Abstract
It is generally accepted that plastids first arose by acquisition of photosynthetic prokaryotic endosymbionts by non-photosynthetic eukaryotic hosts. It is also accepted that photosynthetic eukaryotes were acquired on several occasions as endosymbionts by non-photosynthetic eukaryote hosts to form secondary plastids. In some lineages, secondary plastids were lost and new symbionts were acquired, to form tertiary plastids. Most recent work has been interpreted to indicate that primary plastids arose only once, referred to as a ‘monophyletic’ origin. We critically assess the evidence for this. We argue that the combination of Ockham's razor and poor taxon sampling will bias studies in favour of monophyly. We discuss possible concerns in phylogenetic reconstruction from sequence data. We argue that improved understanding of lineage-specific substitution processes is needed to assess the reliability of sequence-based trees. Improved understanding of the timing of the radiation of present-day cyanobacteria is also needed. We suggest that acquisition of plastids is better described as the result of a process rather than something occurring at a discrete time, and describe the ‘shopping bag’ model of plastid origin. We argue that dinoflagellates and other lineages provide evidence in support of this.
Keywords: endosymbiosis, polyphyly, monophyly, plastid, phylogenetics, dinoflagellate
1. Introduction
There is no longer serious doubt about the idea that chloroplasts first arose through symbiosis between a free-living photosynthetic organism (ancestral to present-day cyanobacteria) and a non-photosynthetic host, as first proposed by Schimper (1883) and developed by Mereschkowsky (1905; Martin & Kowallik 1999). It was Mereschkowsky who originally raised the possibility that lineages with different photosynthetic pigment composition might owe their origins to distinct endosymbioses with photosynthetic organisms with different pigment compositions. He suggested that because ‘there are green, yellow and red cyanophytes, as is also the case for the direct antecedents of the cyanophytes… the green, the brown and the red algae could have thus originated independently’ (translation by Martin & Kowallik 1999). The idea of multiple independent primary origins of chloroplasts (more generally referred to as plastids) with different pigment types was supported by others (e.g. Raven 1970). However, particularly with the advent of sequence-based trees, the idea of multiple primary origins of plastids has fallen from favour, so that by 2000, for example, Palmer (2000) suggested that a ‘single primary origin of plastids now seems certain’. However, a number of workers have urged caution over, or argued against, this conclusion (recent examples including Nozaki et al. 2007 and Stiller 2007). The formation of plastids, and ultimately all eukaryotic algae and plants, has had an enormous impact on the evolution of life on the Earth, and it is therefore important to understand the evidence for and against the idea that there was a single primary formation of these organelles. In this paper, we critically assess the evidence for a single or multiple primary origins of plastids. Some aspects of this evidence have been reviewed by us and others (Stiller 2003; Larkum et al. 2007), so we discuss those in less detail. We also discuss the idea (Larkum et al. 2007) that formation of an endosymbiosis should be regarded as a process involving a number of partners, rather than a single event involving two partners. We discuss the reason why one particular algal lineage, the dinoflagellates, appears to have been involved in multiple episodes of plastid loss and gain.
2. Plastid types
Plastids are the photosynthetic organelles of plant and algal cells. The term also includes non-photosynthetic organelles that are derived from them by development (such as carotenoid-containing chromoplasts) or evolution (such as the remnant plastid of Apicomplexa; Wilson 2005). The term chloroplasts can be used to refer to plastids of green plants and algae containing chlorophylls a and b, or to plastids more generally. We use the term plastid throughout to refer to all forms of the organelle to avoid any confusion. Plastids can be divided into three categories: primary, secondary and tertiary. The primary plastids are bounded by two membranes and represent the acquisition of a photosynthetic prokaryote by a non-photosynthetic eukaryotic host. Broadly, three groups of organisms contain primary plastids. These are the green plants and green algae (containing chlorophylls a and b as their photosynthetic pigments), the red algae (containing chlorophyll a and phycobiliprotein) and the glaucophyte algae (exemplified by Cyanophora paradoxa and also containing chlorophyll a and phycobiliprotein). Other possible examples of primary plastids have been proposed, most recently the nitrogen-fixing ‘spheroid bodies’ in the diatom Rhopalodia (Prechtl et al. 2004) and the cyanobacterium-like ‘chromatophore’ of the amoeba Paulinella (Marin et al. 2005). Because gene loss from symbiont to host is arguably a defining feature of a true endosymbiont (Theissen & Martin 2006), here we do not include the symbionts of Rhopalodia and Paulinella as primary plastids, as there are no indications of gene loss from symbiont to host in these organisms (Yoon et al. 2006).
Secondary and tertiary plastids are bounded by more than two membranes and represent the acquisition of a photosynthetic eukaryote by a non-photosynthetic eukaryotic host followed by varying levels of simplification. Such simplification includes reduction in number of membranes surrounding the secondary plastid and a loss of the nucleus of the photosynthetic eukaryote. (In some lineages, the cryptophytes and the chlorarachniophytes, it is retained as a nucleomorph between two of the membranes surrounding the plastid; Douglas et al. 2001; Gilson et al. 2006.) Tertiary plastids are generated by the loss of secondary plastids from a photosynthetic eukaryote and their replacement by plastids from another source. Examples of plastid types are given in table 1. There is no dispute that secondary and tertiary plastids have arisen on a number of different occasions, i.e. that they are polyphyletic. The question at issue is whether the plastids of glaucophytes, red algae and green plants and algae all have a single origin (monophyly) or separate origins (polyphyly).
Table 1.
Examples of different plastid types, modified from Larkum et al. (2007).
| group | primary, secondary or tertiary | presence of chlorophylls and/or phycobiliprotein | no. of surrounding membranes |
|---|---|---|---|
| green plants/algae | 1° | chlorophyll a, b | 2 |
| red algae | 1° | chlorophyll a, phycobiliprotein | 2 |
| Glaucophyta | 1° | chlorophyll a, phycobiliprotein | 2 |
| Cryptophyta | 2° | chlorophyll a, c phycobiliprotein | 4 |
| Chlorarachniophyta | 2° | chlorophyll a, b | 4 |
| Ochrophyta and heterokonts | 2° | chlorophyll a, c | 4 |
| Haptophyta | 2° | chlorophyll a, c | 4 |
| Euglenophyta | 2° | chlorophyll a, b | 3 |
| dinoflagellates | 2°, 3° | mostly chlorophyll a, c | 3 |
| Apicomplexa | 2° | none | 4 |
3. Can we actually prove (or disprove) anything?
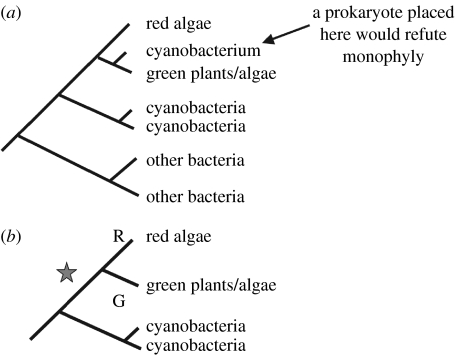
A monophyletic hypothesis of plastid origin predicts that the red, green and glaucophyte plastids form a single group to the exclusion of oxygenic photosynthetic prokaryotes, either as a group within those prokaryotes or as a sister group. Finding a prokaryote lineage that broke up the monophyletic plastid group would therefore refute the monophyletic hypothesis (figure 1a). If we failed to find such a prokaryote lineage, that could be because (i) we had not yet sampled the right prokaryotic lineage or (ii) the monophyletic hypothesis was true. If the prokaryote lineage that could have broken up the group had become extinct, we would never be able to disprove monophyly even if the hypothesis were false. (This extinction could have been part of the mass extinctions of prokaryotic and other lineages occurring as a result of global snowballs or other extreme environmental events.) Thus, as we increase the number of prokaryote lineages sampled without finding one that breaks up the monophyletic group, we may increasingly prefer the monophyletic hypothesis, but if extinctions have occurred we cannot ‘prove’ the hypothesis conclusively.
