Abstract
As the field of glycobiology grows, important roles for glycolipids and glycoproteins in neurological disorders are being increasingly appreciated. However, few studies have explored the involvement of these molecules in the pathology of psychiatric illnesses. We investigated molecular differences related to glycobiology in subjects with schizophrenia by analyzing gene expression profiles using a focused glycogene chip, a custom-designed, oligonucleotide array containing genes encoding proteins related to glycobiology, including glycosyltransferases, carbohydrate-binding proteins, proteoglycans and adhesion molecules. We measured expression profiles in prefrontal cortical (BA46) samples from schizophrenic subjects and matched controls. We find differential expression of genes particularly related to glycosphingolipid/sphingolipid metabolism and N- and O-linked glycan biosynthesis in subjects with schizophrenia. Expression decreases of seven genes associated with these pathways, UGT8, SGPP1, GALC, B4GALT6, SPTLC2, ASAH1 and GAL3ST1, were validated by quantitative PCR in schizophrenic subjects with short-term illness. Only one of these genes, SPTLC2, showed differential expression in chronic schizophrenic subjects, although an increase in expression was observed. Covariate analysis found that the expression of five of these genes was significantly positively correlated with age in schizophrenic, but not control, subjects. These changing patterns of expression could represent an adaptive response to pathology with disease progression or a compensatory effect of antipsychotic medication, although no significant correlations between gene expression levels and drug doses were observed. Disruption of sphingolipid metabolism early in illness could result in widespread downstream effects encompassing diverse pathological deficits already described in schizophrenia, especially those involving myelination and oligodendrocyte function; hence this system may represent an important link in schizophrenia pathology.
Keywords: microarray, gene expression glycolipid, schizophrenia, glycosylation
Introduction
A growing interest in the field of glycobiology, defined in a broad sense as the study of sugars and their roles in biology, has emerged in recent years. Glycoconjugates, which include three broad groups, proteoglycans, glycoproteins, and glycolipids, are ubiquitously distributed in the cells, bodily fluids, tissues and organs, including the brain, of all animals, and are primarily, localized on the cell surface. Research in this area has uncovered many roles for both glycolipids and glycoproteins in the basic functioning of the brain. Further, disruptions in these systems have been linked to several neurological disorders, such as Alzheimer's disease (Grimm et al., 2005; Katsel et al., 2007; Tamboli et al., 2005) and Huntington's disease (Desplats et al., 2007; Katsel et al. 2007). In addition, lysosomal storage disorders, such as Gaucher disease, Tay-Sachs and Sandhoff disease, are specifically characterized by abnormalities in the degradation of glycosphingolipids (Kolter and Sandhoff 2006).
Glycoconjugates are critical determinants of various cellular functions, including signal transduction and cell-to-cell interactions. They further play important roles in CNS development (Rutishauser 2008), in particular in synapse formation, maturation and modulation (Yamaguchi 2002) and immune function (Axford 2001). Dysfunctions in these systems have been implicated in the pathophysiology of schizophrenia (Nawa and Takei 2006; Rapoport et al., 2005; Saetre et al., 2007; Thompson et al., 2004; Woo and Crowell 2005), a devastating, heterogeneous psychiatric disorder with largely unknown etiology (Harrison 1999). However, aside from a few early reports, none have explored the potential involvement of glycoconjugates in psychiatric disease mechanisms. Studies from the 1980s have found that, based on the levels of glycoconjugates excreted in the urine, metabolism of glycoproteins and glycosaminoglycans is abnormal in schizophrenic patients (Brunngraber et al., 1981). Other studies have demonstrated altered levels of GM3 and GD3 gangliosides, the primary glycolipids on neurons, in erythrocyte membranes of schizophrenic patients (Haselhorst et al., 1988). A further association between glycan biology and psychiatric disorders comes from accounts that some of the lysosomal storage disorders present with psychiatric manifestations (Navon 1991; Rosebush et al., 1995; Zelnik et al., 2000).
