Abstract
Umami is one of the 5 basic taste qualities. The umami taste of L-glutamate can be drastically enhanced by 5′ ribonucleotides and the synergy is a hallmark of this taste quality. The umami taste receptor is a heteromeric complex of 2 class C G-protein-coupled receptors, T1R1 and T1R3. Here we elucidate the molecular mechanism of the synergy using chimeric T1R receptors, site-directed mutagenesis, and molecular modeling. We propose a cooperative ligand-binding model involving the Venus flytrap domain of T1R1, where L-glutamate binds close to the hinge region, and 5′ ribonucleotides bind to an adjacent site close to the opening of the flytrap to further stabilize the closed conformation. This unique mechanism may apply to other class C G-protein-coupled receptors.
Keywords: glutamate, G protein-coupled receptors, IMP, T1R
Humans can detect at least 5 basic taste qualities, including sweet, umami, bitter, salty, and sour. Umami, the savory taste of L-glutamate, was first discovered in 1908 by K. Ikeda, but only recently accepted as a basic taste quality by the general public. The most unique characteristic of umami taste is synergism. Purinic ribonucleotides, such as IMP and GMP, can strongly potentiate the umami taste intensity (1). In human taste tests, 200 μM of IMP, which does not elicit any umami taste by itself, can increase one's umami taste sensitivity to glutamate by 15-fold (2).
Among the 5 basic taste qualities, sweet, umami, and bitter taste are mediated by G protein-coupled receptors (GPCRs) (3). Receptors for umami taste and sweet taste are closely related to each other. The 3 subunits of the T1R family form 2 heteromeric receptors: umami (T1R1/T1R3) (2, 4) and sweet (T1R2/T1R3) (2, 5). T1R receptors belong to the class C GPCRs, along with metabotropic glutamate receptors (mGluRs), γ-aminobutyric acid receptor B (GABABR), calcium sensing receptors (CaSR), and so forth. The defining motif in these receptors is an outer membrane N-terminal Venus flytrap (VFT) domain that consists of 2 globular subdomains, the N-terminal upper lobe and the lower lobe, that are connected by a 3-stranded flexible hinge. The VFT domain of C-GPCRs contains the ligand-binding site (6), as demonstrated by studies on mGluRs, GABABR, and the sweet taste receptor (7). The crystal structures of mGluR VFT domains revealed that the bi-lobed architecture can form an open or closed conformation (8, 9). Glutamate binding stabilizes both the active dimer and the closed conformation. This scheme in the initial receptor activation has been applied generally to other C-GPCRs.
Studies on sweet taste-receptor functional domains revealed multiple binding sites for its structurally diverse ligands. Using human–rat chimeric receptors, we demonstrated the T1R2 VFT domain of the human sweet receptor interacts with 2 structurally related synthetic sweeteners aspartame and neotame, while the transmembrane domain (TMD) of human T1R3 interacts with another sweetener cyclamate and a human sweet-taste inhibitor lactisole (7). Works from several other laboratories indicated potential roles in ligand interaction for the T1R3 VFT domain (10, 11) and Cys-rich region (12).
The ligand interaction domains of the umami receptor remained uncharacterized until now. The unique synergistic effect is particularly intriguing to us. Here we report identification of the ligand binding sites of human umami taste receptors. Using sweet-umami chimeric receptors, mutagenesis analysis and molecular modeling, we illustrate a unique mechanism for the synergy between glutamate and IMP in activating the umami taste receptor.
Results and Discussion
Functional Mapping of Ligand-Interaction Sites.
