Abstract
‘Replaying the tape’ is an intriguing ‘would it happen again?’ exercise. With respect to broad evolutionary innovations, such as photosynthesis, the answers are central to our search for life elsewhere. Photosynthesis permits a large planetary biomass on Earth. Specifically, oxygenic photosynthesis has allowed an oxygenated atmosphere and the evolution of large metabolically demanding creatures, including ourselves. There are at least six prerequisites for the evolution of biological carbon fixation: a carbon-based life form; the presence of inorganic carbon; the availability of reductants; the presence of light; a light-harvesting mechanism to convert the light energy into chemical energy; and carboxylating enzymes. All were present on the early Earth. To provide the evolutionary pressure, organic carbon must be a scarce resource in contrast to inorganic carbon. The probability of evolving a carboxylase is approached by creating an inventory of carbon-fixation enzymes and comparing them, leading to the conclusion that carbon fixation in general is basic to life and has arisen multiple times. Certainly, the evolutionary pressure to evolve new pathways for carbon fixation would have been present early in evolution. From knowledge about planetary systems and extraterrestrial chemistry, if organic carbon-based life occurs elsewhere, photosynthesis—although perhaps not oxygenic photosynthesis—would also have evolved.
Keywords: photosynthesis, carbon fixation, carboxylase, evolution, rubisco, enzyme evolution
1. Introduction
To an economist, solar power may seem like a boutique product next to oil, but to life on Earth it is by far the most important source of energy. Even fossil fuels are traced to solar energy. Today, an average of 342 W m−2 of solar radiation reaches our atmosphere and 198 W m−2 of that reaches the Earth's surface (Nielsen 2005). Thus, it is clear that for organisms inhabiting planets with a source of light such as their Sun, solar power is a functionally limitless source of energy.
While the first life was most probably heterotrophic (reviewed in Fenchel et al. 1998), early on, life on Earth evolved to exploit solar energy. Autotrophy—the biotic production of organic carbon from inorganic sources—allowed life to go beyond a reliance on abiotically produced organic carbon. Using solar radiation as the means to produce the energy for carbon fixation, i.e. photosynthesis, remains a very powerful combination. With the use of H2O as a reductant, the raw materials were available to support aerobic metabolism and energy-intensive species such as ourselves. Today, there is an estimated 600–1000 Gt of living biomass on Earth (Falkowski et al. 2000) supported largely by photosynthesis. Photosynthesis, and possibly oxygenic photosynthesis more specifically, appears critical for maintaining large planetary populations.
It is clear from our vantage point that photosynthesis, especially oxygenic photosynthesis, was the key to the history of life on Earth. But was that the result of contingency—an unpredictable and lucky accident (e.g. Gould 1989)—or, rather, a predictable outcome given the physical, chemical and biological conditions on the early Earth, as might be suggested by Conway Morris (1998)? This question is important to evolutionary biology because it asks whether the field has predictive power; in this case, could we have predicted the evolution of photosynthesis?
The question of whether life exists elsewhere is particularly intriguing, and is thus one of the three fundamental foci of astrobiology. Owing to its importance to life on Earth, the question of whether photosynthesis may—or is likely—to evolve again or elsewhere is fundamental. This question is the focus of this study.
To assess the possibility of photosynthesis arising again requires an inventory of the necessary prerequisites including the following: a carbon-based life form; the presence of inorganic carbon; the availability of reductants; the presence of light; a light-harvesting mechanism to convert the light energy into chemical energy; and carboxylating enzymes. Second, evolutionary pressures should be present, which provide a clear advantage to organisms with this function. Third, there should be, if not data, at least suggestive evidence. This comes in the form of convergence.
2. Autotrophy
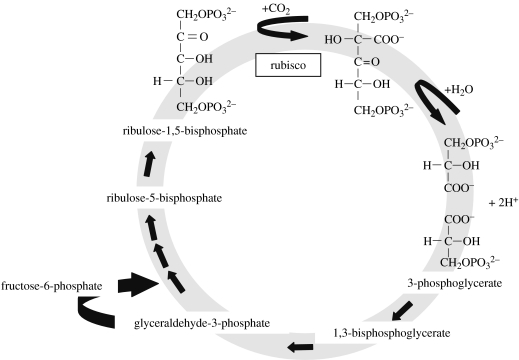
Autotrophs are organisms that can derive some or all of their organic carbon needs from inorganic sources. Autotrophy is widespread among all three domains, including plants, algae, cyanobacteria and even a significant fraction of marine archaea (Ingalls et al. 2006). There are several major pathways for autotrophy (reviewed in Shively & Barton 1991), but the best known and most prevalent one involves the fixation of CO2 by the enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco; EC 4.1.1.39; figure 1).
Figure 1.
Fixation of carbon dioxide by ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco). This step is the first crucial step in the CBB cycle, which is shown in the abridged form.
(a) The origin of autotrophy
When did autotrophy arise? The ‘Great Oxidation Event’ provides a minimum age constraint for oxygenic photosynthesis. This Great Oxidation Event occurred ca 2.4 billion years (Ga), when atmospheric oxygen concentrations rose from less than 10−5 present atmospheric level (PAL) to more than 0.01 PAL or, possibly, more than 0.1 PAL. Geological and theoretical arguments suggest that oxygenic photosynthesis arose at least 300 Myr before the Great Oxidation Event (e.g. Goldblatt et al. 2006; Anbar et al. 2007), although others suggest a greatly compressed interval with cyanobacteria evolving only a million years or so earlier (Kopp et al. 2005). Suggestive of autotrophy arising even earlier, the Archaean fossil record contains cellularly preserved microfossils that are morphologically similar to extant autotrophic organisms, and some are oriented in a way that implies that they were photosynthetic (Walter 1983; Schopf 1992). Stromatolite evidence of erect filaments in shallow isolated basins with insignificant sulphate concentrations in the Tumbiana Formation, Australia, suggests that oxygenic photosynthesis evolved by 2700 Myr ago (Buick 1992). Similarly, the presence of the biological lipids 2α-methylhopanes, characteristic of cyanobacteria, in the Pilbara Craton, Australia, also indicates that oxygenic photosynthesis was well established by 2.7 Ga (Brocks et al. 1999). Filamentous fossils suggesting that either cyanobacteria or other bacteria date to 3.5 Ga (Walsh & Lowe 1985). In the 3.416 Ga Buck Reef Chert of South Africa, there are probably remains of photosynthetic bacteria that used H2 as a reductant (Tice & Lowe 2006). Stable carbon isotope data suggest that carbon fixation evolved by 3.5–3.8 Ga (Schidlowski 1988), and, more recently, carbon isotope data suggesting production by form I rubisco in oxic environments by ca 2900 Myr ago (Nisbet et al. 2007). Rosing & Frei (2004) found organic carbon in a more than 3.7-Ga-old shale from Isua, West Greenland, with a carbon isotopic signature suggesting oxygenic photosynthesis coupled with the presence of oxidized ocean water.
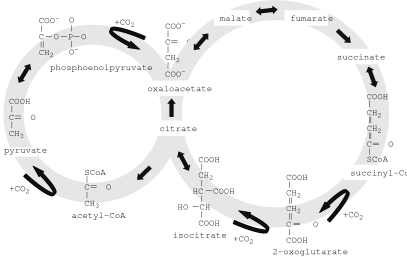
Some have argued that autotrophy is the fundamental process around which life evolved, and thus dates from the origin of life. Wachtershauser (1990) proposed that the primordial metabolism was an autocatalytic cycle that can be derived from the reductive tricarboxylic acid (rTCA) cycle (figure 2) by replacing thioesters by thioacids and by assuming that the required reducing power was obtained from the oxidative formation of pyrite (FeS2). This proposal is appealing because it circumvents the issue of a dilute solution of prebiotic precursors having sufficient concentration to organize into a life form. Smith & Morowitz (2004) pointed out that the rTCA cycle is a network-autocatalytic cycle, which provides the organic building blocks for the synthesis of all major classes of biomolecules. Furthermore, they proposed that the rTCA cycle is favoured relative to other redox reaction pathways under early Earth conditions and that this feature drove its emergence and also accounts for its evolutionary robustness and universality. Thus, subsequent layers of biochemical complexity were built around the rTCA cycle.
Figure 2.
Reverse (reductive) TCA cycle. The rTCA cycle is a carbon dioxide fixation pathway found in autotrophic eubacteria and archaea. It is considered to be a primordial pathway for the production of starting organic molecules for the biosynthesis of sugars, lipids, amino acids, pyrimidines and pyrroles. The rTCA cycle is largely the oxidative, catabolic TCA cycle operating in reverse. Most of the enzymes of the TCA cycle work reversibly and could catalyse both directions. Only three steps are thought to be non-reversible, and thus determine the oxidative or reductive direction of the cycle: the conversion of citrate to oxaloacetate and acetyl-CoA; the conversion of fumarate to succinate; and the conversion of 2-ketoglutarate to isocitrate. The presence of such enzyme activities in autotrophically grown bacteria and archaea is considered indicative of the presence of the rTCA cycle in these organisms.
Persuasive though these proposals might be, only the heterotrophic theory of the origin of life has experimental support because it has been shown that the reduction of CO2 under prebiotic conditions results in the synthesis of organic compounds (Lazcano & Miller 1999). Others (e.g. Orgel 2000) argue that the organization of the rTCA cycle on mineral surfaces makes unreasonable assumptions about the catalytic properties of minerals and the ability of minerals to organize sequences of dissimilar reactions. Clearly, the origin of life on Earth remains highly controversial.
In summary, the origin of oxygenic photosynthesis must have occurred prior to 2.4 Ga, and possibly as far back as the earliest fossil record at 3.7 Ga. Autotrophy has been proposed to be as old as life itself.