In this study, we utilized a custom-designed oligonucleotide array developed and annotated by the Consortium for Functional Glycomics for assessing gene expression changes associated with various aspects of glycobiology, to explore transcriptional differences in the prefrontal cortex of subjects with schizophrenia. This array contains various classes of genes related to glycosyltransferases, carbohydrate-binding proteins, proteoglycans, nucleotide sugar synthesis, adhesion molecules, transporter proteins, growth factors and related genes Our results reveal altered expression of genes encoding glycosyltransferase enzymes in subjects with schizophrenia, suggesting that dysfunction in sphingolipid metabolism is important to disease pathology, particularly early in illness. Given the importance of certain sphingolipids to oligodendrocyte function, these findings support the hypothesis that decreased myelination and disruption of connectivity in schizophrenia might be related to decreases in lipid membrane components.
Materials and Methods
Subjects
Approval for this study was obtained from the Ethics Committee of the Victorian Institute of Forensic Medicine and the North Western Mental Health Program Behavioral and Psychiatric Research and Ethics Committee. Psychiatric diagnoses of schizophrenia for each subject was made according to DSM-IV criteria (American Psychiatric Association, 1994) by consensus between two senior psychiatrists and a psychologist following extensive case history review using a structured diagnostic instrument, the Diagnostic Instrument for Brain Studies (DIBS) (Hill et al., 1996). Subject cohorts were carefully chosen to represent short-term duration of illness (DOI) (within 5 years of diagnosis; n=8 patients, n=8 controls), intermediate-term DOI (7−18 years of illness; n=14 patients, n=14 controls) and long-term DOI (> 28 years of illness; n=8 patients, n=8 controls) (Table 1), as our previous studies have indicated that gene expression levels change with increasing length of illness (Dean et al., 2007; Narayan et al., 2008). DOI was calculated as the time from initial diagnosis to death. To minimize variation in the effects of antipsychotic drugs, all subjects chosen were treated with “typical” antipsychotic drugs. The final recorded dose of antipsychotic drug for each subject was converted to chlorpromazine equivalents, which incorporates lifetime exposure to antipsychotic medications (Bezchlibnyk-Butler and Jeffries 1999). Control subjects were those with no known history of psychiatric illness, and were matched for age and gender (Table 1). No information regarding ethnicity of the subjects was available. In cases where death was witnessed, the time between death and autopsy was taken as the postmortem interval (PMI). Where death was not witnessed, tissue was only taken from individuals who had been seen alive up to 5 hours before being found dead, in these cases PMI was taken as the interval half way between the donors being found dead and last being seen alive. Importantly, in all cases, cadavers were refrigerated within 5 hours of being found and tissue was rapidly frozen to −70°C within 30 minutes of autopsy. The pH of the CNS tissue was measured as described previously (Kingsbury et al., 1995). There were no significant differences in the PMI or pH between controls and schizophrenic subjects.
Table 1.
Demographic and array parameters for all subjects.
| Schizophrenics Short DOI: | Short Controls: | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AGE | DOI | SEX | pH | PMI | 3′/5′ | %P | Drugs | Drug dose* | AGE | SEX | pH | PMI | 3′/5′ | %P | ||
