Abstract
OBJECTIVE—Elevated plasma triglyceride concentration is a component of the insulin resistance syndrome and is commonly associated with type 2 diabetes, obesity, and coronary heart disease. The goal of our study was to perform a genome-wide linkage scan to identify genetic regions that influence variation in plasma triglyceride levels in families that are enriched with individuals with type 2 diabetes.
RESEARCH DESIGN AND METHODS—We used phenotypic and genotypic data from 1,026 individuals distributed across 294 Mexican-American families, who were ascertained for type 2 diabetes, from the Veterans Administration Genetic Epidemiology Study (VAGES). Plasma triglyceride values were transformed, and a variance-components technique was used to conduct multipoint linkage analysis.
RESULTS—After adjusting for the significant effects of sex and BMI, heritability for plasma triglycerides was estimated as 46 ± 7% (P < 0.0001). Multipoint linkage analysis yielded the strongest evidence for linkage of plasma triglycerides near marker D12S391 on chromosome 12p (logarithm of odds [LOD] = 2.4). Our linkage signal on chromosome 12p provides independent replication of a similar finding in another Mexican-American sample from the San Antonio Family Diabetes Study (SAFDS). Combined multipoint linkage analysis of the VAGES and SAFDS data yielded significant evidence for linkage of plasma triglycerides to a genetic location between markers GATA49D12 and D12S391 on 12p (LOD = 3.8, empirical P value = 2.0 × 10−5). This region on 12p harbors the gene-encoding adiponectin receptor 2 (AdipoR2), where we previously have shown that multiple single nucleotide polymorphisms are associated with plasma triglyceride concentrations in the SAFDS. In the present study, we provided suggestive evidence in favor of association for rs929434 with triglyceride concentrations in the VAGES.
CONCLUSIONS—Collectively, these results provide strong evidence for a major locus on chromosome 12p that influences plasma triglyceride levels in Mexican Americans.
The insulin resistance (metabolic) syndrome is characterized by an increase in plasma triglyceride concentration, reduced HDL, visceral obesity, elevated blood pressure, and glucose intolerance (1,2). The prevalence of the insulin resistance syndrome and/or related diseases, including obesity and type 2 diabetes, is rapidly increasing. Although major risk factors for these disorders have been identified, the genetic factors remain largely unknown. Elevated plasma triglyceride concentration is a component of the insulin resistance syndrome and is commonly associated with type 2 diabetes, obesity, and coronary heart disease (1,2). Since knowledge of specific genetic determinants of the common forms of hypertriglyceridemia is limited, we conducted a genome-wide linkage scan to localize genes that influence variation in plasma triglyceride levels in Mexican-American families ascertained for type 2 diabetes from the Veterans Administration Genetic Epidemiology Study (VAGES).
Genome-wide linkage screens for plasma triglyceride levels have been performed in several populations, including our previous study in Mexican Americans from the San Antonio Family Diabetes Study (SAFDS) (3,4). In that study, we reported significant evidence for the linkage of triglycerides to a genetic location on chromosome 15q and found potential evidence for linkage on several chromosomal regions including 12p (3,4). Linkage to chromosomes 2p, 11p, and 11q also have been reported for plasma triglyceride levels in French-Canadian families from the Quebec Family Study (5). In addition, a study performed in families with northern European ancestry demonstrated evidence for linkage to plasma triglyceride levels on chromosome 19 (6). Additionally, a study in a Chinese population from the Hong Kong Family Diabetes Study identified several chromosomes, including 2q, 3q, 6q, 9q, 10q, and 17q, containing genes that influence variation in triglyceride levels (7). Other evidence for the linkage of plasma triglyceride levels has been reported on 8q in African Americans (8), 2q in Hutterites (9), 11q in Old Order Amish (10), and 10p in Finnish families (11). More recently, a meta-analysis of genome-wide linkage studies for plasma triglyceride levels provided suggestive evidence for linkage at chromosome 7p (12). The purpose of this study was to perform a genome-wide linkage scan to identify genetic regions that influence variation in plasma triglyceride levels in Mexican-American individuals from VAGES, in which the families were ascertained for type 2 diabetes.