Figure 1.
(a) A cyanobacterium breaking up the red/green lineage provides a refutation of monophyly. (b) The hypothesis preferred under Ockham's razor for explaining the origins of characters specific to red and green lineages, i.e. a single endosymbiosis, when no cyanobacteria have been sampled that (a) break up the red/green lineage. R and G indicate the origin of characters specific for the red and green lineages; the star indicates endosymbiosis.
A polyphyletic hypothesis predicts the existence of prokaryotic lineages that break up the monophyletic group. A failure to find those lineages could likewise be either because (i) we have not yet sampled the right lineages (perhaps because they are extinct) or (ii) the polyphyletic hypothesis was false.
Limited taxon sampling therefore makes it difficult to distinguish conclusively between monophyletic and polyphyletic models. However, the general principle that we prefer, the explanation that makes fewest assumptions, means that we are likely to prefer a monophyletic model over a polyphyletic one. (This is the principle generally referred to as Ockham's Razor; Spade 1999.) Suppose that (figure 1b) the red and green plastid lineages each have a distinct character state. Although we could invoke two separate endosymbioses with prokaryotes having those character states, unless we can find prokaryotes having the same character states as the plastids, we will prefer a priori a hypothesis that has a single endosymbiosis followed by divergence into the separate character states (figure 1b).
The consequences of limited taxon sampling for our ability to distinguish between monophyly and polyphyly make it important to determine the extent of extinctions that have occurred within the cyanobacterial lineages. In the extreme case, present-day cyanobacteria may represent a recent radiation (after a bottleneck caused by a global snowball) with the evidence that would allow us to distinguish conclusively between monophyly and polyphyly lost. Although there are morphological data indicating that some cyanobacterial lineages, e.g. those with and without akinetes, may have diverged over 2.5 Ga ago (Tomitani et al. 2006), these studies rest on potentially subjective comparison of present-day and fossil material. Estimates of when present-day cyanobacterial lineages diverged will therefore be very valuable. Nevertheless, the combination of Ockham's Razor with limited taxon sampling and mass extinction will implicitly favour the conclusions that (i) evidence favours monophyly over polyphyly but (ii) we cannot be sure.
4. Phylogenetic trees and associated problems
In addition to the problems caused by limited taxon sampling, there are technical difficulties with phylogenetic tree construction. Although a very large number of studies have been carried out to recover phylogenetic trees from sequence data, the results have been contradictory. Many have indicated a monophyletic origin of plastids from within, or as a sister group to, cyanobacteria. However, some have indicated a polyphyletic origin of plastids. For example, Rodriguez-Ezpeleta et al. recently reported an analysis of 50 genes from 16 plastid and 15 cyanobacterial genomes and 143 nuclear genes from 34 eukaryotic lineages, and obtained evidence for a monophyletic origin of plastids (Rodriguez-Ezpeleta et al. 2005). However, a subsequent analysis by Nozaki et al. (2007) using slowly evolving nuclear sequences mainly derived from the dataset of Rodriguez-Ezpeleta et al. provided robust evidence for a polyphyletic origin, with the red algae separate from the glaucophytes and green algae. A number of possible interlinked concerns have been raised over sequence-based trees. Although individual problems have been well addressed in different publications, we attempt here to produce a compilation.
(a) Long-branch attraction
This is a well-recognized tree-building artefact in which some lineages, but not others, have accumulated large amounts of change. The lineages with more change appear as long branches, and these tend to be grouped artificially closely in phylogenetic reconstruction (Hendy & Penny 1989). Longer branches are most often associated with lineages that have an increased number of substitutions per site (Felsenstein 1978), and it has been suggested that this artefact may account for the evidence for monophyly reported by Rodriguez-Ezpeleta et al. (Nozaki et al. 2007). Lockhart & Steel (2005) have discussed an alternative evolutionary scenario to that of Felsenstein, where the rate of substitution is the same at all the sites that can vary, but where there are different numbers of sites that vary in different lineages. They have shown that, if sequences have different proportions of variable sites, tree reconstruction can be misled in a way very similar to that described by Felsenstein. Lineages with greater proportions of variable sites are represented by longer branches, and these tend to join together in tree reconstructions irrespective of the true phylogeny. The Lockhart & Steel (2005) scenario is more worrying than the Felsenstein (1978) scenario, because even the most sophisticated covarion substitution models (Galtier 2001; Huelsenbeck 2002) implemented for tree building require that the proportion of variable sites is constant across lineages. Thus, the problem Lockhart and Steel envisage will affect all tree building methods, not just parsimony or compatibility methods.
Improving our understanding of the spatial patterns of sequence evolution (i.e. how many sites, and which, are varying) is important. There is evidence that differences in the spatial substitution patterns of sequences make a significant contribution to the phylogenetic structure in analyses of oxygenic photosynthetic organisms (Lockhart et al. 1998, 2000, 2006; Shalchian-Tabrizi et al. 2006). If the fact that two lineages share a common set of variable sites to the exclusion of other lineages is a reflection of their actual evolutionary relationship, this phenomenon will reinforce the ‘correct’ phylogenetic signal in a tree. However, if the distribution of variable sites does not reflect evolutionary relationships, artefactual signals will arise.
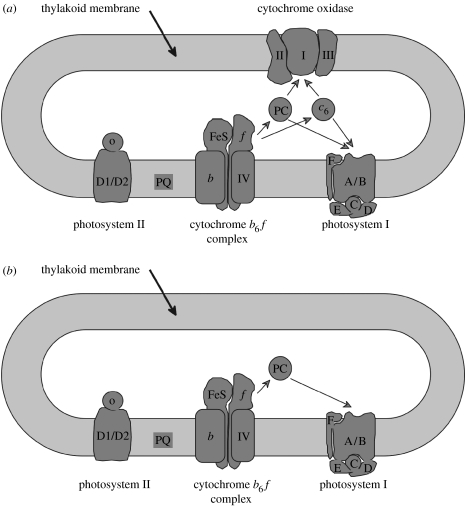
Recognizing which is the case may require detailed understanding of the biochemistry and structural biology of the proteins we are considering, and it is possible to envisage situations where similar distributions of variable sites may arise convergently in different lineages. For example, in oxygenic photosynthetic bacteria the complexes carrying out the light reactions of photosynthesis are located in the same compartment (the thylakoid membrane) as the respiratory cytochrome oxidase complex. The redox proteins plastocyanin or cytochrome c6 that accept electrons from the cytochrome b6f complex in photosynthetic electron transfer may pass them on either to cytochrome oxidase or photosystem I (Hart et al. 2005). However, plastids do not retain the conventional cytochrome oxidase complex, and the plastocyanin or cytochrome c6 (where present) no longer need to interact with cytochrome oxidase (figure 2). Thus, sites on plastocyanin or cytochrome c6 that are involved with cytochrome oxidase interaction in photosynthetic bacteria may be free to vary in plastid-containing lineages. This variability would be a consequence of the loss of cytochrome oxidase from the lineages, and not necessarily their evolutionary relationship.
