Abstract
The circadian clock controls daily oscillations of gene expression at the cellular level. We report the development of a high-throughput circadian functional assay system that consists of luminescent reporter cells, screening automation, and a data analysis pipeline. We applied this system to further dissect the molecular mechanisms underlying the mammalian circadian clock using a chemical biology approach. We analyzed the effect of 1,280 pharmacologically active compounds with diverse structures on the circadian period length that is indicative of the core clock mechanism. Our screening paradigm identified many compounds previously known to change the circadian period or phase, demonstrating the validity of the assay system. Furthermore, we found that small molecule inhibitors of glycogen synthase kinase 3 (GSK-3) consistently caused a strong short period phenotype in contrast to the well-known period lengthening by lithium, another presumed GSK-3 inhibitor. siRNA-mediated knockdown of GSK-3β also caused a short period, confirming the phenotype obtained with the small molecule inhibitors. These results clarify the role of GSK-3β in the period regulation of the mammalian clockworks and highlight the effectiveness of chemical biology in exploring unidentified mechanisms of the circadian clock.
Keywords: screening, small molecule library, kinase
Genetic networks of regulated transcription and protein turnover lie at the core of circadian regulation in all organisms. Mutation in key nodes of the circadian networks causes changes in overt behavioral and physiological rhythms (1, 2). For example, familial advanced sleep phase syndrome with early sleep times and early-morning awakening is attributed to missense mutations of human PER2 and CSNK1D genes (3, 4). The clock genes constitute the transcription/translation-based negative feedback loop of the core oscillator; CLOCK/BMAL1 heterodimers activate transcription of Per and Cry genes, and PER and CRY proteins in turn inhibit their own transcription (5). In addition to transcriptional regulation, posttranslational modifications of clock proteins by phosphorylation, ubiquitination, and acetylation play essential roles in the oscillator mechanism (6, 7).
The molecular clock machinery resides at the cellular level, and each single cell shows circadian rhythmicity in a cell-autonomous manner (8–10). At the organismal level, the cellular oscillators are organized in a hierarchy, in which the suprachiasmatic nucleus (SCN) constitutes the central circadian pacemaker (11). In the SCN, the cellular clocks are synchronized to form a coherent oscillator through intracellular coupling (12), making the SCN clock more robust against genetic and environmental perturbations than peripheral oscillators (13). Therefore, a cell-based assay system using cultured fibroblasts that lack intercellular coupling (9, 10) will provide a particularly responsive system to characterize the circadian clockwork through an unbiased, phenotype-driven screening (14, 15). Perturbations may be revealed in such a cell-based approach that might otherwise be masked via coupling in the SCN and thus missed using behavioral genetic screens.
Although many clock genes forming the core oscillatory loop have been identified, evidence suggests the existence of more additional unknown clock components and modulators (16). Chemical biology methods use small molecules as proof-of-concept probes for biological systems and can be effective in discovering biological mechanisms (14, 17). The approach can complement the limitations of classical forward and reverse genetic screens associated with lethality, pleiotropy, and functional overlapping of closely related proteins. Chemical probes can be applied in a dose-dependent and reversible manner at multiple levels of biological organization. A set of compounds that potently affect the circadian clock function will lead to the identification of clock components and form the basis for therapeutic strategies directed toward circadian disorders. In this study, we developed a robust cell-based circadian screening paradigm for the identification of compounds. To test the screening pipeline, we used a structurally diverse chemical library [Library of Pharmacologically Active Compounds (LOPAC)] containing 1,280 pharmacologically active compounds that span a broad range of biological pathways. Among them, we successfully identified 11 compounds causing reproducible period changes. Therefore, the assay system is competent for large-scale compound screening to discover new chemical probes for dissecting circadian pathways.
Results and Discussion
Development of a High-Throughput Circadian Assay System.