RESEARCH DESIGN AND METHODS
VAGES.
The study participants for whom both genetic and phenotypic data were available consisted of 1,026 Mexican Americans distributed across 294 mostly nuclear families from VAGES. Of 294 families, however, 41 were found to be represented by single individuals with triglyceride data. These 41 single, unrelated individuals were considered for the analysis because they contribute to the evaluation of covariate effects. The remaining 985 individuals from 253 families generated 1,715 relative pairs that were distributed across nine relative-pair categories (Table 1). The VAGES families were ascertained on at least two siblings affected with type 2 diabetes and only one parent with type 2 diabetes. Metabolic, anthropometric, demographic, and medical history information were obtained on all examined individuals. All procedures were approved by the institutional review board of the University of Texas Health Science Centre at San Antonio, and all subjects gave informed written consent before their participation.
TABLE 1.
Distribution of relative pairs by category in the VAGES and SAFDS families
| Relative pair | Relationship coefficient* | Number of VAGES pairs† | Number of SAFDS pairs‡ |
|---|---|---|---|
| Parent-offspring | 0.5 | 416 | 463 |
| Siblings | 0.5 | 1,003 | 586 |
| Grandparent-grandchild | 0.25 | 19 | 123 |
| Avuncular | 0.25 | 184 | 810 |
| Half-siblings | 0.25 | 47 | 88 |
| First and second cousins | 0.15625 | — | 20 |
| Double first cousins | 0.25 | — | 10 |
| Great grandparent-grandchild | 0.125 | — | 12 |
| Grand avuncular | 0.125 | 6 | 204 |
| Half-avuncular | 0.125 | 15 | 106 |
| First cousins | 0.125 | 24 | 793 |
| Double first cousins, once removed | 0.125 | — | 35 |
| First and second cousins, once removed | 0.078125 | — | 22 |
| Great grand avuncular | 0.0625 | — | 2 |
| Half grand avuncular | 0.0625 | — | 5 |
| First cousins, once removed | 0.0625 | 1 | 759 |
| Half first cousins | 0.0625 | — | 77 |
| Double second cousins | 0.0625 | — | 25 |
| Second and third cousins | 0.0390625 | — | 6 |
| First cousins, twice removed | 0.03125 | — | 30 |
| Half first cousins, once removed | 0.03125 | — | 13 |
| Second cousins | 0.03125 | — | 287 |
| Second cousins, once removed | 0.015625 | — | 75 |
| Third cousins | 0.0078125 | — | 8 |
| Total | 1,715 | 4,559 |
The relationship coefficient is 2 × coefficient of kinship of two individuals.
The total data used for the present study were obtained from 1,026 individuals distributed across 294 families. Of 1,026, however, 41 were unrelated individuals (see text for details).
The total data used for the present study were obtained from 537 individuals distributed across 31 families.
Blood samples were obtained after a 12-h fast. Plasma triglyceride concentrations were determined by an enzymatic colorimetric quantification method (Wako Chemicals, Nuess, Germany). After excluding three extreme outliers, plasma triglyceride values were available for 1,023 individuals and were used to conduct the present study, and no values >1 SD from the next lowest value were included. To address the issue of continued nonnormality, plasma triglyceride values were transformed using an inverse-normal transformation.
Genotype data, genetic map, and estimation of identity-by-descent matrices.
DNA was prepared from whole blood (Gentra Systems, Minneapolis, MN). The Center for Inherited Disease Research (CIDR) performed a 10-cM genome scan using the DNA samples from the VAGES participants. We used CIDR genotypic data on 385 highly polymorphic autosomal markers. The program PREST (13) was used to resolve pedigree discrepancies. The genotype data were cleaned for both Mendelian inconsistencies and spurious double recombinants using the program SimWalk2 (14,15). Overall, the blanking rate for errors was <0.22% of the total number of genotypes. The PEDSYS (16) program INFER was used to infer unknown genotypes from the genotypes of relatives, when possible without ambiguity. The genetic maps used for the present analyses were similar to the Marshfield maps. The multipoint identical-by-descent (IBD) matrices given a number of genetic markers (map distance in Haldane cM) were calculated using Markov chain Monte Carlo methods implemented in the program Loki (17,18).