| S1 | 38 | 4 | M | 6.02 | 50 | 1.25 | 31.8 | Haloperidol, Clozapine | 50 mg | Sc1 | 38 | M | 6.42 | 46 | 1.24 | 33.8 |
| S2 | 26 | 2 | M | 6.39 | 52 | 2.3 | 30.2 | Haloperidol | 500 mg | Sc2 | 26 | M | 6.42 | 24 | 1.17 | 29.9 |
| S3 | 19 | 3 | M | 6.22 | 43 | 1.27 | 32 | Haloperidol, Risperidone | 750 mg | Sc3 | 21 | M | 5.82 | 40 | 1.03 | 31.5 |
| S4 | 22 | 3 | M | 6.17 | 37 | 1.48 | 31.8 | Pimozide | 200 mg | Sc4 | 22 | M | 6.39 | 62 | 1.12 | 32.4 |
| S5 | 22 | 3 | M | 6.07 | 37 | 1.29 | 29.2 | Trifluoperazine, Flupenthixol | 450 mg | Sc5 | 25 | M | 6.48 | 50 | 1.19 | 32.4 |
| S6 | 25 | 2 | M | 6.38 | 49 | 1.1 | 31.9 | Trifluoperazine | 200 mg | Sc6 | 25 | M | 6.15 | 35 | 1.2 | 32.6 |
| S7 | 21 | 2 | F | 6.24 | 56 | 2.45 | 23.3 | Haloperidol | NA | Sc7 | 21 | F | 6.03 | 58 | 1.55 | 30.4 |
| S8 | 36 | 4 | F | 6.28 | 45 | 1.12 | 31.9 | Haloperidol | 160 mg | Sc8 | 38 | F | 6.14 | 50 | 1.19 | 31.4 |
| AVG = 330 ± 92.68 | ||||||||||||||||
| Schizophrenics Intermediate DOI: | Intermediate Controls: | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | DOI | Sex | pH | PMI | 3′/5′ | %P | Drugs | Drug dose* | Age | Sex | pH | PMI | 3′/5′ | %P | ||
| Int1 | 53 | 7 | M | 6.23 | 25 | 1.35 | 27.8 | Cholorpromazine | 200 mg | Ic1 | 53 | M | 6.43 | 12 | 1.23 | 30.6 |
| Int2 | 38 | 11 | M | 6.44 | 36 | 1.28 | 28 | Fluphenazine | 200 mg | Ic2 | 38 | M | 6.19 | 44 | 1.2 | 27.3 |
| Int3 | 41 | 11 | M | 6.2 | 31 | 1.13 | 29.5 | Fluphenazine | 500 mg | Ic3 | 42 | M | 6.21 | 26 | 1.08 | 32 |
| Int4 | 26 | 11 | F | 6.11 | 41 | 1.3 | 28.6 | Remoxipride | 750 mg | Ic4 | 29 | F | 6.38 | 34 | 1.19 | 30.1 |
| Int5 | 36 | 12 | M | 6.04 | 38 | 2.15 | 27.3 | Fluphenazine | 200 mg | Ic5 | 36 | M | 6.46 | 42 | 1.26 | 29.2 |
| Int6 | 45 | 12 | M | 6.33 | 68 | 1.09 | 31.6 | Stelazine | 300 mg | Ic6 | 48 | M | 6 | 52 | 1.3 | 29.2 |
| Int7 | 42 | 15 | M | 6.26 | 34.5 | 1.13 | 32.9 | Haloperidol | 670 mg | Ic7 | 44 | M | 6.38 | 44 | 1.34 | 27.7 |
| Int8 | 32 | 15 | M | 6.05 | 17 | 1.2 | 32.7 | Fluphenthixol | 610 mg | Ic8 | 34 | M | 6.46 | 15.5 | 1.24 | 31.5 |
| Int9 | 41 | 15 | M | 6.64 | 52 | 1.22 | 31.1 | Fluphenazine | 25 mg | Ic9 | 42 | M | 6.34 | 63 | 1.23 | 30.7 |
| int10 | 33 | 14 | M | 6.54 | 36 | 1.21 | 30.5 | None | Ic10 | 37 | M | 6.4 | 47 | 1.06 | 32.1 | |
| Int11 | 65 | 18 | F | 6.35 | 50 | 1.38 | 28.2 | Fluphenazine, Haloperidol | 550 mg | Ic11 | 66 | F | 6.37 | 43 | 1.33 | 30.5 |
| AVG = 400.5 ± 77.6 | ||||||||||||||||
| Schizophrenics Long DOI: | Long Controls: | |||||||||||||||
| AGE | DOI | SEX | pH | PMI | 3′/5′ | %P | Drugs | Drug dose* | AGE | SEX | pH | PMI | 3′/5′ | %P | ||
| L1 | 57 | 28 | M | 6.06 | 24 | 1.22 | 31.2 | Fluphenazine | 150 mg | Lc1 | 57 | M | 6.43 | 27 | 1.19 | 28.8 |
| L2 | 65 | 35 | M | 6.57 | 41 | 1.36 | 31.4 | Fluphenazine | 150 mg | Lc2 | 68 | M | 6.06 | 41 | 1.34 | 31.8 |
| L3 | 65 | 36 | M | 6.29 | 42 | 1.23 | 30.5 | Trifluphenazine, Haloperidol | 460 mg | Lc3 | 65 | M | 6.47 | 20.5 | 1.19 | 31.2 |
| L4 | 55 | 33 | M | 6.1 | 25 | 1.14 | 32.7 | Thioridazine | 400 mg | Lc4 | 50 | M | 6.43 | 69 | 1.38 | 30.2 |
| L5 | 61 | 38 | M | 6.46 | 37.5 | 1.19 | 29 | Fluphenazine | 745 mg | Lc5 | 60 | M | 6.11 | 27.5 | 1.05 | 30.4 |
| L6 | 71 | 53 | M | 6.45 | 48 | 1.35 | 30.3 | Thioridazine | 150 mg | Lc6 | 72 | M | 6.21 | 39 | 1.65 | 29.6 |
| L7 | 81 | 40 | F | 6.31 | 25 | 1.11 | 30.1 | Trifluphenazine | 100 mg | Lc7 | 80 | F | 6.28 | 55 | 1.54 | 30.6 |
| L8 | 67 | 36 | M | 6.46 | 21 | 1.15 | 33.9 | Fluphenazine | 75 mg | Lc8 | 65 | M | 6.56 | 41 | 1.24 | 30.7 |