We used the circadian luciferase reporter Bmal1-dluc (18) for monitoring circadian rhythms in cultured cells and developed a 384-well plate-based assay system to screen compound libraries. Among all cell lines tested, a human U2OS cell line showed prominent rhythmicity with high luminescence intensity. We established several clonal U2OS lines stably expressing the Bmal1-dluc reporter and selected one clone with high amplitude and a low damping rate rhythm for further study. In parallel, we developed a curve fitting program for the analysis of large amounts of luminescence data to obtain rhythm parameters such as period length (CellulaRhythm; see Materials and Methods).
By optimizing cell culture and luminescence measurement conditions, we obtained highly consistent rhythms from each well in a 384-well format [supporting information (SI) Fig. S1]. More than 97% of the wells are within the period range of mean ± 0.5 h, which is similar to and better than a system recently reported (15) that used a U2OS reporter line, 384-well plate assays, and kinetic luminescence measurements to perform siRNA-based perturbations to the circadian machinery. Furthermore, treatment of the cells with D4476, a CKIδ/ε inhibitor known to lengthen the circadian period (19), caused a long period phenotype in a dose-dependent manner at final concentrations of 3–20 μM (Fig. S2). These results indicate the validity of the system for the screening of compounds affecting the circadian period.
Screening of LOPAC Chemical Library.
The potential effect of 1,280 compounds on the cellular circadian rhythm was investigated at final concentration of 7 μM (Fig. 1). The screening was repeated twice, and 13 primary “hits” were identified based on period change (lengthening or shortening) of ≥0.5 h in both screening (Table 1). The effect of 13 hit compounds was further investigated at various concentrations. Eleven of the compounds showed dose-dependent period lengthening or shortening (Figs. 2, 3, and Fig. S3), confirming the result of the screening. Interestingly, vincristine, an inhibitor of microtubule assembly, showed a bidirectional effect: period shortening at lower dose and lengthening at higher dose (Fig. 2).
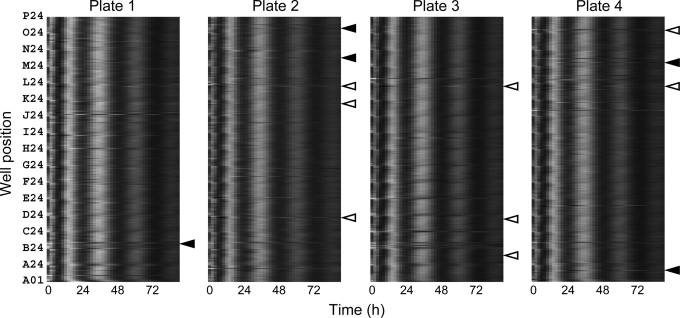
Fig. 1.
Raster plot of the result of LOPAC chemical library screening. Luminescence rhythms of Bmal1-dluc cells were monitored by using ViewLux system in the presence of compounds (final concentration, 7 μM). One screening contained four 384-well plates, and profiles of one 384-well plate are represented in each panel. Each horizontal raster line represents a single well, with elapsed time plotted to the right. Luminescence intensity data from each well are normalized for amplitude, and then indicated by gray scale: peak is white and trough is black. The screening was repeated twice, and the result from the first experiment is shown. Solid and open arrowheads indicate the positions of long and short period compounds in Table 1, respectively. Note that there are many compounds that change the phase of the rhythm without affecting the period.
Table 1.