Variance-components linkage analysis.
We used a pedigree-based multipoint variance-components approach to test for linkage between genetic locations and the transformed triglyceride levels using maximum-likelihood methods (19–21). The variance-components method uses information from all possible biological relationships simultaneously to disentangle the genetic architecture of the quantitative trait. This method specifies the expected genetic covariances between relatives as a function of their IBD relationships at a marker locus (which is hypothesized to be linked to a locus influencing the quantitative trait [QTL]). It allows for locus-specific effects (h2q is heritability attributed to the QTL), residual additive genetic effects (h2 is heritability attributed to the residual genetic effects), and individual-specific random environmental factors [e2 = 1 − (h2q + h2)].
In addition to the variance components, mean ± SD of the phenotype and covariate effects (e.g., age and BMI) were simultaneously estimated by maximum-likelihood techniques. Hypothesis testing was performed by likelihood ratio tests. The hypothesis of no linkage (i.e., additive genetic variance due to the QTL = 0) was tested by comparing the likelihood of this restricted model with that of a model in which the additive genetic variance due to the QTL is estimated. The difference between the two log10 likelihoods produces a logarithm of odds (LOD) score. Twice the difference in the loge likelihoods of these models yields a test statistic that is asymptotically distributed as the ½:½ mixture of χ21 and a point mass at 0, denoted by χ20, where the degree of freedom is equal to the difference in the number of parameters estimated between the two competing models (22). An LOD score of ≥3.0 was considered as strong evidence in support of linkage. For the purpose of discussion, other genetic regions across the genome with LOD scores ≥1.2 and ≥1.9 are considered evidence for potential linkage and suggestive linkage, respectively. Given the initial set of covariates of transformed triglyceride levels considered for an analysis, the program SOLAR was used to evaluate the statistical significance (P < 0.05) of a given covariate using likelihood ratio tests. Each of these tests involved comparison for a given parameter and had one degree of freedom. Given the VAGES family ascertainment criteria, all of our genetic analyses incorporated correction for ascertainment bias by computing the likelihood of a pedigree conditional on the transformed triglyceride value of the proband, using the program SOLAR. Since the VAGES families had more than one proband per family, we randomly chose one of the probands from each family to correct for ascertainment bias since correcting for ascertainment bias in datasets ascertained on multiple affected probands can result in a loss of power to detect linkage (23,24). We performed simulation analysis to determine the empirical P value to verify our major linkage finding. A fully informative marker, not linked to triglyceride, was simulated, IBD information for this simulated marker was calculated, and linkage analysis was performed. The empirical P value was determined based on information obtained from 100,000 replicates. The statistical genetic analytical procedures were implemented in the SOLAR program.
Single nucleotide polymorphism genotyping and association analysis.
We typed rs929434 in AdipoR2 in the VAGES data to perform association analysis with triglyceride concentrations using methods described previously (4). DNA samples for genotyping were available for 907 individuals only.
SAFDS.
The goals and related genetic investigations of SAFDS have been extensively described previously (25–27). Briefly, probands for SAFDS were individuals with type 2 diabetes, and all of the probands’ first-, second-, and third-degree relatives aged ≥18 years were invited to participate in the study. Type 2 diabetes–related phenotypes were collected, and a 10-cM genome scan was performed by CIDR. The SAFDS baseline data used for this study were reported in Table 2, and 537 individuals from 31 complex, multigenerational families generated 4,559 relative pairs that were distributed across 24 relative-pair categories (Table 1).
TABLE 2.
Characteristics of VAGES and SAFDS participants for whom both phenotypic and genotypic data were available
| Variable | VAGES | SAFDS |
|---|---|---|
| n | 1,026 | 537 |
| Age at examination (years) | 49.6 ± 13.1 | 42.1 ± 16.8 |
| Female subjects (%) | 65.9 | 58.3 |
| Diabetic subjects (%) | 68.6 | 24.6 |
| BMI (kg/m2) | 33.2 ± 8.0 | 30.2 ± 6.6 |
| ln BMI* | 3.5 ± 0.2 | 3.4 ± 0.2 |
| Plasma triglycerides (mg/dl)† | 159 ± 136 | 180 ± 135 |
Data are means ± SD or percent.
ln = log transformed.
