dependent and true parthenogens, the second scenario could significantly affect the biogeography of the complex resulting in extinctions of parental populations or species.
In the present study, we focused on the evolution of host switches among gynogenetic lineages of spined loaches from the Cobitis taenia complex sensu (Janko et al. 2005b). These are small bottom-dwelling fish that inhabit non-Mediterranean Europe. Six parapatric species are known to hybridize (Cobitis elongatoides, C. taenia, Cobitis tanaitica, Cobitis taurica, Cobitis strumicae and Cobitis melanoleuca), producing numerous virtually all-female asexual forms that coexist with the parental species (Bohlen & Ráb 2001; reviewed in Janko et al. 2007b). We have identified three primary patterns of hybridization leading to hybrid asexual lineages, i.e. C. elongatoides–taenia, C. elongatoides–tanaitica, C. elongatoides–taurica. These have spread over the whole range of the complex and in some cases have incorporated additional genomes through mating with parental or non-parental species, resulting in a variety of diploid and polyploid biotypes, including tri-genomic hybrids (Vasil'ev et al. 1989; Janko et al. 2005a). Gynogenesis has been proven for C. elongatoides–taenia and C. elongatoides–taenia–melanoleuca hybrid females (Saat 1991; Vasil'ev et al. 2003; Janko et al. 2007a) and is assumed for all hybrid lineages since (i) spined loach females may not spawn without males, (ii) all known hybrids are fixed heterozygotes and (iii) hybrids even co-occur with the sexual species far from the hybrid zone (Saat 1991; Janko et al. 2007a,b). Asexual lineages of these species have evolved as a result of hybridization and subsequent dispersal during the Holocene (Janko et al. 2005a). Two putatively ancient clonal lineages are also known stemming from the hybridization between C. elongatoides and C. tanaitica (the so-called hybrid lineage I) or C. taurica (hybrid lineage II). The former lineage has a remarkably large range that encompasses almost the entire known range of its original parents, including areas where both of its progenitors are absent, namely the Rhine R. and southern Ukraine. This is an abnormal pattern, because all hybrid lineages are generally restricted only to the range of the parental taxa coming from the primary hybridization (Janko et al. 2007b). Similarly, in southern Ukraine, hybrid lineage II occurs exclusively in the absence of its parental species (Janko et al. 2005a,b). In both cases, hybrid females switched their sperm dependence to the local host species C. taenia and there is a high frequency of individuals that carry the C. taenia genome in addition to the original parental genomes (Janko et al. 2007b).
To assess the dispersal potential and evolutionary plasticity of spined loach gynogens and, particularly, the way in which host switches originate, we have combined new information on the distribution of asexual loaches from previously unsampled areas of the lower Danube R. and the Aegean Sea drainage area with the results of Janko et al. (2005a, 2007b). Although we are still far from understanding the evolutionary plasticity of sperm-dependent parthenogens, our data suggest that sperm dependence may not necessarily be as restrictive as previously thought.
2. Material and methods
Four hundred and twenty-four individuals were collected from previously unsampled areas of the lower Danube R. watershed, western Black Sea tributaries and Struma R. (localities no. 1–25 in figure 1 and table 1). In addition, data were included for 385 individuals from previously sampled sites (localities no. 26–44 and part from no. 1 and no. 3–6 in figure 1 and table 1; Janko et al. 2005a, 2007b) to provide complete coverage of the entire range of biotypes of interest. Fin or muscle tissue was taken from each specimen. For each specimen, a tissue was available in both frozen form and preserved in ethanol for allozyme and mtDNA analyses, respectively. In order to unambiguously determine the taxonomical status of each sample (i.e. sexual species, interspecific hybrid of given genomic composition), we used three previously published diagnostic allozyme loci (GPI-A, sAAT and sMDH-A; Šlechtová et al. 2000; Janko et al. 2007b). Previously analysed specimens (Janko et al. 2007b) were used as electrophoretic mobility standards. For individuals from localities no. 14 and 15 in the Struma R., for which no frozen material was available, we used the first intron of the nuclear S7 gene (primers: S7RPEX1F, TGGCCTCTTCCTTGGCCGTC; S7RPEX2R, AACTCGTCTGGCTTTTCGCC; Chow & Hazama 1998) to identify the sample material: sequences were compared with the previously published haplotypes of C. elongatoides, C. taenia, C. tanaitica, C. taurica (GenBank accessions EF675687–EF675698), as described in Janko et al. (2007b). Hybridized genomes contain diagnostic heterozygous sites that allow the identification of hybridizing Cobitis genomes. Only C. tanaitica and C. taurica share common alleles (Janko et al. 2007b). All hybrid individuals were subjected to flow cytometry (FC) analysis to determine their ploidy according to the protocol of Lamatsch et al. (2000).
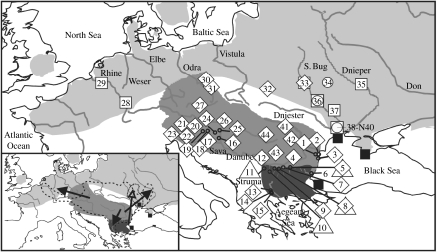
Figure 1.
Map of the known distribution area of Cobitis sexual species and hybrid biotypes of interest in Europe relevant to the recent studies of Janko et al. (2005a), Bohlen et al. (2006), Culling et al. (2006) and Janko et al. (2007b) and to new sampling. Numbers inside the symbols correspond with numbers of localities given in table 1. Map in the inset corresponds with the main map and the arrows represent the expected directions of the range expansions of the primary C. elongatoides–tanaitica (filled arrows) and C. elongatoides–taurica (open arrow) gynogenetic lineages. The southern population of C. taurica was recently described as a new species, Cobitis pontica (Vasil'eva & Vasil'ev 2006). Grey, C. elongatoides; light grey, C. taenia; dark grey, C. strumicae; black, C. taurica and C. pontica; diamonds, C. elongatoides–tanaitica; triangles, C. elongatoides–tanaitica–strumicae; squares, C. elongatoides–tanaitica–taenia; circles, C. elongatoides–taurica–taenia; no. 1–4 and 12, C. tanaitica; no. 6, C. elongatoides–strumicae; dashed line, distribution of hybrid lineage I; dotted line, distribution of hybrid lineage II.