Long and short period compounds identified from LOPAC chemical library screening
| Compound name | Function | Period change, h |
Confirmation | |
|---|---|---|---|---|
| Exp1 | Exp2 | |||
| Roscovitine | Inhibitor of CDK | +1.1 | +1.4 | Figs. 2 and 3 |
| SP600125 | Inhibitor of JNK | +1.0 | +1.4 | Fig. 2 |
| SB 202190 | Inhibitor of p38 MAPK | +1.0 | +1.2 | Fig. S3 |
| DRB | Inhibitor of CK2 | +0.7 | +1.0 | Fig. 2 |
| Vincristine | Inhibitor of microtuble | +0.6 | +1.0 | Fig. 2 |
| Etoposide | Inducer of DNA damage | −3.5 | −1.5 | Fig. 2 |
| Mitoxantrone | Inducer of DNA damage | −1.5 | −2.0 | Fig. S3 |
| PMA | Activator of PKC | −1.9 | −1.6 | Fig. 2 |
| SKF-96365 | Inhibitor of Ca2+ entry | −1.3 | −2.1 | Fig. 2 |
| Indirubin-3′-oxime | Inhibitor of CDK and GSK-3 | −1.0 | −0.7 | Fig. 2 |
| Kenpaullone | Inhibitor of CDK and GSK-3 | −1.1 | −0.5 | Fig. 3 |
| Ethamivan | Respiratory stimulant | −2.5 | −0.6 | No period effect |
| 2-Methoxyestradiol | Estrogen derivative | −1.2 | −1.6 | No period effect |
Luminescence rhythms of Bmal1-dluc cells were monitored by using ViewLux system in the presence of compounds (final concentration, 7 μM). Each luminescence profile was fitted with damped cosine curve to obtain period parameter. The compounds that caused ≥0.5-h period changes in both screening were listed with period change (in hours) relative to the median period of DMSO control wells. Positive and negative values represent period lengthening and shortening, respectively. Ethamivan and 2-methoxyestradiol did not show period changing effect in confirmation assay (data not shown).
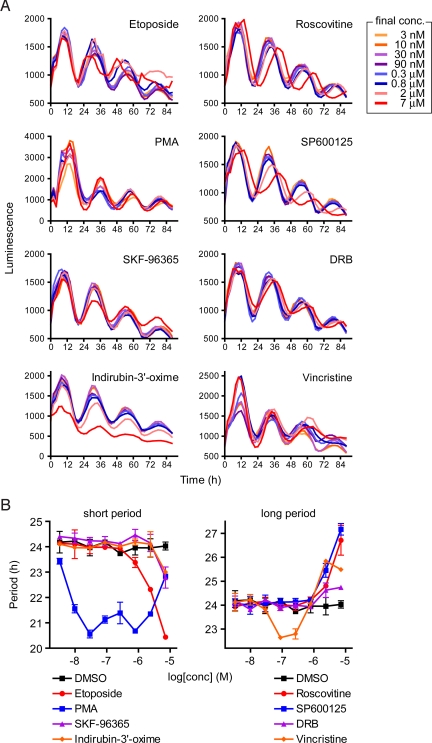
Fig. 2.
Dose-dependent effect of hit compounds. Luminescence rhythms of Bmal1-dluc cells were monitored by using ViewLux system in the presence of various concentrations of compounds (8 points of 3-fold dilution series; final concentrations, 3 nM to 7 μM). (A) Luminescence profiles are indicated for each compound. Data are the representative of duplicate experiment. (B) Period parameter was obtained by the curve fitting and plotted against final concentration of the compound. Data are the mean with variation of duplicate experiment.
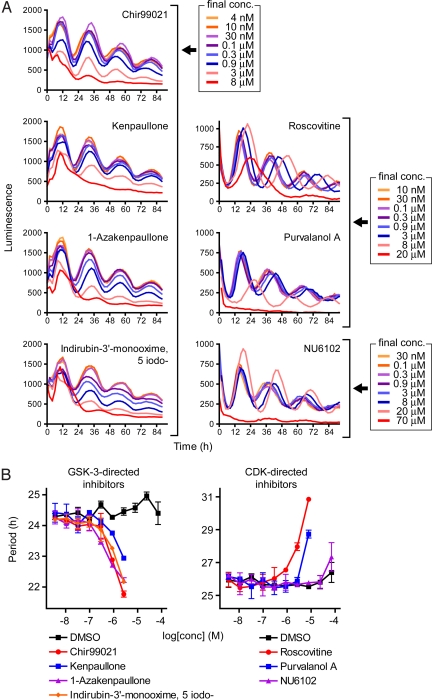
Fig. 3.