Plasma triglyceride values were transformed for the genetic analysis using inverse normal transformation.
RESULTS
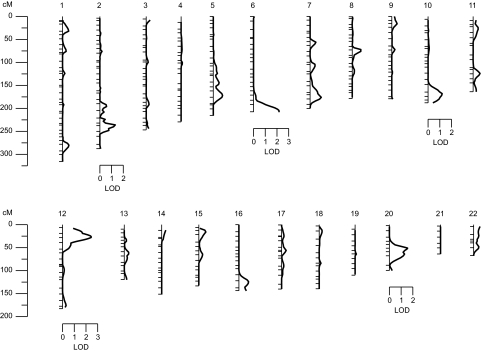
The characteristics of the VAGES participants are reported in Table 2. After accounting for the significant covariate effects of sex and log-transformed BMI (ln BMI), the overall polygenic heritability (h2) for transformed plasma triglyceride values was estimated to be 46 ± 7%, with high statistical significance (P < 0.0001). It should be noted that normality was maintained approximately, after adjustment for covariate influences (i.e., the skewness and kurtosis coefficients of the residuals is −0.01 and −0.11, respectively). The initial set of covariates considered for evaluation of statistical significance included sex, age, age2, age × sex, age2 × sex, and ln BMI. Following the determination of the heritability for triglycerides, the multipoint linkage analysis was conducted, and the results of our genome-wide linkage screen (i.e., peak LOD score by chromosome) are summarized in Fig. 1.
FIG. 1.
Summary of the genome-wide linkage scan for genes influencing (transformed) plasma triglyceride levels in VAGES.
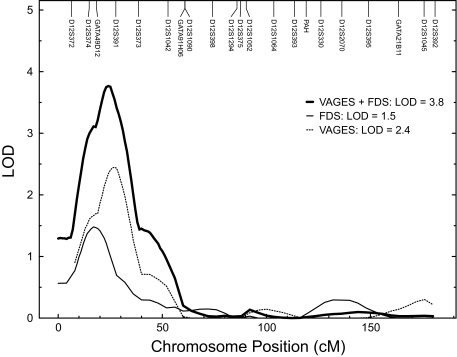
The strongest evidence for linkage (LOD = 2.4) of triglycerides occurred on chromosome 12 at ∼26 cM near marker D12S391. The multipoint LOD scores obtained for transformed plasma triglyceride were plotted against map positions on chromosome 12 (Fig. 2). In addition to the evidence for suggestive triglyceride linkage on chromosome 12 near marker D12S391, four other regions on four separate chromosomes exhibited potential evidence for linkage, with LOD scores ≥1.2 (Table 3).
FIG. 2.
Multipoint linkage results for (transformed) plasma triglyceride levels at the chromosome 12 region in the VAGES, SAFDS, and combined VAGES and SAFDS datasets.
TABLE 3.
Chromosomal regions potentially linked (LOD ≥1.2) to (transformed) plasma triglyceride levels
| Nearest marker | Distance (cM)* | Chromosome location | LOD |
|---|---|---|---|
| D2S434 | 216 | 2q | 1.3 |
| D6S1027 | 187 | 6q | 2.2 |
| D10S1656 | 149 | 10q | 1.2 |
| D20S477 | 47 | 20p | 1.5 |
Marshfield data.