(b) Pathways for autotrophy
There are four main pathways for autotrophic carbon fixation: the Calvin–Benson–Bassham (CBB) cycle; the rTCA cycle; the 3-hydroxypropionate cycle; and the reductive acetyl-CoA pathway. In addition, there are two pathways for the assimilation of one-carbon organic compounds: the ribulose monophosphate cycle and the serine pathway. The eukaryotes use only the CBB cycle, whereas, at the other extreme, the Crenarchaeota use three or possibly four autotrophic pathways (Hügler et al. 2003a,b). This diversity of pathways demonstrates that autotrophy has arisen multiple times on the Earth, increasing the likelihood that it would arise elsewhere.
(i) Calvin–Benson–Bassham cycle
The CBB cycle is the most widespread route for autotrophic carbon fixation on Earth. It is the cycle used by all photosynthetic eukaryotes, cyanobacteria, chemoautotrophs such as purple sulphur and non-sulphur bacteria (Shively et al. 1998), and most anoxygenic photolithotrophs. The incorporation of CO2 into cellular materials requires a carboxylase, as well as energy and a reductant, in the case of the CBB cycle, ATP and NADPH, which are obtained from the ‘light reactions’.
The CBB cycle contains three unique enzymes. Ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) fixes CO2 onto the five-carbon sugar, ribulose-1,5-bisphosphate, yielding two molecules of 3-phosophoglycerate through an unstable six-carbon intermediate (figure 1). While the CO2 usually comes from the environment, it can also come from methanol, formaldehyde (e.g. Beijerinckia mobilis: Dedysh et al. 2005) or CO2 produced by CO oxidation. Functional rubisco has been found in all the three domains of life. The other unique enzymes in the CBB cycle are sedoheptulose bisphosphatase (SBPase), which catalyses the dephosphorylation of sedoheptulose-1,7-bisphosphate, and phosphoribulokinase (PRK), which phosphorylates ribulose-5-phosphate (RuMP).
A key to the evolution of rubisco may be found by studying the form IV subfamily of the rubisco large subunit. Although similar in sequence to forms I, II and III, form IV lacks multiple conserved active-site residues, and thus none of these enzymes has been demonstrated to have robust rubisco activity, even when recombinant rubisco-like protein (RLP) was synthesized in E. coli (Hanson & Tabita 2001). Such RLPs have been found in the green sulphur bacteria Chlorobium tepidum and Chlorobium limicola, organisms that use the rTCA cycle for carbon fixation, as well as in Bacillus subtilis and Archaeoglobus fulgidus. Although the RLPs do not appear to fix carbon, the loss of the gene in C. tepidum resulted in a series of effects including a decreased production of bacteriochlorophyll c, a decrease in photoautotrophic growth rate, a decrease in carbon fixation, the accumulation of sulphur grains and the accumulation of two oxidative stress proteins. Hanson & Tabita (2001) hypothesized that these effects were attributable to a defect in the oxidation of sulphur compounds that provide the reducing power for carbon fixation in C. tepidum.
Ashida et al. (2005) studied the RLP of the non-photosynthetic bacterium B. subtilis and found that it was the 2,3-diketo-5-methylthiopentyl-1-phosphate enolase in the methionine salvage pathway. They suggested that photosynthetic rubiscos evolved from RLPs. Ashida et al. (2003) rescued a RLP-deficient B. subtilis with the gene for the photosynthetic rubisco large subunit from Rhodospirillum rubrum. Yet, the question remains: are RLPs precursors to bona fide functional rubisco or do they represent descendant states (Hanson & Tabita 2001)?
(ii) Reductive TCA cycle
The rTCA cycle (reverse Krebs cycle or reverse citric acid cycle) is used by some bacteria to produce carbon compounds from carbon dioxide and water, sometimes using hydrogen or sulphates as electron donors. The reaction is approximately the citric acid cycle run in reverse (figure 2). The reaction is one of the possible candidates for prebiotic early Earth reactions and so is of interest in the origin of life research. Some of the steps can be catalysed by minerals. Among the bacteria, the rTCA cycle is known for at least some Aquificales, Chlorobiales and Proteobacteria including magnetotactic proteobacteria (Williams et al. 2006). The rTCA cycle may predominate over the CBB cycle among autotrophs in hydrothermal vents (Campbell & Cary 2004) and possibly other microaerophilic environments, suggesting that the rTCA cycle is a key route for carbon fixation on the Earth even today.
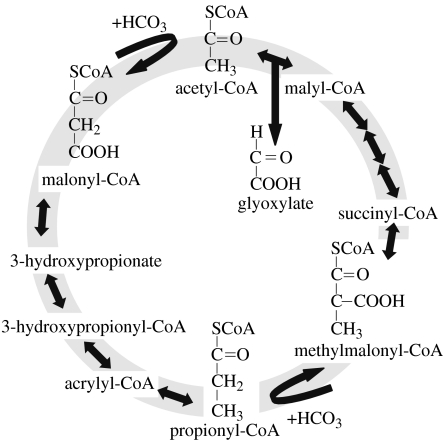
(iii) 3-Hydroxypropionate cycle
The 3-hydroxypropionate cycle (figure 3) is proposed as the pathway for autotrophic carbon fixation in Chloroflexus aurantiacus (Ivanovsky et al. 1993) and for some acidophilic archaebacteria of the phylum Crenarchaeota, such as Acidianus brierleyi (Ishii et al. 1997) and Sulfolobus metallicus (Hügler et al. 2003a,b). For each turn of the 3-hydroxypropionate cycle, two molecules of are fixed into one molecule of glyoxylate (Alber & Fuchs 2002). The thermophilic acidophile Metallosphaera sedula uses a modified 3-hydroxypropionate cycle. In M. sedula, there are ATP-dependent carboxylations of acetyl-CoA and propionyl-CoA, both of which are catalysed by one large enzyme, acetyl-CoA/propionyl-CoA carboxylase (Hügler et al. 2003a,b). The enzyme contains biotin carboxylase and carboxytransferase, and a small biotin carrier protein (Hügler et al. 2003a,b).
Figure 3.
Proposed 3-hydroxypropionate cycle of autotrophic CO2 fixation in the phototrophic green non-sulphur bacterium Chloroflexus aurantiacus. Carboxylating enzymes in the 3-hydroxypropionate cycle include acetyl-CoA carboxylase and propionyl-CoA carboxylase. After Alber & Fuchs (2002).
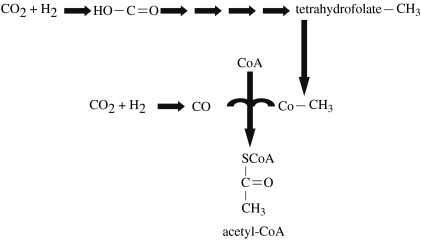
(iv) Reductive acetyl-CoA
The reductive acetyl-CoA pathway is found among many anaerobes in the archaea and bacteria including acetogens, such as Clostridium thermoaceticum, methanogens and most autotrophic sulphate reducers. In the reductive acetyl-CoA pathway, two CO2 are ultimately combined and reduced to form acetyl-CoA (figure 4). In this pathway, one CO2 is captured on tetrahydrofolate and reduced to a methyl group, while the other CO2 is reduced to a carbonyl group (C=O), and then added to the methyl group (Fuchs 1986).
Figure 4.
Reductive acetyl-CoA pathway (Ljungdahl–Wood pathway). Note that there are three carboxylation steps, including the final one where carbon monoxide is added to form acetyl-CoA by acetyl-CoA carboxylase, also known as acetyl-CoA synthase complex. Pathway after http://www.bact.wisc.edu/microtextbook/index.php?name=sections&req=viewarticle&artid=63&allpages=1&theme=printer.
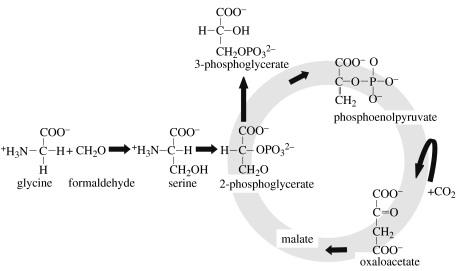
(v) Ribulose monophosphate (RuMP) cycle and serine pathway
There are two known pathways that are used by bacteria for the assimilation of formaldehyde or methane: the serine pathway (figure 5) and the RuMP cycle. Since these C1 compounds are not oxidized to CO2 before assimilation, their fixation is not strictly autotrophic. However, as both are also produced abiotically and even found in space in hot molecular cores and comets (Ehrenfreund & Charnley 2000), these pathways could be considered a way to get abiotically produced organic carbon into the biosphere. In the RuMP cycle, fixation begins with the condensation of formaldehyde and RuMP to form hexulose-6-phosphate (HuMP), which in turn is converted to fructose-6-phosphate (FMP). The FMP is then cleaved to form two glyceraldehyde-3-phosphates. This overall pattern is certainly reminiscent of the CBB cycle using rubisco. 3-Hexulose-6-phosphate synthase (HPS) and 6-phospho-3-hexuloisomerase (PHI) are the only enzymes that are unique to this cycle. Overall, three molecules of formaldehyde are assimilated forming a three-carbon intermediate of central metabolism. The RuMP cycle has been found in chemolithoautotrophs including several methanotrophs.
Figure 5.
Serine pathway (assimilation of formaldehyde) is used by some chemolithoautotrophs, for example several methanotrophs. Methanotrophic bacteria oxidize methane and methanol to formaldehyde that condenses with glycine to form serine that can be assimilated to form intermediates of the central metabolic pathways. During this stage, phosphoenolpyruvate is carboxylated by phosphoenolpyruvate carboxylase to form oxaloacetate. The intermediate compounds are then used for biosynthesis. The net balance of this cycle is the fixation of 2 mol of formaldehyde and 1 mol of CO2 into 1 mol of 3-phosphoglycerate, which is used for biosynthesis, at the expense of 2 mol of ATP and the oxidation of 2 mol of NAD(P)H.