| AVG = 278.8 ± 83.2 | ||||||||||||||||
Chlorpromazine equivalents; DOI, duration of illness; PMI, post-mortem interval; 3′/5′, GAPDH 3′/5′ ratio; %P, percent Present call. The average (AVG) cumulative drug doses ± S.E.M. for each schizophrenic cohort are shown.
RNA Preparation and Array Hybridizations
Total RNA was extracted from prefrontal cortical samples (Brodmann Area 46; left hemisphere; ∼100 mg) using the Queen Mini kit according to the manufacturer's suggested protocol (Queen, Inc., Valencia, CA). All samples were treated with DNAseI to eliminate genomic DNA contamination. RNA quantification was determined by spectrophotometer readings. The ratio of OD260/OD280 was used to evaluate the purity of the nucleic acid samples and the quality of the extracted total RNA was determined by Agilent Bioanalyzer scans. RNA integrity (RIN) numbers were not available at the time of RNA extraction, but Bioanalyzer traces of each RNA sample showed no evidence for degradation products. RNA from each preparation was amplified and biotin-labeled using the GeneChip® One-Cycle Target Labeling and Control Reagents kit (Affymetrix, Inc., Santa Clara, CA). Biotin labeled samples were then hybridized to the GLYCOv2 array using standard Affymetrix protocol as described in Lockhart et al, 1996.
The GLYCOv2 array is a custom Affymetrix GeneChip containing 1900 transcripts designed for the Consortium for Functional Glycomics. On this array, each gene is represented by at least three different probesets, which were optimized based on the hybridization signals from the GLYCOv1 (version 1) array. The average signal (geometric mean) from the multiple probesets was used for calculating the overall signal for each gene, hence providing a more accurate representation of the expression levels per gene. A complete description and annotation for the GLYCOv2 array is available at http://www.scripps.edu/researchservices/dna_array/glyco_genelist.xls.
Chips were scanned using the Affymetrix ScanArray 3000 using default settings and a target intensity of 250 for scaling. Present and Absent calls were determined with the Affymetrix algorithm (GCOS, Affymetrix, Santa Clara, CA). All Marginal calls were treated as Absent. The quality controls of the hybridization for each array were assessed by the percent Present calls and GAPDH 3’/5’ ratios for each array (see Table 1 for array parameters of each sample used). Because the GLYCOv2 array contains probesets directed against both mouse and human sequences, % Present calls are approximately one-half of that expected from a human-specific array. Although we find that our human samples do hybridize to mouse sequences, these were ignored in the final analyses. Samples with GAPDH 3’/5’ ratios of >2.5, or % Present calls of <23%, were omitted (n=4). The matched control samples for the schizophrenic samples with high GAPDH 3’/5’ ratios were also excluded, resulting in 54 total samples (n=27 schizophrenic and n=27 control) for statistical analysis. We further performed Pearson's analyses to determine if there were any correlations between the pH and PMI of each sample versus percent Present call and GAPDH 3’/5’ratios for each corresponding array. No significant correlations were detected. All raw and normalized microarray data files can be found at Consortium for Functional Glycomics website (https://www.functionalglycomics.org/). In previous studies, most of these same samples (49 of the 54 samples) were hybridized to the Affymetrix Human Genome U133 Plus 2.0 array, as described in a separate manuscript (Narayan et al. 2008).