Table 1.
Geographical origin and identification of samples. (Country codes: BG, Bulgaria; CZE, Czech Republic; D, Germany; HR, Croatia; HUN, Hungary; PL, Poland; RO, Romania; SLO, Slovenia; UA, Ukraine. Biotypes: EE, C. elongatoides; NN, C. tanaitica; SS, C. strumicae; TT, C. taenia; CC, C. taurica; EN, C. elongatoides–tanaitica; ENS, C. elongatoides–tanaitica–strumicae; ENT, C. elongatoides–tanaitica–taenia; ECT, C. elongatoides–taurica–taenia; ES, C. elongatoides–strumicae. N, number of sequenced hybrids.)
| noa | site | taxon (number of individuals) | taxon-mtDNA haplotype (N)b |
|---|---|---|---|
| 1 | Ghiol Lake, RO | NN(5); EN(17) | EN-E30(2) |
| 2 | Raselm Lake, RO | NN(1); EN(11) | EN-E29(1) |
| 3 | Sinoe Lake, RO | NN(67); EN(10) | EN-E29(2) |
| 4 | Oltenica, Danube R., RO | EE(1); NN(5); EN(20) | EN-E29(1), E30(3) |
| 5 | Jantra R., BG | SS(8); EN(7); ENS(17) | EN-E30(1); ENS-E29(3), E30(1), En(1) |
| 6 | Kamchya R., BG | EE(1); SS(3); ES(1) | ES-E13(1) |
| 7 | Kozarevska R., BG | EE(10); EN(1) | EN-E30(1) |
| 8 | Katunechka R., BG | EE(1); SS(7); EN(3); ENS(4) | EN-E29(1); ENS-E29(1), E30(2) |
| 9 | Shugavica R., BG | EN(11) | EN-E30(1) |
| 10 | Cibrica R., BG | SS(7); ENS(5) | ENS-E30(4) |
| 11 | Archar R., BG | SS(4); ENS(4) | ENS-E29(2) |
| 12 | Koshava, Danube R., BG | NN(1); EN(3) | EN-E29(1) |
| 13 | Struma R., BG | SS(19); EN(2); ENS(1) | EN-E29(3) |
| 14 | Struma R., BG | EN(7) | EN-E29(5) |
| 15 | Struma R., BG | SS(3); EN(8) | EN-E29(6) |
| 16 | Vuka R., HR | EE(26); EN(1) | EN-E30(1) |
| 17 | Kupa R., HR | EE(21); EN(3) | EN-E30(1) |
| 18 | Ilova R., HR | EE(7); EN(2) | EN-E30(2) |
| 19 | Glogovnica R., HR | EE(11); EN(16) | EN-E29(1), E30(1) |
| 20 | Drava R., SLO | EE(7); EN(15) | EN-E29(2) |
| 21 | Krka R., SLO | EE(22); EN(6) | EN-E29(1), E30(1) |
| 22 | Ledava R., SLO | EE(10); EN(18) | EN-E30(3) |
| 23 | N. Gyongyos R., HUN | EE(22); EN(3) | EN-E30(1) |
| 24 | Koppany R., HUN | EE(15); EN(10) | EN-E30(1) |
| 25 | Tisza R., HUN | EE(5); EN(20) | EN-E30 (1) |
| 26 | Szodrakosz Creek, HUN | EE(8); EN(23) | EN-E1(2), E29(1), E30(2) |
| 27 | Dyje R., CZE | EE(2); EN(42) | EN-E13(2), E31(1) |
| 28 | Rhine R., D | TT(3); ENT(11) | ENT-E30(2) |
| 29 | Rhine R., D | TT(3); ENT(5) | ENT-E30(2) |
| 30 | Polska Woda R., PL | EE(4); EN(6) | EN-E31(1) |
| 31 | Storawa R., PL | EE(4); NN(1); EN(3) | EN-E31(2) |
| 32 | Western Bug R., UA | EN(3); ENT(1) | ENT-E31(1) |
| 33 | Slutch R., UA | TT(8); EN(3); ENT(3) | EN-E32(2); ENT-E32(3) |
| 34 | Slutch R., UA | TT(19); EN(1); ECT(5) | ECT-E23(4) |
| 35 | Irpien R., UA | TT(8); EN(3); ENT(3) | ENT-E29(1), E30(1), E31(1) |
| 36 | Southern Bug R., UA | TT(5); ENT(3); ECT(12) | ENT-E29(1); ECT-E24(1) |
| 37 | Savranka R., Bug, UA | TT(9); ENT(1) | ENT-E29(1) |
| 38 | Southern Bug R., UA | CC(15); ECT(1) | ECT-E28(1) |
| 39 | Southern Bug R., UA | TT(3); ENT(2); ECT(3) | ENT-E29(2); ECT-E23(1), E25(1), E26(1) |
| 40 | Kodyma, Bug R., UA | TT(19); ECT(1) | ECT-E27(1) |
| 41 | Prut R., RO | EE(3); EN(10) | EN-E14(1), E29(2) |
| 42 | Birlat R, RO | EE(2); EN(10) | EN-E29(3) |
| 43 | Comana, Danube R., RO | EE(2); EN(11) | EN-E1(1), E29(1) |
| 44 | Mures R., RO | EE(9); EN(1) | EN-E1(1) |
In addition, 1088 bp of the cytochrome b gene were sequenced for a subset of hybrid individuals from previously unsampled areas following Janko et al. (2005a). Sequences were edited in Seqman II v. 5.05 (Dnastar software package) and compared with the previously identified haplotypes (GenBank accessions AY706159–AY706203, Janko et al. 2005a). Collapse v. 1.2 (Posada 2004, http://darwin.uvigo.es) was used to identify unique haplotypes among the newly obtained sequences and previously published ones. Phylogenetic relationships among haplotypes were determined using a haplotype network constructed under the statistical parsimony criterion implemented in TCS v. 1.21 (Clement et al. 2000).