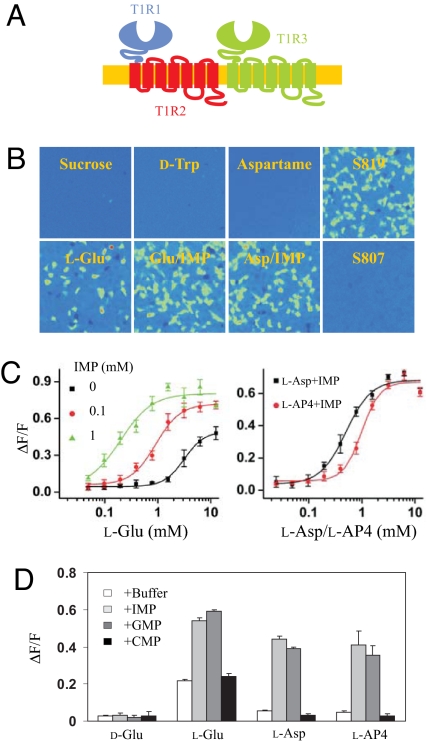
The sweet taste receptor and the umami taste receptor share a common subunit T1R3 but recognize different types of taste stimuli. The unique subunits, T1R1 and T1R2, are probably responsible for ligand binding, most likely via the VFT domains. To test this hypothesis, we generated chimeric receptors of T1R1 and T1R2. T1R2–1 consists of the T1R2 N-terminal extracellular domain and T1R1 transmembrane domain (Fig. 1A). Conversely, T1R1–2 consists of the T1R1 N-terminal extracellular domain and T1R2 transmembrane domain (Fig. 2A). By characterizing the ligand specificity of these chimeric receptors, we can map the ligand-binding domains of umami and sweet taste receptors. To elevate the hybrid receptor activity to measurable level, we coexpressed human T1R1–2 with rat T1R3 instead of human T1R3. Several compounds identified in our high throughput-screening program interact with the TMDs of the T1R taste receptors. S807 (N-(heptan-4-yl)benzo[d][1,3]dioxole-5-carboxamide) (13) is an umami compound that interacts with T1R1 TMD [see supporting information (SI) Fig. S1], while S819 [1-((1H-pyrrol-2-yl)methyl)-3-(4-isopropoxyphenyl)thiourea] (14) is a sweet compound that interacts with T1R2 TMD (Fig. S2). These compounds are expected to activate the hybrid receptors according to the identity of the TMDs and are used as internal controls for the chimeric-receptor activities.
Fig. 1.
The sweet-umami chimeric receptor. (A) A schematic of sweet-umami chimeric receptor. Human T1R1 sequence is in blue, T1R2 is in red, and T1R3 is in green. (B) Responses of sweet-umami chimeric receptor to different sweet and umami stimuli. HEK293 cells stably expressing hT1R2–1/hT1R3 and G16gust25 were assayed for intracellular calcium increase in response to sucrose (200 mM), D-tryptophan (10 mM), aspartame (2.5 mM), S819 (25 μM), cyclamate (80 mM), L-glutamate/IMP (100/10 mM), L-aspartate/IMP (100/10 mM), and S807 (6 μM). (C) Dose-dependent responses of the hT1R2–1/hT1R3 stable cell line to sucralose and aspartame in the absence and presence of IMP (10 mM) and cyclamate (10 mM). The EC50s (mM) are 0.96, 0.97 (with IMP), and 0.41 (with cyclamate) for sucralose, and 2.5, 2.6 (with IMP), and 1.6 (with cyclamate) for aspartame. (D) Responses of the hT1R2–1/hT1R3 stable cell line to more sweet and umami taste stimuli in the absence and presence of IMP (10 mM) and cyclamate (10 mM). Concentrations: aspartame (2.5 mM), D-tryptophan (10 mM), sucrose (200 mM), fructose (200 mM), sucralose (1 mM), saccharin (2 mM), neotame (0.1 mM), acesulfame K (2.5 mM), S807 (3 μM), L-glutamate (100 mM). The values in C to D represent the mean ± SD of ΔF/F for 4 independent responses recorded using Fluorescence Imaging Plate Reader (FLIPR).
Fig. 2.
The umami-sweet chimeric receptor. (A) A schematic of umami-sweet chimeric receptor. (B) Responses of umami-sweet chimeric receptor to different sweet and umami stimuli. HEK293 cells stably expressing hT1R1–2/rT1R3 and G16gust44 were assayed for intracellular calcium increase in response to sucrose (200 mM), D-tryptophan (10 mM), aspartame (2.5 mM), S819 (25 μM), L-glutamate (100 mM), L-glutamate/IMP (100/10 mM), L-aspartate/IMP (100/10 mM), and S807 (6 μM). (C) Dose-dependent responses of the umami-sweet chimeric receptor. The EC50s (mM) for L-glutamate are 3.2, 0.9, and 0.2 in the presence of 0, 0.1 and 1 mM IMP respectively. The EC50s (mM) for L-AP4 and L-aspartate are 1.0 and 0.5 in the presence of 10 mM IMP. (D) Responses of hT1R1–2/rT1R3 stable cell line to umami taste stimuli in the absence and presence of 10 mM IMP or GMP. D-glutamate was used as a negative control for L-glutamate, and CMP was used as a negative control for IMP and GMP. The values in C to D represent the mean ± SD of ΔF/F for four independent responses recorded using FLIPR.