Figure 2.
Organization of the electron transfer chain of (a) cyanobacteria and (b) chloroplasts. The figure shows that chloroplasts lack a cytochrome oxidase complex and (at least for green chloroplasts) a cytochrome c6. Figure courtesy of D. S. Bendall.
(b) Nucleotide compositional bias
Plastid genomes typically have high AT contents, e.g. Nicotiana tabacum, Porphyra purpurea and Odontella sinensis have, respectively, plastid genomes with 62, 67 and 68% AT content compared with 52% for the cyanobacterium Synechocystis sp. PCC 6803 (Howe et al. 2003). The reason(s) for the high AT content of plastid genomes remain unclear, but it is a common feature of reduced genomes in bacterial symbionts (Canback et al. 2004) as well as plastids. The biased nucleotide composition can lead to plastid genomes being grouped on the basis of their high AT content, and to the exclusion of sequences with a lower AT content (Lockhart et al. 1992; Barbrook et al. 1998). If two lineages share a high AT content as a result of common ancestry, this effect will reinforce the historical signal in the data. If they have independently acquired a high AT content, the effect may override the ability to recover the true tree topology. The problem is worst when internal tree branches are very short (Jermiin et al. 2004). Methods such as the LogDet transformation (Lockhart et al. 1994) have been developed to attempt to deal with the problem of non-random nucleotide composition. However, our ability to deal with the problem depends on having a good understanding of the spatial pattern of substitution in sequences and, as mentioned above, we still lack this knowledge.
(c) Lateral transfer
Lateral gene transfer may be responsible for the unexpected tree topology obtained with the rbcLS sequences from plastids. Analyses based on these sequences indicated a common ancestry of the red and brown plastid rbcLS sequences with those from beta purple photosynthetic bacteria, rather than with the rbcLS sequences of cyanobacteria as expected. The green chloroplast and glaucophyte sequences shared most recent common ancestry with the cyanobacterial sequences (Morden et al. 1992). One possible explanation for this is that the rbcLS sequences of red plastids were acquired by lateral transfer from a purple photosynthetic bacterium. Although lateral transfer events can lead to phylogenetic trees that do not reflect the history of other genes, they may themselves be useful indicators of evolutionary history. Thus, if a particular gene in all green, red and glaucophyte plastids appeared to have been acquired by lateral transfer, this could be interpreted as evidence of a monophyletic origin as the chance of the same lateral transfer happening independently in all the lineages would be low. Examples of the use of lateral transfer events as phylogenetic markers are discussed in more detail below.
(d) Plastid impact hypothesis
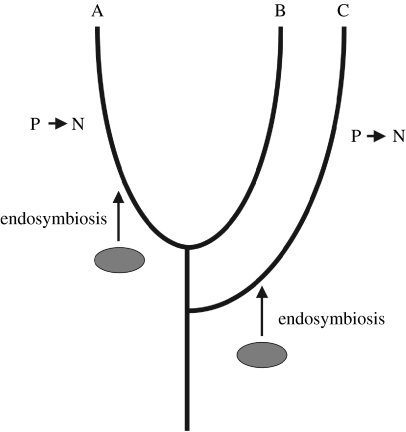
In organisms with multiple plastids there is believed to be a continuous flux of DNA from plastid to nucleus (reviewed by Barbrook et al. 2006a). A significant fraction of nuclear genes—not just those encoding plastid proteins—are of endosymbiont origin (Martin et al. 2002), although the extent of this in different lineages has been questioned (Reyes-Prieto et al. 2007). Thus, host genes can be replaced by plastid homologues. Consider a situation where primary plastids originated separately in two different host lineages, but in both cases with a cyanobacterial endosymbiont (figure 3). A phylogenetic tree based on a host (i.e. nuclear) gene would be expected to show two independent origins of plastids. However, if the host gene were replaced independently in both lineages with the counterpart from the endosymbiont, a tree constructed from the sequence would group the plastid-containing lineages to the exclusion of the others, indicating a monophyletic origin. This potential artefact was described by Stiller (2007) and is referred to as the ‘plastid impact’ hypothesis.
Figure 3.
The plastid impact hypothesis. Two lineages (A and C) acquire a plastid by endosymbiosis of closely related organisms. The plastid gene subsequently replaces a nuclear counterpart in both lineages (P→N). Thus, phylogenetic trees based on that gene will group A and C to the exclusion of B.
(e) Conflicting signals within the data
Many studies use concatenated sequences, on the assumption that this will provide greater phylogenetic resolution and more reliable tree topologies. However, this is not always so. Where a systematic error, such as long-branch attraction, results in the recovery of an incorrect tree topology, the addition of further sequences subject to the same systematic error will not affect the topology recovered. It will simply increase the statistical confidence placed on the incorrect topology. In other circumstances, different subsets of sequences may give support for different tree topologies, for reasons discussed above. The strongest signal will be likely to mask weaker ones, but may not necessarily be correct. The approach of spectral analysis, where different phylogenetic signals supporting different topologies are extracted from the data, may allow conflicting signals to be identified. For example, Lockhart et al. (1999), reanalysing a concatenated dataset of 45 proteins with regard to the placement of Odontella in relation to sequences from red and green lineages, demonstrated a different phylogenetic signal from the three RNA polymerase genes within the dataset from the remaining 42 sequences. It will be important to understand better how sequences encoding components of different complexes, such as the RNA polymerase, the ribosomes and the photosystems, behave in phylogenetic reconstruction.
5. Relationships inferred from other characters
A number of other characters have been used to infer phylogenetic relationships among lineages with primary plastids. We have reviewed these recently (Larkum et al. 2007) and will not discuss them in detail here.
(a) Gene content and organization
The retention of a similar set of genes in plastid genomes of different lineages could be explained in terms of large-scale transfer in an ancestral lineage, i.e. in support of monophyly. However, it has also been argued that the similarity in genes retained in different lineages is no greater than would be expected by chance (Stiller et al. 2003) especially as similar selective factors may be operating to retain genes in the chloroplast in independent lineages. Particular genes may be retained in order to allow them to be controlled in response to the redox or other biochemical needs of the organelle (Allen 2003; Barbrook et al. 2006a).
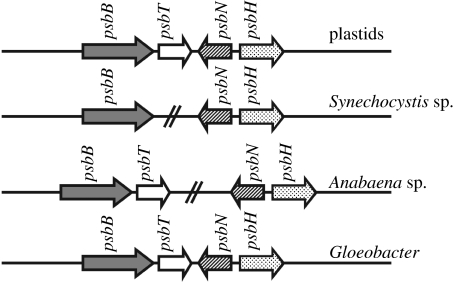
The presence of gene clusters, which are not observed in prokaryotes sampled, in different plastid lineages may also be taken as an indication of monophyly. However, this interpretation depends on adequate sampling of prokaryote lineages. Thus, although it had been supposed for some time that the psbB, psbT, psbN and psbH gene cluster united plastids to the exclusion of cyanobacteria, the same cluster was then found in the Gloeobacter genome sequence, as shown in figure 4 (Reith & Munholland 1995; Nakamura et al. 2003).
Figure 4.
The psbBTNH cluster. This was initially thought to be unique to plastids, indicating a monophyletic origin. However, a similar cluster was then found to exist in the cyanobacterium Gloeobacter.