The serine pathway is similar to the RuMP pathway in that the initial substrate is formaldehyde, which itself may be derived from methane or methanol in methanotrophic bacteria. It also contains a carboxylation step. Some of the 2-phosphoglycerate produced is converted to phosphoenolpyruvate that is carboxylated by phosphoenolpyruvate carboxylase to oxaloacetate. Similar to the RuMP cycle, the serine pathway has been found in chemolithoautotrophs including several methanotrophs.
Both the RuMP and serine pathways oxidize CO or organic compounds including methanol and formate to CO2 prior to fixation. This suggests that CO2 fixation is the primitive substrate and the oxidation of the initial substrate was added later in evolution.
(vi) Variations on a theme
There are organisms known that have variations on these well-known pathways. For example, the thermophilic hydrogen-oxidizing bacterium Hydrogenobacter thermophilus fixes CO2 by the rTCA cycle (Shiba et al. 1985). However, pyruvic carboxylase catalyses the carboxylation of pyruvic to oxaloacetate instead of phosphoenolpyruvate catalysing the carboxylation of phosphoenolpyruvate to oxaloacetate as is often done in the rTCA cycle.
(vii) Which came first?
The most direct pathway imaginable for an autotroph is to condense two molecules of CO2, as acetogens do using the acetyl-CoA pathway and the rTCA cycle. This mode of autotrophy is used by anaerobes, which evolved prior to aerobes. In fact, formate dehydrogenase, which catalyses the reversible reduction of CO2 or , is extremely oxygen sensitive. H2, CO2, CO and metals such as cobalt, nickel, iron, tungsten, molybdenum and selenium, which are involved in the catalysis of the pathways, were present on the early Earth. For these reasons, Wood & Ljungdahl (1991) suggested that these might be considered as earliest autotrophic pathways. Some bacteria and archaea, such as Chlorobium tepidum, use the rTCA cycle but their genome encodes an orthologue of the large subunit of rubisco, although the purified protein has no detectable carbon-fixation activity (Hanson & Tabita 2001; Eisen et al. 2002).
Type III rubiscos, found only in the archaea, have rubisco activity although a functional CBB pathway has not been identified in that domain (Sato et al. 2007).
(c) Why is rubisco dominant today?
Why did rubisco-based carbon fixation come to dominate the biosphere? The enzyme itself is not particularly efficient (e.g. Tcherkez et al. 2006). One possibility is that the cyanobacteria used the CBB pathway, and thus this pathway was ‘fixed’ in all eukaryotes since carbon fixation is the result of the chloroplast, an evolved cyanobacterian. As eukaryotes came to dominate the biosphere, so did rubisco. In turn, cyanobacteria use the CBB pathway because they are oxygenic, and the acetyl-CoA and rTCA cycles are oxygen sensitive, as discussed above. Thus, even with its oxygenase activity, rubisco is superior in an oxygenated environment. Similarly, the 3-hydroxypropionate cycle has a narrow taxonomic distribution, and is found in a few thermophiles and acidophiles. Again, this suggests that, in cooler and aerobic environments, rubisco is superior.
Using a different approach, McFadden (1973) postulated that the acquisition of rubisco and PRK by ancient heterotrophic bacteria would have given huge selective advantage if hexoses or intermediates of the CBB cycle were limiting in the environment, assuming that all other enzymes of the CBB cycles were already present. He hypothesized that rubisco could have evolved from phosphofructokinase, noting that fructose-1,6-disphosphate is a good competitive inhibitor for rubisco.
3. Photosynthesis, a specialized case of autotrophy
Photosynthesis is a special case of autotrophy in that it exploits light as its source of energy. Photosynthesis can be aerobic (oxygenic), where water is used as a source of electrons and thus oxygen is produced as a by-product. Alternatively, it may be anaerobic (anoxygenic), i.e. a reductant other than water is used. Reductants for anaerobic photosynthesis are listed in table 1. Photosynthesis is found among the eukaryotes, notably the plants and algae, and six phyla of bacteria including cyanobacteria, Chlorobi, Proteobacteria, Chloroflexi, Firmicutes and Acidobacteria (Bryant et al. 2007).
Table 1.
Examples of reductants used by autotrophs.
| reductants for CO2 | type of photosynthesis |
|---|---|
| H2O | aerobic (oxygenic) |
| H2 | anaerobic (anoxygenic) |
| anaerobic (anoxygenic) | |
| anaerobic (anoxygenic) | |
| S° | anaerobic (anoxygenic) |
| H2S | anaerobic (anoxygenic) |
| CO | anaerobic (anoxygenic) |
| CH4 | anaerobic (anoxygenic) |
| Mn2+ | anaerobic (anoxygenic) |
| Fe2+ | anaerobic (anoxygenic) |
How probable is photosynthesis, specifically, to arise again? Once again, the diversity of reductants and even pathways, some of which have more than one step where carbon fixation occurs, increases the probability of it arising again.
Some cyanobacteria and algae have carbon-concentrating mechanisms ranging from active influx of CO2 or , passive influx of CO2, to prefixation of the inorganic carbon on a three-carbon sugar prior to release and re-fixation (Giordano et al. 2005). Among the plants, there are three major variations in photosynthetic pathways: the C3 pathway (no carbon concentration); the C4 pathway; and Crassulacean acid metabolism (CAM). Over 95% of the plants on Earth use the C3 pathway, in which inorganic carbon is incorporated directly into the CBB cycle. In C4 and CAM, there is an initial fixation of inorganic carbon with a subsequent decarboxylation prior to incorporation into the CBB cycle, an adaptation that aids in water conservation. In C4 plants, a distinct leaf anatomy keeps the two steps physically separated. Inorganic carbon is fixed to a three-carbon sugar, phosphoenolpyruvate, in the mesophyll cells to form oxaloacetate, which is converted to malate and then transported to the bundle-sheath cell where it is decarboxylated thus delivering CO2 to the CBB cycle enzymes. CAM is an adaptation for arid conditions. Similar to C4, there is an initial fixation of CO2 and a subsequent release to the CBB cycle, but in this case the two processes are temporally separated with CO2 entering the stomata during the night, and decarboxylation occurring during the day when the stomata are closed, thus minimizing water loss.
4. Could photosynthesis evolve again?
In order to estimate the probability of photosynthesis evolving again on Earth or elsewhere requires that the prerequisites for photosynthesis be met and selective pressures to act. When these components have arisen multiple times, this increases the likelihood of photosynthesis arising again.
(a) Prerequisites for the evolution of biological carbon fixation
There are at least six prerequisites for the evolution of biological carbon fixation as stated earlier. Each are now explored in order to assess the probability of the evolution of carbon fixation.
(i) Carbon-based life form
Organic carbon forms the basis for life on Earth, and is likely to do so if life arose again on Earth or elsewhere. Carbon is the fourth most abundant element in the Universe and forms the basis for a complex and varied chemistry. It may be twice as common relative to hydrogen in our Solar System than elsewhere (Snow & Witt 1995), but even at half the amount of our Solar System, it is still abundant. While silicon is also common (though not nearly as common as carbon in the Universe as a whole) and can form interesting polymers (Benner et al. 2004), its flexibility pales in comparison with organic chemistry, particularly in the ability of carbon to form polymers. Most stunning of all, organic compounds—including amino acids and nucleotide bases—have been detected in the interstellar medium (Irvine 1998; Ehrenfreund & Charnley 2000; Allamandola & Hudgins 2003). Even Earth, which is composed of a substantial quantity of silicates and thus should be biased towards silicon-based life, harbours carbon-based life.
(ii) The presence of inorganic carbon
While organic carbon is quite common in the Universe, the predominant form of carbon is inorganic. Carbon is formed by the triple collision of alpha particles (helium nuclei) within the core of a giant or supergiant star, and is made available for life when it disperses from its parent star during a supernova. Today's Earth is estimated to contain approximately 38 000 Gt inorganic carbon (Falkowski et al. 2000).
(iii) Availability of reductants
Much of the photosynthesis on Earth today is oxygenic; that is, the reducing power is derived from H2O. Yet many reductants, from elemental sulphur to hydrogen, are used by prokaryotes that do not produce oxygen (table 1). Some can use multiple electron donors for photosynthesis, such as Chlorobium tepidum, which can use both sulphide and thiosulphate (Eisen et al. 2002).
(iv) The presence of light
Any planet, comet or asteroid near enough to a parent star will receive sufficient radiation for photosynthesis on its surface. ‘Near enough’ is surprisingly low, as only a small photon flux is needed for photosynthesis, with an absolute minimum thought to be 0.01 μmol of photons m−2 s−1 (Raven & Cockell 2006). By contrast, midday sunlight in summer in the continental USA is approximately 2000 μmol of photons m−2 s−1 (L. J. Rothschild 2008, unpublished). We have shown that interstitial microbial mats found just below the sand surface where they receive a tiny fraction of the surface solar flux can still fix approximately 41 mg C m−2 d−1 (Rothschild & Giver 2003). Nisbet et al. (1995) proposed that even the presence of solar radiation may not be required for the evolution of the light-harvesting pigment chlorophyll as it could have arisen for thermal detection in order for organisms to properly orient with respect to hydrothermal vents.
Which wavelengths are necessary? On Earth, most organisms use solar radiation between 400 and 700 nm. Much below, ultraviolet radiation damage becomes of increasing concern, a problem with any organic carbon-based life form. At the other extreme, longer wavelengths are more benign but are also of much less energy, hence may not be energetic enough to drive photosynthesis. However, there are prokaryotes that can access slightly longer wavelengths. For example, the cyanobacterium Acaryochloris marina is unique among phototrophs in that it uses chlorophyll d as its main light-harvesting pigment instead of chlorophyll a (Kühl et al. 2005). The peak absorption for chlorophyll d is 700–720 nm (Kühl et al. 2005). Some anoxygenic prokaryotes use even higher wavelengths into the near infrared (IR) because bacteriochlorophylls a and b have secondary absorption peaks at 770 and 795 nm, respectively. The purple bacteria are able to access solar radiation even further into the IR, with the LH1 complex absorbing as high as 963 nm (Permentier et al. 2001).