The results are consistent with our hypothesis. T1R2–1/T1R3 has similar ligand specificity as the sweet taste receptor T1R2/T1R3. It recognizes a number of natural and synthetic sweeteners, with the exception of cyclamate (Fig. 1 B and D). T1R2–1/T1R3 does not respond to glutamate and IMP cannot enhance the activity of the sweeteners (see Fig. 1 B–D). In contrast, T1R1–2/T1R3 has similar ligand specificity as the umami taste receptor T1R1/T1R3 (Fig. 2B). The chimeric receptor recognizes glutamate, and more importantly, the activity can be strongly enhanced by IMP or GMP (Fig. 2 C and D). No activity was observed with sweeteners. As predicted, S807 activates the T1R2–1/T1R3 receptor, while S819 does the T1R1–2/T1R3 receptor (see Figs. 1B and 2B). The hybrid receptors can respond to stimuli of different taste qualities through different VFT and TM domains. These observations indicate that the N-terminal extracellular domains of T1R1 and T1R2 determine the ligand specificity of umami and sweet taste receptors, and that the same domain of T1R1 is also crucial for the enhancement activity of IMP and GMP.
Cyclamate, a synthetic sweetener that interacts with the human T1R3 TMD, has a different effect on the sweet and umami taste receptors (7). It is an agonist of the sweet taste receptor but an enhancer of the umami taste receptor. Interestingly, cyclamate did not activate the T1R2–1/T1R3 receptor like other sweeteners. Instead, it enhances the receptor activity (see Fig. 1 C and D). Our data suggest that the TMD of T1R1 is required for the enhancement activity, and the TMD of T1R2 is important for the agonist activity. Recently, a mechanism of intersubunit rearrangement has been proposed for the activation of class C GPCRs (15). We propose that cyclamate induces conformation changes in the T1R3 TMD, which in turn leads to intersubunit rearrangement between the two TMDs. The rearrangement is sufficient to activate T1R2/T1R3 but not T1R1/T1R3, possibly because of differences in the energy barrier of activation of these 2 receptors. Further experiments are needed to fully elucidate the mechanisms.
Critical Residues for Glutamate Activity.
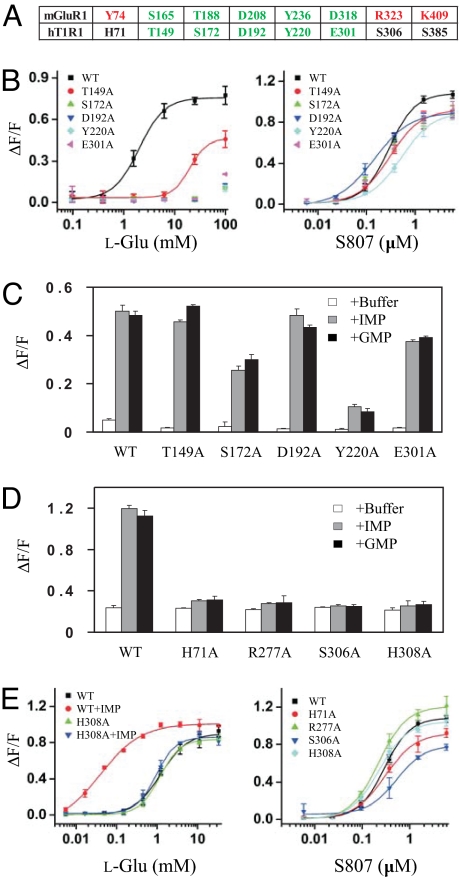
The T1R N-terminal extracellular fragment consists of a large VFT domain followed by a small Cys-rich region. Based on the mapping data alone, we cannot rule out the involvement of the Cys-rich region. To further define the ligand-binding sites, we performed mutagenesis on the human T1R1 VFT domain using the crystal structure of mGluR1 as guidance. In the ligand-binding pocket of mGluR1, glutamate makes direct contact with 8 residues, among which 5 interact with the α-amino acid moiety, and the remaining 3 interact with the glutamate side chain carboxylate (Fig. 3A). Tellingly, all 5 residues that interact with the glutamate α-amino acid moiety are conserved in T1R1, suggesting a similar binding mechanism in these receptors.
