Abstract
The Balbiani body is an evolutionarily conserved asymmetric aggregate of organelles that is present in early oocytes of all animals examined, including humans. Although first identified more than 150 years ago, genes acting in the assembly of the Balbiani body have not been identified in a vertebrate. Here we show that the bucky ball gene in the zebrafish is required to assemble this universal aggregate of organelles. In the absence of bucky ball the Balbiani body fails to form, and vegetal mRNAs are not localized in oocytes. In contrast, animal pole-localized oocyte markers are expanded into vegetal regions in bucky ball mutants, but patterning within the expanded animal pole remains intact. Interestingly, in bucky ball mutants an excessive number of cells within the somatic follicle cell layer surrounding the oocyte develop as micropylar cells, an animal-pole specific cell fate. The single micropyle permits sperm to fertilize the egg in zebrafish. In bucky ball mutants, excess micropyles cause polyspermy. Thus bucky ball provides the first genetic access to Balbiani body formation in a vertebrate. We demonstrate that bucky ball functions during early oogenesis to regulate polarity of the oocyte, future egg and embryo. Finally, the expansion of animal identity in oocytes and somatic follicle cells suggests that somatic cell fate and oocyte polarity are interdependent.
Keywords: animal-vegetal, polarity, Balbiani body, oocyte, follicle cells, micropyle, fertilization, mRNA localization, mitochondrial cloud, zebrafish
INTRODUCTION
The complex body plan of the vertebrate embryo begins with the establishment of the embryonic axes. Formation of both the dorsal-ventral and left-right axes depends on the prior establishment of the animal-vegetal axis, which itself gives rise to the anterior-posterior axis. Although the first axis to form, specification of the animal-vegetal axis is significantly less well understood than the other axes, in large part due to its establishment during oogenesis making it less accessible to experimental manipulation. In frogs and fish, the animal-vegetal axis is apparent in primary oocytes. In mouse, this axis is evident during oocyte maturation, although earlier oocyte asymmetries also exist. However, the relationship between polarity of the oocyte and the embryonic axes of the mouse zygote is not fully understood and remains an active area of study (reviewed in (Gardner, 2005; Hiiragi et al., 2006; Prodon et al., 2004; Zernicka-Goetz, 2005; Zernicka-Goetz, 2006)).
The first asymmetry evident in vertebrate oocytes is a prominent aggregate of organelles that includes endoplasmic reticulum (ER), golgi, and mitochondria, called the Balbiani body or mitochondrial cloud (Billett and Adam, 1976; Heasman et al., 1984; Marinos and Billett, 1981; Pepling et al., 2007) and reviewed in (Guraya, 1979; Kloc et al., 2004a)). The Balbiani body is transiently present in all vertebrates examined, including mammals. In mammals, the Balbiani body has been postulated to select the healthiest mitochondria for the future zygote (Cox and Spradling, 2003; Guraya, 1979). In frogs and fish the Balbiani body is thought to be a component of a vegetal pole RNA transport pathway (Kloc et al., 2001; Kloc and Etkin, 1995; Kloc et al., 1996; Wilk et al., 2005) and reviewed in (Kloc et al., 2004b).
Three pathways are postulated to sort RNA along the animal-vegetal (AV) axis in Xenopus. Two pathways direct transcripts to the vegetal pole that together comprise the Xenopus message transport organizer (METRO) pathway, while the third less understood pathway governs animal pole targeting (Kloc and Etkin, 1995). The early vegetal-directed pathway utilizes RNA binding proteins and ER to “trap” germ plasm transcripts within aggregates that associate with the Balbiani body (King et al., 1999; Kloc et al., 2001; Mowry and Cote, 1999). In contrast, vegetal localization in the late pathway requires intact microtubules rather than Balbiani body association (King et al., 1999). Transient overlap between early and late components suggests the two pathways intersect, the early pathway likely regulates the assembly of cytoskeletal “tracks” required for the later process (King et al., 2005; Kloc et al., 2001; Kloc et al., 1996).
The bucky ball (buc) mutant was identified based on its maternal-effect egg phenotype, whereby cytoplasm segregates radially around the yolk rather than to the animal pole to form the blastodisc (Dosch et al., 2004). In addition, the animal pole marker cyclinB and vegetal marker bruno-like are not localized in eggs from bucky ball mutant mothers (Dosch et al., 2004). Here we show that bucky ball is required to localize animal and vegetal transcripts during oogenesis to generate oocyte polarity, and to assemble the evolutionarily conserved Balbiani body in primary oocytes. Furthermore, our studies reveal an additional role for Bucky ball in the zebrafish ovary in restricting the number of animal-pole specific micropylar cells in the surrounding somatic follicle cell layer, suggesting interdependent patterning between the oocyte and follicle cell layer.
MATERIALS AND METHODS
In situ hybridization and histology
Whole mount in situ hybridization was performed as described (Thisse and Thisse, 1998). Stained oocytes were cleared in benzoyl benzoate/Canada balsam or embedded in JB4 plus resin; infiltration was for 30 minutes. 5 µm plastic sections were cut using a microtome. Sections were coated with permount and then a coverslip was applied. Images were captured with a Leica MZ 12-5 and Cool Snap/Cool Snap cf using Openlab (Improvision), Iplab (Scanalytics), or a Zeiss AXIOSKOP and Prog/Res/3012 (Kontron Elektronic) control software on Apple Macintosh Computers. Images were processed in Abobe Photoshop and Illustrator. In situ hybridization on slides was essentially the same, except 10 µm cryo-sections (using a Leica Cryotome) of tissue tek (Meyers) embedded ovaries were stained.
Hematoxylin & Eosin staining. Ovaries were dissected from anesthetized adult females and fixed in 4% PFA or 3.7% formaldehyde overnight. Following three 30-minute washes in PBS, the ovaries were transferred to MeOH, then embedded in JB4 plus plastic resin and sectioned, as described above. Slides were stained in Hematoxylin for 20 minutes, washed with water, then stained with Eosin for 30 minutes, washed with water, and cleared in 95% EtOH. Dried slides were coated with permount solution and then cover slipped.
Oocytes were staged according to Selman et al., 1993, based on oocyte size or other distinctive morphological features of each stage.
Immunostaining, confocal microscopy
Ovaries were dissected from adult or 6 week old wild-type and buc fish, fixed in 4% PFA, and stained as described (Topczewski et al., 2001), using FITC-Phalloidin or the Gasz primary antibody (1:200 dilution) (Yan et al., 2004) with 1:500 diluted anti-rabbit Alexa Fluor 488 or 546 secondary (Molecular Probes), Vectashield (Vector labs) containing DAPI was used to label the nuclei. Hermes and Mitotracker red staining were done as described in Kosaka et al., 2007, except Alexa Fluor 546 secondary antibody (Molecular Probes) was used to detect Hermes and oocytes were stained as wholemounts rather than on sections. Mitochondria and ER were also visualized with the vital dye, DiOC6 (3) (Molecular Probes D-273) (0.5µg/mL in M199 media (Gibco 11150)), for 10minutes, followed by 6×5 minute washes in M199 media. Images were acquired using a Zeiss LSM 510 Laser Scanning Inverted Microscope.
RTPCR
Ovaries were dissected from sibling and bucp106re mutant female fish. 50 stage IV oocytes were manually singled out for RNA isolation. For stage I/II oocytes, 3 ovaries dissected from juvenile zebrafish were used. RNA was isolated using Trizol. RNA and cDNA concentrations were measured using a spectrophotometer (Beckman Coulter DU 640). PCR was performed on a dilution series prepared from a stock concentration of 0.4µg/µl (0.2µg/µl, 0.1µg/µl, 0.05µg/µl). Primers: mago nashi 5’-TACCGCGCACAGTTACCGCGCACAGTTATGTCCACGAG-3’ and 5’-GGCCG ATGAGA CTGAATACGAGGCA-3’ based on (Pozzoli et al., 2004). For brl, 5’-AAGCACAAAATGCCCTTCAC-3’ and 5’-GCACTCCTCGATTTGACCAT-3’ based on (Suzuki et al., 2000). For EF1α, primers 5’-AGCCTGGTATGGTTGTGACCTTCG-3’ and 5’-CCAAGTTGTTTTCCTTTCCTGCG-3’. All primers were synthesized by IDT, Inc. Images acquired with BioRad Geldoc.
For QRTPCR: cDNA was prepared as stated above from manually sorted Stage IV oocytes (50 oocytes per sample). Triplicate reactions (25µl reactions) with SYBR green (Applied Biosystems), were carried out using the DNA Engine Opticon 2 System (MJ Research). The relative mRNA levels of the averaged CT were quantified and normalized to wild-type EF1α or Actin expression in Stage IV oocytes. The relative expression is shown in the graph.
Electron microscopy
Dissected adult or 6 week old wild-type and buc mutant ovaries were fixed in 2.5% glutaraldehyde + 2% paraformaldehyde in 0.1 M Sodium Cacodylate overnight at 4 °C. Samples were then rinsed in 0.1M Sodium Cacodylate buffer, post-fixed with 2% osmium tetroxide, dehydrated in graded ethanol, and embedded in Epon. 70 nm thin sections were obtained by standard EM protocols, and examined with the JEOL JEM 1010 electron microscope after uranyl acetate and bismuth sub-nitrite staining (Hayat, 1986; Hayat, 2000). Imaging was with a Hamamatsu CCD camera and AMT 12-HR software. All supplies were purchased from Electron Microscopy Sciences, Fort Washington, PA.