Our linkage on chromosome 12p provides an independent replication of a similar finding in other Mexican-American data obtained from the SAFDS (3,4). Given that SAFDS has a new genome scan performed by CIDR (26), we reanalyzed the SAFDS data (3) but included all of the available triglyceride information (n = 537 compared with the initial linkage data based on a sample of 418 individuals) (Table 2) and the new (CIDR) chromosome 12 marker data. As in the case of VAGES, the plasma triglyceride values in SAFDS were transformed using the inverse-normal transformation and the analysis incorporated correction for the ascertainment bias given the SAFDS ascertainment scheme (3). After adjusting for the significant covariate effects of age, sex, age × sex, age2, and ln BMI on triglycerides, the reanalysis (the skewness and kurtosis values for residuals were 0.08 and 0.04, respectively) yielded potential evidence for linkage (LOD = 1.5) at ∼17 cM near marker GATA49D12. Following this, we performed a combined VAGES and SAFDS plasma triglyceride linkage analysis using a combined chromosome 12 map and study-specific multipoint IBD matrices. After accounting for the above-said covariates and study as an additional covariate (the skewness and kurtosis coefficients for residuals were 0.07 and 0.02, respectively), the combined multipoint linkage analysis yielded significant evidence for linkage of transformed plasma triglycerides (LOD = 3.8, empirical P value = 2.0 × 10−5) to a genetic location at ∼ 24 cM between markers GATA49D12 (17 cM) and D12S391 (26 cM) on chromosome 12p (Fig. 2). The empirical P value that corresponds to our observed LOD score is similar to our nominal P value, which suggests that our nominal P value has not overstated the evidence of linkage.
As part of the present study, we genotyped (n = 907) the most strongly associated single nucleotide polymorphisms (SNPs) (rs929434) with triglycerides observed in the SAFDS (P = 0.00016) (4), in the VAGES data and found suggestive evidence in favor of association (P = 0.06505). The rare allele was associated with decreased triglyceride concentrations in both the SAFDS and VAGES datasets. We obtained the combined P value (0.00013) using Fisher's combined P value approach, in turn finding a slight improvement in overall evidence for association. In the VAGES data, however, the observed suggestive association explains only a small portion (∼5%) of the initial linkage signal.
DISCUSSION
In this study, we report the results from a genome-wide linkage scan undertaken in VAGES to identify genes that influence variation in plasma triglyceride levels. We found that plasma triglyceride levels were suggestively linked to a genetic location near marker D12S391 on chromosome 12. Potential evidence for linkage also was observed on chromosomes 2q, 6q, 10q, and 20p. The linkage on chromosome 12 was an independent replication of our previously (or presently) reported triglyceride linkage finding in Mexican Americans from SAFDS, where we found modest evidence for linkage of plasma triglyceride levels at 12p (3,4). Our combined linkage analysis of VAGES and SAFDS increased our power to detect statistically significant evidence for linkage at 12p (LOD = 3.8, empirical P value = 2.0 × 10−5). Taken together, these results provide strong evidence for a major susceptibility locus on 12p that influences plasma triglyceride levels in Mexican Americans.
Other genome-wide linkage screens in different populations reported evidence of linkage for plasma triglyceride levels on chromosomes 2p, 2q, 3q, 6q, 8q, 9q, 10p, 10q, 11p, 11q, 17q, and 19q (5–11). With the exception of 2q, 6q, and 10q, the majority of these loci appear to have no strong effects in the Mexican Americans. Consistent with our findings, a genome-wide linkage scan of plasma triglyceride concentrations in the Framingham Heart Study identified modest evidence for linkage on 12p (28). In contrast, the linkage of 12p to plasma triglyceride levels has not been replicated in the studies mentioned above (5–11), including a meta-analysis of genome-wide linkage studies for plasma triglyceride levels (12). This lack of replication is not surprising and may arise due to factors such as different study designs, sample size, choice or power of the statistical methods used, and genuine genetic heterogeneity. In addition, replication of a true linkage in a phenotype with complex inheritance patterns (e.g., oligogenic inheritance) appears to be difficult since it is likely to require a larger number of families than is required for initial detection (29). As can be seen from our present study, however, the combined VAGES and SAFDS linkage analysis provided stronger evidence for linkage of triglyceride levels on chromosome 12p compared with the independent linkage information provided by VAGES and SAFDS separately. In addition to these genome-wide linkage findings, more recently, genome-wide association studies have also identified additional loci/SNPs associated with triglyceride levels including GCKR, LPL, APOA5, and TRIB1 (30–33). These reported loci/SNPs do not overlap with the chromosomal regions that we have identified in the present study.