There are ways to distinguish between the active dispersal and range expansion of hybrid linages and host switches that occur after the local extinction of the parental species. Under the first hypothesis, a signature of demographic growth in the lineage would be predicted. Using the 98 sequences obtained from the previous study (Janko et al. 2005a) as well as those from the current study, we employed three approaches to infer the demographic growth in lineages I and II. We used the DnaSP software (Rozas & Rozas 1999) to assess the distribution of the number of pairwise mutation differences between sequences (the mismatch distribution), which is typically unimodal in recently expanded populations, but irregular in shape in stationary populations (Rogers & Harpending 1992). Then, using the same software, we applied the Tajima's D test of neutrality, which is expected to result in significantly negative values under a selective sweep or population growth and/or bottleneck, whereas balancing selection or population structuring typically result in positive values (Ramos-Onsins & Rozas 2002). Finally, we computed maximum likelihood (ML)-based estimates of theta (θML) and exponential growth rate s(g) (as characterized by ) using the software Fluctuate v. 1.4 (Kuhner et al. 1998). This method takes into account the phylogenetic relationship among haplotypes, and samples the genealogical space by the Markov Chain Monte Carlo procedure. We used a transition/transversion ratio of 10 and Watterson's (1975) estimator of θ. The initial run included default settings and a starting value of 0.01 for the growth parameter g. Several subsequent runs were performed with the best estimates of g obtained from previous runs (Kuhner 2003). The final run was performed using five short chains of 8000 steps and two long chains of 200 000 steps with a sampling increment of 20. A lineage was considered to have undergone expansion when the log likelihood of the zero value of g was more than two units lower than the log likelihood for the best estimate of the g (Kuhner 2003).
3. Results
(a) Nuclear genotype analysis
The species-diagnostic allozyme markers (Janko et al. 2007b) identified three sexual species (i.e. C. elongatoides, C. tanaitica and C. strumicae) among the new samples (tables 1 and 2). In addition, two types of interspecific hybrids were also identified within the study area, which generally coexisted with either of the sexual species (table 1, figure 1): C. elongatoides–tanaitica and C. elongatoides–tanaitica–strumicae. All biotypes determined as hybrids were fixed heterozygotes for alleles of more than one distinct species and carried the species-specific alleles in all diagnostic loci (Janko et al. 2003, 2007b). Furthermore, the gene dose effect (Vrijenhoek 1975) or tri-allelic constitutions in some loci suggested that most of these individuals (167 out of 189) were polyploids. In two cases, we identified ‘pure’ hybrid populations, but due to low sample sizes it is probable that the local sexual species was unsampled. Two individuals from locality no. 7 carrying C. elongatoides-specific alleles at all loci were heterozygous at sMDH-A for C. tanaitica-like allele 060, suggesting that they may be introgressive hybrids. These individuals were discarded from subsequent analyses and shall be studied in detail in a further project aiming to describe the level of introgression between hybridizing loach species.
Table 2.
Species-specific allozyme alleles at three loci diagnostic among parental species.
| Cobitis species | locus | ||
|---|---|---|---|
| GPI-A | sAAT | sMDH-A | |
| C. elongatoides | 098a, 100ab, 113ab | 085ab, 087a, 100ab | 040ab, 070ab, 100ab |
| C. tanaitica | 087ab | 071ab, 078a, 100a | 060ab, 100ab |
| C. strumicae | 060b, 074ab, 080ab | 085ab, 095b | 064ab |
Alleles published by Janko et al. (2007b).
Alleles found in the present study.
Using sequences of the first intron of the S7 nuclear ribosomal gene, three individuals were identified as C. strumicae (GenBank accessions EU295700–EU295702), confirming its different morphology, while 15 individuals expressed heterozygous positions diagnostic for C. elongatoides, and either C. tanaitica or C. taurica (GenBank accessions EU295703–EU295717). Since the previous analyses did not suggest the presence of C. taurica genome in this area (Janko et al. 2005a; Bohlen et al. 2006; Janko et al. 2007b), we conclude that these hybrids are C. elongatoides–tanaitica (table 1).
Only 21 out of 139 hybrid specimens provided reliable results in FC. However, all individuals identified as polyploids by FC were also identified as polyploids by allozyme analysis, providing support for the results of the latter method. For reasons of simplicity and our interest in the distributions of the primary hybrid lineages, and in the presence/absence of specific genomes in hybrid biotypes, we did not assess the ploidy level of all hybrid specimens.
(b) Maternal ancestry and mtDNA diversity of hybrids
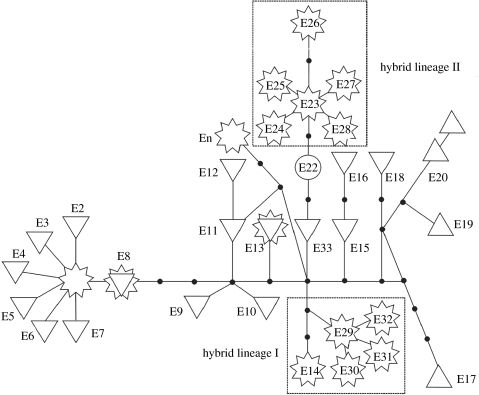
We sequenced 1088 bp of the cytochrome b gene for 59 hybrids. All individuals possessed C. elongatoides-type haplotypes (table 1, figure 2), namely E13, E29 and E30 (GenBank accessions AY706198, AY706202, AY706199), as described in Janko et al. (2005a), and one previously unidentified haplotype, here referred to as ‘En’. Haplotype En has been deposited under the GenBank accession no. EU262736. A detailed phylogeographic study of parental species (and their haplotypes) will be the subject of future publications. However, none of the parental haplotypes clustered in ‘hybrid clades’ I or II, supporting our previous conclusion (Janko et al. 2005a) that both clades represent assemblages of clonal lineages of monophyletic ancient origin.
Figure 2.
Unrooted tree constructed by a statistical parsimony corresponding to the sampled mtDNA haplotypes. Haplotype numbers correspond to Janko et al. (2005a). Dotted lines delimit the ‘hybrid’ clades I (haplotypes E14, E29–E31) and II (E23–E28). Black dots represent missing (unobserved) haplotypes. Down triangles, C. elongatoides; circles, C. taurica (sens. str. C. pontica); up triangles, C. tanaitica; stars, hybrids.