RESULTS
bucky ball is required for animal-vegetal pole mRNA localization during oogenesis
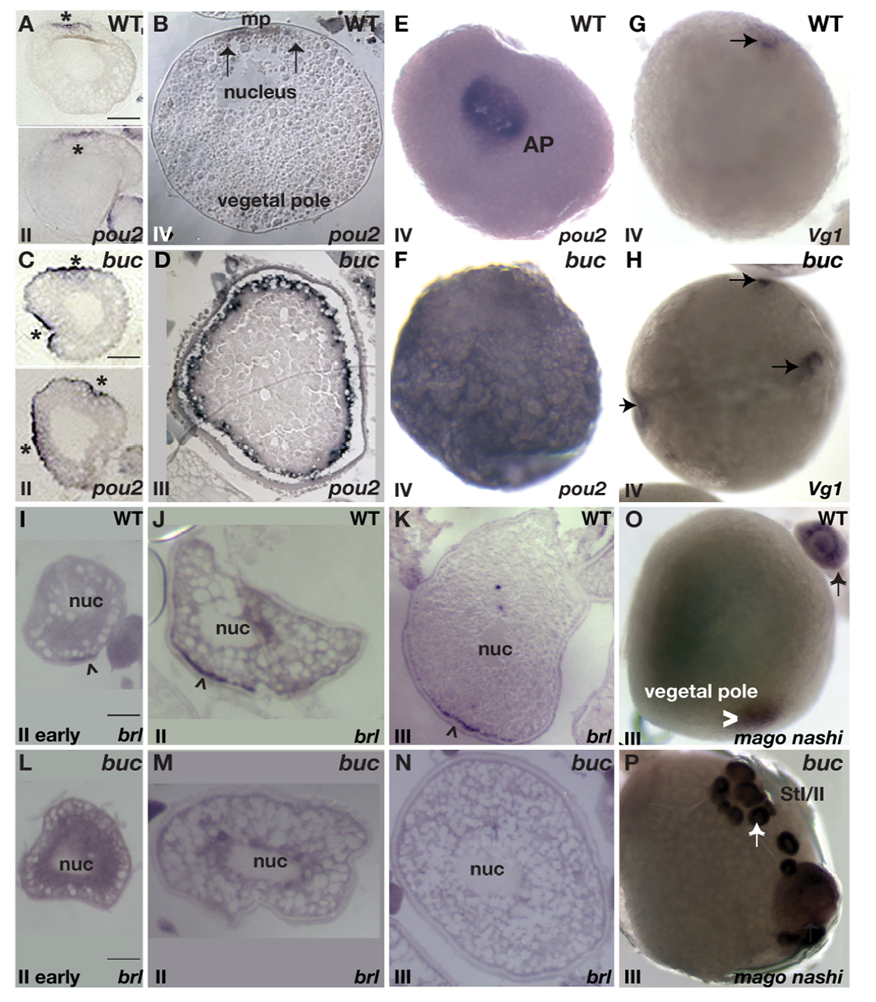
To determine if the transcript mislocalization seen previously in the eggs of homozygous maternal-effect bucky ball mutant females (Dosch et al., 2004) was due to abnormal radial cytoplasmic segregation following egg activation or an earlier defect in transcript localization, we examined RNA localization during oogenesis in bucky ball mutants. The animal pole marker pou2 was initially present throughout the cytoplasm of stage I wild type and bucky ball mutant oocytes (data not shown, n>100 for each). In stage II wild-type oocytes, pou2 transcripts became asymmetrically localized (Fig 1A n=52), localizing to a single animal pole domain in stage III wild-type oocytes (Fig. 1B,E n=31) (Howley and Ho, 2000). In stage II bucky ball oocytes, pou2 occupied multiple domains at the cortex (Fig 1C) and was found radially throughout the cortex of stage III bucky ball mutant oocytes (Fig. 1D, F n>20), indicating a defect in oocyte polarity or animal pole mRNA localization during oogenesis.
Figure 1. bucky ball is required to localize vegetal pole and restrict animal pole localized oocyte transcripts.
Animal pole localization of pou2 in in situ hybridizations on sections of wild-type stage II (A) and stage IV (B) oocytes. C) in situ hybridizations on sections shows pou2 enriched at multiple cortical sites in buc stage II oocytes, and D) radially expanded at the cortex in buc stage IV oocytes E) Animal pole (AP) view of pou2 localization in a wild-type stage IV oocyte. F) In buc mutant oocytes pou2 transcripts are not asymmetrically localized. G) Vg1 localization (arrow) at the animal pole beneath the micropyle in wild type, and H) in multiple, ectopic domains in buc mutant oocytes. In situ hybridization on sections reveals brl enrichment at the vegetal cortex (arrowhead) in wild-type early stage II (I) and late stage II (J) oocytes. At stage III of oogenesis, brl transcripts are discretely localized to the vegetal cortex in wild type (K). In stage II bucky ball mutant oocytes, brl remains throughout the oocyte (L,M) and is also not localized in stage III (N) oocytes. O) mago nashi transcripts localize to the vegetal pole in wild-type stage III oocytes (arrowhead). P) Although present in primary oocytes (arrow, also in (O)), mago nashi is not detectable by in situ hybridization in stage III buc oocytes. A–D, I–N) Hybridizations on slides containing 10µM ovary sections. E-H and O–P) Whole mount in situ hybridization images. Stage is shown in lower, left of each panel. Bars are 50µm.
In stage I and II wild-type and bucky ball oocytes, Vg1 transcripts are not asymmetrically localized (data not shown). In contrast to their vegetal pole localization in Xenopus, Vg1 transcripts in zebrafish are localized to an animal pole domain just below the micropylar follicle cell in late stage oocytes (Bally-Cuif et al., 1998) (Fig. 1G). In contrast to wild-type oocytes, which display a single Vg1 localization domain, buc mutant oocytes display multiple discrete foci of Vg1 localization around the oocyte cortex (Fig.1H; n=43). Therefore, Bucky ball is required to limit animal pole transcripts to a discrete animal pole domain at the cortex during oogenesis.
We next investigated if vegetally-localized transcripts were affected in bucky ball mutants. bruno-like (brl) transcripts were initially detected throughout primary oocytes from wild-type and bucky ball mutants. brl progressively accumulated during stage II of oogenesis at the vegetal pole cortex (Fig. 1I (early), J (late); n=19), and is asymmetrically localized in stage III oocytes (Fig. 1K; n=42) (Hashimoto et al., 2006; Suzuki et al., 2000). In bucky ball mutants brl was not asymmetrically localized in stage II oocytes (Fig. 1L,M; n=22), and remained unlocalized in stage III oocytes (Fig.1N; n=23).
The zebrafish mago nashi transcript segregates to the animal pole blastodisc during early cleavage stages (Pozzoli et al., 2004). Here we show that mago nashi transcripts are localized to the vegetal pole of wild-type stage III oocytes (Fig. 1O n=21), but are not asymmetrically localized in stage I or II oocytes. Similar to wild-type, mago nashi was present throughout early bucky ball oocytes, but was not apparent by in situ hybridization in stage III bucky ball oocytes (Fig. 1P). To determine if our inability to detect mago nashi transcripts was due to their lack of stability or maintenance when not localized, we tested if they could be detected by RT-PCR. mago nashi transcripts were clearly detectable by RT-PCR in early and late stage oocytes, and QRT-PCR confirmed that the abundance of brl and mago nashi were indistinguishable from wild-type (supplemental Fig.1). Thus, our inability to visualize mago nashi transcripts by in situ hybridization was not due to their absence, but due to their unlocalized nature and likely low concentration when distributed throughout large, late stage oocytes. These results show that Bucky ball functions during oogenesis to mediate the localization of mRNAs to distinct animal and vegetal domains of the oocyte.