(b) Light-harvesting machinery
Plants and green algae contain a set of membrane-intrinsic light-harvesting proteins, designated the light-harvesting complex (LHC) family, that are not found in cyanobacteria. Where cyanobacteria do have a chlorophyll b, like that found in plants and green algae, it is bound by a protein that is not part of the LHC family. However, red algae contain polypeptides with similarity to the LHC family, consistent with a monophyletic origin of the red and green lineages (Wolfe et al. 1994; Durnford et al. 1999). Furthermore, the glaucophyte Cyanophora paradoxa contains a polypeptide immunochemically related to this family (Rissler & Durnford 2005). Assuming that this represents genuine homology (and the same authors inferred structural and functional differences between the Cyanophora protein and conventional LHCs so that assumption needs to be made with caution), this indicates monophyly of all three groups. Such an interpretation is subject to the assumptions that lateral transfer has not occurred, and that the present-day cyanobacteria that have been sampled are adequately representative of the plastid ancestors.
(c) Metabolic pathways
The enzymes for individual steps in a metabolic pathway could come from one of three sources: the plastid endosymbiont, the host (whether from the nucleus or mitochondrion), or something else by lateral transfer. Obornik & Green (2005) analysed sequences of enzymes involved in haem biosynthesis. They showed that green algae and plants, the red alga Cyanidioschyzon and the diatom Thalassiosira pseudonana all had a porphobilinogen deaminase of mitochondrial origin and a glutamyl-tRNA synthetase of nuclear origin. This supports monophyly of the green and red lineages, although the possibility of convergent evolution cannot be excluded.
Richards et al. (2006) recently analysed the origin of aro enzymes for the shikimate pathway. The aroA gene provided support for a monophyletic origin of red and green chloroplasts, as the gene for both these lineages showed most recent common ancestry with the beta- and gamma-proteobacteria, rather than the cyanobacteria. This indicates a similar lateral gene transfer event in both lineages. However, not all genes supported a monophyletic origin. For example, aroK/L placed the green lineage separately from Cyanidioschyzon and Thalassiosira within the cyanobacteria.
(d) Import pathways
Plastids need a machinery for the import of nuclear-encoded proteins. Elements of this (such as Toc75) were probably derived from the endosymbiont itself, as there are homologues in cyanobacterial genomes. At least two proteins (Tic110 and Toc34) are present in red algae and the green lineage (and in the case of Tic110 also in glaucophytes) but do not have obvious homologues in cyanobacterial genomes (McFadden & van Dooren 2004; Steiner et al. 2005). These data therefore support monophyly. However, this conclusion is subject to the concern over how representative present-day cyanobacteria are of the ancestral organisms, especially as independent losses of some proteins involved in import have taken place in a number of lineages (McFadden & van Dooren 2004). Thus, it might be that ancestral cyanobacteria had Tic110 and Toc34 proteins, which have been lost from present-day cyanobacteria. The recent observation that an Arabidopsis mitochondrial outer membrane protein (mtOM64) and the chloroplast outer envelope receptor protein TOC64 share sequence similarity (Lister et al. 2007) also highlights the danger in inferring a common origin of organelles based on a limited number of shared homologous import proteins.
6. What really happens? The plastid as a shopping bag
Taken together, the available data provide some support for a monophyletic origin of primary plastids. However, as outlined above, the interpretation of the data makes assumptions about the reliability of tree reconstruction, how representative present-day cyanobacteria are and the extent of convergent evolution. The discussion so far has regarded endosymbiosis as the product of a single host organism taking up a single symbiont and establishing a long-term relationship with gene transfer from the symbiont to the host, whether happening on a single occasion or on multiple independent ones. This may well be an oversimplification of what actually happens. It seems unlikely that the stable symbiont ultimately acquired by the host cell would be the first one it had ever acquired. The acquisition would almost certainly have been preceded by the uptake of other photosynthetic organisms. Some of these would have been lysed very quickly. Others might have persisted for a while and succeeded in dividing a few times. Although the plant plastid division system is of bacterial origin (Aldridge et al. 2005; and presumably deriving from the symbiont) it is unlikely that there would have been close synchrony of division with the host at an early stage. The Nephroselmis symbiont of the flagellate Hatena, which is suggested to be an endosymbiont in the process of establishment, shows a similar lack of synchrony (Okamoto & Inouye 2005). So in many cases, the first few rounds of division would have ended with the loss of the would-be symbiont, probably with lysis of a number of symbiont cells. We know that there is a remarkably high rate of transfer of DNA from organelle to host (Huang et al. 2003; Stegemann et al. 2003), and even in the apparent absence of selection for its retention inserted DNA can persist for significant periods of time, of the order of a million years (Huang et al. 2005; Matsuo et al. 2005). Mutation and rearrangement of inserted sequences occurs (Noutsos et al. 2005) and this can lead to functional activation in the nucleus of genes carrying plastid promoters (Stegemann & Bock 2006) and acquisition of plastid-targeting sequences (Ueda et al. 2006). The transfer of DNA from plastids to nucleus appears to be mediated by plastid lysis (Lister et al. 2003) as organisms that have a single essential plastid have a much lower rate of transfer of plastid DNA to the nucleus. Furthermore, uptake of DNA into the nucleus from the cytosol seems to be a typical feature of eukaryotic cells. So it seems highly probable that the early rounds of failed endosymbiosis, with some would-be endosymbionts eventually lysing and liberating DNA into the cytosol, would result in integration of endosymbiont DNA into the nuclear genome (a process known as ‘endosymbiotic gene transfer’; Dagan & Martin 2006). This DNA would have persisted in the nucleus for a period of time, even if there were no longer functional symbionts in the host cytoplasm. If, finally, a symbiont were able to establish a balanced relationship with the host, the reservoir of sequences in the host nucleus that were derived from previous photosynthetic organisms would have provided a pool of sequences to encode proteins to be imported into the newly established plastid (in addition to sequences transferred to the nucleus from the newly established plastid itself).
The final plastid would therefore have a hybrid origin. Although at least one of the membranes surrounding it was derived from a single organism that entered the host at a defined time, the protein complement would have had mixed origins. The majority of proteins would have originated from the successful symbiont, but some would have originated from its unsuccessful predecessors. It is therefore misleading to refer to the plastid as being derived from a single endosymbiont. We have previously coined the terms ‘polysymbiosis’ (Larkum 2007) or the ‘shopping bag’ model to describe this proposal (Larkum et al. 2007). A shopping bag may have come from a particular store, and the same may be true for most of the contents. However, some of the articles (in this case, the genes) have a different origin and we cannot say that the shopping collectively came from a single place.
7. Predictions of the shopping bag model
The shopping bag model makes a number of predictions. Gene acquisition by the nucleus begins before the symbiont becomes ‘locked in’ to the host. So we predict that organisms that do not currently have stable endosymbionts will nevertheless have genes in their nucleus from failed would-be symbionts. These nuclear genes of symbiont origin may be from organisms still present in the host cytoplasm (but which may eventually be lost) or from organisms that were lost some time ago. The report that the nuclear genome of the sea slug Elysia crispata contains a gene for the fucoxanthin- and chlorophyll-binding protein from the algae that the slug eats (and whose chloroplasts remain functional in gut cells for some time) is consistent with the predictions of the model (Pierce et al. 2003). We predict that organisms such as Paulinella and Rhopalodia will have chromatophore- and spheroid body-derived sequences in their nuclear genomes, even though the symbionts probably retain their own complete genomes. Note that although foreign genes might be expected in the nuclei of phagotrophic organisms by simple lateral transfer, under our model the foreign genes would predominantly be derived from would-be symbionts.