Many genes that could plausibly play a role in the regulation of plasma triglyceride levels reside within the critical linkage region at 12p. One of these positional candidate genes is adiponectin receptor 2 (AdipoR2, 12p13.33). AdipoR2 is one of two receptors that mediate the effects of adiponectin, an important regulator of energy homeostasis and glucose and lipid metabolism (34). As we reported previously (4), multiple SNPs in AdipoR2 were significantly associated with decreased plasma triglyceride levels (4). In this same study, we demonstrated that one of these SNPs (rs929434) accounted for 47% of the linkage in the previously reported genome-wide linkage scan performed in a subset of SAFDS (3). In the present study, we found suggestive evidence in favor of association with rs929434 (P = 0.06505) in VAGES, and the observed suggestive association explains only a small portion (∼5%) of the initial linkage signal. Following these findings, we have started typing a large battery of SNPs in AdipoR2 to further confirm the association between genetic variation in AdipoR2 and triglyceride levels.
In addition to our own findings, several other groups have reported that polymorphisms in AdipoR2 are associated with type 2 diabetes, obesity, and other components of the insulin resistance syndrome (35–37). Interestingly, one of these association studies reported that AdipoR2 polymorphisms were associated with decreased plasma triglyceride levels (35). Another gene of interest in this region of 12p is the wingless-type MMTV integration site family, member 5B (WNT5B). An association study in a Japanese population identified WNT5B as a new candidate for conferring susceptibility to type 2 diabetes (38). Their results suggest that the WNT5B gene plays an important role in the regulation of adipocyte function and might contribute to the pathogenesis of type 2 diabetes (38).
In conclusion, we found evidence for a major susceptibility locus on chromosome 12p influencing plasma triglyceride levels in Mexican Americans. The findings of the present study, combined with the SNP association studies performed in AdipoR2 (4,35), indicate that this locus appears to have substantial influence on the phenotypic variation in plasma triglyceride levels in Mexican Americans. Further fine-structure mapping will be required to identify the remaining potential functional variants in this region contributing to the linkage signal.
Acknowledgments
This work was supported by a Veterans Administration Epidemiologic grant and supported in part by grants from the National Institutes of Health (DK42273, DK47482, DK53889, and DK70746). This work was also supported by an American Heart Association National Scientist Development grant (to D.K.C.). We thank CIDR for providing a genome scan using the VAGES data.
No potential conflicts of interest relevant to this article were reported.
We thank Marcel J. Fourcaudot and Lenore M. Rodriguez for excellent technical assistance. We thank our nurses (James King, John Kincaid, Rose Kaminski-Graham, and Norma Diaz) for their excellent care of the patients throughout the study. We thank the participants of VAGES and are grateful for their participation and cooperation.
Published ahead of print at http://diabetes.diabetesjournals.org on 17 October 2008.