IR radiation contains less energy than visible radiation, so it is difficult to understand why bacteriochlorophyll should use this portion of the spectrum. Nisbet et al. (1995) have suggested that bacteriochlorophylls arose as heat (i.e. near IR) sensing devices for early organisms that lived near hydrothermal vents. In practice, niche partitioning of different species with different suites of primary and secondary pigments occurs so that each organism can access as much solar radiation as possible (e.g. Stomp et al. 2004).
(v) A light-harvesting mechanism to convert the light energy into chemical energy
The synthesis of chlorophyll/bacteriochlorophyll arose only once in evolution (Xiong & Bauer 2002). But chlorophyll biosynthesis is closely related to the widespread haem, the former being a magnesium porphyrin and the latter an iron porphyrin. Haem is extremely widespread as it is the prosthetic group for such proteins as myoglobin, haemoglobin, catalase, peroxidase and cytochrome c. Thus, it seems plausible biochemically that a chlorophyll-like molecule could have arisen multiple times in multiple taxa.
In addition, several types of photoreceptor molecules are known, some of which convert light energy into chemical energy, as in photosynthesis, or use the light energy for signal transduction. Béjá et al. (2000) presented evidence that bacterial rhodopsin could be the photoreceptor driving photoheterotrophy or even photosynthesis in a variety of marine bacteria.
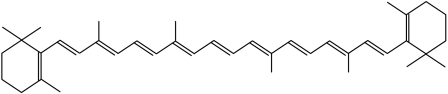
Photopigments include carotenoids, bilins, chlorophylls, flavins and pterins (table 2). There may be a few other photopigments such as parahydroxycinnamic acid that may act as the photoactive pigment in association with green fluorescent protein (Sancar 2000). A common theme in the molecular structure of light-harvesting compounds is the network of alternating single and double bonds called polyenes that makes them particularly effective photoreceptors. Chlorophyll is a substituted tetrapyrrole, with the four nitrogen atoms of the pyrroles coordinated to a magnesium atom. Carotenoids, used as secondary compounds in photosynthesis, also contain polyenes (figure 6). Rhodopsin, the photosensitive molecule in rods, contains opsin (a protein) and 11-cis-retinal, a prosthetic group that contains alternating polyenes.
Table 2.
Photoreactive pigments (see Sancar 2000 for more details). (The actual absorption depends on both the pigment and the chemical environment in which it is found.)
| class of pigment | absorption range | description | examples |
|---|---|---|---|
| carotenoids | ∼400–550 | photoantenna (accessory) pigments in the photosystems and the catalytic pigments in animal and bacterial rhodopsins | xanthophylls, carotenes such as β-carotene, retinal |
| bilins | 400–500 nm, 600–700 nm | photoantenna pigments in the photosystems and the chromophore of the plant photoreceptor phytochrome | |
| chlorophylls | 350–450 nm, 600–700 nm, >700 for chlorophyll d | photoantenna pigments in the photosystems and the primary electron donors in the reaction centre of photosystems | chlorophylls a, b, c and d |
| flavins | 360 nm in the two-electron reduced form; 370 and 440 nm in the two-electron oxidized form; 380, 480, 580, 625 nm in the one-electron reduced (blue neutral radical) form | photoactive cofactor in the photolyase/blue-light photoreceptor family | flavin adenine dinucleotide (photoactive cofactor in photolyase/blue-light photoreceptor family), deazoriboflavin |
| pterins | 360–420 nm | photoantenna in the majority of photolyase/cryptochrome blue-light photoreceptors | 5,10-methenyltetrahydrofolate (MTHF) |
Figure 6.
β-Carotene, a light-harvesting pigment with peak absorptions of 455 and 480 nm. It is an effective photoreceptor owing to the alternating single and double bonds.
Not all of these molecules originated as light-harvesting compounds. For example, it is thought that the carotenoids arose in archaea as lipids reinforcing cell membranes (Vershinin 1999). However, their structure contains a number of properties that pre-adapted them to the various functions they serve today ranging from light harvesting to quenching singlet oxygen, while still maintaining their ability to reinforce membrane structure as they do today in mycoplasms, some fungi and some animals such as molluscs. Cryptochromes may have evolved from photolyases, but no longer possess the photolyase function (Lin & Shalitin 2003).
Often, such as in the case of the bilins, the photoreceptor is covalently attached to a protein. Grossman et al. (1995) hypothesized that the phycobiliprotein subunits were derived from a common precursor protein that already had the amino acid residues needed for photoreceptor attachment and stability. The integral membrane proteins called opsins bind retinal (vitamin A aldehyde) forming rhodopsins. Rhodopsins are capable of generating a chemiosmotic membrane potential in response to light. Rhodopsins belong to two distinct protein families: the visual rhodopsins and archaeal and bacterial rhodopsins (Béjá et al. 2000). Although they have no significant sequence similarity and may have different origins, they have identical topologies (Béjá et al. 2000).
(vi) Carbon-fixation enzymes
Carbon fixation, in the broadest sense, is a process where inorganic carbon is converted or incorporated into organic compounds. Carbon dioxide is a ‘featureless molecule’ (Tcherkez et al. 2006) as is oxygen, leading to confusion in catalysis with at least rubisco, resulting in both a carboxylase and oxygenase function. Carboxylases are usually associated with autotrophs only, particularly phototrophs, but, in fact, multiple enzymes found in all taxa have evolved to fix carbon (table 3). Different types of compounds can be carboxylated from sugars to proteins to assorted other compounds.
Table 3.
Carboxylating enzymes.
| name | alternate name | number | reaction catalysed | cofactor |
|---|---|---|---|---|
| pyruvic-malic carboxylase | malate dehydrogenase (oxaloacetate-decarboxylating) | 1.1.1.38 | (S)-malate+NAD+↔pyruvate+CO2+NADH | |
| pyruvic-malic carboxylase | malate dehydrogenase (decarboxylating) | 1.1.1.39 | (S)-malate+NAD+↔pyruvate+CO2+NADH | |
| pyruvic-malic carboxylase | malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+) | 1.1.1.40 | (S)-malate+NADP+↔pyruvate+CO2+NADPH | |
| β-ketoglutaric-isocitric carboxylase | isocitrate dehydrogenase (NAD+) | 1.1.1.41 | isocitrate+NAD+↔2-oxoglutarate+CO2+NADH | manganese or magnesium |
| triphosphopyridine nucleotide-linked isocitrate dehydrogenase-oxalosuccinate carboxylase | isocitrate dehydrogenase (NADP+) | 1.1.1.42 | isocitrate+NADP+↔2-oxoglutarate+CO2+NADPH | |
| 6-phosphogluconic carboxylase | phosphogluconate dehydrogenase (decarboxylating) | 1.1.1.44 | 6-phospho-d-gluconate+NADP+↔d-ribulose-5-phosphate+CO2+NADPH | |
| 2-oxopropyl-CoM reductase (carboxylating) | 2-oxopropyl-CoM reductase (carboxylating) | 1.8.1.5 | 2-mercaptoethanesulphonate+acetoacetate+NADP+↔2-(2-oxopropylthio)ethanesulphonate+CO2+NADPH | |
| α-ketoacid carboxylase | pyruvate decarboxylase | 4.1.1.1 | a 2-oxo acid↔an aldehyde+CO2 | thiamine diphosphate |
| phosphoribosylaminoimidazole carboxylase | 4.1.1.21 | 5-amino-1-(5-phospho-d-ribosyl)imidazole-4-carboxylate →5-amino-1-(5-phospho-d-ribosyl)imidazole+CO2 | ||
| phosphoenolpyruvate carboxylase | phosphoenolpyruvate carboxykinase (GTP) | 4.1.1.32 | GTP+oxaloacetate→GDP+phosphoenolpyruvate+CO2 | |
| phosphopyruvate carboxylase | phosphoenolpyruvate carboxykinase (diphosphate) | 4.1.1.38 | diphosphate+oxaloacetate→phosphate+phosphoenolpyruvate+CO2 | |
| ribulose-1,5-bisphosphate carboxylase/oxygenase | carboxydismutase | 4.1.1.39 | 2 3-phospho-d-glycerate+2H+→d-ribulose-1,5-bisphosphate+CO2+H2O | |
| phosphoenolpyruvate carboxylase | methylmalonyl-CoA decarboxylase | 4.1.1.41 | ATP+oxaloacetate→ADP+phosphoenolpyruvate+CO2 | |
| urea carboxylase | urea amidolyase | 6.3.4.6 | ATP+urea+→ADP+phosphate+urea-1-carboxylate→→ 2NH3+2CO2 | biotin |
| biotin carboxylase | 6.3.4.14 | ATP+biotin-carboxyl-carrier protein+CO2→ADP+phosphate+carboxybiotin-carboxyl-carrier protein | ||
| pyruvate carboxylase | 6.4.1.1 | ATP+pyruvate+→ ADP+phosphate+oxaloacetate | biotin, manganese or zinc | |
| acetyl-CoA carboxylase | 6.4.1.2 | ATP+acetyl-CoA+→ ADP+phosphate+malonyl-CoA | biotin | |
| propionyl-CoA carboxylase | 6.4.1.3 | ATP+propionyl-CoA+→ ADP+phosphate+(S)-methylmalonyl-CoA | biotin | |
| methylcrotonyl-CoA carboxylase | 6.4.1.4 | ATP+3-methylcrotonyl-CoA+→ ADP+phosphate+3-methylglutaconyl-CoA | biotin | |
| geranoyl-CoA carboxylase | 6.4.1.5 | ATP+geranoyl-CoA+→ ADP+phosphate+3-(4-methylpent-3-en-1-yl)pent-2-enedioyl-CoA | biotin | |
| acetone carboxylase | 6.4.1.6 | Acetone+CO2+ATP+2H2O→ acetoacetate+AMP+2 phosphate | magnesium | |
| 2-oxoglutarate carboxylase | oxalosuccinate synthetase | 6.4.1.7 | ATP+2-oxoglutarate+→ ADP+phosphate+oxalosuccinate | biotin, magnesium |
| γ-glutamyl carboxylase | carboxylation of specific glutamate residues in vitamin K-dependent proteins to Gla in the presence of carbon dioxide, oxygen and reduced vitamin K; in the process, vitamin K is converted to vitamin K epoxide, which is subsequently converted to vitamin K by vitamin K epoxide reductase and used in the carboxylation reaction |
Examples of a carboxylating enzyme that is not used in autotrophy are the γ-carboxylation enzymes. These enzymes are important in blood clotting in vertebrates, but have recently been found in some invertebrates including Drosophila and the marine cone snail Conus, where they have a role in the production of venom peptides (Bandyopadhyay et al. 2002). γ-Carboxylation enzymes are integral proteins of the endoplasmic reticulum, which, in the presence of carbon dioxide, oxygen and reduced vitamin K, results in the carboxylation of specific glutamate residues in vitamin K-dependent proteins. The general topology of the enzyme from all the three taxa are similar, as was the sequence, especially in the transmembrane portion of the carboxylase and the putative substrate-binding pocket (Bandyopadhyay et al. 2002).