Effect of CDK and GSK-3 inhibitors on the cellular circadian rhythm. Luminescence rhythms of Bmal1-dluc cells were monitored by using ViewLux system (for GSK-3-directed inhibitors) or Tecan luminometer (for CDK-directed inhibitors) in the presence of various concentrations of compounds (10 points of 3-fold dilution series; final concentrations, 4 nM to 70 μM). (A) Luminescence profiles are indicated for each compound. Data are the representative of duplicate experiment. (B) Period parameter was obtained by the curve fitting and plotted against final concentration of the compound. Data are the mean with variation of duplicate experiment. Similar result was obtained from another experiment. Note that obtained period and phase of the basal rhythm and luminescence intensity are different between ViewLux system and Tecan luminometer, because of the setup difference that did not affect the effect of compounds (data not shown).
The hit compounds can generally be classified as inhibitors/activators of protein kinases, inhibitors of microtubule assembly, inhibitors of Ca2+ entry, and inducers of DNA damage (Table 1). Many of the compounds are related to the pathways already known to affect the circadian clock function in a variety of organisms and tissues as follows. Roscovitine (a CDK inhibitor), SP600125 (a JNK inhibitor), and SB 203580 (an analog of p38 MAPK inhibitor SB 202190) cause period lengthening in cultured Aplysia eye (20), mouse tissues (21), and chicken pineal gland (22), respectively. Decreased activity of CK2 causes long period behavioral rhythms in Drosophila (23, 24). Furthermore, supporting the short period phenotypes of PMA (an activator of PKC), etoposide, and mitoxantrone (DNA damage inducers) in our assay (Table 1), PMA treatment and DNA damage by γ-radiation have been reported to cause phase advances but not delays of the rhythms in cultured hamster SCN and free-running mice, respectively (25, 26). Taken together, the identification of these previously known compounds and pathways validates our screening paradigm and suggests highly conserved mechanism of the circadian clock across species and tissue types.
Period Shortening by Inhibition of GSK-3β.
Period-shortening compounds indirubin-3′-oxime and kenpaullone (Table 1) are known to inhibit both CDK and GSK-3 (27). In contrast, roscovitine, that inhibits CDK but not GSK-3 (27), caused a long period phenotype (Table 1). To clarify the difference of the effect of CDK inhibition and GSK-3 inhibition on the period length, we tested additional CDK and GSK-3 inhibitors (Fig. S4). We found that GSK-3-directed inhibitors including Chir99021 (28) and 1-azakenpaullone (29) shortened the period, but CDK-directed inhibitors such as purvalanol A (27) and NU6102 (30) did not (Fig. 3). Interestingly, the period shortening effect of GSK-3-directed inhibitors is opposite to the well-known period lengthening effect of lithium (31), which has been proposed to act through GSK-3 inhibition (32). We also observed prominent period lengthening by LiCl in our assay system (Fig. S5).