Fig. 3.
Mutagenesis analysis. (A) Critical ligand-binding residues in mGluR1 VFT domain and equivalent residues in human T1R1. mGluR1 residues that interact with the carboxylate side chain are in red, and those that interact with the α-amino acid moiety are in green. All 5 residues in green are conserved in T1R1. (B) The dose responses of mutant umami receptors that affect L-glutamate activity. (C) Mutant receptors that affect L-glutamate activity can still be enhanced by IMP and GMP. The L-glutamate concentrations used in the test are 0.4 (WT), 1.0 (T149A), and 100 mM (S172A, D192A, Y220A, and E301A). IMP and GMP concentration is 10 mM. (D) Human T1R1 mutations that affect the enhancement activity of IMP and GMP. The L-glutamate concentrations (mM) used in the test are 0.4 (WT), 10 (H71A), 3.0 (S306A), and 1.0 (R277A and H308A), roughly EC10 based on their respective dose-response curves. IMP and GMP concentration is 10 mM. (E) The dose responses of human T1R1 mutant receptors that affect the enhancement activity of IMP and GMP. The EC50s of WT receptor are 1.3 and 0.02 mM in the absence and presence of 1 mM IMP, respectively. The EC50s of H308A mutant receptor are 1.2 and 0.9 mM in the absence and presence of 1 mM IMP, respectively. All 4 mutants responded to S807 similarly as the wild type. The values in B to D represent the mean ± SD of ΔF/F for 4 independent responses recorded using FLIPR.
Results from mutagenesis analysis confirmed the hypothesis: 4 of the 5 conserved residues are essential for glutamate recognition. S172A, D192A, Y220A and E301A mutant receptors showed no detectable response to glutamate up to 100 mM, while mutation of the fifth residue, T149, resulted in partially reduced activity (Fig. 3B). Importantly, all 5 mutants can still be significantly enhanced by IMP and GMP (Fig. 3C). S807, which interacts with the TMD of T1R1, served as a nice internal control for these T1R1 VFT mutants. None of the 5 mutations had significant effect on the activity of S807 (see Fig. 3B), indicating the diminished glutamate response is not caused by altered protein expression, folding, or surface targeting. In contrast, among the 3 nonconserved residues, mutations of S306 and S385 showed little direct effect on glutamate-induced activity, while mutation of H71 resulted in partially reduced activity (Fig. S3).
Molecular Modeling.
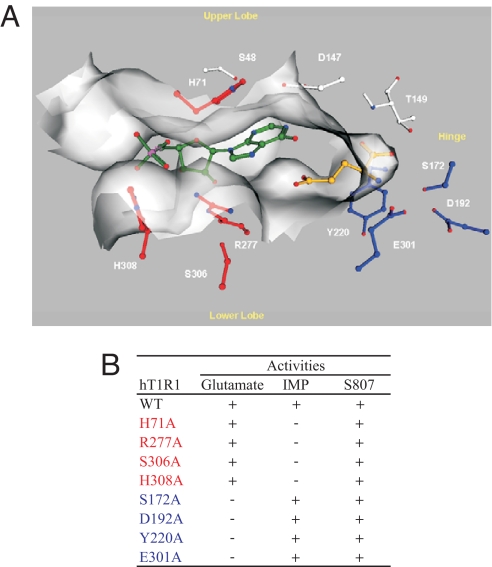
We built homology models of the T1R1 VFT domain in open and closed forms using available structures of mGluR1, mGluR3, mGluR7 from the Protein Databank (8, 9). Despite low overall sequence identity (≈30%) of T1R1 and T1R3 to mGluR1, residues near the hinge of the VFT domain that connect the 2 lobes and that coordinate the zwitterionic backbone of glutamate in the mGluR family of proteins (16) are conserved. Our assumption is therefore that the glutamate binding position in T1R1 closely follows that in the mGluR family of proteins. In our T1R1 model, the alpha carboxylate group of glutamate makes hydrogen bonds with a group of residues close to the hinge of the flytrap and conserved in mGluRs, specifically to the backbone nitrogens of S172 and T149 and to the serine side-chain of S172 (Fig. 4). The alpha amine group of glutamate is coordinated by upper lobe S172 and lower lobe E301. The ring of lower lobe Y220 makes cation-pi interactions with the amine group of glutamate. Notably, the charged residues that form both direct contacts to the side chain of glutamate and stabilizing contacts between the lobes in the closed form of mGluR1 are largely absent in T1R1, which could help explain T1R1's lower affinity for glutamate than mGluRs.