Degradation of urea is widely accomplished by the action of urease, but in algae, yeast and some bacteria, the degradation occurs through the action of the enzyme complex urea amidolyase (Kanamori et al. 2004). Bicarbonate is phosphorylated by ATP, with the subsequent transfer of the CO2 to biotin. Through the urea carboxylase activity, the urea is carboxylated leading to its degradation to two molecules each of NH3 and CO2.
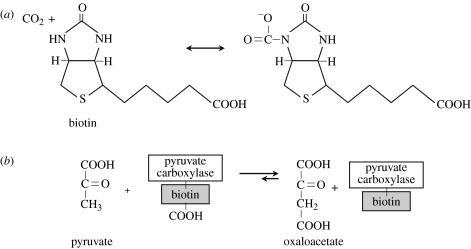
Biotin, a water-soluble member of the B complex of vitamins, is a mobile carrier of activated CO2 (figure 7). Biotin is a cofactor for many carboxylating, decarboxylating and transcarboxylating enzymes, for example pyruvate carboxylase (table 2; Nikolau et al. 2003). For all carboxylase reactions, the carboxyl donor is bicarbonate/ATP (Visser & Kellogg 1978). Biotin-dependent carboxylases share a catalytic mechanism and a structure that consists of biotin carboxylase, biotin-carboxyl-carrier protein and carboxyl transferase. The carboxyl terminus of biotin is linked to the amino group of a specified lysine residue by an amide bond. The biotin itself is carboxylated by biotin carboxylase (EC 6.3.4.14) during the following reaction:
Most coenzymes are ribonucleotides, or at least have a cyclic nitrogenous base in common, except biotin and lipoic acid. Coenzymes and prosthetic groups and tRNA can be seen as relics from the RNA world, except biotin and lipoic acid. This suggests that biotin and lipoic acid arose after the RNA world (Visser & Kellogg 1978).
Figure 7.
Biotin (vitamin B7 or vitamin H) is a cofactor for CO2 transfer. (a) The carboxyl terminus of biotin is linked to the amino group of a specified lysine residue by an amide bond. Carboxylating enzymes using biotin are: acetyl-CoA carboxylase alpha; acetyl-CoA carboxylase beta; methylcrotonyl-CoA carboxylase; propionyl-CoA carboxylase; and pyruvate carboxylase. (b) An example of the use of biotin in the carboxylation of pyruvate by pyruvate carboxylase.
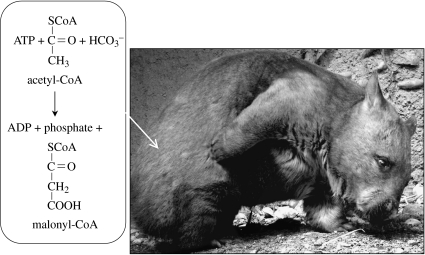
Some carboxylating enzymes are used for autotrophic carbon fixation in some cases and not others. Acetyl-CoA (figure 8) and propionyl-CoA carboxylation are normally used in fatty acid synthesis; however, they are also found in archaea that do not have fatty acids. It is thought that, in these archaea, the enzymes function in autotrophic carbon fixation. The carboxylase enzyme from the archaeans Metallosphaera sedula (Hügler et al. 2003b) and Acidianus brierleyi (Chuakrut et al. 2003) catalyses equally effectively both acetyl-CoA and propionyl-CoA carboxylation. The holoenzyme contains subunits of biotin carboxylase, carboxytransferase and a biotin-carrier protein.
Figure 8.
Acetyl-CoA carboxylase catalyses the first step in fatty acid synthesis. This carboxylation is irreversible. Photo of a wombat by L.J.R.
Acetone degradation in the actinomycete Rhodococcus rhodochrous is accomplished with an initial carboxylation by acetone carboxylase. Carboxylation is increased in the presence of a nucleotide triphosphate, with GTP and IPT particularly effective (Clark & Ensign 1999).
How probable is the evolution of a carboxylase? The wide diversity of carboxylases outlined above and their substrates suggests multiple origins of carboxylases, implying a relative ease of evolutionary origin. Other putative examples of convergence in enzyme function have been identified, including the d- and l-specific 6-hydroxynicotine oxidases, both found in the soil bacterium Arthrobacter nicotinovorans, but unrelated at the DNA and protein levels (Schenk & Decker 1999). Sugar kinases, enzymes that catalyse the phosphorylation of sugars, arose at least three times leading to distinct non-homologous families (Bork et al. 1993), and the lactate dehydrogenase from the amitochondriate protists Trichomonas vaginalis appears to have evolved from the cytosolic malate dehydrogenase of the same species rather than from another lactate dehydrogenase (Wu et al. 1999).
As Pain (1982) pointed out, natural selection should act on the active site of an enzyme rather than its amino acid sequence, and with the featureless nature of the substrate, CO2, again, one should predict relative ease in the evolution of carboxylation activity although, as in the case of rubisco, it may lack in specificity. But, if there are similarities in the active site, is this the result of common ancestry or of a limited number of ways to construct an active site with a particular catalytic activity? Conversely, the subtilisin group of the microbial serine proteases is similar to the mammalian type in their active sites but not conformation, suggesting that this catalytic activity was solved at least twice in evolution with a resulting convergence on the active site (Pain 1982). Furthermore, there is modularity in the evolution of many carboxylases in that the incorporation of biotin allows the rapid evolution of carboxylation.
But biomolecules do not exist in either physical or functional isolation. When an enzyme is part of a metabolic network, changes in components may need compensation in order to keep the entire network functional (Vitkup et al. 2006). Not surprisingly, analyses of the photosynthetic machinery of cyanobacterial gene products that need to interact show genetic linkage suggesting coordinated evolution (Shi et al. 2005). However, rubisco may not be one of them, but rather a ‘frozen accident’ because it allows oxygen to act as a competitive inhibitor of carboxylase activity (Shi et al. 2005). Rather than evolution acting on removing the oxygenase activity during geological time as atmospheric levels of oxygen rose, organisms have responded by evolving carbon-concentrating mechanisms and increasing the production of rubisco.
(b) Selection pressure for photosynthesis
In addition to the prerequisites for the evolution of biological carbon fixation, a selective pressure should be present. An excess of inorganic carbon (e.g. CO2, CO, carbonates, cyanides, cyanates, carbides and thiocyanates) over organic carbon, in most cases a vast surfeit, should provide the selective pressure. Even on today's life-infested Earth, there are approximately 38 000 Gt carbon in contrast to 1000 Gt organic carbon in the oceans (Falkowski et al. 2000).
(c) Proposed evolutionary scenario for the origin of photosynthesis
Given these conditions, it is easy to envision an evolutionary scenario that includes strong selective pressure for organisms that evolve autotrophy, particularly photosynthesis. The evolutionary background includes the following:
Life on Earth is based on organic carbon.
Most of the carbon in the Universe is inorganic.
Life is lazy, or, to put it more scientifically, one must be efficient to maintain the competitive edge.
Evolution uses what is available; ‘evolution as tinkerer’ rather than an engineer (Jacob 1977). Parts are co-opted from within or from others rather than designed, as Ashida et al. (2005) have suggested for the origin of rubisco from RLPs. Another source of novelty are units—from genes to teeth—which replicate and diversify.
Of course, contingency does occur. Asteroids, floods, atmospheric catastrophes and so on may change the rules substantially.
From these rules, I hypothesize a planet-independent scenario for the evolution of life in the Universe.
The first living organisms were built from abiotically produced organic carbon, as the heterotrophic model for the origin of life postulates (e.g. Lazcano & Miller 1999). The struggle for existence began over the remains of the first ‘free lunch’, and life was forced to make its own organic carbon in order to persist. Perhaps this was doubly beneficial because it was more efficient; for example, organic carbon production could be combined with the generation of energy. In either case, the first autotroph was born.
But life conserves energy whenever possible for other uses. Why synthesize organic carbon when it can be delivered in an integrated package? In other words, why be autotrophic when organic carbon is not limiting?
There is experimental evidence to support this point. For example, a mixed community of the red alga Cyanidium caldarium, a green alga described as ‘Chlorella-like’ or ‘Chlorella’ protothecoides var. acidicola and a minor component of the fungus Dactylaria (Belly et al. 1973; Ferris et al. 2005) was tested for gross carbon fixation in the presence and absence of added fixed carbon (L. J. Rothschild 2008, unpublished). Acetate and glucose were added to mat samples, which were then incubated in situ with supplemental 14CO2. While, for the most part, there was little difference with the addition of glucose, the presence of acetate had a powerful inhibitory effect on carbon fixation (figure 9). This suggests that these photosynthetic algae were able to take up a source of fixed carbon; they downregulated their own carbon fixation. Similarly, oceanic photoheterotrophic bacteria metabolize organic carbon from the environment when available, but photosynthesize when organic carbon is sparse (Kolber et al. 2001).