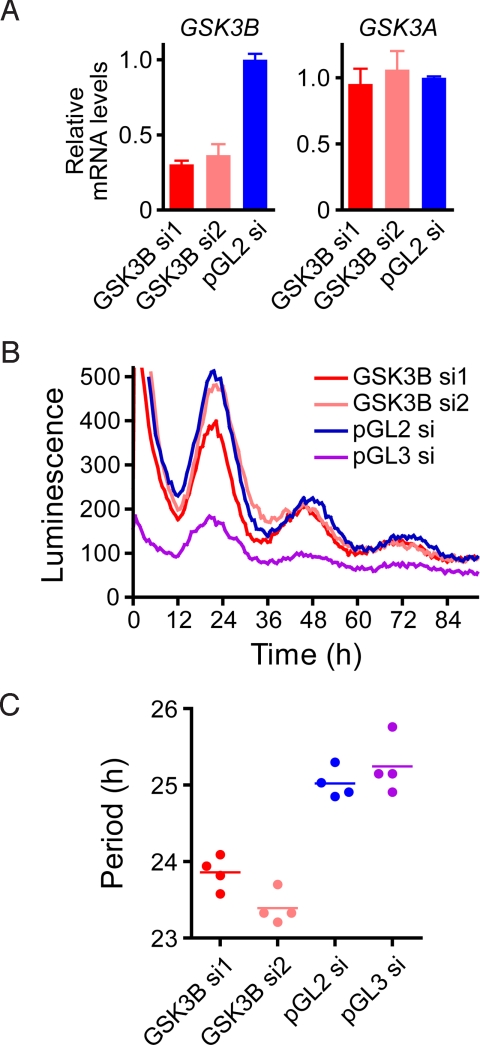
To elucidate the role of GSK-3 in the period regulation of mammalian cells, we performed siRNA-mediated knockdown experiments. Transfection of GSK-3β siRNA (si1 and si2) strongly and specifically reduced endogenous GSK-3β mRNA levels (Fig. 4A) and shortened the circadian period (Fig. 4 B and C). We obtained similar period shortening in mouse primary fibroblasts infected with a lentivirus vector encoding GSK-3β shRNA (Fig. S6). Together, the chemical biology and genomic approaches clarify the role of GSK-3β in the period regulation of the mammalian circadian clock.
Fig. 4.
Effect of GSK-3β knockdown on the circadian rhythm. Bmal1-dluc cells were transfected with GSK-3β siRNA (GSK3B si1 or si2), luciferase siRNA (pGL3 si), or control siRNA (pGL2 si), and luminescence rhythms were monitored by using Tecan luminometer. After 4-d monitoring, the cells were collected for RT-qPCR analysis. (A) Endogenous GSK-3β (GSK3B) and GSK-3α (GSK3A) mRNA levels were analyzed by RT-qPCR and indicated by normalization with GAPDH. Data are the mean with variation of 2 independent experiments. (B) Luminescence profiles are indicated for each siRNA. Data are the representative of 4 independent experiments. (C) Period parameter was obtained by the curve fitting and plotted. Data from each experiment is shown by a circle, and mean period is indicated by a horizontal bar.
Role of GSK-3β in the Circadian Clock Mechanism.
In Drosophila, reduction of GSK-3 activity by genetic manipulation causes period lengthening (33), in contrast to our findings in mammalian cell culture. Of note, GSK-3 phosphorylates TIM protein in Drosophila (33), but there is no tim ortholog in mammals (34). Instead, GSK-3β is known to phosphorylate PER2, CRY2, and Rev-erbα in mammals (35–37). GSK-3β-mediated phosphorylation leads to proteasomal degradation of CRY2 (36) and stabilization of Rev-erbα (37). Considering that Cry2 and Rev-erbα knockout mice show long and short period phenotypes, respectively (38–40), it is possible that the period shortening by GSK-3β inhibition is mediated at least in part by the regulation of CRY2 and Rev-erbα protein levels (i.e., stabilization of CRY2 and degradation of Rev-erbα). The role of GSK-3α is of interest and should be addressed in future studies.
Although lithium lengthens the period of circadian rhythms in a wide range of experimental systems such as unicellular organisms, insects, mice, and humans (31), the exact mode of action is still uncertain. Because lithium inhibits inositol monophosphatase and other phosphomonoesterases and GSK-3 (32), the long period phenotype in mammals might be mediated by lithium-targeted protein(s) other than GSK-3.
Period Changing Compounds from LOPAC Chemical Library.
The LOPAC chemical library contains many drugs currently on the market and in clinical trials. Therefore, our screening results obtained from human cells might have an important implication for the application of such drugs. Roscovitine, vincristine, etoposide, and mitoxantrone are in clinical trials against cancers, and they showed significant period changing effects (Table 1) that may affect the circadian clock of normal, non-dividing tissues.