D.K.C. is currently affiliated with the Center for Metabolic Biology, College of Liberal Arts and Sciences, Arizona State University, Tempe, Arizona.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Reaven GM: Pathophysiology of insulin resistance in human disease. Physiol Rev 75: 473–486, 1995 [DOI] [PubMed] [Google Scholar]
- 2.DeFronzo RA: Insulin resistance: a multifaceted syndrome responsible for NIDDM, obesity, hypertension, dyslipidaemia and atherosclerosis. Neth J Med 50: 191–197, 1997 [DOI] [PubMed] [Google Scholar]
- 3.Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, O'Connell P, Stern MP: A major susceptibility locus influencing plasma triglyceride concentrations is located on chromosome 15q in Mexican Americans. Am J Hum Genet 66: 1237–1245, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richardson DK, Schneider J, Fourcaudot MJ, Rodriguez LM, Arya R, Dyer TD, Almasy L, Blangero J, Stern MP, DeFronzo RA, Duggirala R, Jenkinson CP: Association between variants in the genes for adiponectin and its receptors with insulin resistance syndrome (IRS)-related phenotypes in Mexican Americans. Diabetologia 49: 2317–2328, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Bosse Y, Chagnon YC, Despres JP, Rice T, Rao DC, Bouchard C, Perusse L, Vohl MC: Genome-wide linkage scan reveals multiple susceptibility loci influencing lipid and lipoprotein levels in the Quebec Family Study. J Lipid Res 45: 419–426, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Elbein SC, Hasstedt SJ: Quantitative trait linkage analysis of lipid-related traits in familial type 2 diabetes: evidence for linkage of triglyceride levels to chromosome 19q. Diabetes 51: 528–535, 2002 [DOI] [PubMed] [Google Scholar]
- 7.Ng MC, So WY, Lam VK, Cockram CS, Bell GI, Cox NJ, Chan JC: Genome-wide scan for metabolic syndrome and related quantitative traits in Hong Kong Chinese and confirmation of a susceptibility locus on chromosome 1q21–q25. Diabetes 53: 2676–2683, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Kullo IJ, Ding K, Boerwinkle E, Turner ST, de Andrade M: Quantitative trait loci influencing low density lipoprotein particle size in African Americans. J Lipid Res 47: 1457–1462, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Newman DL, Abney M, Dytch H, Parry R, McPeek MS, Ober C: Major loci influencing serum triglyceride levels on 2q14 and 9p21 localized by homozygosity-by-descent mapping in a large Hutterite pedigree. Hum Mol Genet 12: 137–144, 2003 [DOI] [PubMed] [Google Scholar]
- 10.Pollin TI, Hsueh WC, Steinle NI, Snitker S, Shuldiner AR, Mitchell BD: A genome-wide scan of serum lipid levels in the Old Order Amish. Atherosclerosis 173: 89–96, 2004 [DOI] [PubMed] [Google Scholar]
- 11.Pajukanta P, Terwilliger JD, Perola M, Hiekkalinna T, Nuotio I, Ellonen P, Parkkonen M, Hartiala J, Ylitalo K, Pihlajamäki J, Porkka K, Laakso M, Viikari J, Ehnholm C, Taskinen MR, Peltonen L: Genomewide scan for familial combined hyperlipidemia genes in finnish families, suggesting multiple susceptibility loci influencing triglyceride, cholesterol, and apolipoprotein B levels. Am J Hum Genet 64: 1453–1463, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malhotra A, Elbein SC, Ng MC, Duggirala R, Arya R, Imperatore G, Adeyemo A, Pollin TI, Hsueh WC, Chan JC, Rotimi C, Hanson RL, Hasstedt SJ, Wolford JK: Meta-analysis of genome-wide linkage studies of quantitative lipid traits in families ascertained for type 2 diabetes. Diabetes 56: 890–896, 2007 [DOI] [PubMed] [Google Scholar]
- 13.McPeek MS, Sun L: Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet 66: 1076–1094, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sobel E, Lange K: Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58: 1323–1337, 1996 [PMC free article] [PubMed] [Google Scholar]
- 15.Sobel E, Papp JC, Lange K: Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet 70: 496–508, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyke B: PEDSYS: A Pedigree Data Management System. Population Genetics Laboratory, Department of Genetics, Southwest Foundation for Biomedical Research, San Antonio, 1996. [Tech. rep. no. 2]
- 17.Heath SC: Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet 61: 748–760, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heath SC, Snow GL, Thompson EA, Tseng C, Wijsman EM: MCMC segregation and linkage analysis. Genet Epidemiol 14: 1011–1015, 1997 [DOI] [PubMed] [Google Scholar]
- 19.Amos CI: Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet 54: 535–543, 1994 [PMC free article] [PubMed] [Google Scholar]