bucky ball is required for Balbiani body formation
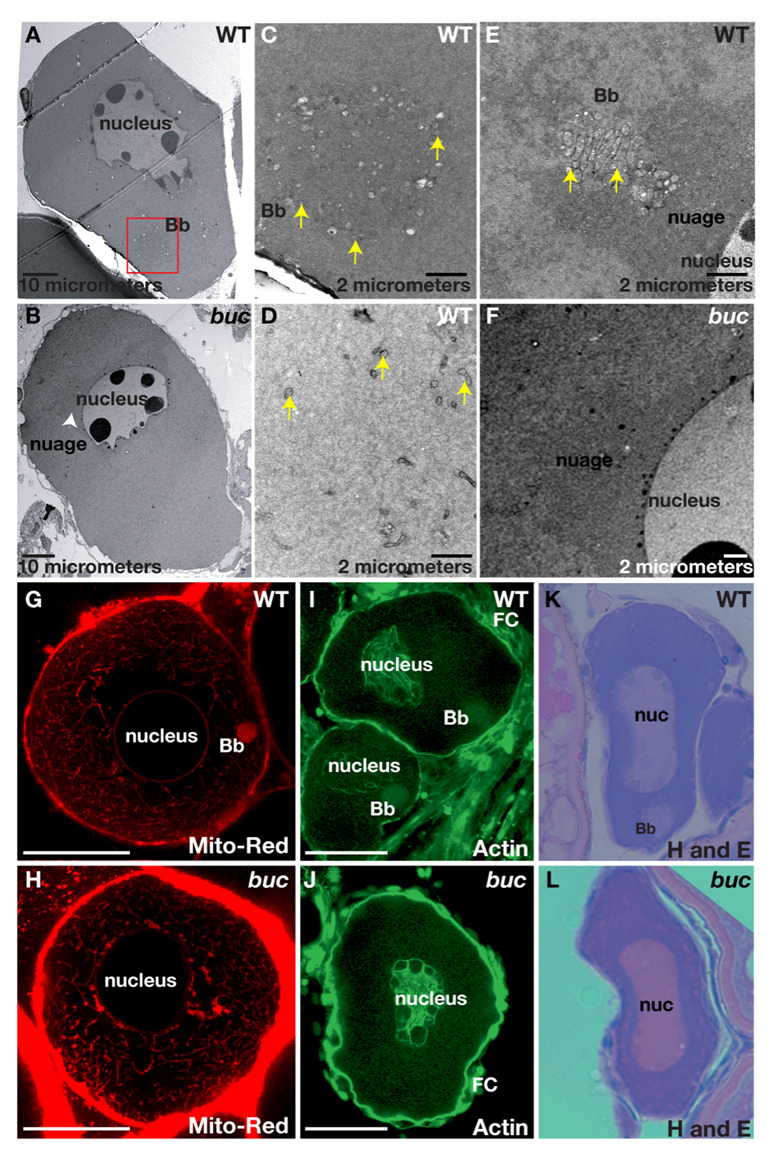
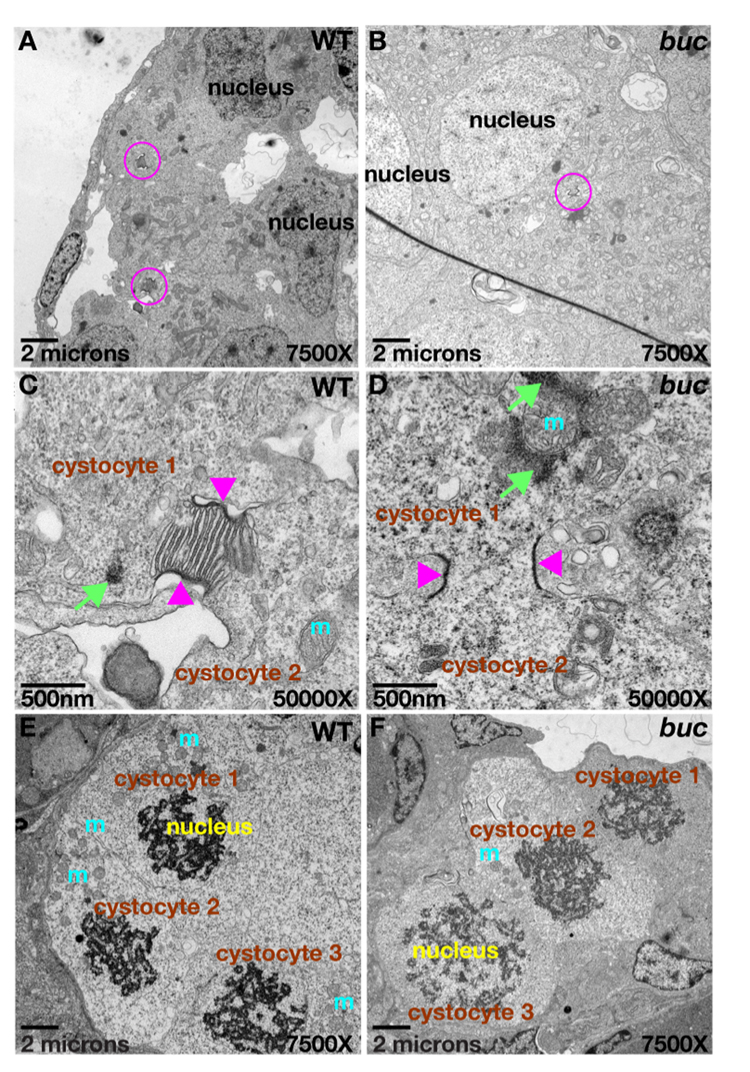
To investigate if bucky ball functions in the establishment of the earliest known polarized structure in zebrafish and Xenopus, the Balbiani body or mitochondrial cloud, we determined if the Balbiani body was affected in bucky ball mutants. The Balbiani body is an aggregate of mitochondria, ER, germinal granules and transcripts, which forms adjacent to the germinal vesicle (GV, the oocyte nucleus) in stage I oocytes. It moves vegetally and dissociates, leaving the germinal granules and vegetally-destined transcripts localized to the vegetal pole. To investigate if the Balbiani body is present in bucky ball oocytes, we used a mitochondrial dye, Mitotracker, which labels the zebrafish Balbiani body (Kosaka et al., 2007). In wild type, labeled mitochondria accumulated around the nucleus in stage Ia oocytes, and subsequently coalesced into a polarized aggregate (Fig.2G n=20 oocytes). In bucky ball mutant stage I oocytes, mitochondria were abundant around the oocyte nucleus, but failed to form a polarized aggregate (Fig.2H n=50 oocytes). Transmission electron microscopy, TEM, analysis of wild-type oocytes revealed an asymmetric aggregate of mitochondria and nuage that likely comprise the Balbiani body (Fig.2 A,C–E; n=10 ovaries, 14 oocytes). Although mitochondria and nuage were present in bucky ball oocytes, they failed to assemble into a polarized aggregate (Fig. 2B,F n=5 ovaries,17 oocytes).
Figure 2. bucky ball is required for Balbiani body assembly.
A,B) 1500X TEM images reveal the Balbiani body (red box) in wild type (A), but not in buc (B). Mitochondria (yellow arrows) are abundant and clustered within the Balbiani body (C), whereas on the opposite side of the nucleus they are sparse (D, yellow arrows). E) TEM reveals mitochondria (yellow arrowheads) and associated nuage in the Balbiani body of another wild-type oocyte. Nuage is adjacent to the nucleus and surrounds the wild-type Balbiani body. In buc stage I oocytes, perinuclear nuage is present (F and white arrowhead in B). G) Mitotracker red labels mitochondria in the Balbiani body (Bb) of wild-type stage I oocytes, and (H) reveals abundant mitochondria in buc stage I oocytes, which fail to aggregate into a Bb. I) Actin localizes to the oocyte nucleus, the somatic cells (FC) and the Bb in wild-type stage I oocytes. J) In buc mutants, Actin localization is like wild-type except for the lack of a Bb aggregate. Hematoxylin and Eosin stained sections of primary oocytes confirms the presence of a Bb in wild-type (K), but not in buc (L). C–F) 10,000X TEM; G–H) 63x Confocal images; (I–J) 40x Confocal images; (K, L) 40x images. Scale bars are 50µm for G–L.
We further investigated whether other aspects of Balbiani body formation are disrupted in bucky ball mutants. In Xenopus oocytes, Actin is found in the Balbiani body (Gard, 1999). In wild-type zebrafish oocytes, we found Actin localized to the oocyte cortex, oocyte nucleus, and enriched at the apical follicle cell surface (the surface closest to the oocyte) (Fig. 2I). Actin also formed a polarized “cloud-like” structure in stage I oocytes (Fig.2I n=30). In contrast, polarized Actin “clouds” were absent in bucky ball mutant oocytes despite normal Actin localization at the oocyte cortex, the oocyte nucleus, and at the apical follicle cell surface (Fig.2J n=63). The RNA rich Balbiani body can also be detected on histological sections stained with the basic dye Hematoxylin, which accumulates in DNA and RNA abundant regions of the cell (Guraya, 1979). The presence of a Balbiani body in wild-type and its absence in bucky ball oocytes was further confirmed by sectioning and Hematoxylin and Eosin (H&E) staining of the ovary (Fig. 2K, n=39; L, n=47).