The specificity of 20 kinase inhibitors including clinical drugs was reported by developing a small molecule-kinase interaction profiling method (41). The kinase-interaction of roscovitine, SP600125, and SB 202190 was investigated, and interestingly, all of the compounds showed strong binding with CKIε (41), the inhibition of which caused a long period phenotype (Fig. S2). It is possible that the period lengthening effect of roscovitine, SP600125, and SB 202190 (Table 1) is mediated via CKIε inhibition and their primary target (CDK, JNK, or p38 MAPK). Further studies are necessary to determine the responsible protein for the circadian effect of these compounds.
Chemical Biology Approach for Circadian Clock Mechanism.
We have successfully miniaturized the automated luminescence monitoring of cellular circadian rhythms to a 384-well format and set up data analysis tools to extract circadian parameters from thousands of datasets, enabling us to identify period altering compounds from LOPAC chemical library screening. To discover clock components through the chemical biology approach, we need a more comprehensive, large-scale approach with many different types of compounds. Such diversity-oriented screening usually requires hundreds of thousands of chemical probes to identify a viable lead compound (42). We are screening a nonproprietary compound collection containing ≈650,000 diverse compounds with predicted drug-like properties. A wide variation of chemical structures has the advantage of probing many classes of potential targets, some of which may lead to the identification of new mechanisms regulating circadian clock functions in many tissues.
Materials and Methods
Compounds.
The 1 mM solution of LOPAC chemical library (Sigma) and its 8-point dilution series were obtained as 384-well format from GNF HTS core. D4476, indirubin-3′-monooxime, 5 iodo-, roscovitine, purvalanol A, and NU6102 were purchased from Calbiochem; SB 202190, mitoxantrone, and kenpaullone were from Sigma; 1-azakenpaullone was from Alexis Biochemicals; and Chir99021 was synthesized. The dilution series of the compounds was made on 384-well plate by using a robotic liquid handling system (MiniTrak, Perkin–Elmer).
Cell-Based Circadian Assay.
A clonal line of human osteosarcoma U2OS cells stably expressing the Bmal1-dluc reporter was established by using the method described in ref. 18 and grown in the culture medium (DMEM supplemented with 10% FBS, 0.29 mg/ml L-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin).
For the luminescence recording, the cells were suspended in the culture medium and plated onto 384-well white solid-bottom plates (Greiner) at 20 μL (2,000 cells) per well by using a microplate dispenser (μFill; BioTek). They were cultured for 2 days to reach confluence. Then, 50 μL of the explant medium [DMEM supplemented with 2% B27 (Invitrogen), 10 mM Hepes (pH 7.2), 0.38 mg/ml sodium bicarbonate, 0.29 mg/ml L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 0.1 mg/ml gentamicin, and 1 mM luciferin (Promega)] was dispensed to each well by using μFill, followed by the application of 500 nL of compounds (dissolved in DMSO; final 0.7% DMSO) by using 384-well head PinTool (GNF Systems). The plate was covered by an optically clear film (USA Scientific) and set to luminescence monitoring system. This procedure is sufficient for the synchronization of the cellular rhythms accompanied by the induction of endogenous PER1 and PER2 genes (data not shown). The luminescence was recorded by using a microplate reader (Infinite M200; Tecan) or a GNF automated robotic system (GNF Systems) equipped with a CCD imager (ViewLux, Perkin–Elmer). The settings for Tecan luminometer are as follows: temperature setting, 35 °C; integration time, 14 sec; interval time, 100 min. The setting for ViewLux system: incubator temperature, 37 °C; exposure time, 120 sec; interval time, 120 min.
Luminescence Data Analysis.