Microarray Analysis
Data were normalized using Robust Multichip Average (RMA) via RMA Express. After filtering for Present calls, a list of 783 genes was used for statistical testing. When all 27 schizophrenia subjects were analyzed together (compared to all 27 controls), the resulting lists of differentially expressed genes showed low absolute log2 ratios and high FDRs (>0.8). This is likely due to the heterogeneous nature of the disease and the wide range of age in our subjects. The use of DOI as a continuous variable in linear regression statistical models was not possible, given that control subjects do not have a DOI. Therefore, in this study, we retained the structure of different cohorts divided according to DOI described above and shown in Table 1. Two-sample t-tests were performed using BRB Array Tools (Radmacher et al., 2002) to identify differentially expressed genes between schizophrenic and control subjects in each cohort separately. Both unpaired and pair-wise comparisons were performed. For unpaired analyses, the geometric mean of the normalized expression values for all schizophrenic subjects in each cohort was compared against the geometric mean of the normalized expression values for all control subjects in each cohort. For the pair-wise analysis, the normalized expression value for each schizophrenic subject was compared against its matched control in a pairwise manner, then the results for all pairs in each cohort were averaged. The FDR was calculated according to Benjamini and Hochberg (Benjamini and Hochberg 1995). The FDRs for the short-term DOI cohort were the lowest (<0.05 for unpaired analysis and 0.13 for the paired analysis), giving high confidence in our resulting lists of differentially expressed genes. The FDRs for the intermediate-term and long-term DOI cohorts were much higher (0.60 and 0.80, respectively). The resulting lists of differentially expressed genes (p-value <0.05) from both analyses were filtered for an absolute log2 ratio >0.270 (i.e. 1.20 fold, increase or decrease). One drawback of such a multiple testing correction is the assumption that each gene varies independently, which in the context of CNS gene expression, is often not the case (Sequeira and Turecki 2006). For instance, genes belonging to a common pathway or associated with a common molecular function would be expected to vary in concert. Therefore, to allow for a more inclusive understanding of potential pathological bases in schizophrenia, for our pathways analysis, we used a less stringent criteria, as discussed previously (Benes et al., 2006), which included genes with FDR<1.0.
Covariate analyses
For all genes validated by qPCR analysis, Pearson Product Moment correlations were performed to assess the effects of age, pH and PMI on gene expression values in all subjects, and the effects of drug dose (in chlorpromazine equivalents) on expression in all subjects with schizophrenia. Student's t test (two-tailed, unpaired) was used to determine sex differences. Before the Pearson's correlations were run, the datasets were tested for a Gaussian distribution using the Kolmogorov-Smirnov method, which confirmed that the expression data was normally distributed. All statistical analyses were performed using GraphPad Prism Software (GraphPad Software Inc, San Diego, CA).
Pathways Analyses
We analyzed biological significance of differentially expressed genes using Ingenuity Pathways Analysis (IPA) (www.ingenuity.com) and the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/), which consists of over 40 annotation categories, including Gene Ontology (GO) terms, protein-protein interactions, protein functional domains, disease associations, KEGG and other bio-pathways. The IPA Functional Analysis Tools calculates a p-value using the right-tailed Fisher Exact Test, in order to determine statistically significant over-representation of genes in known canonical pathways.