- 20.Blangero J, Almasy L: Multipoint oligogenic linkage analysis of quantitative traits. Genet Epidemiol 14: 959–964, 1997 [DOI] [PubMed] [Google Scholar]
- 21.Almasy L, Blangero J: Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62: 1198–1211, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Self SG, Liang K-Y: Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am Stat Assoc 82: 605–610, 1987 [Google Scholar]
- 23.Comuzzie AG, Williams JT: Correcting for ascertainment bias in the COGA data set. Genet Epidemiol 17: 109–114, 1999 [DOI] [PubMed] [Google Scholar]
- 24.Duggirala R, Almasy L, Blangero J, Jenkinson CP, Arya R, DeFronzo RA, Stern MP, O'Connell P, the American Diabetes Association GENNID Study Group: Further evidence for type 2 diabetes susceptibility locus on chromosome 11q. Genet Epidemiol 24: 240–242, 2003 [DOI] [PubMed] [Google Scholar]
- 25.Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, O'Connell P, Stern MP: Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet 64: 1127–1140, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puppala S, Dodd GD, Fowler S, Arya R, Schneider J, Farook VS, Granato R, Dyer TD, Almasy L, Jenkinson CP, Diehl AK, Stern MP, Blangero J, Duggirala R: A genomewide search finds major susceptibility loci for gallbladder disease on chromosome 1 in Mexican Americans. Am J Hum Genet 78: 377–392, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunt KJ, Lehman DM, Arya R, Fowler S, Leach RJ, Göring HH, Almasy L, Blangero J, Dyer TD, Duggirala R, Stern MP: Genome-wide linkage analyses of type 2 diabetes in Mexican Americans: the San Antonio Family Diabetes/Gallbladder Study. Diabetes 54: 2655–2662, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Lin JP: Genome-wide scan on plasma triglyceride and high density lipoprotein cholesterol levels, accounting for the effects of correlated quantitative phenotypes. BMC Genet 31: S47, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suarez BK, Hampe CL, Van Eerdewegh P: Problems of replicating linkage claims in psychiatry. In Genetic Approaches to Mental Disorders. 1st ed. Gershon ES, Cloninger CR, Eds. American Psychiatric, Washington, DC, 1994, p. 23–46
- 30.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, Novartis Institutes of BioMedical Research; Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Boström K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Råstam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjögren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S: Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331–1336, 2007 [DOI] [PubMed] [Google Scholar]
- 31.Kathiresan S, Manning AK, Demissie S, D'Agostino RB, Surti A, Guiducci C, Gianniny L, Burtt NP, Melander O, Orho-Melander M, Arnett DK, Peloso GM, Ordovas JM, Cupples LA: A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet 8: S17, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR: Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40: 161–169, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M: Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet 40: 189–197, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berg AH, Combs TP, Scherer PE: ACRP30/adiponectin: an adipokine regulating glucose and lipid metabolism. Trends Endocrinol Metab 13: 84–89, 2002 [DOI] [PubMed] [Google Scholar]
- 35.Broedl UC, Lehrke M, Fleischer-Brielmaier E, Tietz AB, Nagel JM, Goke B, Lohse P, Parhofer KG: Genetic variants of adiponectin receptor 2 are associated with increased adiponectin levels and decreased triglyceride/VLDL levels in patients with metabolic syndrome. Cardiovasc Diabetol 15: 5–11, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaxillaire M, Dechaume A, Vasseur-Delannoy V, Lahmidi S, Vatin V, Lepretre F, Boutin P, Hercberg S, Charpentier G, Dina C, Froguel P: Genetic analysis of ADIPOR1 and ADIPOR2 candidate polymorphisms for type 2 diabetes in the Caucasian population. Diabetes 55: 856–861, 2006 [DOI] [PubMed] [Google Scholar]
- 37.Damcott CM, Ott SH, Pollin TI, Reinhart LJ, Wang J, O'connell JR, Mitchell BD, Shuldiner AR: Genetic variation in adiponectin receptor 1 and adiponectin receptor 2 is associated with type 2 diabetes in the Old Order Amish. Diabetes 54: 2245–2250, 2005 [DOI] [PubMed] [Google Scholar]
- 38.Kanazawa A, Tsukada S, Sekine A, Tsunoda T, Takahashi A, Kashiwagi A, Tanaka Y, Babazono T, Matsuda M, Kaku K, Iwamoto Y, Kawamori R, Kikkawa R, Nakamura Y, Maeda S: Association of the gene encoding wingless-type mammary tumor virus integration-site family member 5B (WNT5B) with type 2 diabetes. Am J Hum Genet 75: 832–843, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]