A second prediction is that nuclear genes for plastid proteins may come from different sources as determined by phylogenetic analysis. This may be true for many nuclear genes encoding proteins in other compartments as well, as it has been shown that many Arabidopsis genes for proteins functioning in compartments other than the plastid are of cyanobacterial (i.e. symbiont) origin (Martin et al. 2002). Given the time that has elapsed since the origin(s) of primary chloroplasts, and the range of substitution processes occurring during that time (as discussed previously), it might be thought unlikely that there would be sufficient resolution to distinguish reliably the different origins of nuclear sequences for plastid proteins. However, recent reports suggest that some nuclear genes for plastid proteins are of Chlamydial origin (Huang & Gogarten 2007; Tyra et al. 2007). These observations suggest a transient endosymbiosis involving Chlamydia-like bacteria as well as photosynthetic bacteria in the evolution of the chloroplast, in accordance with the shopping bag model.
The best chance of detecting differences in origin of nuclear genes for plastid proteins would probably come with organisms that had acquired a plastid more recently, i.e. a secondary or a tertiary plastid, and retain phagocytosis. One such example comes from the chlorarachniophyte, Bigelowiella natans, which has a green chloroplast related to that of green algae (and with an independent origin from the Euglena plastid; Rogers et al. 2007). Phylogenetic analysis of a collection of cDNAs for plastid-located proteins of Bigelowiella indicated that although the majority of these genes appeared to be of chlorophyte green algal origin, 21% appeared to have a different origin, with the majority of the exceptions (where the origin was identifiable) being from red algae (Archibald et al. 2003). A very small number of the exceptions were identified as being from bacteria. The varied nature of the origin of these genes was interpreted as lateral gene transfer reflecting the phagotrophic nature of Bigelowiella. Under the shopping bag model, some of these instead represent genes transferred from transient endosymbionts. The fact that the majority of these genes are from photosynthetic organisms is consistent with their arising from a transient endosymbiosis rather than simple uptake of DNA from degraded bacterial prey.
Similarly, the genome of the diatom Thalassiosira pseudonana contains several hundred sequences that are homologous to red algal proteins, but not to green plant ones, and vice versa (Armbrust et al. 2004). This is again consistent with the predictions of the shopping bag model.
The dinoflagellate algae also fulfil the predictions of the shopping bag model. This group of organisms has undergone a large number of plastid replacements. The common ancestor of this group and the Apicomplexa (which contains the important pathogens Theileria, Plasmodium and Toxoplasma) was photosynthetic, with a secondary plastid of probable red algal origin (Wilson 2005; Sanchez-Puerta et al. 2007; Stelter et al. 2007; Moore et al. 2008). In the Apicomplexa, the plastid was retained but its genome was greatly reduced following the loss of photosynthesis (Barbrook et al. 2006a). In the dinoflagellates, the plastid has been lost completely on a number of separate occasions, being replaced in as many as five different lineages with plastids derived from a range of photosynthetic eukaryotes including haptophytes and green algae (Watanabe et al. 1987; Nosenko et al. 2006). The reason why so many different lineages of dinoflagellates should have undergone plastid loss is not clear, but it may be due to their unusual plastid genome organization. While the plastid genome in most organisms comprises a hundred or more genes physically linked on the same molecule, in dinoflagellates most of the chloroplast genes have been transferred to the nucleus (Koumandou et al. 2004; Barbrook et al. 2006b). Most of those that are retained encode subunits of the complexes involved in the light reactions of photosynthesis, together with rRNAs and a limited number of tRNAs (table 2; Barbrook et al. 2006b). Rather than being physically linked on the same molecule, the genes are present on small plasmids, known as minicircles, typically in the region of 3 kbp in size, and with a single gene, although in some cases a few genes, on each minicircle (Zhang et al. 1999; Barbrook & Howe 2000; Nisbet et al. 2004; Barbrook et al. 2006b). Each minicircle has a conserved core region that is believed to function as an origin of replication and also transcription (Nisbet et al. 2008). The copy number of the minicircles appears to vary widely with growth phase, ranging from a few molecules per cell during periods of rapid growth, to a hundred or more during periods of very slow growth (table 3; Koumandou & Howe 2007). This may reflect a loose coupling of chloroplast DNA replication to cell division. A consequence of this gene arrangement and loose control of replication may be the relatively frequent loss of genes essential to photosynthesis, and thus photosynthesis itself (Green 2004; Howe et al. 2008).
Table 2.
Genes identified on dinoflagellate chloroplast minicircles, modified from Barbrook et al. (2006b).
| photosystem I | photosystem II | cytochrome b6f complex | ATP synthase | tRNA | rRNA |
|---|---|---|---|---|---|
| psaA | psbA | petB | atpA | f-Met | LSU |
| psaB | psbB | petD | atpB | Pro | SSU |
| psbC | Trp | ||||
| psbD | |||||
| psbE | |||||
| psbI |
Table 3.
Variation in copy number of dinoflagellate chloroplast gene minicircles with growth phase (Koumandou & Howe 2007).
| minicircle | exponential growth | slow growth |
|---|---|---|
| atpB | 4 | 420 |
| petD | 2 | 50 |
| psbB | 4 | 200 |
| psbDEI | 2 | 170 |
| 23S rRNA | 3 | 300 |
As predicted under the shopping bag model, the dinoflagellates Karenia brevis and Karlodinium micrum, which have tertiary plastids, both have plastid-targeted proteins of multiple origins (Nosenko et al. 2006; Patron et al. 2006). Thus, the tertiary plastid of Karlodinium is haptophyte in origin and many nuclear genes for plastid-targeted proteins are most closely related to other haptophyte sequences, indicating their origin from the tertiary endosymbiont. However, there are also many genes for plastid-targeted proteins that are most closely related to those found in other dinoflagellates, indicating an origin from the secondary endosymbiont.
8. Summary and conclusions
To conclude, there is some support for a monophyletic origin of plastids, but some analyses indicate polyphyly. To differentiate more reliably, we need to have better models for the evolution of the sequences used for phylogenetic analysis. These models will necessarily include information from biochemistry and structural biology. We also need to have a better understanding of the evolution of cyanobacteria, and in particular whether present-day examples represent anciently diverged lineages or the results of more recent divergences following population bottlenecks. In some instances, particularly involving secondary and tertiary plastids, it is not accurate to regard the plastid we see as the result of a single endosymbiotic event (albeit followed by cellular remodelling, such as the movement of DNA to the nucleus). Instead, the plastid is a chimaera. Even though the membrane systems delineating them may be traceable to a single ancestor the protein complement reflects the culmination of a series of symbioses, of varying degrees of transience. The same chimeric nature may be true of primary plastids as well.
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council, the Leverhulme Trust, the University of Cambridge Newton Trust and the Biochemical Society for financial support. We thank two anonymous referees and a number of contributors to the discussion for their comments, which have been incorporated into the published version of the manuscript.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘Photosynthetic and atmospheric evolution’.