Real-Time PCR Analysis
Real-time PCR experiments were performed as described previously (Desplats et al., 2006). Specific primers for each sequence of interest were designed using Primer Express 1.5 software and their specificity to bind the desired sequence was analyzed by BLAST analysis (Table 2). Primers sequences for four housekeeping genes, human β-2-microglobulin (B2M), beta-tubulin (TUBB), hypoxanthine guanine phosphoribosyl transferase (HPRT) and porphobilinogen deaminase (PBGD) were synthesized as published previously (Zhang et al., 2005). Standard curves were generated for each primer pair using serial dilutions of human cDNAs to test for efficiency of amplification. Primer efficiencies were calculated by linear regression analysis of the Ct vs log[template] blots using GraphPad Prism 3.0 software and were between 94−115%.
Table 2.
Primer sequences used for real-time PCR analysis.
| Gene ID: | Gene Name: | Accession #: | Forward Primer: | Reverse Primer: |
|---|---|---|---|---|
| B2M | beta-2-microglobulin | NM_004048 | actgaattcacccccactga | cctccatgatgctgcttaca |
| TUBB* | tubulin, beta | NM_178014 | cttcggccagatcttcagac | agagagtgggtcagctggaa |
| ASAH1 | N-acylsphingosine amidohydrolase 1 | NM_004315 | tgagcacacgtctggcctacaga | cagctgcagtgttcggtcacatg |
| B4GALT6 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 | NM_004775 | tggctgtcgtggcaaggtagac | cagccaatcactgacctccactaaga |
| SGPP1 | sphingosine-1-phosphate phosphatase 1 | NM_030791 | gaagtggtgctggaattgcatgtg | gcaatatggcttttccaaacagagtca |
| SPTLC2 | serine palmitoyltransferase, long chain base subunit 2 | NM_004863 | ggccgtgaagaaagaatgtgagtaactg | caagagaggaaagataccttgggaattagg |
| UGT8 | UDP glycosyltransferase 8 | NM_003360 | tggtcaccctgtcaatcgaactatct | gcaaaagcacaaaggcaatatcca |
| GAL3ST1 | galactose-3-O-sulfotransferase 1 | NM_004861 | gaagacgcacaagacggcca | aaggcgaacttgagccggtg |
| GALC | galactosylceramidase | NM_000153 | cctgcatgaagatgagctgttcaca | taggtacttgggaagggctggga |
primer sequence taken from Zhang X et al., BMC Molecular Biology 2005, 6:4.
Real-time PCR experiments were performed using the ABI PRISMs 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). Amplification was performed on a cDNA amount equivalent to 50 ng total RNA with 1 × SYBR Green universal PCR Master mix (Applied Biosystem, Foster City, CA) containing dNTPs, MgCl2, AmpliTaq Gold DNA polymerase, and forward and reverse primers. PCR reactions were performed on two independent sets of cDNA samples. Experimental samples and no-template controls were run in duplicate. The PCR cycling parameters were: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 94°C for 15 s, 60°C for 1 min. We first compared the expression of all four housekeeping genes in samples from control and schizophrenic subjects to assess variability in expression among all subjects and to determine if there were expression differences between control and schizophrenic subjects. In subjects with short DOI and their matched controls, B2M showed the least variation in threshold cycle (Ct) among all samples and showed no significant differences in expression between control and schizophrenic subjects, hence B2M was using as the internal control for subjects with short DOI and their controls. In subjects with long DOI and their matched controls, TUBB showed the least variation in Ct among all subjects and no significant differences in expression between control and schizophrenic subjects, hence TUBB was used for normalization. The amount of cDNA in each sample was calculated using SDS2.1 software by the comparative Ct method and expressed as 2exp(Ct). Significant differences in expression (p<0.05) were determined by Student's t test; because the direction of the changes was predicted based on the microarray data, a one-tailed t test was used. All statistics were performed with GraphPad Prism Software, (San Diego, CA).