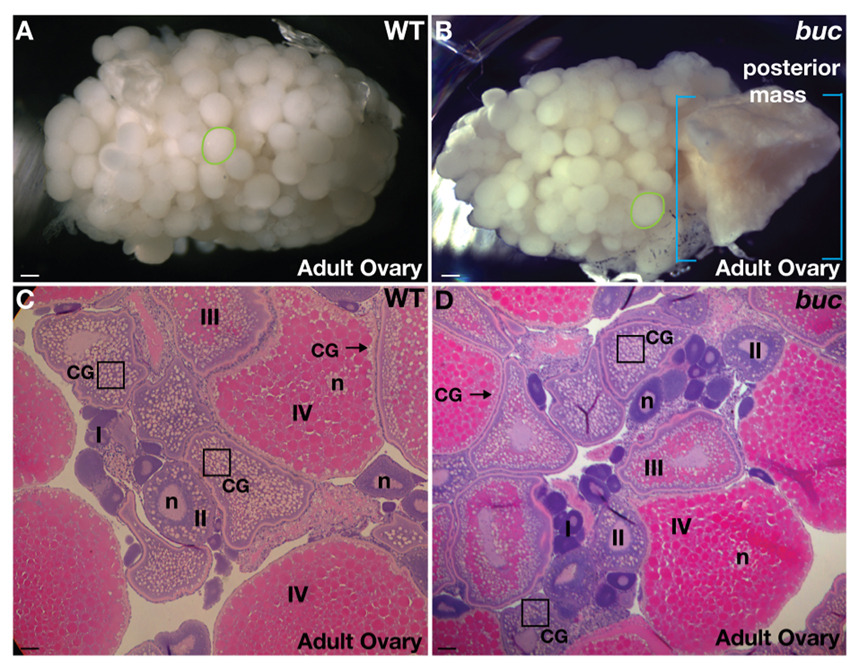
In dissecting ovaries from older bucky ball mutant females, we detected tissue masses frequently in the posterior region of the ovary of females older than 1.5 years (Fig. 3B; 83%, n=46) that are not frequently seen in wild-type (Fig. 3A; 1.9%, n=961) or in young bucky ball mutant females 6–8 weeks old (0/20 in buc mutants; 0/20 in heterozygous siblings). Histological analysis suggests that this tissue is incompletely absorbed oocyte tissue, seen in small amounts in wild-type ovaries, when mature oocytes are not ovulated (data not shown and Bauer and Goetz, 2001). Other aspects of bucky ball mutant ovaries appeared grossly normal both morphologically, as well as by histological analysis (Fig. 3C,D). Homozygous mutant bucky ball males are viable and fertile, with no apparent defects in spermatogenesis (data not shown), indicating that bucky ball plays a sexually dimorphic role in regulating polarity in the gametes of the female gonad.
Figure 3. bucky ball ovaries are grossly normal.
A) Wild-type and (B) buc mutant ovaries from adults are of similar size and composition (green circles outline single late stage III or IV oocytes), except that older buc mutant ovaries often have a mass of tissue in the posterior region of the ovary (blue brackets highlight the extent of the posterior mass). Other aspects of oogenesis (oocyte stages I–IV) appeared normal in buc mutants, as revealed by H and E stained adult ovary sections from (C) wild-type and (D) buc. A,B) Dissecting microscope images. C,D) 10x images. CG indicates cortical granules located centrally in young oocytes (e.g. box) and at the cortex (arrow) in late stage WT and mutant oocytes. n indicates the oocyte nucleus. Scale bars, 200µm (A,B); 50µm (C,D).
Defective protein and mRNA localization to the Balbiani body
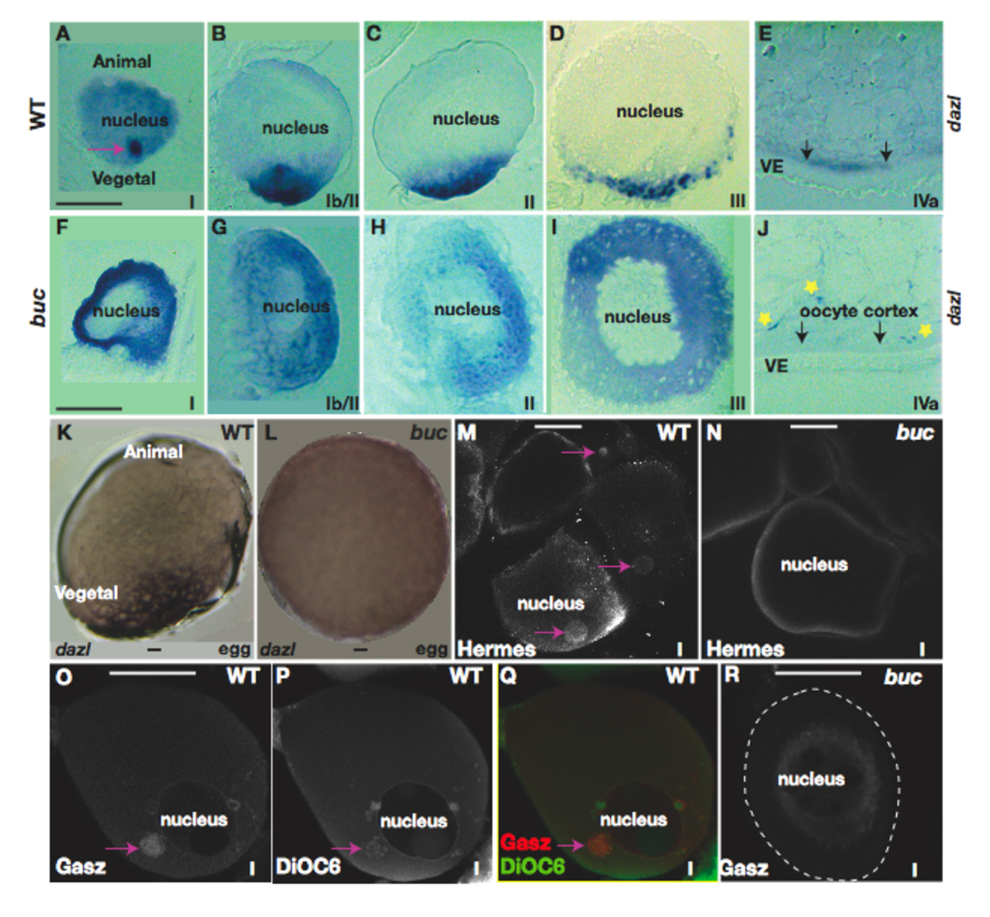
Specific proteins and transcripts also localize to the Balbiani body in Xenopus and zebrafish. To investigate if the localization of these factors is also affected in bucky ball mutants, we examined dazl RNA localization in stage I oocytes. Our analysis revealed a dynamic localization pattern of dazl during oogenesis, as also recently reported (Fig. 4, Kosaka et al., 2007). dazl transcripts first aggregate in the Balbiani body (Fig. 4A), then expand toward the future vegetal pole (Fig. 4B). By stage II of oogenesis, smaller dazl aggregates are positioned near the vegetal pole (Fig. 4C), then are confined to a discrete vegetal pole domain in stage III and IV oocytes (Fig. 4D,E) and in wild-type eggs prior to activation (Fig. 4K). This developmental time course of dazl localization is strongly reminiscent of the events described for the Xenopus message transport (METRO) pathway that localizes transcripts to the vegetal pole (see also Kosaka et al., 2007). Despite abundant transcripts, however, we did not detect a dazl aggregate in stage I bucky ball mutant oocytes (Fig. 4F). Although small unlocalized foci of transcripts were observed in stage II (Fig.4G,H) and III (Fig. 4I) bucky ball mutant oocytes, dazl transcripts did not accumulate at the oocyte cortex (Fig. 4J) or later in bucky ball mutant eggs (Fig. 4L). These results indicate that dazl transcripts localize to the vegetal cortex through a Bucky ball dependent localization mechanism.
Figure 4. Asymmetric localization of dazl and Gasz to the Balbiani body fails in bucky ball mutants.
A) In wild-type primary oocytes dazl mRNA localizes to the Balbiani body, (B) then expands toward the vegetal pole. In stage II (C), III (D), and IV (E) wild-type oocytes, dazl transcripts are localized to the vegetal cortex. F) In wild-type eggs before activation, dazl is localized to the vegetal pole. G) In bucky ball mutants dazl fails to localize to the Balbiani body in primary oocytes and H) remains dispersed throughout early stage II oocytes. In stage II (I), early stage III (J), and stage IV bucky ball oocytes, dazl mRNA is not localized, although small foci of transcripts are present (yellow asterisks in K) at all stages of oogenesis Arrows indicate the cortex in J, K. dazl mRNA remains unlocalized in mutant eggs (L). M) Hermes protein in wild-type stage I oocytes (pink arrows indicate the Balbiani body). Hermes does not localize in a cytoplasmic aggregate in bucky ball mutant primary oocytes (N). O) Gasz protein localizes to an aggregate in stage I wild-type oocytes, colocalizing with mitochondria labeled with DiOC6 (P) in the Balbiani body (Q, merged; Gasz (red), DiOC6 (green)). R) Gasz protein is not asymmetrically localized in bucky ball stage I oocytes. The oocyte stage is shown at lower right of panels. VE-vitelline envelope. A–E, G–K, are 5 µM sections following wholemount in situ hybridization. F,L) are wholemounts. M–R) Confocal images (63x objective). Bars=50µm.
Hermes, a conserved RNA binding protein localizes to the Balbiani body in Xenopus and zebrafish stage I oocytes (Fig. 4M; n=18/30, Kosaka et al., 2007; Zearfoss et al., 2004), colocalizing with mitochondria (data not shown). In contrast, Hermes protein is not localized in a cytoplasmic aggregate in bucky ball mutant oocytes (Fig. 4N; n=50), indicating that Hermes protein either requires the Bucky ball protein and/or the Balbiani body for its localization.