We did not observe any significant signal of population expansion in hybrid lineage I, for which Tajima's D was slightly negative, but non-significant (−0.74), the likelihood surface for the parameter g was flat and the ML value low, although the mismatch distribution was unimodal. By contrast, clear evidence of population expansion was observed in lineage II for all three tests: unimodal mismatch distributions and a highly significant negative Tajima's D value (−1.85) were indicative of population growth. In addition, the ML estimate of g was of the order of 104 and the log-likelihood values of the ML estimates of g were approximately five units from the log-likelihood value for no growth (0 versus −5.4), although the likelihood surface was relatively flat.
4. Discussion
(a) Host-sperm parasite switch
Although, in natural populations, the species that provides sperm for the sperm-dependent asexual is generally the parental species involved in the primary hybridization (Alves et al. 1997), several more recent genetic studies have identified some cases of gynogens and hybridogens relying on sperm from non-parental species. We went through the lists of recent studies on sperm-dependent parthenogenetic animals by Beukeboom & Vrijenhoek (1998) and Simon et al. (2003) and referred to the original literature to identify cases of asexuals that occur in the absence of parental species. Although we do not claim that this is an exhaustive search, we found several documented cases of host switches among asexual vertebrates (see §1 for the list of genera and original citations). Among the invertebrates, we found only one case (the sperm-dependent planarian Schmidtea polychroa) where the asexual form lived outside the range of the sexual species (see Pongratz et al. 2003), but due to the simultaneous production of eggs and sperm in this asexual hermaphrodite, we did not consider this to be a host switch.
Janko et al. (2005a) documented the ability of both ancient asexual lineages of Cobitis (i.e. hybrid lineages I and II) to use a non-parental species as a sperm donor, but current data suggest that even an unrelated species such as C. strumicae from a different Cobitis subgenus (Bohlen et al. 2006) may serve as a host. The presence of a hybrid female with a previously unknown haplotype not belonging to either of the putatively ancient asexual lineages that coexist with C. strumicae and that has incorporated the C. strumicae genome (see table 1 and figure 2) suggests that the ability to incorporate unrelated genomes and host switches is more common among spined loaches than previously thought. Our study adds to the growing body of evidence supporting the Schultz's (1967) hypothesis that sperm from more non-parental species may be used in gynogenetic reproduction. Given the assumed gynogenetic nature of Cobitis hybrids (Saat 1991; Vasil'ev et al. 2003; Janko et al. 2007a), our data are also in agreement with the premise of Beukeboom & Vrijenhoek (1998) that gynogens are more likely to be able to switch sperm hosts than hybridogens that, in addition to the compatibility of gametes, rely on the compatibility of hybridizing genomes.
(b) Escaping from parents or becoming orphans?
It is unclear whether host switches in general arise by an active expansion of hybrids outside of its parental taxon (hypothesis 1) or via local parental extinction followed by a host switch (hypothesis 2). To differentiate between these alternatives, one would not only need to identify parental species of asexuals but also have knowledge about the history of clonal lineages. This would require a thorough phylogeographic analysis of the complex. Unfortunately, available data are ambiguous in this respect. In some cases, local ‘orphan’ asexual populations that are putatively dependent on sperm from a non-parental species are fully encompassed within the ranges of one or both parental species (Niemeitz et al. 2002; Schlupp et al. 2002; Mateos & Vrijenhoek 2005), preventing the distinction between models.
Gynogenetic females of silver crucian carp (Carassius auratus gibelio) are known to have expanded in Europe after human-mediated introduction (Holčík & Žitňan 1978; Kalous et al. 2007) used the sperm from various cyprinid species for reproduction (Peňáz et al. 1979; Yang et al. 2001). This case seems to support the ‘escape from parental species’ hypothesis. However, recent discoveries from silver crucian carp males in Europe (Lusková et al. 2004) raise doubts about such an interpretation. In theory, both sexuals and gynogens might have been introduced to Europe, and males might have remained undetected for a long time due to their low frequency in the population, which is otherwise typical for gynogens (Vasil'ev et al. 1989; Zhou et al. 2000; Janko et al. 2007b).
The contrasting scenario of local parental extinction followed by a host switch (hypothesis 2) seems supported by the hybridogenetic Squalius alburnoides complex in the Iberian Peninsula, in which the paternal species is assumed to be extinct, and asexual hybrids occur in the absence of the maternal progenitor in the northern parts of their range, relying therein on an unrelated species as a host (Alves et al. 2001). mtDNA-based phylogeographic analysis (Cunha et al. 2004) suggests multiple origins for asexual lineages endemic to several river basins with putative extinction of a maternal progenitor in the rivers of northern Spain. However, this explanation has been questioned on the grounds that mtDNA introgression was discovered in asexual S. alburnoides lineages, making the hypothesis of a single origin followed by subsequent dispersion over the Iberian Peninsula even more parsimonious (Sousa-Santos et al. 2006). The apparent ‘absence’ of paternal species was also recently hypothesized as an artefact of divergence of the nuclear S. alburnoides genome from existing paternal species (Sousa-Santos et al. 2007).
Similarly, European hybridogenetic water frogs of the Rana esculenta complex contain populations that might provide evidence of a host switch. Here, the Rana lessonae genome is replaced by Rana perezi in the hybridogen R. kl. grafi. In addition, some populations of R. kl. esculenta are entirely independent of a sperm donor due to the existence of hybridogenetic males and females (Günther 1979). It still remains unclear whether R. kl. grafi is a primary hybrid (Graf et al. 1977) or whether it is derived from an expanding R. kl. esculenta that has changed host species (e.g. Arano et al. 1995), and we do not know how the orphan R. kl. esculenta arose.
To our knowledge, the best analysis to date of host switch was performed in the sperm-dependent asexuals of mole salamanders of the genus Ambystoma. Spolsky et al. (1992) reported Ambystoma jeffersonianum–laterale hybrids dependent on sperm from Ambystoma texanum that exist outside the north-western range border of the continuous distribution of both original parental species. The presence of fragmented relict populations of A. jeffersonianum in this area led the authors to suggest that before switching to A. texanum, hybrid salamanders expanded into this area together with the parental species A. jeffersonianum, which subsequently retreated to the south (Lowcock 1989).