Fig. 4.
A molecular model of T1R1 VFT domain. (A) Key residues for L-glutamate (blue) and for IMP (red) binding. The VFT domain is oriented so that the opening to solvent is horizontal, with the upper lobe on the top and the lower lobe on the bottom. The flytrap hinge region is to the right, and the flytrap opening is to the left. L-glutamate (golden) is located deep inside the VFT domain near the hinge region, while IMP (green) is located close to the opening of the VFT domain. (B) A summary of the mutagenesis data of the key residues. Residues important for L-glutamate activity are in blue, while those for IMP are in red.
Energetic analysis of agonist and antagonist binding in mGluR crystal structures showed that ligands bind strongly to the upper lobe (9, 17). In fact, crystal structures exist of the mGluR1 VFT domain in an open conformation, showing glutamate bound to the upper lobe only (9). The dynamics of flytrap closure has been measured for the VFT domain of ionotropic glutamate receptor iGluR2, revealing fast (≈μs) binding of glutamate (docking) followed by slow (≈ms) stabilization of the closed form (locking) (18). A 2-step mechanism of flytrap closure was proposed (19). In the first step, glutamate binds to the upper lobe and lowers the entropic barrier to form the closed conformation. In a second step, the lobes close up and facilitate interactions between glutamate and the lower lobe, as well as interactions between the upper lobe and the lower lobe. The same mechanism likely persists in T1R1. We refer to critical interacting residues of opposing lobes that are brought together on closure as pincer residues. In the closed conformation, pincer residues of both lobes can energetically interact with each other or with ligands. To further investigate these interactions, we performed normal mode analysis on the VFT domain (20). Of possible movements of the lobes relative to one another, only selected movements result in stabilization of the closed form and likely propagation of signal.
Given the importance of the phosphate group of IMP, the cluster of positively charged residues located near the opening of T1R1 VFT domain provided us with an anchor for positioning IMP in the cleft of VFT domain. An initial model was constructed by placing the phosphate of IMP adjacent to the cluster of positively charged residues described above, which were later confirmed by site-directed mutagenesis. The position and orientation of IMP relative to glutamate are further constrained by the shape of the active-site cleft of the flytrap domain and by the nature of residues lining the active-site cleft. In our model, glutamate and IMP are positioned deeper in the active-site cleft, providing additional stabilization interaction between the upper and lower lobes. The enhancement activity of IMP may be sufficiently explained by the binding of IMP adjacent to glutamate, stabilizing the closed form of the T1R1 VFT domain through electrostatic interactions. It is, however, possible to consider direct additional interaction between IMP and glutamate.
Critical Residues for IMP Activity.
Based on our homology model of T1R1 VFT, we selected 38 residues for mutagenesis studies. While most of the 38 mutations do not affect the synergy (data not shown), residues H71, R277, S306, and H308 are found to be critical for IMP and GMP activities (Fig. 3D). All 4 mutant receptors respond well to glutamate and S807, but the activities could not be enhanced by IMP or GMP up to 10 mM (Fig. 3E and Fig. S4). The set of mutants that affected glutamate activity is essentially separate from the set that affected IMP activity, suggesting separate binding sites for glutamate and the enhancers within the same T1R1 VFT domain.
According to our model, the negatively charged phosphate group coordinates the positively charged pincer residues to stabilize the closed conformation of T1R1 VFT. Therefore, it can be predicted that reversing the charge on 1 of the 2 lobes could stabilize the closed conformation by adding a salt-bridging interaction between the pincer residues, creating a receptor more responsive to glutamate by in part mimicking the enhancement mechanism of IMP. We created 3 such T1R1 mutants: H71E, R277E, and H308E. Indeed, 1 of the 3 mutations, H308E, resulted in a receptor more active than the wild type (Fig. 5A). When coexpressed with rat T1R3, H308E showed an EC50 of 0.8 mM for glutamate, more than 3-fold lower than the wild type. The maximal activity is also increased by nearly 50% over that of wild type. The effect was even more dramatic when H308E was coexpressed with human T1R3 (Fig. 5C). In contrast, S807 activity was not affected by the mutation (Fig. 5 B and D). As with H308A (see Fig. 3E), the H308E/T1R3 mutant receptor activity could not be further enhanced by IMP (Fig. 5C) or GMP (data not shown). This observation reinforced the important function of the positively charged pincer residues and the validity of our model.