Gasz, a Germ cell-specific gene encoding a protein with four Ankyrin repeats, a Sterile-α motif, and a basic leucine Zipper, is expressed in spermatocytes and oocytes of mammals, several fish species including zebrafish, and in Xenopus its protein localizes to a Balbiani body-like aggregate. Here we report that Gasz protein, but not mRNA, is present in an aggregate (Fig. 4O) that colocalizes with mitochondria in the Balbiani body in wild-type primary zebrafish oocytes (Fig. 4P,Q). In contrast, Gasz was not localized in an asymmetric aggregate in bucky ball mutants (Fig. 4R). Based on the lack of a mitochondrial aggregate in bucky ball primary oocytes by Mitotracker staining and TEM analysis, lack of asymmetric aggregation of dazl, Hermes, and Gasz, and lack of detection of the Balbiani body by histological analysis, we conclude that the Balbiani body does not assemble in bucky ball mutants and therefore, that Bucky Ball is required for Balbiani body formation.
Cystoblast cytokinesis is synchronous and incomplete in zebrafish
We next investigated if earlier stages of oogenesis were altered in bucky ball mutants. During oogenesis, a germ line stem cell gives rise to cystoblasts, which undergo mitosis, generating a nest of primary oocytes called cystocytes. It has been shown in several animals that cystoblast cytokinesis is incomplete, generating cystocytes that are connected by intercellular bridges. These cytoplasmic connections are thought to facilitate synchronous cyst development and to limit the total number of cells that will survive or develop as oocytes in Drosophila and the mouse (Deng and Lin, 2001; Lin and Spradling, 1995; Lin et al., 1994; Mowry and Cote, 1999; Pepling and Spradling, 1998; Snapp et al., 2004; Storto and King, 1989) and reviewed in (Kloc et al., 2004a). In Drosophila, polarity and oocyte specification depends on intact cyst architecture (Grieder et al., 2000; Lin and Spradling, 1995; Ohlmeyer and Schupbach, 2003) (Bolivar et al., 2001). A conserved cyst organization in Xenopus has led to the hypothesis that cyst architecture also plays a similar role in limiting cystoblast divisions, and conferring oocyte polarity by regulating the inheritance of Balbiani body precursor materials in vertebrates (Kloc et al., 2004a). In the teleost fish medaka, female gametes form interconnected cysts that also undergo synchronous divisions during their differentiation (Saito et al., 2007).
Although primary oocyte nests have been identified in zebrafish, their cytokinesis has been suggested to be complete, since intercellular bridges have not been identified (Matova and Cooley, 2001; Selman et al., 1993). Since most animals examined exhibit incomplete cystocyte divisions, we reasoned that zebrafish may also have cystocytes with intercellular bridges. If so, then it was possible that perturbed cytoplasmic bridges could underlie the absence of the Balbiani body and oocyte polarity in bucky ball oocytes. To investigate if zebrafish cystocytes share cytoplasmic connections, we examined wild-type and bucky ball juvenile ovaries by TEM. Our analysis clearly demonstrates that zebrafish cystocytes undergo incomplete mitotic divisions, retaining ring canal-like interconnections (Fig. 5A,C). In wild-type cystocytes we observed intercellular bridges both with and lacking ER structures within the lumen (Fig. 5C and data not shown; n=5 ovaries). When multiple bridges of a single cyst were observed within the same section (the maximum observed in a single section was three), all the bridges displayed or lacked the ER structures. This suggests that the cysts are either in different stages or that the ER structures are only apparent in distinct planes of section. The nuclei of cystocytes within a cyst were prominent and synchronous in their progression through mitosis and synaptonemal complexes were evident (Fig. 5E). In the bucky ball mutant, cystocyte nests also developed synchronously, with well-formed intercellular bridges between cystocytes, as well as synaptonemal complexes and abundant mitochondria (Fig. 5B,D,F; n=3 ovaries). Synchronous development within cysts was also confirmed by DAPI staining in wild type and bucky ball mutants (data not shown). Therefore, in zebrafish cytoplasmic bridges do form between cystocytes, likely synchronizing their development, as also found in Drosophila, mouse, Xenopus, and humans, and they are not perturbed in bucky ball mutants.
Figure 5. Cytoplasmic bridges connect cystocytes in zebrafish.
Ring canal-like cytoplasmic bridges (pink circles) connect primary oocytes in a cyst (cystocytes) in A) wild-type and B) buc mutants. C,D) 50,000X TEM images of cysts from (A) and (B), Mitochondrial cement (green arrows) and mitochondria (m) are near the cytoplasmic bridges (pink arrowheads) in wild-type and mutants. E,F) Synchronous cyst division in wild-type and buc mutant cysts; 7500x TEM images.
bucky ball is required to restrict an animal-pole follicle cell layer cell fate
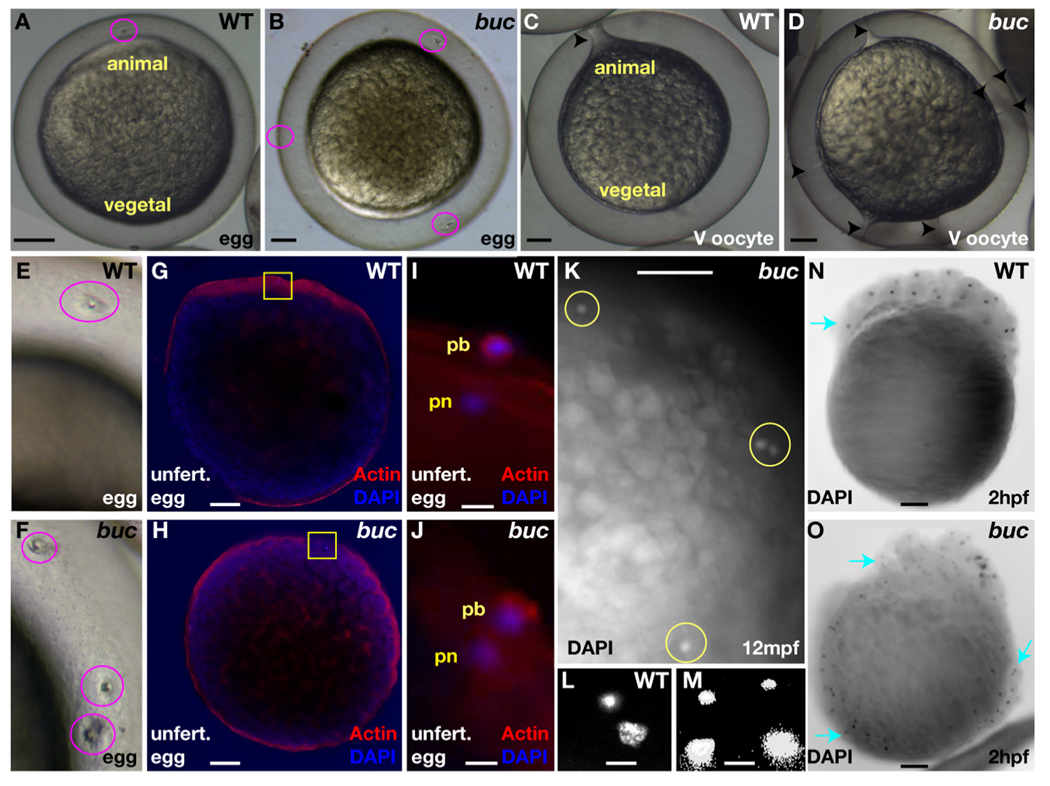
Both the oocyte and the surrounding follicular epithelium display AV polarity in the zebrafish. In zebrafish, a specialized follicle cell adjacent to the animal pole of the oocyte makes the single micropyle through which the sperm fertilizes the egg. To determine if the animal pole specific micropylar cell fate within the follicular epithelium is affected, we counted the number of micropyles in wild-type and mutant oocytes and eggs. In contrast to wild-type eggs, which possess a single micropyle at the animal pole (Fig. 6A,E), as many as 12 micropyles were found on bucky ball chorions all around the surface (average = 6; n > 200 eggs; Fig. 6B,F). Very interestingly, in examining activation of dissected stage V wild-type and mutant oocytes, we observed a striking cytoplasmic extension from the egg plasma membrane to the micropyle in the chorion (Fig. 6C). Egg activation and formation of these cytoplasmic extensions did not depend on fertilization and instead was initiated by contact of the egg with natural spawning medium. In contrast to wild-type, bucky ball mutant stage V oocytes exhibited multiple cytoplasmic extensions from the egg plasma membrane to each of the micropyles (Fig. 6D), suggesting that these ectopic structures are functional micropyles.
Figure 6. bucky ball limits the micropylar follicle cell fate preventing polyspermy.
A,E) The single micropyle (pink circle) at the animal pole of a wild-type chorion. B,F) Multiple micropyles on buc chorions (pink circles). C) An oocyte cytoplasmic projection extends from the plasma membrane to the micropyle in activated wild-type oocytes (arrowhead). D) Multiple projections in activated buc mutant oocytes (arrowheads). G) In activated wild-type eggs, Actin is enriched at the animal pole cortex. H) In activated buc eggs, Actin is uniform around the cortex due to the lack of animal-vegetal polarity. I) Actin and DAPI label the extruded polar body (pb), while DAPI alone marks the female pronucleus (pn), indicating the completion of meiosis in unfertilized wild-type. J) Like wild-type a single pronucleus and polar body are observed in activated buc mutants. (G,H) yellow boxed areas indicate region depicted in I and J. L) Female and male pronuclei in a wild-type egg 10 mpf. K,M) Polyspermy is indicated by excess pronuclei (yellow circles in K) in buc mutant eggs 10 mpf. N) DAPI staining showing cleavage occurs at the animal pole in wild-type (blue arrow), while nuclear cleavage O) is circumferential in buc progeny (blue arrows). A–F) Live dissecting microscope images. G,H) 10X confocal objective. I,J) Confocal images 63x objective. K) Dissecting microscope image; L,M) 40x acquired images. N,O) Images 5x objective. Scale bars are 200µm except for (I,J,L,M) which are 10µm.