By using the R-project computing environment (www.r-project.org), we have developed an automated analysis algorithm for curve fitting and data display (“CellulaRhythm,” see SI Appendix for code). Raw luminescence data were fit to a damped cosine curve using nonlinear least squares to the following equation:
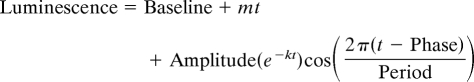 |
where m = Slope, k = Damping rate, and t = Time. Due to transient luminescence changes upon the medium change, the first 20 h data were excluded from the analysis. The quality of the curve fitting was inspected manually, and the data with poor fitting were filtered out. In many case, the poor fitting arose from experimental problem such as toxic effect of the compound. Local effects such as edge effect on the rhythm were evaluated by visual inspection of the raster plot of luminescence data.
siRNA-Mediated Knockdown.
siRNAs were purchased from Qiagen (GSK3B si1, Hs_GSK3B_7; GSK3B si2, Hs_GSK3B_8), and 3 pmol of each siRNA was spotted onto 96-well white solid-bottom plates (Corning). A total of 60 μL of Opti-MEM (Invitrogen) containing 0.2 μL of Lipofectamine 2000 (Invitrogen) was dispensed onto each well and incubated for 20 min at room temperature. Then, 60 μL of the cells in DMEM supplemented with 20% FBS was dispensed (6,000 cells per well). The cells were cultured overnight, and the medium was changed to 180 μL of the culture medium. They were cultured for another 2 days to reach confluence. Then, the medium was changed to 180 μL of the explant medium, and the plate was covered by the optically clear film. The luminescence was recorded by using Tecan luminometer with the following settings: temperature setting, 35 °C; integration time, 20 sec; interval time, 36 min. To obtain the period parameter, the luminescence data (10–60 h) were analyzed by using MultiCycle software (Actimetrics), in which polynomial order was set at 1 for background subtraction.
The cells were collected after 4 days of recording for RT-qPCR analysis. cDNA sample was prepared by using RNeasy kit (Qiagen) and SuperScript III Reverse Transcriptase (Invitrogen). qPCR was performed on 7900HT Fast Real-Time PCR System (Applied Biosystems) by using SYBR Green PCR Master Mix (Applied Biosystems). The sequences of qPCR primers were as follows: GAPDH-F, TGCAC CACCA ACTGC TTAGC; GAPDH-R, ACAGT CTTCT GGGTG GCAGT G; GSK3B-F, TGGAA TCTGC CATCG GGATA; GSK3B-R, ATTGG GTTCT CCTCG GACCA; GSK3A-F, TGATG AACTG CGATG TCTGG; and GSK3A-R, TGAGA GACGG TTGGA TGGAG.
Supplementary Material
Acknowledgments.
We thank Achim Brinker, Michael Garcia, Jason Matzen, Paul Anderson, Jeff Janes, Charles Cho, Xu Wu, and Richard Glynne for their help on chemical screening and Eric Zhang for critical reading of the manuscript. This work was supported by National Institutes of Health Grants R01 GM074868 and R01 MH051573 (to S.A.K.) and grants from the Skaggs Institute for Chemical Biology (to P.G.S.). This is manuscript no. 081103 of Genomics Institute of the Novartis Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811410106/DCSupplemental.