(c) Dispersal and range expansion in the genus Cobitis
By contrast, we have good reason to believe that the current distribution of Cobitis hybrid lineages reflects the opposite scenario to the pattern described in Ambystoma (hypothesis 1, i.e. the independent expansion of hybrids beyond the distribution of original parents). First, the monophyly of both hybrid lineages I and II implies geographically restricted origins probably somewhere in the mid-Danubian basin (Janko et al. 2005a), and that their present positions (including the areas outside the original parental ranges) are the result of recent dispersal. Second, it is very unlikely that the original parental species occurs undetected in those areas where both lineages coexist with C. taenia; if it did exist, then our thorough sampling of central Europe and the southern Bug R., where C. elongatoides and C. taenia occur, would have allowed us to detect it (Bohlen & Ráb 2001; Janko et al. 2007b). Third, unlike in the cases discussed above, the areas where both hybrid lineages coexist with non-parental species lay outside the ranges of original parents, mostly in different river systems, where no relict parental populations have been documented. Fourth, the new host's genomes were incorporated into expanding lineages on multiple occasions and the resulting tri-genomics are not found in the territories of the original parents. Finally, the demographic history of hybrid lineage II suggests that it underwent a recent population expansion, most probably linked to the invasion of new host populations.
The lack of evidence for population expansion in lineage I is difficult to explain if we accept the previous argument. The signal of population expansion (mismatch distribution and Tajima's D) may be an artefact from the process of accumulation of deleterious mutations, especially in ancient asexual lineages (Gordo et al. 2002). On the other hand, the inferred signal in lineage I might have been distorted by the fact that its demographic history may be more complex, as it is distributed throughout the refugial areas and is fragmented among several river systems.
The alternative scenario of host switching following local parental extinction (hypothesis 2) is a less parsimonious explanation than the escape of gynogens from their parents, since it assumes three independent extinctions of local populations of the original parental species in addition to the independent expansion events into these areas. Furthermore, in Bulgaria and southern Ukraine, the orphan gynogenetic populations are found at the centres of the ranges of C. strumicae and C. taenia, which are supposed to have Aegean (Bohlen et al. 2006) and Ponto-Caspian (Culling et al. 2006) origins, respectively. Furthermore, the ages of these species substantially predate the origin of both gynogenetic lineages (Janko et al. 2005a).
It is possible that the hybrid lineages and either of their parental species might have been present in this area before C. taenia settled there. The application of phylogeographic inference methods enabling sophisticated joint estimates analyses of the ages of population splits and their interconnections via migrations (Hey & Nielsen 2004) would allow us to distinguish between the alternative hypotheses. We regret that we have not collected a sufficient number of C. taenia samples from Ukrainian rivers, apart from southern Bug R., to perform such an analysis. However, the C. taenia population from the lower Bug R. does not appear to have undergone population expansion (Culling et al. 2006), suggesting long-term stability. Hybrid lineage II probably expanded in the recent past (as evidenced by the form of the mismatch distribution), which supports the invasion of hybrid gynogens to a new host territory.
Our data further demonstrate surprising, and perhaps unprecedented, genome compatibility among hybridizing loach species. We documented the ability of hybrid lineages to incorporate the genome of C. strumicae, which split from the original parental species during the Middle Miocene (Bohlen et al. 2006). Furthermore, previous data suggest that hybrid lineages of C. elongatoides–taenia origin (Janko et al. 2005a) form successful tetraploid clones that incorporate the C. melanoleuca genome (Vasil'ev et al. 1989), which probably diverged from both parental species during the Oligocene (Bohlen et al. 2006). If we assume the evidence of expansion of loach gynogens independent of their parental species to be true, then our data suggest that sperm dependence is not necessarily as restrictive to geographical expansion when compared with true parthenogenesis as previously thought.
Whereas apomixis in plants is often assumed to be under genetic control of one or several candidate genes (reviewed in Savidan 2000), the mechanisms responsible for the origin of asexuality are very poorly understood in animals. Among the various studies (for review see Schlupp 2005), gynogenetic systems allow the study of the interaction of hybridizing genomes and its effect on meiosis. When ‘suitable’ species meet, asexual hybrids are rather the rule than exception (review in Dawley 1989). Here, we have shown that once such gynogenetic lineages arise, they do not necessarily depend on the original parental species and are surprisingly tolerant of the incorporation of quite distantly related genomes. This gives us a possibility to study more general questions, i.e. whether it is the genetic distance between hybridizing species (as hypothesized by Moritz et al. 1989) and/or the control of specific genes and/or even a single allele that decides whether or not the hybrid shall be asexual, or whether the presence of some particular forms (genomes) is required to trigger gynogenesis as proposed by Hotz et al. (1985) for the hybridogenetic R. esculenta complex.
Acknowledgments
‘Valid Animal Use Protocols’ were in force at IAPG AS CR, Libechov.
We thank Milen Vasilev, Zdeněk Lajbner, Alena Šedivá and Štěpánka Hulová for their help in the field, Věra Šlechtová for determination help, Jana Kopecká and Šárka Pelikánová for their technical support, Jana Lokajová for personal support and Ann Puschell and Paul Bloor for their language corrections. We gratefully acknowledge the grant support provided by Grant Agency of Charles University in Prague (GAUK) no. 148407 and partially by GAUK no. 187/2005 B-BIO, GAČR 206/05/P586, GAČR 206/06/1763, MŠMT ČR LC06073, MŽP VaV-SM/6/3/05, the Academy of Sciences of the Czech Republic IRP IAPG ASCR AV0Z50450515 and the Joint Research Project no. 6 between ASCR and Bulgarian Academy of Sciences for the period 2005–2007.
Footnotes
One contribution of 16 to a Theme Issue ‘Hybridization in animals: extent, processes and evolutionary impact’.