To test directly if these ectopic structures are functional micropyles, we examined the number of DAPI stained pronuclei in the absence of, or following, fertilization of bucky ball mutant eggs. In zebrafish, egg activation induces the completion of meiosis and polar body extrusion (Wolenski and Hart, 1988). In unfertilized, but activated wild-type and bucky ball mutant eggs, the female pronucleus and a single extruded polar body surrounded by Actin was observed (Fig.6G–J, n=17 for wild-type, n=38 for buc), indicating the completion of meiosis in both wild-type and bucky ball mutant eggs. In fertilized wild-type eggs, both the male and female pronuclei and the female polar body were evident near the oocyte cortex at the animal pole (Fig. 6L). If the ectopic micropyles are functional in bucky ball eggs and the single micropyle is the mechanism that blocks polyspermy in zebrafish (Hart and Donovan, 1983), then we expect to see ectopic male pronuclei, in addition to the female pronucleus. In fertilized bucky ball eggs, multiple DAPI-labeled pronuclei were evident around the egg circumference (Fig. 6K,M, n>19). Since unfertilized bucky ball eggs exhibit only a single pronucleus (Fig. 6J), these results indicate that multiple sperm fertilized bucky ball eggs. Thus, we conclude that bucky ball mutant eggs contain multiple functional micropyles, and that the block to polyspermy in zebrafish is the presence of a single micropyle, consistent with previous studies (Hart and Donovan, 1983).
Embryos derived from bucky ball mutant females attempt to undergo cell division. However, in contrast to wild-type embryos in which cellular cleavages are restricted to the animal pole (Fig. 6N), cell division appeared to occur radially around the circumference of bucky ball mutant progeny (Fig. 6O; n>200). These data together with the circumferential expression of animal pole localized mRNAs in bucky ball mutant oocytes demonstrate that animal pole identity is expanded in the oocyte, the follicle cell layer, and later in embryos from bucky ball mutant mothers. Furthermore, we found that the ectopic domains of Vg1 in bucky ball mutant oocytes coincide with the positions of ectopic micropylar follicle cells (not shown), suggesting interdependence between the specification of this follicle cell fate and the establishment of the Vg1 animal pole oocyte domain.
DISCUSSION
bucky ball is the first gene in a vertebrate known to function in Balbiani body formation. In Xenopus and zebrafish, vegetal pole destined transcripts co-localize with the Balbiani body, leading to the hypothesis that it functions early in a vegetal pole localization pathway (Kloc et al., 2004b; Kosaka et al., 2007). Individual vegetal pole localized mRNAs have been depleted (Heasman et al., 2001; Kloc et al., 2005) (reviewed in (Kloc et al., 2004b; Kloc and Etkin, 2005), but only at stages after the Balbiani body has formed and dispersed. Thus far, it has not been possible to eliminate the Balbiani body or to identify genes essential for its formation. Based on our analysis of bucky ball mutants, we postulate that Bucky ball regulates animal-vegetal polarity and directs mRNA localization along the animal-vegetal oocyte axis in zebrafish, in part through mediating Balbiani body formation.
The Balbiani body has been postulated to select the healthiest mitochondria for the prospective zygote (Cox and Spradling, 2003; Cox and Spradling, 2006; Guraya, 1979; Kloc et al., 2004b; Pepling et al., 2007). In bucky ball mutants, mitochondria are abundant and mitochondrial cement, implicated in mitochondrial expansion, is present. Although a population of more active mitochondria may associate with the Balbiani body and co-localize with germ plasm constituents, mitochondrial function in bucky ball mutants is sufficient to permit normal oocyte growth in the absence of the Balbiani body. Based on the abundant mitochondria, normal oocyte growth, and completion of meiosis in bucky ball mutants, we conclude that the Balbiani body is not generally required for mitochondrial function or amplification in the zebrafish during oogenesis.
The failure to localize vegetal markers and the circumferential expansion of animal pole markers in bucky ball mutant oocytes indicates that Bucky ball is required to localize vegetal transcripts and exclude animal transcripts from the vegetal pole. Animal and vegetal pole transcripts may localize by an interdependent mechanism, where Bucky ball specifies vegetal identity and thereby restricts animal pole identity to generate polarity in the early oocyte. Alternatively, Bucky ball could function independently to establish both animal and vegetal pole oocyte territories, as is known for Staufen in Drosophila (Ferrandon et al., 1994; St Johnston et al., 1991). Bucky ball may restrict animal pole identity in both the oocyte and follicle cell layer by acting cell non-autonomously from one tissue to the other or autonomously in each tissue. Such relationships between the somatic and germ line cells are known to influence follicle cell fate and oocyte pattern in Drosophila, but were not known in vertebrate oocytes. In either case, in the absence of bucky ball, the failure to generate the animal-vegetal oocyte axis leads to a radial expansion of animal identity.
Our results are consistent with the model proposed in Xenopus that a late vegetal-pole localization pathway depends on an earlier Balbiani body-dependent vegetal localization pathway. The vegetal markers bruno-like and mago nashi do not localize to the Balbiani body in stage I oocytes in zebrafish, and instead only localize to the vegetal cortex during stage II and III of oogenesis, respectively (Fig 1, (Kosaka et al., 2007; Suzuki et al., 2000)). These temporal differences in vegetal transcript localization may reflect distinct pathways that localize transcripts to the vegetal pole, as demonstrated in Xenopus and proposed in zebrafish (Heasman et al., 2001) (King et al., 2005; Kloc et al., 2004b; Kloc and Etkin, 1998; Kloc and Etkin, 2005; Kloc et al., 1998; Kloc et al., 1996; Kosaka et al., 2007; Mowry and Cote, 1999; Wilk et al., 2005). In bucky ball mutants, the Balbiani body is absent and the late localizing vegetal transcripts bruno-like and mago nashi are not localized. This result is consistent with the late localization of vegetal pole transcripts, not associated with the Balbiani body in stage I, also depending on the earlier Balbiani body-mediated vegetal pole localization pathway. Alternatively, Bucky ball could function independently at stage I and at stage II/III to localize transcripts to the vegetal pole.
Zebrafish dazl localizes to the Balbiani body and then to the vegetal pole during stage I of oogenesis. Recent studies of the zebrafish dazl 3’UTR identified distinct localization elements that mediate its association with the Balbiani body and later with the vegetal pole (Kosaka et al., 2007). Our results demonstrate that Bucky ball function is essential for dazl localization to the Balbiani body and the oocyte vegetal pole. In agreement with this finding, Kosaka and colleagues (2007) demonstrated that failure to localize to the Balbiani body abolishes all other steps of dazl localization.
Interestingly, the animal pole localized marker Vg1 was present in multiple restricted domains in bucky ball mutants. The limited size of the multiple, ectopic Vg1 domains in bucky ball mutant oocytes (Fig. 1H) compared to the more broadly expanded pou2 animal pole transcript (Fig. 1B,D) shows that some patterning remains intact within the expanded animal domain. The co-localization of Vg1 below the ectopic micropylar cells in bucky ball mutants suggests that interactions between the oocyte animal surface and the micropyle cell govern the localization of Vg1 to the most animal pole region of the oocyte, in a manner distinct from the localization of pou2. Consistent with the possibility of distinct mechanisms localizing Vg1 versus pou2, is their distinct temporal periods of localization. pou2 localizes to the animal pole as early as stage II of oogenesis (Howley and Ho, 2000), whereas Vg1 localizes animally later in late stage III or early stage IV oocytes (Bally-Cuif et al., 1998). Although Vg1 is abundant in stage II oocytes, it does not become localized, consistent with a mechanism localizing pou2 that cannot localize Vg1 at this stage. The broad expansion of pou2 transcripts in bucky ball mutants, together with their early localization at stage II, just following vegetal pole transcript localization and disassembly of the Balbiani body at the end of stage I of oogenesis, suggests that the Balbiani body or a vegetally-localized factor dependent on the Balbiani body restricts the animal pole domain of pou2. Alternatively, bucky ball may also act at this later stage to restrict pou2 animally.
Vg1 localization and specification of the micropylar follicle cell fate, in contrast, display both Bucky ball dependent and independent mechanisms of localization. We show that bucky ball is required to restrict the micropylar follicle cell fate to just one cell of up to 12 (Fig. 6). Since all follicle cells do not adopt a micropylar cell fate in bucky ball mutants and those that do are generally well spaced from the next micropylar cell, additional interactions between the follicle cells themselves or between the oocyte and follicle cell layer independent of Bucky ball likely contribute to limiting the Vg1 domain and the micropylar cell fate to just one at the animal pole. Bucky ball could act autonomously within the oocyte, and cell non-autonomously to restrict follicle cell fate or vice versa. Such relationships between the somatic and germ line cells are known to influence follicle cell fate and oocyte pattern in Drosophila, but were not known in vertebrate oocytes. Further investigation is required to determine if a single or multiple signaling events occur between the oocyte and the follicle cells to limit the micropyle follicle cell fate and the Vg1 domain within the oocyte. Finally, it is also possible that Bucky ball functions uniquely and autonomously to limit animal identity in the germ line and somatic tissues, in which case other signaling mechanisms would align the micropyle and the oocyte animal pole.