References
- 1.Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toh KL, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 5.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 6.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 7.Belden WJ, Dunlap JC. SIRT1 is a circadian deacetylase for core clock components. Cell. 2008;134:212–214. doi: 10.1016/j.cell.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 9.Welsh DK, et al. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagoshi E, et al. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 12.Aton SJ, Herzog ED. Come together, right.now: Synchronization of rhythms in a mammalian circadian clock. Neuron. 2005;48:531–534. doi: 10.1016/j.neuron.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu AC, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu AC, Lewis WG, Kay SA. Mammalian circadian signaling networks and therapeutic targets. Nat Chem Biol. 2007;3:630–639. doi: 10.1038/nchembio.2007.37. [DOI] [PubMed] [Google Scholar]
- 15.Vollmers C, Panda S, DiTacchio L. A high-throughput assay for siRNA-based circadian screens in human U2OS cells. PLoS ONE. 2008;3:e3457. doi: 10.1371/journal.pone.0003457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi JS. Finding new clock components: Past and future. J Biol Rhythms. 2004;19:339–347. doi: 10.1177/0748730404269151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding S, Schultz PG. A role for chemistry in stem cell biology. Nat Biotechnol. 2004;22:833–840. doi: 10.1038/nbt987. [DOI] [PubMed] [Google Scholar]
- 18.Liu AC, et al. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reischl S, et al. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms. 2007;22:375–386. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- 20.Sankrithi N, Eskin A. Effects of cyclin-dependent kinase inhibitors on transcription and ocular circadian rhythm of Aplysia. J Neurochem. 1999;72:605–613. doi: 10.1046/j.1471-4159.1999.0720605.x. [DOI] [PubMed] [Google Scholar]
- 21.Chansard M, et al. c-Jun N-terminal kinase inhibitor SP600125 modulates the period of mammalian circadian rhythms. Neuroscience. 2007;145:812–823. doi: 10.1016/j.neuroscience.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi Y, et al. p38 mitogen-activated protein kinase regulates oscillation of chick pineal circadian clock. J Biol Chem. 2003;278:25166–25171. doi: 10.1074/jbc.M212726200. [DOI] [PubMed] [Google Scholar]
- 23.Lin JM, et al. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- 24.Akten B, et al. A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci. 2003;6:251–257. doi: 10.1038/nn1007. [DOI] [PubMed] [Google Scholar]
- 25.Schak KM, Harrington ME. Protein kinase C inhibition and activation phase advances the hamster circadian clock. Brain Res. 1999;840:158–161. doi: 10.1016/s0006-8993(99)01787-4. [DOI] [PubMed] [Google Scholar]
- 26.Oklejewicz M, et al. Phase resetting of the mammalian circadian clock by DNA damage. Curr Biol. 2008;18:286–291. doi: 10.1016/j.cub.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 27.Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: An update. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ring DB, et al. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes. 2003;52:588–595. doi: 10.2337/diabetes.52.3.588. [DOI] [PubMed] [Google Scholar]
- 29.Kunick C, et al. 1-Azakenpaullone is a selective inhibitor of glycogen synthase kinase-3 beta. Bioorg Med Chem Lett. 2004;14:413–416. doi: 10.1016/j.bmcl.2003.10.062. [DOI] [PubMed] [Google Scholar]
- 30.Davies TG, et al. Structure-based design of a potent purine-based cyclin-dependent kinase inhibitor. Nat Struct Biol. 2002;9:745–749. doi: 10.1038/nsb842. [DOI] [PubMed] [Google Scholar]
- 31.Engelmann W. In: Chronobiology and Psychiatric Disorders. Halaris A, editor. Amsterdam: Elsevier Science; 1987. pp. 263–289. [Google Scholar]
- 32.Quiroz JA, Gould TD, Manji HK. Molecular effects of lithium. Mol Interv. 2004;4:259–272. doi: 10.1124/mi.4.5.6. [DOI] [PubMed] [Google Scholar]
- 33.Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- 34.Gotter AL, et al. A time-less function for mouse timeless. Nat Neurosci. 2000;3:755–756. doi: 10.1038/77653. [DOI] [PubMed] [Google Scholar]
- 35.Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem. 2005;280:29397–29402. doi: 10.1074/jbc.M503526200. [DOI] [PubMed] [Google Scholar]
- 36.Harada Y, et al. Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J Biol Chem. 2005;280:31714–31721. doi: 10.1074/jbc.M506225200. [DOI] [PubMed] [Google Scholar]
- 37.Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- 38.Thresher RJ, et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 39.van der Horst GT, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 40.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 41.Fabian MA, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 42.Mullin R. Drug Discovery: As high-throughput sreening draws fire, researchers leverage science to put automation into perspective. Chem Eng News. 2004;82:23–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.