Figure 9.
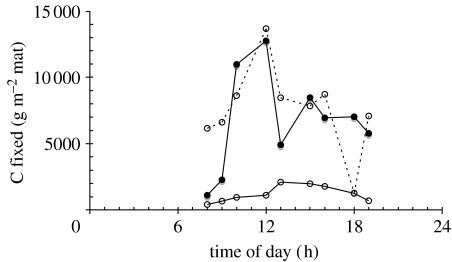
Effect of exogenous fixed carbon on carbon-fixation rates in Nymph Creek (N 44°45.178′, W 110°43.439′), a natural red and green algal ecosystem in Yellowstone National Park, WY, USA. Carbon fixation was measured by the acid-stable incorporation of in situ, as described by Rothschild & Mancinelli (1990) and Rothschild (1991). The incubation mix was stream water (control: filled circle, solid line) or stream water supplemented with 25 mM acetate (open circle, solid line) or 25 mM glucose (open circle, dashed line) and 1 μCi ml−1 14C-bicarbonate (New England Nuclear, Wilmington, DE, USA, catalogue number NEC 086H). Algal mat cores (surface area=17 cm2) were placed in Whirlpack bags, incubation mix was added and the bags were incubated in Nymph Creek. Experiments were conducted in duplicate, and all data points were averaged. Dark controls were performed by wrapping the incubation vessel in foil. The dissolved inorganic carbon of Nymph Creek water was determined using a CO2 probe (Lazar Research Laboratories, Los Angeles, CA, USA) calibrated with aqueous solutions of . Incubation times were 15 min in order to minimize the impact of remineralization of the fixed carbon and to be sensitive to light levels that are constantly changing in nature.
And thus, herbivory arose. Of course, this required various modifications such as sufficient size and structure. Organic ‘slurp’ was still available for those unable to feed on autotrophs, but unlike the first life forms, the soup course was now biologically generated; a satisfying broth for countless members of all the three domains of life. Even today, it is fashionable to acquire an in-house source of organic carbon through symbiosis, a strategy that has been brilliantly exploited on Earth by a variety of protists and animals.
Assimilating fixed carbon is metabolically less expensive than fixing carbon, and if the fixed carbon comes with proteins, nucleic acids and other nutrients, so much the better. The autotrophs provided a new biological source of organic carbon for heterotrophs (more specifically saprotrophs) by excreting organic compounds. More significantly, a new type of heterotrophy that could not be supported previously arose: herbivory. Many microbes have remained mixotrophic (e.g. Kolber et al. 2001). Raven's (1997) excellent analysis of the costs and benefits of phagotrophy points out that for all the versatility of mixotrophy, that is, retaining both the ability to photosynthesize and consume, there are costs. The most obvious cost is the necessity of maintaining two sets of machinery, which he estimates as approximately 50% for the photosynthetic apparatus and approximately 10% to maintain phagotrophic capability, although there can be some regulation in the amount of machinery based on environmental conditions. A possible secondary problem is that a phagotrophic cell must regulate volume. A photosynthetic eukaryote must make sure that all of the components for the photosynthetic apparatus including genetic instructions are inherited properly.
(d) The danger of oxygenic photosynthesis
Oxygenic photosynthesis is tremendously advantageous owing to the wide availability of water. But oxygenic photosynthesis creates hazards in the form of reactive oxygen species that can damage nucleic acids, proteins and lipids. Oxygen, water, superoxide and hydrogen peroxide are not particularly reactive, but hydrogen peroxide can be reduced to the hydroxyl radical that is extremely reactive.
During the course of the production of O2 from H2O in photosystem II, oxygen of the ground (triplet) state (3O2) may be excited to singlet state (1O2) as a side or back reaction that generates triplet chlorophyll. Fortunately, 1O2 is rapidly quenched by water (Asada 2006). Photoreduction of oxygen to hydrogen peroxide (H2O2) occurs in photosystem I. The primary reduced product is superoxide anion (), and its enzymatically catalysed disproportionation products are H2O2 and O2 (Asada 2006). In isolated thylakoids, H2O2 accumulates, but not in intact chloroplasts owing to enzymatic degradation. However, the prerequisite for oxygenic photosynthesis is the ability to tolerate oxygen, and aerobic metabolism also generates reactive oxygen species during the reduction of O2 to H2O. Thus, oxygenic photosynthesis is fraught with biochemical risks that must be addressed with antioxidants.
5. Photosynthesis, a universal phenomenon?
The preceding argument is that given certain prerequisites—a carbon-based life form, the presence of inorganic carbon, the availability of reductants, the presence of light, a light-harvesting mechanism to convert the light energy into chemical energy and carboxylating enzymes—and evolutionary pressure, photosynthesis is likely to evolve again, whether on Earth or elsewhere.
Oxygenic photosynthesis, however, carries with it the danger of reactive oxygen species but the advantage of, at least on Earth, a widely available source of reducing power relative to other reductants. It is unclear what the relative costs to benefits are, but these must always be accompanied by a cautionary note. By contrast, Lovelock (1975) assumed that oxygenic photosynthesis would evolve multiple times, and thus the detection of O3 on other planets would be an indication of life. A thorough and thoughtful analysis of the likelihood of oxygenic photosynthesis arising elsewhere has been presented by Wolstencroft & Raven (2002).
Leaving oxygenic photosynthesis aside, are the prerequisites for the evolution of any form of photosynthesis available elsewhere? The argument was advanced above that life anywhere in the Universe is likely to be based on organic carbon, and that the inorganic and organic carbon is widely available in the Universe. Even in our Solar System, Europa and Enceladus are thought to contain substantial quantities of organic carbon at least on their surface, and the Huygens probe confirmed that Titan has large quantities of organics. Meteorites, comets and dust particles rain organic carbon through the Solar System, so it is unclear why we have yet to detect it on Mars unless it is being oxidized. Mars does have a large inventory of CO2. All these bodies have water and other compounds that could be used as reductants.
Light becomes increasingly scarce with distance from the Sun, decreasing with the inverse of the square of the distance. Mars at 1.6 AU has approximately 44% of the terrestrial solar radiation flux. Of the bodies mentioned, Saturn's moons Titan and Enceladus are the farthest from the Sun at approximately 9.2 AU, which means they should receive approximately 1.1% of the terrestrial solar radiation flux, a flux sufficient to support photosynthesis on Earth. Even beyond our Solar System, Tarter et al. (2007) showed that an M dwarf emits enough light for photosynthesis. From a star of Teff=4000 K (slightly hotter than a M0 star), photosynthetically active radiation (400–700 nm) incident at the top of the atmosphere would be roughly a third that of the Earth, and the range that penetrates clear ocean water (450–550 nm) would be about a quarter that incident on the Earth. And even if the average wavelength is shifted towards the IR, there should be enough useful radiation for photosynthesis (Raven 2007). But note that if a planet is tidally locked, then any stationary organism, such as terrestrial plants, would always see light from the same angle, so less of the canopy would be able to access it.
The wide diversity of photoreceptors and carboxylation enzymes on Earth and their multiple origin argue for a relative ease in the evolution of these mechanisms. If so, and the selection pressure arose with regard to a dearth of organic carbon relative to biotic demands, photosynthesis would arise.
Acknowledgments
I am indebted to Euan and Ellen Nisbet for the invitation to participate in this meeting, and especially to Euan for his ever-stimulating suggestions and perspectives. This manuscript has benefited by the insightful suggestions of John Ellis, Robert Tabita and two anonymous reviewers, and the sharp eyes of Derek Bendall. This work was supported by grants from NASA's Astrobiology Program, including the NASA Astrobiology Institute.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘Photosynthetic and atmospheric evolution’.