The interdependence of the oocyte and the somatic cells that surround it in forming a functional follicle has long been appreciated. The early events of follicle formation are similar between zebrafish and mammals. In mammals, however, the follicle cells proliferate more extensively forming multiple layers surrounding the oocyte within a pre-antral follicle. In the antral follicle an asymmetric fluid filled cavity forms generating distinct populations of pre-antral derived follicle cells, the mural and cumulus cells. Formation of these distinct cell types requires signaling from the oocyte (Diaz et al., 2007; Eppig, 2001; Hussein et al., 2005; Hussein et al., 2006; Plancha et al., 2005; Su et al., 2004). Whether there is a relationship between the formation of these distinct cell types, the asymmetric antral cavity, and the asymmetric Balbiani body present in early mouse oocytes, like the micropylar follicle cell in the zebrafish, remains to be determined.
In conclusion, we show that the bucky ball gene is required to assemble the evolutionarily conserved asymmetric Balbiani body and to generate animal-vegetal oocyte polarity. We do not know if loss of vegetal identity and AV transcript localization are secondary defects resulting from a primary failure to assemble the Balbiani body or if bucky ball function is required independently in some or all of these processes. We also show that bucky ball is required to properly pattern the follicle cell layer, specifically to limit the number of animal pole micropylar cells, which is crucial for preventing polyspermy in zebrafish. Lastly, our studies provide the first genetic evidence that oocyte and follicle cell pattern are interrelated in zebrafish.
Supplementary Material
Supplemental Figure 1. RTPCR reveals vegetal marker presence in early (A) and late (B) stage oocytes in wild-type and buc (mut). C) QRTPCR shows that the relative expression levels of bruno-like and mago nashi transcripts are similar in wild-type and mutant oocytes corroborating the notion that these vegetal mRNAs are present but not localized in bucky ball mutant oocytes.
ACKNOWLEDGEMENTS
We would like to thank Frank Cotelli, Kunio Inoue, and Benjamin Feldman for gifts of probes. Martin Matzuk for generously providing the Gasz antibody, and Malgorzata Kloc for kindly providing the Hermes antibody. We are grateful to Ashley Tai for assisting with pou2 in situ hybridization and RTPCR, and Neelima Shah for her assistance and EM expertise. We thank Tripti Gupta for thoughtful reading and comments on the manuscript. This work was funded by NIH grant HD050901 to MCM. FLM was supported by the Damon Runyon Cancer Research Foundation DRG 1826-04.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bally-Cuif L, Schatz WJ, Ho RK. Characterization of the zebrafish Orb/CPEB-related RNA binding protein and localization of maternal components in the zebrafish oocyte. Mech Dev. 1998;77:31–47. doi: 10.1016/s0925-4773(98)00109-9. [DOI] [PubMed] [Google Scholar]
- Bauer MP, Goetz FW. Isolation of gonadal mutations in adult zebrafish from a chemical mutagenesis screen. Biol Reprod. 2001;64:548–554. doi: 10.1095/biolreprod64.2.548. [DOI] [PubMed] [Google Scholar]
- Billett FS, Adam E. The structure of the mitochondrial cloud of Xenopus laevis oocytes. J Embryol Exp Morphol. 1976;36:697–710. [PubMed] [Google Scholar]
- Bolivar J, Huynh JR, Lopez-Schier H, Gonzalez C, St Johnston D, Gonzalez-Reyes A. Centrosome migration into the Drosophila oocyte is independent of BicD and egl, and of the organisation of the microtubule cytoskeleton. Development. 2001;128:1889–1897. doi: 10.1242/dev.128.10.1889. [DOI] [PubMed] [Google Scholar]
- Cox RT, Spradling AC. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development. 2003;130:1579–1590. doi: 10.1242/dev.00365. [DOI] [PubMed] [Google Scholar]
- Cox RT, Spradling AC. Milton controls the early acquisition of mitochondria by Drosophila oocytes. Development. 2006;133:3371–3377. doi: 10.1242/dev.02514. [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. Asymmetric germ cell division and oocyte determination during Drosophila oogenesis. Int Rev Cytol. 2001;203:93–138. doi: 10.1016/s0074-7696(01)03005-4. [DOI] [PubMed] [Google Scholar]
- Diaz FJ, Wigglesworth K, Eppig JJ. Oocytes determine cumulus cell lineage in mouse ovarian follicles. J Cell Sci. 2007;120:1330–1340. doi: 10.1242/jcs.000968. [DOI] [PubMed] [Google Scholar]
- Dosch R, Wagner DS, Mintzer KA, Runke G, Wiemelt AP, Mullins MC. Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Dev Cell. 2004;6:771–780. doi: 10.1016/j.devcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Eppig JJ. Oocyte control of ovarian follicular development and function in mammals. Reproduction. 2001;122:829–838. doi: 10.1530/rep.0.1220829. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Elphick L, Nusslein-Volhard C, St Johnston D. Staufen protein associates with the 3'UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Gard DL. Confocal microscopy and 3-D reconstruction of the cytoskeleton of Xenopus oocytes. Microsc Res Tech. 1999;44:388–414. doi: 10.1002/(SICI)1097-0029(19990315)44:6<388::AID-JEMT2>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Gardner RL. The case for prepatterning in the mouse. Birth Defects Res C Embryo Today. 2005;75:142–150. doi: 10.1002/bdrc.20038. [DOI] [PubMed] [Google Scholar]
- Grieder NC, de Cuevas M, Spradling AC. The fusome organizes the microtubule network during oocyte differentiation in Drosophila. Development. 2000;127:4253–4264. doi: 10.1242/dev.127.19.4253. [DOI] [PubMed] [Google Scholar]
- Guraya SS. Recent advances in the morphology, cytochemistry, and function of Balbiani's vitelline body in animal oocytes. Int Rev Cytol. 1979;59:249–321. doi: 10.1016/s0074-7696(08)61664-2. [DOI] [PubMed] [Google Scholar]
- Hart NH, Donovan M. Fine structure of the chorion and site of sperm entry in the egg of Brachydanio. Journal of Experimental Zoology. 1983;227:277–296. [Google Scholar]
- Hashimoto Y, Suzuki H, Kageyama Y, Yasuda K, Inoue K. Bruno-like protein is localized to zebrafish germ plasm during the early cleavage stages. Gene Expr Patterns. 2006;6:201–205. doi: 10.1016/j.modgep.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Hayat MA. Basic Techniques for transmission Electron Microscopy. San Diego and London: Academic Press; 1986. [Google Scholar]
- Hayat MA. Principles and Techniques of Electron Microscopy. Biological Applications. 2000 doi: 10.1017.S1431927601010066. [DOI] [PubMed] [Google Scholar]
- Heasman J, Quarmby J, Wylie CC. The mitochondrial cloud of Xenopus oocytes: the source of germinal granule material. Dev Biol. 1984;105:458–469. doi: 10.1016/0012-1606(84)90303-8. [DOI] [PubMed] [Google Scholar]
- Heasman J, Wessely O, Langland R, Craig EJ, Kessler DS. Vegetal localization of maternal mRNAs is disrupted by VegT depletion. Dev Biol. 2001;240:377–386. doi: 10.1006/dbio.2001.0495. [DOI] [PubMed] [Google Scholar]
- Hiiragi T, Alarcon VB, Fujimori T, Louvet-Vallee S, Maleszewski M, Marikawa Y, Maro B, Solter D. Where do we stand now? Mouse early embryo patterning meeting in Freiburg, Germany (2005) Int J Dev Biol. 2006;50:581–586. doi: 10.1387/ijdb.062181th. discussion 586-7. [DOI] [PubMed] [Google Scholar]
- Howley C, Ho RK. mRNA localization patterns in zebrafish oocytes. Mech Dev. 2000;92:305–309. doi: 10.1016/s0925-4773(00)00247-1. [DOI] [PubMed] [Google Scholar]
- Hussein TS, Froiland DA, Amato F, Thompson JG, Gilchrist RB. Oocytes prevent cumulus cell apoptosis by maintaining a morphogenic paracrine gradient of bone morphogenetic proteins. J Cell Sci. 2005;118:5257–5268. doi: 10.1242/jcs.02644. [DOI] [PubMed] [Google Scholar]
- Hussein TS, Thompson JG, Gilchrist RB. Oocyte-secreted factors enhance oocyte developmental competence. Dev Biol. 2006;296:514–521. doi: 10.1016/j.ydbio.2006.06.026. [DOI] [PubMed] [Google Scholar]