Supplementary Material
References
- Aldridge C, Maple J, Møeller S.G. The molecular biology of plastid division in higher plants. J. Exp. Bot. 2005;56:1061–1077. doi: 10.1093/jxb/eri118. doi:10.1093/jxb/eri118 [DOI] [PubMed] [Google Scholar]
- Allen J.F. The function of genomes in bioenergetic organelles. Phil. Trans. R. Soc. B. 2003;358:19–37. doi: 10.1098/rstb.2002.1191. doi:10.1098/rstb.2002.1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald J.M, Rogers M.B, Toop M, Ishida K.-I, Keeling P.J. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc. Natl Acad. Sci. USA. 2003;100:7678–7683. doi: 10.1073/pnas.1230951100. doi:10.1073/pnas.1230951100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust E.V, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. doi:10.1126/science.1101156 [DOI] [PubMed] [Google Scholar]
- Barbrook A.C, Howe C.J. Minicircular plastid DNA in the dinoflagellate Amphidinium operculatum. Mol. Gen. Genet. 2000;263:152–158. doi: 10.1007/s004380050042. doi:10.1007/s004380050042 [DOI] [PubMed] [Google Scholar]
- Barbrook A.C, Lockhart P.J, Howe C.J. Phylogenetic analysis of plastid origins based on secA sequences. Curr. Genet. 1998;34:336–341. doi: 10.1007/s002940050404. doi:10.1007/s002940050404 [DOI] [PubMed] [Google Scholar]
- Barbrook A.C, Howe C.J, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006a;11:101–108. doi: 10.1016/j.tplants.2005.12.004. doi:10.1016/j.tplants.2005.12.004 [DOI] [PubMed] [Google Scholar]
- Barbrook A.C, Santucci N, Plenderleith L.J, Hiller R.G, Howe C.J. Comparative analysis of dinoflagellate chloroplast genomes reveals rRNA and tRNA genes. BMC Genomics. 2006b;7:297. doi: 10.1186/1471-2164-7-297. doi:10.1186/1471-2164-7-297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canback B, Tamas I, Andersson S.G.E. A phylogenomic study of endosymbiotic bacteria. Mol. Biol. Evol. 2004;21:1110–1122. doi: 10.1093/molbev/msh122. doi:10.1093/molbev/msh122 [DOI] [PubMed] [Google Scholar]
- Dagan T, Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. doi:10.1186/gb-2006-7-10-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S, et al. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410:1091–1096. doi: 10.1038/35074092. doi:10.1038/35074092 [DOI] [PubMed] [Google Scholar]
- Durnford D.G, Deane J.A, Tan S, McFadden G.I, Gantt E, Green B.R. A phylogenetic assessment of the eukaryotic light-harvesting antenna proteins, with implications for plastid evolution. J. Mol. Evol. 1999;48:59–68. doi: 10.1007/pl00006445. doi:10.1007/PL00006445 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Sys. Zool. 1978;27:401–410. doi:10.2307/2412923 [Google Scholar]
- Galtier N. Maximum-likelihood phylogenetic analysis under a covarion-like model. Mol. Biol. Evol. 2001;18:866–873. doi: 10.1093/oxfordjournals.molbev.a003868. [DOI] [PubMed] [Google Scholar]
- Gilson P.R, Su V, Slamovits C.H, Reith M.E, Keeling P.J, McFadden G.I. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature's smallest nucleus. Proc. Natl Acad. Sci. USA. 2006;103:9566–9571. doi: 10.1073/pnas.0600707103. doi:10.1073/pnas.0600707103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B.R. The chloroplast genome of dinoflagellates—a reduced instruction set? Protist. 2004;155:23–31. doi: 10.1078/1434461000161. doi:10.1078/1434461000161 [DOI] [PubMed] [Google Scholar]
- Hart S.E, Schlarb-Ridley B.G, Bendall D.S, Howe C.J. Terminal oxidases of cyanobacteria. Biochem. Soc. Trans. 2005;33:832–835. doi: 10.1042/BST0330832. doi:10.1042/BST0330832 [DOI] [PubMed] [Google Scholar]
- Hendy M.D, Penny D. A framework for the quantitative study of evolutionary trees. Sys. Zool. 1989;38:297–309. doi:10.2307/2992396 [Google Scholar]
- Howe C.J, Barbrook A.C, Koumandou V.L, Nisbet R.E.R, Symington H.A, Wightman T.F. Evolution of the chloroplast genome. Phil. Trans. R. Soc. B. 2003;358:99–106. doi: 10.1098/rstb.2002.1176. doi:10.1098/rstb.2002.1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe C.J, Nisbet R.E.R, Barbrook A.C. The remarkable chloroplast genome of dinoflagellates. J. Exp. Bot. 2008;59:1035–1045. doi: 10.1093/jxb/erm292. doi:10.1093/jxb/erm292 [DOI] [PubMed] [Google Scholar]
- Huang J, Gogarten J.P. Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol. 2007;8:R99. doi: 10.1186/gb-2007-8-6-r99. doi:10.1186/gb-2007-8-6-r99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.Y, Ayliffe M.A, Timmis J.N. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature. 2003;422:72–76. doi: 10.1038/nature01435. doi:10.1038/nature01435 [DOI] [PubMed] [Google Scholar]
- Huang C.Y, Gruenheit N, Ahmadinejad N, Timmis J.N, Martin W. Mutational decay and age of chloroplast and mitochondrial genomes transferred recently to angiosperm nuclear chromosomes. Plant Physiol. 2005;138:1723–2733. doi: 10.1104/pp.105.060327. doi:10.1104/pp.105.060327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J.P. Testing a covariotide model of DNA substitution. Mol. Biol. Evol. 2002;19:698–707. doi: 10.1093/oxfordjournals.molbev.a004128. [DOI] [PubMed] [Google Scholar]
- Jermiin L.S, Ho S.Y.W, Ababneh F, Robinson J, Larkum A.W.D. The biasing effect of compositional heterogeneity on phylogenetic estimates may be underestimated. Sys. Biol. 2004;53:639–643. doi: 10.1080/10635150490468648. doi:10.1080/10635150490468648 [DOI] [PubMed] [Google Scholar]
- Koumandou V.L, Howe C.J. The copy number of chloroplast gene minicircles changes dramatically with growth phase in the dinoflagellate Amphidinium operculatum. Protist. 2007;158:89–103. doi: 10.1016/j.protis.2006.08.003. doi:10.1016/j.protis.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Koumandou V.L, Nisbet R.E.R, Barbrook A.C, Howe C.J. Dinoflagellate chloroplasts—where have all the genes gone? Trends Genet. 2004;20:261–267. doi: 10.1016/j.tig.2004.03.008. doi:10.1016/j.tig.2004.03.008 [DOI] [PubMed] [Google Scholar]
- Larkum, A. W. D. 2007 Evolution of the reaction centers and photosystems. In Primary processes of photosynthesis: principles and apparatus, vol. 2 (ed. G. Renger), pp. 489–521. Cambridge, UK: Royal Society of Chemistry.