References
- Alber B.E, Fuchs G. Propionyl-coenzyme A synthase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroypropionate cycle for autotrophic CO2 fixation. J. Biol. Chem. 2002;277:12 137–12 143. doi: 10.1074/jbc.M110802200. doi:10.1074/jbc.M110802200 [DOI] [PubMed] [Google Scholar]
- Allamandola L, Hudgins D. From interstellar polycyclic aromatic hydrocarbons and ice to astrobiology. In: Pirronello V, Krelowski J, editors. Proc. NATO ASI entitled “Solid State Astrochemistry”. Kluwer; Dordrecht, The Netherlands: 2003. pp. 251–316. [Google Scholar]
- Anbar A.D, et al. A whiff of oxygen before the great oxidation event? Science. 2007;317:1903–1906. doi: 10.1126/science.1140325. doi:10.1126/science.1140325 [DOI] [PubMed] [Google Scholar]
- Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. doi:10.1104/pp.106.082040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Salto Y, Kojima C, Kobayashi K, Ogasawara N, Yokota A. A functional link between rubisco-like protein of Bacillus and photosynthetic rubisco. Science. 2003;302:286–290. doi: 10.1126/science.1086997. doi:10.1126/science.1086997 [DOI] [PubMed] [Google Scholar]
- Ashida H, Danchin A, Yokota A. Was photosynthetic rubisco recruited by acquisitive evolution from rubisco-like proteins involved in sulfur metabolism? Res. Microbiol. 2005;156:611–618. doi: 10.1016/j.resmic.2005.01.014. doi:10.1016/j.resmic.2005.01.014 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay P.K, Garrett J.E, Shetty R.P, Keate T, Walker C.S, Olivera B.M. γ-Glutamyl carboxylation: an extracellular posttranslational modification that antedates the divergence of mollusks, arthropods, and chordates. Proc. Natl Acad. Sci. USA. 2002;99:1264–1269. doi: 10.1073/pnas.022637099. doi:10.1073/pnas.022637099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béjá O, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. doi:10.1126/science.289.5486.1902 [DOI] [PubMed] [Google Scholar]
- Belly R.T, Tansey M.R, Brock T.D. Algal excretion of 14C-labeled compounds and microbial interactions in Cyanidium caldarium mats. J. Phycol. 1973;9:123–127. doi:10.1111/j.1529-8817.1973.tb04067.x [Google Scholar]
- Benner S.A, Ricardo A, Carrigan M.A. Is there a common chemical model for life in the universe? Curr. Opin. Chem. Biol. 2004;8:672–689. doi: 10.1016/j.cbpa.2004.10.003. doi:10.1016/j.cbpa.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A. Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 1993;2:31–40. doi: 10.1002/pro.5560020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocks J.J, Logan G.A, Buick R, Summons R.E. Archean molecular fossils and the early rise of eukaryotes. Science. 1999;285:1033–1036. doi: 10.1126/science.285.5430.1033. doi:10.1126/science.285.5430.1033 [DOI] [PubMed] [Google Scholar]
- Bryant D.A, et al. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic acidobacterium. Science. 2007;317:523–526. doi: 10.1126/science.1143236. doi:10.1126/science.1143236 [DOI] [PubMed] [Google Scholar]
- Buick R. The antiquity of oxygenic photosynthesis: evidence from stromatolites in sulphate-deficient Archaean lakes. Science. 1992;255:74–77. doi: 10.1126/science.11536492. doi:10.1126/science.11536492 [DOI] [PubMed] [Google Scholar]
- Campbell B.J, Cary S.C. Abundance of reverse tricarboxylic acid cycle genes in free-living microorganisms at deep-sea hydrothermal vents. Appl. Environ. Microbiol. 2004;70:6282–6289. doi: 10.1128/AEM.70.10.6282-6289.2004. doi:10.1128/AEM.70.10.6282-6289.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuakrut S, Arai H, Ishii M, Igarashi Y. Characterization of a bifunctional archaeal acyl coenzyme A carboxylase. J. Bacteriol. 2003;185:938–947. doi: 10.1128/JB.185.3.938-947.2003. doi:10.1128/JB.185.3.938-947.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D.D, Ensign S.A. Evidence for an inducible nuceotide-dependent acetone carboxylase in Rhodococcus rhodochrous B276. J. Bacteriol. 1999;181:2752–2758. doi: 10.1128/jb.181.9.2752-2758.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway Morris S. Oxford University Press; Oxford, UK: 1998. The crucible of creation: the Burgess Shale and the rise of animals. p. 272. [Google Scholar]
- Dedysh S.N, Smirnova K.V, Khmelenina V.N, Suzina N.E, Liesack W, Trotsenko Y.A. Methylotrophic autotrophy in Beijerinckia mobilis. J. Bacteriol. 2005;187:3884–3888. doi: 10.1128/JB.187.11.3884-3888.2005. doi:10.1128/JB.187.11.3884-3888.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenfreund P, Charnley S.B. Organic molecules in the interstellar medium, comets, and meteorites: a voyage from dark clouds to the early Earth. Annu. Rev. Astron. Astrophys. 2000;38:427–483. doi:10.1146/annurev.astro.38.1.427 [Google Scholar]
- Eisen J.A, et al. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl Acad. Sci. USA. 2002;99:9509–9514. doi: 10.1073/pnas.132181499. doi:10.1073/pnas.132181499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski P, et al. The global carbon cycle: a test of our knowledge of Earth as a system. Science. 2000;290:291–296. doi: 10.1126/science.290.5490.291. doi:10.1126/science.290.5490.291 [DOI] [PubMed] [Google Scholar]
- Fenchel T, King G.M, Blackburn T.H. 2nd edn. Academic Press; San Diego, CA: 1998. Bacterial biogeochemistry: the ecophysiology of mineral cycling. [Google Scholar]
- Ferris M.J, Sheehan K.B, Kühl M, Cooksey K, Wigglesworth-Cooksey B, Harvey R, Henson J.M. Algal species and light microenvironment in a low-pH, geothermal microbial mat community. Appl. Environ. Microbiol. 2005;71:7164–7171. doi: 10.1128/AEM.71.11.7164-7171.2005. doi:10.1128/AEM.71.11.7164-7171.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G. CO2 fixation in acetogenic bacteria: variations on a theme. FEMS Microbiol. Lett. 1986;39:181–213. doi:10.1111/j.1574-6968.1986.tb01859.x [Google Scholar]
- Giordano M, Beardall J, Raven J.A. CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu. Rev. Plant Biol. 2005;56:99–131. doi: 10.1146/annurev.arplant.56.032604.144052. doi:10.1146/annurev.arplant.56.032604.144052 [DOI] [PubMed] [Google Scholar]
- Goldblatt C, Lenton T.M, Watson A.J. Bistability of atmospheric oxygen and the great oxidation. Nature. 2006;443:683–686. doi: 10.1038/nature05169. doi:10.1038/nature05169 [DOI] [PubMed] [Google Scholar]
- Gould S.J. W. W. Norton; New York, NY: 1989. Wonderful life: the Burgess Shale and the nature of history. p. 323. [Google Scholar]
- Grossman A.R, Bhaya D, Apt K.E, Kehoe D.M. Light-harvesting complexes in oxygenic photosynthesis: diversity, control, and evolution. Annu. Rev. Genet. 1995;29:231–288. doi: 10.1146/annurev.ge.29.120195.001311. doi:10.1146/annurev.ge.29.120195.001311 [DOI] [PubMed] [Google Scholar]
- Hanson T.E, Tabita F.R. A ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc. Natl Acad. Sci. USA. 2001;98:4397–4402. doi: 10.1073/pnas.081610398. doi:10.1073/pnas.081610398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hügler M, Huber H, Stetter K.O, Fuchs G. Autotrophic CO2 fixation pathways in archaea (Crenarchaeota) Arch. Microbiol. 2003a;179:160–173. doi: 10.1007/s00203-002-0512-5. [DOI] [PubMed] [Google Scholar]
- Hügler M, Krieger R.S, Jahn M, Fuchs G. Characterization of the acetyl-CoA/priopionyl-CoA carboxylase in Metallosphaera sedula. Eur. J. Biochem. 2003b;270:736–744. doi: 10.1046/j.1432-1033.2003.03434.x. doi:10.1046/j.1432-1033.2003.03434.x [DOI] [PubMed] [Google Scholar]
- Ingalls A.E, Shah S.R, Hansman R.L, Aluwihare L.I, Santos G.M, Druffel E.R.M, Person A. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc. Natl Acad. Sci. USA. 2006;103:6442–6447. doi: 10.1073/pnas.0510157103. doi:10.1073/pnas.0510157103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine W.M. Extraterrestrial organic matter: a review. Orig. Life Evol. Biosph. 1998;28:365–383. doi: 10.1023/a:1006574110907. doi:10.1023/A:1006574110907 [DOI] [PubMed] [Google Scholar]
- Ishii M, Miyake T, Satoh T, Sugiyama H, Oshima Y, Kodama T, Igarashi Y. Autotrophic carbon dioxide fixation in Acidianus brierleyi. Arch. Microbiol. 1997;166:368–371. doi: 10.1007/BF01682981. doi:10.1007/s002030050397 [DOI] [PubMed] [Google Scholar]
- Ivanovsky R.N, Krasilnikova E.N, Fal Y.I. A pathway of the autotrophic CO2 fixation in Chloroflexus aurantiacus. Arch. Microbiol. 1993;159:257–264. doi:10.1007/BF00248481 [Google Scholar]
- Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. doi:10.1126/science.860134 [DOI] [PubMed] [Google Scholar]
- Kanamori T, Kanou N, Atomi H, Imanaka T. Enzymatic characterization of prokaryotic urea carboxylase. J. Bacteriol. 2004;186:2532–2539. doi: 10.1128/JB.186.9.2532-2539.2004. doi:10.1128/JB.186.9.2532-2539.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolber Z.S, et al. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science. 2001;292:2492–2495. doi: 10.1126/science.1059707. doi:10.1126/science.1059707 [DOI] [PubMed] [Google Scholar]
- Kopp R.E, Kirschvink J.L, hilburn I.A, Nash C.Z. The Paleoproterozoic Snowball Earth: a climate disaster triggered by the evolution of oxygenic photosynthesis. Proc. Natl Acad. Sci. USA. 2005;102:11 131–11 136. doi: 10.1073/pnas.0504878102. doi:10.1073/pnas.0504878102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühl M, Chen M, Ralph P.J, Schreiber U, Larkum A.W.D. A niche for cyanobacteria containing chlorophyll d. Nature. 2005;433:820. doi: 10.1038/433820a. doi:10.1038/433820a [DOI] [PubMed] [Google Scholar]