References
- Alves M.J, Coelho M.M, Collares-Pereira M.J, Dowling T.E. Maternal ancestry of the Rutilus alburnoides complex (Teleostei, Cyprinidae) as determined by analysis of cytochrome b sequences. Evolution. 1997;51:1584–1592. doi: 10.1111/j.1558-5646.1997.tb01481.x. doi:10.2307/2411210 [DOI] [PubMed] [Google Scholar]
- Alves M.J, Coelho M.M, Collares-Pereira M.J. Evolution on action through hybridization and polyploidy in an Iberian freshwater fish: a genetic review. Genetica. 2001;111:375–385. doi: 10.1023/a:1013783029921. doi:10.1023/A:1013783029921 [DOI] [PubMed] [Google Scholar]
- Arano B, Llorente G.A, Herrero P, Sanchiz B. Current studies on Iberian water frogs. Zool. Pol. 1995;39:365–375. [Google Scholar]
- Arnold M.L. Oxford University Press; Oxford, UK: 1997. Natural hybridization and evolution. [Google Scholar]
- Baker H.G. Self-compatibility and establishment after ‘long-distance’ dispersal. Evolution. 1955;9:347–348. doi:10.2307/2405656 [Google Scholar]
- Baker H.G. Characteristics and modes of origin of weeds. In: Baker H.G, Stebbins G.L, editors. The genetics of colonizing species. Academic Press; New York, NY: 1965. pp. 147–168. [Google Scholar]
- Beukeboom L.W, Vrijenhoek R.C. Evolutionary genetics of sperm-dependent parthenogenesis. J. Evol. Biol. 1998;11:755–782. doi:10.1007/s000360050117 [Google Scholar]
- Bohlen J, Ráb P. Species and hybrid richness in spined loaches of the genus Cobitis (Teleostei: Cobitidae), with a checklist of European forms and suggestions for conservation. J. Fish Biol. 2001;59:75–89. doi:10.1006/jfbi.2001.1751 [Google Scholar]
- Bohlen J, Perdices A, Doadrio I, Economidis P.S. Vicariance, colonisation, and fast local speciation in Asia Minor and the Balkans as revealed from the phylogeny of spined loaches (Osteichthyes; Cobitidae) Mol. Phylogenet. Evol. 2006;39:552–561. doi: 10.1016/j.ympev.2005.12.007. doi:10.1016/j.ympev.2005.12.007 [DOI] [PubMed] [Google Scholar]
- Chow S, Hazama K. Universal PCR primers for S7 ribosomal protein gene introns in fish. Mol. Ecol. 1998;7:1247–1263. doi:10.1046/j.1365-294x.1998.00406.x [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall K.A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. doi:10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Culling M.A, Janko K, Boron A, Vasil'ev V.P, Cote I.M, Hewitt G.M. European colonization by the spined loach (Cobitis taenia) from Ponto-Caspian refugia based on mitochondrial DNA variation. Mol. Ecol. 2006;15:173–190. doi: 10.1111/j.1365-294X.2005.02790.x. doi:10.1111/j.1365-294X.2005.02790.x [DOI] [PubMed] [Google Scholar]
- Cunha C, Coelho M.M, Carmona J.A, Doadrio I. Phylogeographical insights into the origins of the Squalius alburnoides complex via multiple hybridization events. Mol. Ecol. 2004;13:2807–2817. doi: 10.1111/j.1365-294X.2004.02283.x. doi:10.1111/j.1365-294X.2004.02283.x [DOI] [PubMed] [Google Scholar]
- Dawley R.M. An introduction to unisexual vertebrates. In: Dawley R.M, Bogart J.P, editors. Evolution and ecology of unisexual vertebrates. New York State Museum; Albany, NY: 1989. pp. 1–18. [Google Scholar]
- Doncaster C.P, Pound G.E, Cox S.J. The ecological cost of sex. Nature. 2000;404:281–285. doi: 10.1038/35005078. doi:10.1038/35005078 [DOI] [PubMed] [Google Scholar]
- Engeler B, Reyer H.-U. Choosy females and indiscriminate males: mate choice in mixed populations of the water frogs Rana lessonae and Rana esculenta. Behav. Ecol. 2001;12:600–606. doi:10.1093/beheco/12.5.600 [Google Scholar]
- Gordo I, Navarro A, Charlesworth B. Muller's ratchet and the pattern of variation at a neutral locus. Genetics. 2002;161:835–848. doi: 10.1093/genetics/161.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J.D, Karch F, Moreillon M.C. Biochemical variation in the Rana esculenta complex: a new hybrid form related to Rana perezi and Rana ridibunda. Experientia. 1977;33:1582–1584. doi: 10.1007/BF01934010. doi:10.1007/BF01934010 [DOI] [PubMed] [Google Scholar]
- Günther R. General remarks on the evolutionary genetics of the European water frog complex. Mitt. Zool. Mus. Berlin. 1979;55:7–11. [Google Scholar]
- Hedges S.B, Bogart J.P, Maxon L.R. Ancestry of unisexual salamanders. Nature. 1992;356:708–710. doi: 10.1038/356708a0. doi:10.1038/356708a0 [DOI] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. doi:10.1534/genetics.103.024182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holčík J, Žitňan R. On the expansion and origin of Carassius auratus in Czechoslovakia. Folia Zool. 1978;27:279–288. [Google Scholar]
- Hotz H, Mancino G, Bucci-Innocenti S, Ragghianti M, Berger L, Uzzell T. Rana ridibunda varies geographically in inducing clonal gametogenesis in interspecific hybrids. J. Exp. Zool. 1985;236:199–210. doi:10.1002/jez.1402360210 [Google Scholar]
- Janko K, Kotlík P, Ráb P. Evolutionary history of asexual hybrid loaches (Cobitis: Teleostei) inferred from phylogenetic analysis of mitochondrial DNA variation. J. Evol. Biol. 2003;16:1280–1287. doi: 10.1046/j.1420-9101.2003.00627.x. doi:10.1046/j.1420-9101.2003.00627.x [DOI] [PubMed] [Google Scholar]
- Janko K, Kotlík P, Culling M.A, Ráb P. Ice age cloning—comparison of Quaternary evolutionary histories of sexual and clonal forms of European loaches (Cobitis; Teleostei) using the analysis of mitochondrial DNA variation. Mol. Ecol. 2005a;14:2991–3004. doi: 10.1111/j.1365-294X.2005.02583.x. doi:10.1111/j.1365-294X.2005.02583.x [DOI] [PubMed] [Google Scholar]