- Larkum A.W.D, Lockhart P.J, Howe C.J. Shopping for plastids. Trends Plant Sci. 2007;12:189–195. doi: 10.1016/j.tplants.2007.03.011. doi:10.1016/j.tplants.2007.03.011 [DOI] [PubMed] [Google Scholar]
- Lister D.L, Bateman J.M, Purton S, Howe C.J. DNA transfer from chloroplast to nucleus is much rarer in Chlamydomonas than in tobacco. Gene. 2003;316:33–38. doi: 10.1016/s0378-1119(03)00754-6. doi:10.1016/S0378-1119(03)00754-6 [DOI] [PubMed] [Google Scholar]
- Lister R, Carrie C, Duncan O, Ho L.H.M, Howell K.A, Murcha M.W, Whelan J. Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell. 2007;19:3739–3759. doi: 10.1105/tpc.107.050534. doi:10.1105/tpc.107.050534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart P.J, Steel M.A. A tale of two processes. Syst. Biol. 2005;54:948–951. doi: 10.1080/10635150500234682. doi:10.1080/10635150500234682 [DOI] [PubMed] [Google Scholar]
- Lockhart P.J, Howe C.J, Bryant D.A, Beanland T.J, Larkum A.W.D. Substitutional bias confounds inference of cyanelle origins from sequence data. J. Mol. Evol. 1992;34:153–162. doi: 10.1007/BF00182392. doi:10.1007/BF00182392 [DOI] [PubMed] [Google Scholar]
- Lockhart P.J, Steel M.A, Hendy M.D, Penny D. Recovering evolutionary trees under a more realistic model of sequence evolution. Mol. Biol. Evol. 1994;11:605–612. doi: 10.1093/oxfordjournals.molbev.a040136. [DOI] [PubMed] [Google Scholar]
- Lockhart P.J, Steel M.A, Barbrook A.C, Huson D.H, Charleston M.A, Howe C.J. A covariotide model explains apparent phylogenetic structure of oxygenic photosynthetic lineages. Mol. Biol. Evol. 1998;15:1183–1188. doi: 10.1093/oxfordjournals.molbev.a026025. [DOI] [PubMed] [Google Scholar]
- Lockhart P.J, Howe C.J, Barbrook A.C, Larkum A.W.D, Penny D. Spectral analysis, systematic bias, and the evolution of chloroplasts. Mol. Biol. Evol. 1999;16:573–576. [Google Scholar]
- Lockhart P.J, Huson D, Maier U, Fraunholz M.J, Van de Peer Y, Barbrook A.C, Howe C.J, Steel M.A. How molecules evolve in eubacteria. Mol. Biol. Evol. 2000;17:835–838. doi: 10.1093/oxfordjournals.molbev.a026363. [DOI] [PubMed] [Google Scholar]
- Lockhart P, Novis P, Milligan B.G, Riden J, Rambaut A, Larkum T. Heterotachy and tree building: a case study with plastids and eubacteria. Mol. Biol. Evol. 2006;23:40–45. doi: 10.1093/molbev/msj005. doi:10.1093/molbev/msj005 [DOI] [PubMed] [Google Scholar]
- Marin B, Nowack E.C.M, Melkonian M. A plastid in the making: evidence for a second primary endosymbiosis. Protist. 2005;156:425–432. doi: 10.1016/j.protis.2005.09.001. doi:10.1016/j.protis.2005.09.001 [DOI] [PubMed] [Google Scholar]
- Martin W, Kowallik K.V. Annotated English translation of Mereschkowsky's 1905 paper ‘Uber Natur und Ursprung der Chromatophoren im Pflanzenreiche’. Eur. J. Phycol. 1999;34:287–295. [Google Scholar]
- Martin W, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl Acad. Sci. USA. 2002;99:12 246–12 251. doi: 10.1073/pnas.182432999. doi:10.1073/pnas.182432999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M, Ito Y, Yamauchi R, Obokata J. The rice nuclear genome continuously integrates, shuffles, and eliminates the chloroplast genome to cause chloroplast–nuclear DNA flux. Plant Cell. 2005;17:665–675. doi: 10.1105/tpc.104.027706. doi:10.1105/tpc.104.027706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden G.I, van Dooren G. Evolution: red algal genome affirms a common origin of all plastids. Curr. Biol. 2004;14:R514–R516. doi: 10.1016/j.cub.2004.06.041. doi:10.1016/j.cub.2004.06.041 [DOI] [PubMed] [Google Scholar]
- Mereschkowsky C. Ueber Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol. Centralbl. 1905;25:593–604. [Google Scholar]
- Morden C.W, Delwiche C.F, Kuhsel M, Palmer J.D. Gene phylogenies and the endosymbiotic origin of plastids. BioSystems. 1992;28:75–90. doi: 10.1016/0303-2647(92)90010-v. doi:10.1016/0303-2647(92)90010-V [DOI] [PubMed] [Google Scholar]
- Moore R.B, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–963. doi: 10.1038/nature06635. doi:10.1038/nature06635 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, et al. Complete genome structure of Gloeobacter violaceus PCC7421, a cyanobacterium that lacks thylakoids. DNA Res. 2003;10(Suppl.):181–201. doi: 10.1093/dnares/10.4.181. doi:10.1093/dnares/10.4.181 [DOI] [PubMed] [Google Scholar]
- Nisbet R.E.R, Koumandou V.L, Barbrook A.C, Howe C.J. Novel plastid gene minicircles in the dinoflagellate Amphidinium operculatum. Gene. 2004;331:141–147. doi: 10.1016/j.gene.2004.02.008. doi:10.1016/j.gene.2004.02.008 [DOI] [PubMed] [Google Scholar]
- Nisbet R.E.R, Hiller R.G, Barry E.R, Skene P, Barbrook A.C, Howe C.J. Transcript analysis of dinoflagellate plastid gene minicircles. Protist. 2008;159:31–39. doi: 10.1016/j.protis.2007.07.002. doi:10.1016/j.protis.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Nosenko T, Lidie K.L, Van Dolah F.M, Lindquist E, Cheng J.-F, Bhattacharya D. Chimeric plastid proteome in the Florida ‘Red Tide’ dinoflagellate Karenia brevis. Mol. Biol. Evol. 2006;23:2026–2038. doi: 10.1093/molbev/msl074. doi:10.1093/molbev/msl074 [DOI] [PubMed] [Google Scholar]
- Noutsos C, Richly E, Leister D. Generation and evolutionary fate of insertions of organelle DNA in the nuclear genomes of flowering plants. Genome Res. 2005;15:616–628. doi: 10.1101/gr.3788705. doi:10.1101/gr.3788705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki H, Iseki M, Hasegawa M, Misawa K, Nakada T, Sasaki N, Watanabe M. Phylogeny of primary photosynthetic eukaryotes as deduced from slowly evolving nuclear genes. Mol. Biol. Evol. 2007;24:1592–1595. doi: 10.1093/molbev/msm091. doi:10.1093/molbev/msm091 [DOI] [PubMed] [Google Scholar]
- Obornik M, Green B.R. Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol. Biol. Evol. 2005;22:2343–2353. doi: 10.1093/molbev/msi230. doi:10.1093/molbev/msi230 [DOI] [PubMed] [Google Scholar]
- Okamoto N, Inouye I. A secondary symbiosis in progress? Science. 2005;310:287. doi: 10.1126/science.1116125. doi:10.1126/science.1116125 [DOI] [PubMed] [Google Scholar]
- Palmer J.D. A single birth of all plastids? Nature. 2000;405:32–33. doi: 10.1038/35011184. doi:10.1038/35011184 [DOI] [PubMed] [Google Scholar]
- Patron N.J, Waller R.F, Keeling P.J. A tertiary plastid uses genes from two endosymbionts. J. Mol. Biol. 2006;357:1371–1382. doi: 10.1016/j.jmb.2006.01.084. doi:10.1016/j.jmb.2006.01.084 [DOI] [PubMed] [Google Scholar]
- Pierce S.K, Massey S.E, Hanten J.J, Curtis N.E. Horizontal transfer of functional nuclear genes between multicellular organisms. Biol. Bull. 2003;204:237–240. doi: 10.2307/1543594. doi:10.2307/1543594 [DOI] [PubMed] [Google Scholar]