Fig. 5.
Mutations of pincer residues to reverse the charge. (A) The L-glutamate dose responses of 3 mutants with reversed charges. The EC50s are 2.6 (WT), 22 (H71E), 12 (R277E), and 0.8 mM (H308E). (B) The S807 dose responses of 3 mutants with reversed charges. The EC50s are 1.3 (WT), 1.1 (H71E), 0.6 (R277E), and 1.2 μM (H308E). (C) The L-glutamate dose responses of H308E in the absence and presence of 1 mM IMP. Gα15 cells were transiently transfected with human T1R1 WT or H308E mutant together with human T1R3. The EC50s are 4.3 and 4.0 mM for the mutant receptor in the absence and presence of IMP. (D) The S807 dose responses of the H308E mutant. Gα15 cells were transiently transfected with human T1R1 WT or H308E mutant together with human T1R3. The EC50s are 0.6 μM for WT and 0.5 μM for H308E mutant receptor. All values represent the mean ± SD of ΔF/F for 4 independent responses recorded using FLIPR.
In summary, we mapped the glutamate and IMP binding sites to the VFT domain of T1R1. Molecular modeling revealed a unique mechanism for a positive allosteric modulator. In our model, IMP exhibits its effect by binding adjacent to glutamate and stabilizes the closed conformation of the VFT by coordinating the positively charged pincer residues. The model is confirmed by mutagenesis data.
There are very few examples of naturally occurring allosteric modulators for GPCRs. Previously, Ca2+ was shown to be required for high affinity γ-aminobutyric acid binding at GABAB receptor (21), and L-amino acids were shown to potentiate the response of CaSR to Ca2+ in transfected HEK cells (22). Allosteric modulators are attracting more and more interest from the pharmaceutical industry as drug candidates because of their obvious advantages over traditional agonists. For example, in cases of closely related receptors that share the same ligand, such as mGluRs, it is more likely to achieve subtype selectivity with allosteric modulators, as they target sites different from the conserved ligand-binding pocket. Furthermore, positive allosteric modulators only work when the natural ligand is present in vivo, reducing potential side effects of constitutively active agonists. Several positive allosteric modulators for members of family C GPCRs have been identified (23) by screening synthetic chemical libraries. In contrast to IMP and GMP, they all bind to the transmembrane domain and their activities are very weak. IMP and GMP interact with the VFT domain of T1R1, representing a unique mechanism of positive regulation of family C GPCRs. Furthermore, this mechanism of synergism, through cooperative binding to the VFT domain, also applies to the sweet taste receptor (X.L., unpublished data).
Materials and Methods
Stable Cell Lines.
hT1R2–1/hT1R3 stable cell lines were generated by transfecting linearized pCDNA3.1/Neo-derived T1R2-T1R1 and pCDNA3.1/Zeo-derived (Invitrogen) T1R3 vectors into G16gust25 cell, an HEK293 line stably expressing the chimeric G16gust25 protein. Cells were selected with G418 (Invitrogen, 0.4 mg/ml) and zeocin (Invitrogen, 0.1 mg/ml) in low glucose DMEM. hT1R1–2/rT1R3 stable cell lines were generated by transfecting linearized pEAK10-derived T1R1-T1R2 and pCDNA3.1/Zeo-derived (Invitrogen) T1R3 vectors into G16gust44 cell, a HEK293 line stably expressing the chimeric G16gust44 protein (24). Cells were selected with puromycin (Calbiochem, 0.5 μg/ml) and zeocin (Invitrogen, 50 μg/ml) in glutamine-free DMEM supplemented with GlutaMAX. Resistant colonies were expanded, and their responses to sweet- and umami-taste stimuli were evaluated by calcium imaging.
Constructs.