- Janko K, Vasil'ev V.P, Ráb P, Rábová M, Šlechtová V, Vasil'eva E.D. Genetic and morphological analyses of 50-chromosome spined loaches (Cobitis, Cobitidae, Pisces) from the Black Sea basin that are morphologically similar to C. taenia, with the description of a new species. Folia Zool. 2005b;54:405–420. [Google Scholar]
- Janko K, Bohlen J, Lamatsch D, Flajšhans M, Kotlík P, Ráb P, Šlechtová V. Evidence for gynogenesis as the reproductive mode of hybrid loaches (Cobitis: Teleostei): on the evolution of polyploidy in asexual vertebrates. Genetica. 2007a;131:185–194. doi: 10.1007/s10709-006-9130-5. doi:10.1007/s10709-006-9130-5 [DOI] [PubMed] [Google Scholar]
- Janko K, et al. Diversity of European spined loaches (genus Cobitis L.): an update of the geographic distribution of the Cobitis taenia hybrid complex with a description of new molecular tools for species determination. J. Fish. Biol. 2007b;71(Suppl.):387–408. doi:10.1111/j.1095-8649.2007.01663.x [Google Scholar]
- Kalous L, Šlechtová V, Jr, Bohlen J, Petrtýl M, Švátora M. First European record of Carassius langsdorfii from the Elbe basin. J. Fish Biol. 2007;70:132–138. doi:10.1111/j.1095-8649.2006.01290.x [Google Scholar]
- Kirkendall L.R. Sperm is a limiting resource in the pseudogamous bark beetle Ips acumiatus (Scolytidae) Oikos. 1990;57:80–87. doi:10.2307/3565740 [Google Scholar]
- Kuhner M.K. LAMARC: estimating population genetic parameters from molecular data. In: Salemi M, Vandamme A, editors. The phylogenetic handbook: a practical approach to DNA and protein phylogeny. Cambridge University Press; Cambridge, UK: 2003. pp. 379–399. ( doi:10.2277/052180390X) [Google Scholar]
- Kuhner M.K, Yamato J, Felsenstein J. Maximum likelihood estimation of population growth rates based on the coalescent. Genetics. 1998;149:429–434. doi: 10.1093/genetics/149.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamatsch D.K, Steinlein C, Schmid M, Schartl M. Noninvasive determination of genome size and ploidy level in fishes by flow cytometry: detection of triploid Poecilia formosa. Cytometry. 2000;39:91–95. doi: 10.1002/(sici)1097-0320(20000201)39:2<91::aid-cyto1>3.0.co;2-4. doi:10.1002/(SICI)1097-0320(20000201)39:2<91::AID-CYTO1>3.0.CO;2-4 [DOI] [PubMed] [Google Scholar]
- Lowcock L.A. Biogeography of hybrid complexes of Ambystoma: interpreting unisexual–bisexual genetic data in space and time. In: Dawley R.M, Bogart J.P, editors. Evolution and ecology of unisexual vertebrates. New York State Museum; Albany, NY: 1989. pp. 180–208. [Google Scholar]
- Lusková V, Halačka K, Vetešník L, Lusk S. Changes of ploidy and sexuality status of ‘Carassius auratus’ population in the drainage area of the River Dyje (Czech Republic) Ecohydrol. Hydrobiol. 2004;4:165–171. [Google Scholar]
- Mateos M, Vrijenhoek R.C. Ancient versus reticulate origin of a hemiclonal lineage. Evolution. 2002;56:985–992. doi: 10.1111/j.0014-3820.2002.tb01410.x. doi:10.1554/0014-3820(2002)056[0985:AVROOA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Mateos M, Vrijenhoek R.C. Independent origins of allotriploidy in the fish genus Poeciliopsis. J. Hered. 2005;96:1–8. doi: 10.1093/jhered/esi010. doi:10.1093/jhered/esi010 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. Cambridge University Press; Cambridge, UK: 1978. The evolution of sex. [Google Scholar]
- Moritz C, Brown W.M, Densmore L.D, Wright J.W, Vyas D, Donnellan S, Adams M, Baverstock P. Genetic diversity and the dynamics of hybrid parthenogenesis in Cnemidophorus (Teiidae) and Heteronotia (Gekkonidae) In: Dawley R.M, Bogart J.P, editors. Evolution and ecology of unisexual vertebrates. New York State Museum; Albany, NY: 1989. pp. 268–280. [Google Scholar]
- Niemeitz A, Kreutzfeldt R, Schartl M, Parzefall J, Schlupp I. Male mating behaviour of a molly, Poecilia latipunctata: a third host for the sperm-dependent Amazon molly, Poecilia formosa. Acta Ethol. 2002;5:45–49. doi:10.1007/s10211-002-0065-2 [Google Scholar]
- Peňáz M, Ráb P, Prokeš M. Cytological analysis, gynogenesis and early development of Carassius auratus gibelio. Acta Sci. Nat. Brno. 1979;13:1–33. [Google Scholar]
- Pongratz N, Storhas M, Carranza S, Michiels N.K. Phylogeography of competing sexual and parthenogenetic forms of a freshwater flatworm: patterns and explanations. BMC Evol. Biol. 2003;3:1–15. doi: 10.1186/1471-2148-3-23. doi:10.1186/1471-2148-3-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pound G.E, Doncaster C.P, Cox S.J. A Lotka–Volterra model of coexistence between a sexual population and multiple asexual clones. J. Theor. Biol. 2002;217:535–545. doi: 10.1006/jtbi.2002.3040. doi:10.1006/jtbi.2002.3040 [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins S.E, Rozas J. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 2002;19:2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- Rogers A.R, Harpending H. Population-growth makes waves in the distribution of pairwise genetic-differences. Mol. Biol. Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. doi:10.1093/bioinformatics/15.2.174 [DOI] [PubMed] [Google Scholar]