- Prechtl J, Kneip C, Lockhart P, Wenderoth K, Maier U.-G. Intracellular spheroid bodies of Rhopalodia gibba have nitrogen-fixing apparatus of cyanobacterial origin. Mol. Biol. Evol. 2004;21:1477–1481. doi: 10.1093/molbev/msh086. doi:10.1093/molbev/msh086 [DOI] [PubMed] [Google Scholar]
- Raven P.H. A multiple origin for plastids and mitochondria. Science. 1970;169:641–646. doi: 10.1126/science.169.3946.641. doi:10.1126/science.169.3946.641 [DOI] [PubMed] [Google Scholar]
- Reith M, Munholland J. Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol. Biol. Rep. 1995;13:333–335. doi:10.1007/BF02669187 [Google Scholar]
- Reyes-Prieto A, Weber A.P.M, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Ann. Rev. Genet. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. doi:10.1146/annurev.genet.41.110306.130134 [DOI] [PubMed] [Google Scholar]
- Richards T.A, Dacks J.B, Campbell S.A, Blanchard J.L, Foster P.G, McLeod R, Roberts C.W. Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot. Cell. 2006;5:1517–1531. doi: 10.1128/EC.00106-06. doi:10.1128/EC.00106-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissler H.M, Durnford D.G. Isolation of a novel carotenoid-rich protein in Cyanophora paradoxa that is immunologically related to the light-harvesting complexes of photosynthetic eukaryotes. Plant Cell Physiol. 2005;46:416–424. doi: 10.1093/pcp/pci054. doi:10.1093/pcp/pci054 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Brinkmann H, Burey S.C, Roure B, Burger G, Loffelhardt W, Bohnert H.J, Philippe H, Lang B.F. Monophyly of primary photosynthetic eukaryotes: green plants, red algae and glaucophytes. Curr. Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. doi:10.1016/j.cub.2005.06.040 [DOI] [PubMed] [Google Scholar]
- Rogers M.B, Gilson P.R, Su V, McFadden G.I, Keeling P.J. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol. Biol. Evol. 2007;24:54–62. doi: 10.1093/molbev/msl129. doi:10.1093/molbev/msl129 [DOI] [PubMed] [Google Scholar]
- Sanchez-Puerta M.V, Lippmeier J.C, Apt K.E, Delwiche C.F. Plastid genes in a non-photosynthetic dinoflagellate. Protist. 2007;158:105–117. doi: 10.1016/j.protis.2006.09.004. doi:10.1016/j.protis.2006.09.004 [DOI] [PubMed] [Google Scholar]
- Schimper A.F.W. Ueber die entwickelung der chlorophyllkoerner und farbkoerper. Bot. Zeit. 1883;41:105–113. [Google Scholar]
- Shalchian-Tabrizi K, Skanseng M, Ronquist F, Klaveness D, Bachvaroff T.R, Delwiche C.F, Botnen A, Tengs T, Jakobsen K.S. Heterotachy processes in rhodophyte-derived secondhand plastid genes: implications for addressing the origin and evolution of dinoflagellate plastids. Mol. Biol. Evol. 2006;23:1504–1515. doi: 10.1093/molbev/msl011. doi:10.1093/molbev/msl011 [DOI] [PubMed] [Google Scholar]
- Spade P.V, editor. The cambridge companion to ockham. Cambridge University Press; Cambridge, UK: 1999. [Google Scholar]
- Stegemann S, Bock R. Experimental reconstruction of functional gene transfer from the tobacco plastid genome to the nucleus. Plant Cell. 2006;18:2869–2878. doi: 10.1105/tpc.106.046466. doi:10.1105/tpc.106.046466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann S, Hartmann S, Ruf S, Bock R. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc. Natl Acad. Sci. USA. 2003;100:8828–8833. doi: 10.1073/pnas.1430924100. doi:10.1073/pnas.1430924100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner J.M, Yusa F, Pompe J.A, Loeffelhardt W. Homologous protein import machineries in chloroplasts and cyanelles. Plant J. 2005;44:646–652. doi: 10.1111/j.1365-313X.2005.02559.x. doi:10.1111/j.1365-313X.2005.02559.x [DOI] [PubMed] [Google Scholar]
- Stelter K, El-Sayed N.M, Seeber F. The expression of a plant-type ferredoxin redox system provides molecular evidence for a plastid in the early dinoflagellate Perkinsus marinus. Protist. 2007;158:119–130. doi: 10.1016/j.protis.2006.09.003. doi:10.1016/j.protis.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Stiller J.W. Weighing the evidence for a single origin of plastids. J. Phycol. 2003;39:1283–1285. doi:10.1111/j.0022-3646.2003.03-084.x [Google Scholar]
- Stiller J.W. Plastid endosymbiosis, genome evolution and the origin of green plants. Trends Plant Sci. 2007;12:391–396. doi: 10.1016/j.tplants.2007.08.002. doi:10.1016/j.tplants.2007.08.002 [DOI] [PubMed] [Google Scholar]
- Stiller J.W, Reel D.C, Johnson J.C. A single origin of plastids revisited: convergent evolution in organellar genome content. J. Phycol. 2003;39:95–105. doi:10.1046/j.1529-8817.2003.02070.x [Google Scholar]
- Theissen U, Martin W. The difference between organelles and endosymbionts. Curr. Biol. 2006;16:R1016–R1017. doi: 10.1016/j.cub.2006.11.020. doi:10.1016/j.cub.2006.11.020 [DOI] [PubMed] [Google Scholar]
- Tomitani A, Knoll A.H, Cavanaugh C.M, Ohno T. The evolutionary diversification of cyanobacteria: molecular-phylogenetic and paleontological perspectives. Proc. Natl Acad. Sci. USA. 2006;103:5442–5447. doi: 10.1073/pnas.0600999103. doi:10.1073/pnas.0600999103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyra H.M, Linka M, Weber A.P.M, Bhattacharya D. Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biol. 2007;8:R212. doi: 10.1186/gb-2007-8-10-r212. doi:10.1186/gb-2007-8-10-r212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Fujimoto M, Arimura S, Tsutsumi N, Kadowaki K. Evidence for transit peptide acquisition through duplication and subsequent frameshift mutation of a preexisting protein gene in rice. Mol. Biol. Evol. 2006;23:2405–2412. doi: 10.1093/molbev/msl112. doi:10.1093/molbev/msl112 [DOI] [PubMed] [Google Scholar]
- Watanabe M.M, Takeda Y, Sasa T, Inouye I, Suda S, Sawaguchi T, Chihara M. A green dinoflagellate with chlorophylls A and B: morphology, fine structure of the chloroplast and chlorophyll composition. J. Phycol. 1987;23:382–389. doi:10.1111/j.1529-8817.1987.tb04148.x [Google Scholar]
- Wilson R.J.M. Parasite plastids: approaching the end-game. Biol. Rev. 2005;80:129–153. doi: 10.1017/s1464793104006591. doi:10.1017/S1464793104006591 [DOI] [PubMed] [Google Scholar]
- Wolfe G.R, Cunningham F.X, Durnford D, Green B.R, Gantt E. Evidence for a common origin of chloroplasts with light-harvesting complexes of different pigmentation. Nature. 1994;367:566–568. doi:10.1038/367566a0 [Google Scholar]
- Yoon H.S, Reyes-Prieto A, Melkonian M, Bhattacharya D. Minimal plastid genome evolution in the Paulinella endosymbiont. Curr. Biol. 2006;16:R670–R672. doi: 10.1016/j.cub.2006.08.018. doi:10.1016/j.cub.2006.08.018 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Green B.R, Cavalier-Smith T. Single gene circles in dinoflagellate chloroplast genomes. Nature. 1999;400:155–159. doi: 10.1038/22099. doi:10.1038/22099 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.