As previously described, T1R chimeras were constructed by introducing an XhoI site with a silent mutation at human T1R2 amino acid 560. T1R mutants were generated using standard PCR-based mutagenesis protocol. All T1Rs, chimeras and mutants used in HEK293 cell-based assay were cloned into pEAK10 expression vector (EdgeBio). Human T1R TMD constructs for GTPγS binding assay were designed as described for mGlu5a receptor (25). The boundaries for the deletion were determined by alignment of amino acid sequences of hT1R1, hT1R2, and hT1R3 with mGluR5a. The necessary DNA fragments were obtained by PCR. The final plasmids encoded for proteins possessing the signal peptides of hT1Rs (amino acids 1–45) followed by the TMDs and the intracellular C termini (amino acids 553–841 for hT1R1, 552–839 for hT1R2 and 555–852 for hT1R3). All constructs also included the C-terminal myc tag. The fragments were cloned into a pFastBac-1 vector (Invitrogen).
HEK Cell-Based Assay.
Calcium imaging assay was performed as previously described (2, 7). Assays using FLIPR were performed using 384-well plates (≈20,000 cells per well). Transient transfections were performed in suspension using Mirus TransIt-293 (Invitrogen). Briefly, ≈107 cells were mixed with 25 μg of DNA lipid complex, incubated at room temperature for 20 min, and seeded onto 384-well plates.
GTPγS Binding Assay.
Insect Spodoptera frugiperda Sf9 cells were infected for 48 h at 2 × 106 cells/ml with the appropriate baculoviruses at multiplicity of infection 1–2. Cell membrane preparation and GTPγS-binding assay were carried out as previously described (26). Expression of all hT1R-TMD proteins was confirmed by Western blot with anti-myc mAb (Upstate Biotech). Under the assay conditions, the final GTPγS occupancy of transducin in any reaction did not exceed 35%, with basal binding to transducin in the absence of any membranes of ≈0.7% (80 fmol).
Mutagenesis Studies.
Mutagenesis was performed using standard PCR-based method. To map the IMP/GMP binding site, the following 38 residues in human T1R1 (GenBank accession number BK000153) were mutated individually to Ala: H47, S48, R54, R56, R64, E70, H71, R80, R117, H126, D147, R151, K179, R180, R187, K242, F247, R255, S276, R277, Q278, R281, K295, S306, R307, H308, R329, K335, R365, H373, K377, H398, H401, R413, H433, K482, H486, K488. Mutant receptor activities were tested using transiently transfected Gα15 cells (27) and FLIPR. Human T1R1 WT or mutants were cotransfected with rat T1R3 unless otherwise specified.
Molecular Modeling.
Homology models of the T1R1 and T1R3 monomers were constructed in open and closed form using available structures of the metabotropic glutamate receptors mGluR1, mGluR3, mGluR7 from the Protein Databank (8, 9), using the program Homology (Accelrys). Ligands were introduced into the model using the interactive program Insight II (Accelrys) and Biodock (BioPredict). Resulting complexes were subjected to minimization and molecular dynamics-based simulated annealing, both with and without explicit water molecules, using the program Discover (Accelrys). Normal modes were computed using the program ElNemo (28). Visualization was performed using the BioSight (BioPredict).
Supplementary Material
Acknowledgments.
We thank J. Kang for his scientific contribution in molecular modeling, and B. Moyer and G. Servant for critical reading of this manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810174106/DCSupplemental.
References
- 1.Yamaguchi S, Ninomiya K. Umami and food palatability. J Nutr. 2000;130:921S–926S. doi: 10.1093/jn/130.4.921S. [DOI] [PubMed] [Google Scholar]
- 2.Li X, et al. Human receptors for sweet and umami taste. Proc Natl Acad Sci USA. 2002;99:4692–4696. doi: 10.1073/pnas.072090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- 4.Nelson G, et al. An amino-acid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- 5.Nelson G, et al. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 6.Pin JP, Galvez T, Prezeau L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol Ther. 2003;98:325–354. doi: 10.1016/s0163-7258(03)00038-x. [DOI] [PubMed] [Google Scholar]