- Saat T.V. Reproduction of the diploid and polyploid spinous loach (Cobitis, Teleostei). Oocyte maturation and fertilization in the triploid form. Sov. J. Dev. Biol. 1991;22:332–338. [Google Scholar]
- Savidan Y. Apomixis: genetics and breeding. Plant Breed. Rev. 2000;18:13–86. [Google Scholar]
- Schley D, Doncaster C.P, Sluckin T. Population models of sperm-dependent parthenogenesis. J. Theor. Biol. 2004;229:559–572. doi: 10.1016/j.jtbi.2004.04.031. doi:10.1016/j.jtbi.2004.04.031 [DOI] [PubMed] [Google Scholar]
- Schlupp I. The evolutionary ecology of gynogenesis. Annu. Rev. Ecol. Evol. Syst. 2005;36:399–417. doi:10.1146/annurev.ecolsys.36.102003.152629 [Google Scholar]
- Schlupp I, Parzefall J, Schartl M. Biogeography of the Amazon molly, Poecilia formosa. J. Biogeogr. 2002;29:1–6. doi:10.1046/j.1365-2699.2002.00651.x [Google Scholar]
- Schultz R.J. Gynogenesis and triploidy in the viviparous fish Poeciliopsis. Science. 1967;157:1564–1567. doi: 10.1126/science.157.3796.1564. doi:10.1126/science.157.3796.1564 [DOI] [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. doi:10.1016/j.tree.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Semlitsch R.D, Hotz H, Guex G.-D. Competition among tadpoles of coexisting hemiclones of hybridogenetic Rana esculenta: support for the frozen niche variation model. Evolution. 1997;51:1249–1261. doi: 10.1111/j.1558-5646.1997.tb03972.x. doi:10.2307/2411054 [DOI] [PubMed] [Google Scholar]
- Simon J.C, Delmotte F, Rispe C, Crease T. Phylogenetic relationships between parthenogens and their sexual relatives: the possible routes to parthenogenesis in animals. Biol. J. Linn. Soc. 2003;79:151–163. doi:10.1046/j.1095-8312.2003.00175.x [Google Scholar]
- Šlechtová V, Lusková V, Šlechta V, Lusk S, Halačka K, Bohlen J. Genetic differentiation of two diploid–polyploid complexes of spined loach, genus Cobitis (Cobitidae), in the Czech Republic, involving C. taenia, C. elongatoides and C. spp.: allozyme interpopulation and interspecific differences. Folia Zool. 2000;49(Suppl.):67–78. [Google Scholar]
- Sousa-Santos C, Collares-Pereira M.J, Almada V.C. Evidence of extensive mitochondrial introgression with nearly complete substitution of the typical Squalius pyrenaicus-like mtDNA of the Squalius alburnoides complex (Cyprinidae) in an independent Iberian drainage. J. Fish Biol. 2006;68(Suppl. B):292–301. doi:10.1111/j.1095-8649.2006.01081.x [Google Scholar]
- Sousa-Santos C, Collares-Pereira M.J, Almada V. Reading the history of a hybrid fish complex from its molecular record. Mol. Phylogenet. Evol. 2007;45:981–996. doi: 10.1016/j.ympev.2007.05.011. doi:10.1016/j.ympev.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Spolsky C, Phillips C.A, Uzzell T. Gynogenetic reproduction in hybrid mole salamanders (genus Ambystoma) Evolution. 1992;46:1935–1944. doi: 10.1111/j.1558-5646.1992.tb01179.x. doi:10.2307/2410041 [DOI] [PubMed] [Google Scholar]
- Vandel A. La parthénogénèse geographique. Contribution à l'étude biologique et cytologique de la parthénogénèse naturelle. Bull. Biol. Fr. Belg. 1928;62:164–281. [Google Scholar]
- Vasil'ev V.P, Vasil'eva K.D, Osinov A.G. Evolution of a diploid–triploid–tetraploid complex in fishes of the genus Cobitis (Pisces, Cobitidae) In: Dawley R.M, Bogart J.P, editors. Evolution and ecology of unisexual vertebrates. New York State Museum; Albany, NY: 1989. pp. 153–169. [Google Scholar]
- Vasil'ev V.P, Akimova N.V, Emel'yanova N.G, Pavlov D.A, Vasil'eva E.D. Reproductive capacities in the polyploid males of spined loaches from the unisexual–bisexual complex, occurred in the Moscov River. Folia Biol. (Krakow) 2003;51:67–73. [PubMed] [Google Scholar]
- Vasil'eva E.D, Vasil'ev V.P. Cobitis pontica sp. nova—a new spined loach species (Cobitidae) from the Bulgarian waters. J. Ichthyol. 2006;46:15–20. doi:10.1134/S003294520610002X [Google Scholar]
- Vrijenhoek R.C. Gene dosage in diploid and triploid unisexual fishes (Poeciliopsis, Poeciliidae) In: Markert C.L, editor. Isozymes IV genetics and evolution. Academic Press; New York, NY: 1975. pp. 463–476. [Google Scholar]
- Vrijenhoek R.C. Factors affecting clonal diversity and coexistence. Am. Zool. 1979;19:787–797. [Google Scholar]
- Vrijenhoek R.C. Genetic and evolutionary constraints on the origin and establishement of unisexual vertebrates. In: Dawley R.M, Bogart J.P, editors. Evolution and ecology of unisexual vertebrates. New York State Museum; Albany, NY: 1989. pp. 24–31. [Google Scholar]
- Vrijenhoek R.C. Clonal organisms and the benefits of sex. In: Carvalho G.R, editor. Advances in molecular ecology. IOS Press; Amsterdam, The Netherlands: 1998. pp. 151–172. [Google Scholar]
- Watterson G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. doi:10.1016/0040-5809(75)90020-9 [DOI] [PubMed] [Google Scholar]
- Yang L, Shu-Ting Y, Xue-Hong W, Gui J.F. Genetic diversity among different clones of the gynogenetic silver crucian carp Carassius auratus gibelio, revealed by transferrin and isozyme markers. Biochem. Genet. 2001;39:213–225. doi: 10.1023/a:1010297426390. doi:10.1023/A:1010297426390 [DOI] [PubMed] [Google Scholar]
- Zhou L, Wang Y, Gui J.F. Genetic evidence for gonochoristic reproduction in gynogenetic silver crucian carp (Carassius auratus gibelio Bloch) as revealed by RAPD assays. J. Mol. Evol. 2000;51:498–506. doi: 10.1007/s002390010113. doi:10.1007/s002390010113 [DOI] [PubMed] [Google Scholar]