- King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- King ML, Zhou Y, Bubunenko M. Polarizing genetic information in the egg: RNA localization in the frog oocyte. Bioessays. 1999;21:546–557. doi: 10.1002/(SICI)1521-1878(199907)21:7<546::AID-BIES3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Chan AP, Allen LH, Zearfoss NR, Etkin LD. RNA localization and germ cell determination in Xenopus. Int Rev Cytol. 2001;203:63–91. doi: 10.1016/s0074-7696(01)03004-2. [DOI] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Dougherty MT, Brey EM, Etkin LD. Formation, architecture and polarity of female germline cyst in Xenopus. Dev Biol. 2004a;266:43–61. doi: 10.1016/j.ydbio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Etkin LD. The Balbiani body and germ cell determinants: 150 years later. Curr Top Dev Biol. 2004b;59:1–36. doi: 10.1016/S0070-2153(04)59001-4. [DOI] [PubMed] [Google Scholar]
- Kloc M, Etkin LD. Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development. 1995;121:287–297. doi: 10.1242/dev.121.2.287. [DOI] [PubMed] [Google Scholar]
- Kloc M, Etkin LD. Apparent continuity between the messenger transport organizer and late RNA localization pathways during oogenesis in Xenopus. Mech Dev. 1998;73:95–106. doi: 10.1016/s0925-4773(98)00041-0. [DOI] [PubMed] [Google Scholar]
- Kloc M, Etkin LD. RNA localization mechanisms in oocytes. J Cell Sci. 2005;118:269–282. doi: 10.1242/jcs.01637. [DOI] [PubMed] [Google Scholar]
- Kloc M, Larabell C, Chan AP, Etkin LD. Contribution of METRO pathway localized molecules to the organization of the germ cell lineage. Mech Dev. 1998;75:81–93. doi: 10.1016/s0925-4773(98)00086-0. [DOI] [PubMed] [Google Scholar]
- Kloc M, Larabell C, Etkin LD. Elaboration of the messenger transport organizer pathway for localization of RNA to the vegetal cortex of Xenopus oocytes. Dev Biol. 1996;180:119–130. doi: 10.1006/dbio.1996.0289. [DOI] [PubMed] [Google Scholar]
- Kloc M, Wilk K, Vargas D, Shirato Y, Bilinski S, Etkin LD. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development. 2005;132:3445–3457. doi: 10.1242/dev.01919. [DOI] [PubMed] [Google Scholar]
- Kosaka K, Kawakami K, Sakamoto H, Inoue K. Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis. Mech Dev. 2007;124:279–289. doi: 10.1016/j.mod.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC. Fusome asymmetry and oocyte determination in Drosophila. Dev Genet. 1995;16:6–12. doi: 10.1002/dvg.1020160104. [DOI] [PubMed] [Google Scholar]
- Lin H, Yue L, Spradling AC. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 1994;120:947–956. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
- Marinos E, Billett FS. Mitochondrial number, cytochrome oxidase and succinic dehydrogenase activity in Xenopus laevis oocytes. J Embryol Exp Morphol. 1981;62:395–409. [PubMed] [Google Scholar]
- Matova N, Cooley L. Comparative aspects of animal oogenesis. Dev Biol. 2001;231:291–320. doi: 10.1006/dbio.2000.0120. [DOI] [PubMed] [Google Scholar]
- Mowry KL, Cote CA. RNA sorting in Xenopus oocytes and embryos. Faseb J. 1999;13:435–445. doi: 10.1096/fasebj.13.3.435. [DOI] [PubMed] [Google Scholar]
- Ohlmeyer JT, Schupbach T. Encore facilitates SCF-Ubiquitin-proteasome-dependent proteolysis during Drosophila oogenesis. Development. 2003;130:6339–6349. doi: 10.1242/dev.00855. [DOI] [PubMed] [Google Scholar]
- Pepling ME, Spradling AC. Female mouse germ cells form synchronously dividing cysts. Development. 1998;125:3323–3328. doi: 10.1242/dev.125.17.3323. [DOI] [PubMed] [Google Scholar]
- Pepling ME, Wilhelm JE, O'Hara AL, Gephardt GW, Spradling AC. Mouse oocytes within germ cell cysts and primordial follicles contain a Balbiani body. Proc Natl Acad Sci U S A. 2007;104:187–192. doi: 10.1073/pnas.0609923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plancha CE, Sanfins A, Rodrigues P, Albertini D. Cell polarity during folliculogenesis and oogenesis. Reprod Biomed Online. 2005;10:478–484. doi: 10.1016/s1472-6483(10)60824-3. [DOI] [PubMed] [Google Scholar]
- Pozzoli O, Gilardelli CN, Sordino P, Doniselli S, Lamia CL, Cotelli F. Identification and expression pattern of mago nashi during zebrafish development. Gene Expr Patterns. 2004;5:265–272. doi: 10.1016/j.modgep.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Prodon F, Pruliere G, Chenevert J, Sardet C. [Establishment and expression of embryonic axes: comparisons between different model organisms] Med Sci (Paris) 2004;20:526–538. doi: 10.1051/medsci/2004205526. [DOI] [PubMed] [Google Scholar]
- Saito D, Morinaga C, Aoki Y, Nakamura S, Mitani H, Furutani-Seiki M, Kondoh H, Tanaka M. Proliferation of germ cells during gonadal sex differentiation in medaka: Insights from germ cell-depleted mutant zenzai. Dev Biol. 2007;310:280–290. doi: 10.1016/j.ydbio.2007.07.039. [DOI] [PubMed] [Google Scholar]
- Selman K, Wallace R, Sarka A, Qi X. Stages of oocyte development in the Zebrafish, Brachydanio rerio. Journal of Morphology. 1993;218:203–224. doi: 10.1002/jmor.1052180209. [DOI] [PubMed] [Google Scholar]
- Snapp EL, Iida T, Frescas D, Lippincott-Schwartz J, Lilly MA. The fusome mediates intercellular endoplasmic reticulum connectivity in Drosophila ovarian cysts. Mol Biol Cell. 2004;15:4512–4521. doi: 10.1091/mbc.E04-06-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- Storto PD, King RC. The role of polyfusomes in generating branched chains of cystocytes during Drosophila oogenesis. Dev Genet. 1989;10:70–86. doi: 10.1002/dvg.1020100203. [DOI] [PubMed] [Google Scholar]
- Su YQ, Wu X, O'Brien MJ, Pendola FL, Denegre JN, Matzuk MM, Eppig JJ. Synergistic roles of BMP15 and GDF9 in the development and function of the oocyte-cumulus cell complex in mice: genetic evidence for an oocyte-granulosa cell regulatory loop. Dev Biol. 2004;276:64–73. doi: 10.1016/j.ydbio.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Maegawa S, Nishibu T, Sugiyama T, Yasuda K, Inoue K. Vegetal localization of the maternal mRNA encoding an EDEN_BP/Bruno-like protein in zebrafish. Mech Dev. 2000;1–2:205–209. doi: 10.1016/s0925-4773(00)00270-7. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. Zebrafish Science Monitor. Vol. 5 1998. High Resolution Whole-Mount in situ Hybridization. [Google Scholar]
- Topczewski J, Sepich DS, Myers DC, Walker C, Amores A, Lele Z, Hammerschmidt M, Postlethwait J, Solnica-Krezel L. The Zebrafish Glypican Knypek Controls Cell Polarity during Gastrulation Movements of Convergent Extension. Developmental Cell. 2001;1:251–264. doi: 10.1016/s1534-5807(01)00005-3. [DOI] [PubMed] [Google Scholar]
- Wilk K, Bilinski S, Dougherty MT, Kloc M. Delivery of germinal granules and localized RNAs via the messenger transport organizer pathway to the vegetal cortex of Xenopus oocytes occurs through directional expansion of the mitochondrial cloud. Int J Dev Biol. 2005;49:17–21. doi: 10.1387/ijdb.041906kw. [DOI] [PubMed] [Google Scholar]
- Wolenski JS, Hart NH. Effects of cytochalasins B and D on the fertilization of zebrafish (Brachydanio) eggs. Journal of Experimental Zoology. 1988;246:202–215. doi: 10.1002/jez.1402460213. [DOI] [PubMed] [Google Scholar]
- Yan W, Ma L, Zilinski CA, Matzuk MM. Identification and characterization of evolutionarily conserved pufferfish, zebrafish, and frog orthologs of GASZ. Biol Reprod. 2004;70:1619–1625. doi: 10.1095/biolreprod.103.024778. [DOI] [PubMed] [Google Scholar]
- Zearfoss NR, Chan AP, Wu CF, Kloc M, Etkin LD. Hermes is a localized factor regulating cleavage of vegetal blastomeres in Xenopus laevis. Dev Biol. 2004;267:60–71. doi: 10.1016/j.ydbio.2003.10.032. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M. Developmental cell biology: cleavage pattern and emerging asymmetry of the mouse embryo. Nat Rev Mol Cell Biol. 2005;6:919–928. doi: 10.1038/nrm1782. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M. The first cell-fate decisions in the mouse embryo: destiny is a matter of both chance and choice. Curr Opin Genet Dev. 2006;16:406–412. doi: 10.1016/j.gde.2006.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. RTPCR reveals vegetal marker presence in early (A) and late (B) stage oocytes in wild-type and buc (mut). C) QRTPCR shows that the relative expression levels of bruno-like and mago nashi transcripts are similar in wild-type and mutant oocytes corroborating the notion that these vegetal mRNAs are present but not localized in bucky ball mutant oocytes.