- 7.Xu H, et al. Different functional roles of T1r subunits in the heteromeric taste receptors. Proc Natl Acad Sci USA. 2004;101:14258–14263. doi: 10.1073/pnas.0404384101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunishima N, et al. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 9.Muto T, Tsuchiya D, Morikawa K, Jingami H. Structures of the extracellular regions of the group ii/iii metabotropic glutamate receptors. Proc Natl Acad Sci USA. 2007;104:3759–3764. doi: 10.1073/pnas.0611577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nie Y, Vigues S, Hobbs JR, Conn GL, Munger SD. Distinct contributions of T1r2 and T1r3 taste receptor subunits to the detection of sweet stimuli. Curr Biol. 2005;15:1948–1952. doi: 10.1016/j.cub.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 11.Koizumi A, et al. Taste-modifying sweet protein, neoculin, is received at human T1r3 amino terminal domain. Biochem Biophys Res Commun. 2007;358:585–589. doi: 10.1016/j.bbrc.2007.04.171. [DOI] [PubMed] [Google Scholar]
- 12.Jiang P, et al. The cysteine-rich region of T1r3 determines responses to intensely sweet proteins. J Biol Chem. 2004;279:45068–45075. doi: 10.1074/jbc.M406779200. [DOI] [PubMed] [Google Scholar]
- 13.Tachdjian C, et al. PCT Int. Appl. 2005 WO 2005041684. [Google Scholar]
- 14.Tachdjian C, et al. PCT Int. Appl. 2006 WO 2006084184. [Google Scholar]
- 15.Brock C, et al. Activation of a dimeric metabotropic glutamate receptor by intersubunit rearrangement. J Biol Chem. 2007;282:33000–33008. doi: 10.1074/jbc.M702542200. [DOI] [PubMed] [Google Scholar]
- 16.Acher FC, Bertrand HO. Amino acid recognition by Venus flytrap domains is encoded in an 8-residue motif. Biopolymers. 2005;80:357–366. doi: 10.1002/bip.20229. [DOI] [PubMed] [Google Scholar]
- 17.Tsuchiya D, Kunishima N, Kamiya N, Jingami H, Morikawa K. Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+ Proc Natl Acad Sci USA. 2002;99:2660–2665. doi: 10.1073/pnas.052708599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abele R, Keinanen K, Madden DR. Agonist-induced isomerization in a glutamate receptor ligand-binding domain. A kinetic and mutagenetic analysis. J Biol Chem. 2000;275:21355–21363. doi: 10.1074/jbc.M909883199. [DOI] [PubMed] [Google Scholar]
- 19.Ahmed AH, Loh AP, Jane DE, Oswald RE. Dynamics of the S1s2 glutamate binding domain of Glur2 measured using 19f Nmr spectroscopy. J Biol Chem. 2007;282:12773–12784. doi: 10.1074/jbc.M610077200. [DOI] [PubMed] [Google Scholar]
- 20.Suhre K, Sanejouand YH. On the potential of normal-mode analysis for solving difficult molecular-replacement problems. Acta Crystallogr D Biol Crystallogr. 2004;60:796–799. doi: 10.1107/S0907444904001982. [DOI] [PubMed] [Google Scholar]
- 21.Galvez T, et al. Ca(2+) requirement for high-affinity gamma-aminobutyric acid (Gaba) binding at Gaba(B) receptors: involvement of serine 269 of the Gaba(B)R1 subunit. Mol Pharmacol. 2000;57:419–426. doi: 10.1124/mol.57.3.419. [DOI] [PubMed] [Google Scholar]
- 22.Conigrave AD, Quinn SJ, Brown EM. L-amino acid sensing by the extracellular Ca2+-sensing receptor. Proc Natl Acad Sci USA. 2000;97:4814–4819. doi: 10.1073/pnas.97.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brauner-Osborne H, Wellendorph P, Jensen AA. Structure, pharmacology and therapeutic prospects of family C G-protein coupled receptors. Curr Drug Targets. 2007;8:169–184. doi: 10.2174/138945007779315614. [DOI] [PubMed] [Google Scholar]
- 24.Ueda T, Ugawa S, Yamamura H, Imaizumi Y, Shimada S. Functional interaction between T2r taste receptors and G-protein alpha subunits expressed in taste receptor cells. J Neurosci. 2003;23:7376–7380. doi: 10.1523/JNEUROSCI.23-19-07376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goudet C, et al. Heptahelical domain of metabotropic glutamate receptor 5 behaves like rhodopsin-like receptors. Proc Natl Acad Sci USA. 2004;101:378–383. doi: 10.1073/pnas.0304699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pronin AN, Tang H, Connor J, Keung W. Identification of ligands for two human bitter T2r receptors. Chem Senses. 2004;29:583–593. doi: 10.1093/chemse/bjh064. [DOI] [PubMed] [Google Scholar]
- 27.Chandrashekar J, et al. T2rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 28.Suhre K, Sanejouand YH. Elnemo: A normal mode Web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32:W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.