- Lazcano A, Miller S.L. On the origin of metabolic pathways. J. Mol. Evol. 1999;49:424–431. doi:10.1007/PL00006565 [PubMed] [Google Scholar]
- Lin C, Shalitin D. Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. doi:10.1146/annurev.arplant.54.110901.160901 [DOI] [PubMed] [Google Scholar]
- Lovelock J.E. Thermodynamics and the recognition of alien biospheres. Proc. R. Soc. B. 1975;189:167–181. doi:10.1098/rspb.1975.0051 [Google Scholar]
- McFadden B.A. Autotrophic CO2 fixation and the evolution of ribulose diphosphate carboxylase. Bacteriol. Rev. 1973;37:289–319. doi: 10.1128/br.37.3.289-319.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, R. 2005 Solar radiation. See http://home.iprimus.com.au/nielsens/
- Nikolau B.J, Ohlrogge J.B, Wurtele E.S. Plant biotin-containing carboxylases. Arch. Biochem. Biophys. 2003;414:211–222. doi: 10.1016/s0003-9861(03)00156-5. doi:10.1016/S0003-9861(03)00156-5 [DOI] [PubMed] [Google Scholar]
- Nisbet E.G, Carr J.R, Van Dover C.L. Origins of photosynthesis. Nature. 1995;373:479–480. doi:10.1038/373479a0 [Google Scholar]
- Nisbet E.G, Grassineau N.V, Howe C.J, Abell P.I, Regelous M, Nisbet R.E.R. The age of rubisco: the evolution of oxygenic photosynthesis. Geobiology. 2007;5:311–335. doi:10.1111/j.1472-4669.2007.00127.x [Google Scholar]
- Orgel L.E. Self-organizing biochemical cycles. Proc. Natl Acad. Sci. USA. 2000;97:12 503–12 507. doi: 10.1073/pnas.220406697. doi:10.1073/pnas.220406697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain R.H. The evolution of enzyme activity. Nature. 1982;299:486–487. doi: 10.1038/299486a0. doi:10.1038/299486a0 [DOI] [PubMed] [Google Scholar]
- Permentier H.P, Neerken S, Overmann J, Amesz J. A bacteriochlorophyll a antenna complex from purple bacteria absorbing at 963 nm. Biochemistry. 2001;40:5573–5578. doi: 10.1021/bi0024308. doi:10.1021/bi0024308 [DOI] [PubMed] [Google Scholar]
- Raven J.A. Phagotrophy in phototrophs. Limnol. Oceanogr. 1997;42:198–205. [Google Scholar]
- Raven J.A. Photosynthesis in watercolours. Nature. 2007;448:418. doi: 10.1038/448418a. doi:10.1038/448418a [DOI] [PubMed] [Google Scholar]
- Raven J.A, Cockell C.S. Influence on photosynthesis of starlight, moonlight, planetlight, and light pollution (reflections on photosynthetically active radiation in the universe) Astrobiology. 2006;6:668–675. doi: 10.1089/ast.2006.6.668. doi:10.1089/ast.2006.6.668 [DOI] [PubMed] [Google Scholar]
- Rosing M.T, Frei R. U-rich Archaean sea-floor sediments from Greenland indications of >3700 Ma oxygenic photosynthesis. Earth Planet. Sci. Lett. 2004;217:237–244. doi:10.1016/S0012-821X(03)00609-5 [Google Scholar]
- Rothschild L.J. A model for diurnal patterns of carbon fixation in a Precambrian microbial mat based on a modern analog. Biosystems. 1991;25:13–23. doi: 10.1016/0303-2647(91)90009-a. doi:10.1016/0303-2647(91)90009-A [DOI] [PubMed] [Google Scholar]
- Rothschild L.J, Giver L.J. Photosynthesis below the surface in a cryptic microbial mat. Int. J. Astrobiol. 2003;1:295–304. doi:10.1017/S1473550403001320 [Google Scholar]
- Rothschild L.J, Mancinelli R.L. Model of carbon fixation in microbial mats from 3,500 Myr ago to the present. Nature. 1990;345:710–712. doi: 10.1038/345710a0. doi:10.1038/345710a0 [DOI] [PubMed] [Google Scholar]
- Sancar A. Crytochrome: the second photoactive pigment in the eye and its role in circadian photoreception. Annu. Rev. Biochem. 2000;69:31–67. doi: 10.1146/annurev.biochem.69.1.31. doi:10.1146/annurev.biochem.69.1.31 [DOI] [PubMed] [Google Scholar]
- Sato T, Atomi H, Imanaka T. Archaeal type III rubiscos function in a pathway for AMP metabolism. Science. 2007;315:1003–1006. doi: 10.1126/science.1135999. doi:10.1126/science.1135999 [DOI] [PubMed] [Google Scholar]
- Schenk S, Decker K. Horizontal gene transfer involved in the convergent evolution of the plasmid-encoded enantioselective 6-hydroxynicotine oxidases. J. Mol. Evol. 1999;48:178–186. doi: 10.1007/pl00006456. doi:10.1007/PL00006456 [DOI] [PubMed] [Google Scholar]
- Schidlowski M. A 3,800-million-year isotopic record of life from carbon in sedimentary rocks. Nature. 1988;333:313–318. doi:10.1038/333313a0 [Google Scholar]
- Schopf J.W. Paleobiology of the Archean. In: Schopf J.W, Klein C, editors. The Proterozoic biosphere. Cambridge University Press; Cambridge, UK: 1992. pp. 25–39. [Google Scholar]
- Shi T, Bibby T.S, Jiang L, Irwin A.J, Falkowski P.G. Protein interactions limit the rate of evolution of photosynthetic genes in cyanobacteria. Mol. Biol. Evol. 2005;22:2179–2189. doi: 10.1093/molbev/msi216. doi:10.1093/molbev/msi216 [DOI] [PubMed] [Google Scholar]
- Shiba H, Kawasumi T, Igarashi Y, Kodama T, Minoda Y. The CO2 assimilation via the reductive tricarboxylic acid cycle in an autotrophic, aerobic hydrogen-oxidizing bacterium Hydrogenobacter thermophilus. Arch. Microbiol. 1985;141:198–203. doi:10.1007/BF00408058 [Google Scholar]
- Shively J.M, Barton L.L. Variations in autotrophic life. Academic Press; London, UK: 1991. p. 346. [Google Scholar]
- Shively J.M, van Keulen G, Meijer W.G. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 1998;52:191–230. doi: 10.1146/annurev.micro.52.1.191. doi:10.1146/annurev.micro.52.1.191 [DOI] [PubMed] [Google Scholar]
- Smith E, Morowitz H.J. Universality in intermediary metabolism. Proc. Natl Acad. Sci. USA. 2004;101:13 168–13 173. doi: 10.1073/pnas.0404922101. doi:10.1073/pnas.0404922101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow T.P, Witt A.N. The interstellar carbon budget and the role of carbon in dust and large molecules. Science. 1995;270:1455–1460. doi: 10.1126/science.270.5241.1455. doi:10.1126/science.270.5241.1455 [DOI] [PubMed] [Google Scholar]
- Stomp M, Huisman J, de Jongh F, Veraart A.J, Gerla D, Rijkeboer M, Ibelings B.W, Wollenzien U.I.A, Stal L.J. Adaptive divergence in pigment composition promotes phytoplankton biodiversity. Nature. 2004;432:104–107. doi: 10.1038/nature03044. doi:10.1038/nature03044 [DOI] [PubMed] [Google Scholar]
- Tarter J.C, et al. A re-appraisal of the habitability of planets around M dwarf stars. Astrobiology. 2007;7:30–65. doi: 10.1089/ast.2006.0124. doi:10.1089/ast.2006.0124 [DOI] [PubMed] [Google Scholar]
- Tcherkez G.G.B, Farquhar G.D, Andrews T.J. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl Acad. Sci. USA. 2006;203:7246–7251. doi: 10.1073/pnas.0600605103. doi:10.1073/pnas.0600605103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice M.M, Lowe D.R. Hydrogen-based carbon fixation in the earliest photosynthetic organisms. Geology. 2006;34:37–40. doi:10.1130/G22012.1 [Google Scholar]
- Vershinin A. Biological functions of carotenoids—diversity and evolution. BioFactors. 1999;10:99–104. doi: 10.1002/biof.5520100203. [DOI] [PubMed] [Google Scholar]
- Visser C.M, Kellogg R.M. Biotin. Its place in evolution. J. Mol. Evol. 1978;11:171–187. doi: 10.1007/BF01733892. doi:10.1007/BF01733892 [DOI] [PubMed] [Google Scholar]
- Vitkup D, Kharchenko P, Wagner A. Influence of metabolic network structure and function on enzyme evolution. Genome Biol. 2006;7:R39. doi: 10.1186/gb-2006-7-5-r39. doi:10.1186/gb-2006-7-5-r39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtershauser G. Evolution of the first metabolic cycles. Proc. Natl Acad. Sci. USA. 1990;87:200–204. doi: 10.1073/pnas.87.1.200. doi:10.1073/pnas.87.1.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M.M, Lowe D.R. Filamentous microfossils from the 3,500-Myr-old Onverwacht Group, Barberton Mountain Land, South Africa. Nature. 1985;314:530–532. doi: 10.1016/0301-9268(92)90074-x. doi:10.1038/314530a0 [DOI] [PubMed] [Google Scholar]
- Walter M.R. Archean stromatolites: evidence of the Earth's earliest benthos. In: Schopf J.W, editor. Earth's earliest biosphere. Princeton University Press; Princeton, NJ: 1983. pp. 187–213. [Google Scholar]
- Williams T.J, Zhang C.L, Scott J.H, Bazylinski D.A. Evidence for autotrophy via the reverse tricarboxylic acid cycle for the marine magnetotactic coccus strain MC-1. Appl. Environ. Microbiol. 2006;72:1322–1329. doi: 10.1128/AEM.72.2.1322-1329.2006. doi:10.1128/AEM.72.2.1322-1329.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstencroft R.D, Raven J.A. Photosynthesis: likelihood of occurrence and possibility of detection on Earth-like planets. Icarus. 2002;157:535–548. doi:10.1006/icar.2002.6854 [Google Scholar]
- Wood H.G, Ljungdahl L.G. Autotrophic character of the acetogenic bacteria. In: Shively J.M, Barton L.L, editors. Variations in autotrophic life. Academic Press; London, UK: 1991. pp. 201–250. [Google Scholar]
- Wu G, Fiser A, ter Kuile B, Sali A, Müller M. Convergent evolution of Trichomonas vaginalis lactate dehydrogenase from malate dehydrogenase. Proc. Natl Acad. Sci. USA. 1999;96:6285–6290. doi: 10.1073/pnas.96.11.6285. doi:10.1073/pnas.96.11.6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Bauer C.E. Complex evolution of photosynthesis. Annu. Rev. Plant Biol. 2002;53:503–521. doi: 10.1146/annurev.arplant.53.100301.135212. doi:10.1146/annurev.arplant.53.100301.135212 [DOI] [PubMed] [Google Scholar]