Results
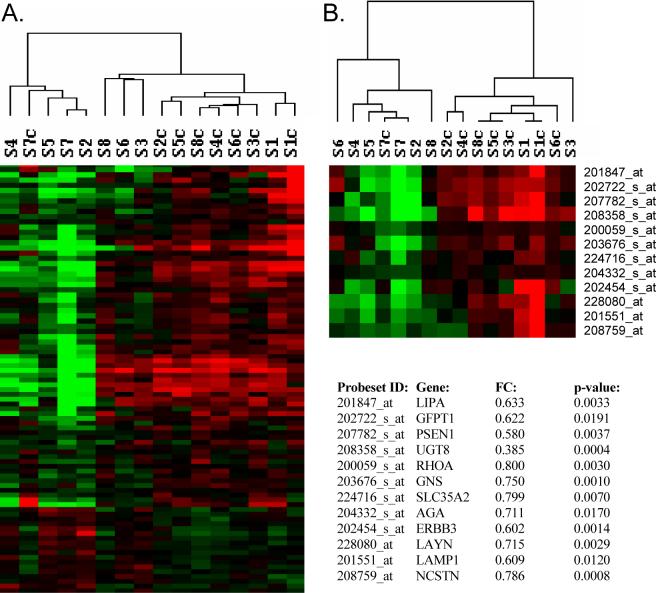
We analyzed mRNA expression data from schizophrenic subjects at three stages of illness, along with their age- and sex- matched controls using a microarray focused on genes related to glycobiology. The highest numbers of differentially expressed genes, (n=57 and n=35 for paired and unpaired analyses, respectively) were found in subjects with short-term illness, defined by death within 5 years of diagnosis. A large majority of these changes were decreases in expression (79% and 91% for paired and unpaired analyses, respectively). 10 of these genes were also found to be differentially expressed in subjects with intermediate- and long-term illness (data not shown). The differentially expressed probe sets (at p<0.01 and p<0.05) were subjected to two-way hierarchal clustering; this revealed that most subjects clustered according to their respective diagnoses (Figure 1), with the exception of control #7, who clustered with the schizophrenic subjects, and schizophrenic #1, who clustered with the controls. Notably, schizophrenic #1 was the only schizophrenic subject who had been treated with the atypical neuroleptic, clozapine. The top 12 differentially expressed genes are shown in Figure 1, with the largest magnitude of change being a 2.63-fold decrease in the expression of UDP glycosyltransferase 8 (UGT8), similar to that reported previously (Aston et al., 2005).
Figure 1.
Hierarchical clustering of gene expression changes. Heatmaps show the top differentially expressed genes at p<0.05, (A), and p<0.01, (B), in schizophrenic subjects with short-term illness and their controls. Log2-transformed expression values were subjected to two-dimensional hierarchical clustering as shown above the heatmaps. Each colored pixel represents an individual gene expression value from a single subject. Relative decreases in gene expression are indicated by green and increases by red. Schizophrenic subjects: S1-S8 and their controls, S1c-S8c, are shown. FC, fold-change. The top 12 differentially expressed genes are shown as indicated by their UniGene gene symbol.
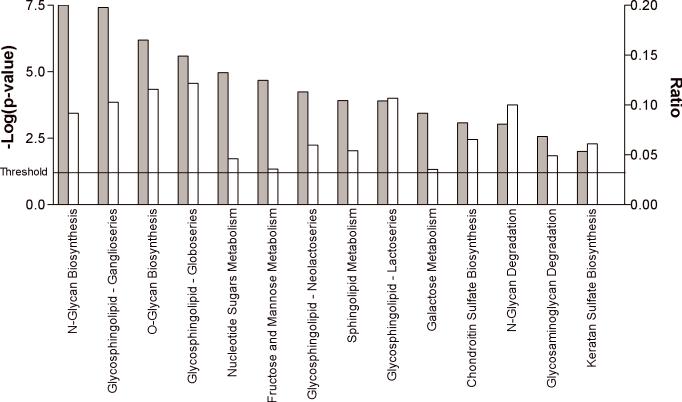
The lists of differentially expressed genes from the paired and unpaired analyses were evaluated for biological relevance using Ingenuity Systems Pathways analysis. While the GLYCOv2 array was designed to study various aspects of glycobiology, there is, nonetheless, a vast scope to this topic. Within the realm of glycobiology, significantly enriched “Functions”, included post-translational modification, carbohydrate metabolism, cancer, cell-to-cell interaction and lipid metabolism (Table 3). Canonical Pathways Analysis revealed top pathways of N- and O-linked glycan biosynthesis, which included several genes encoding glycan transferases (MAN2A2, MGAT3, ST6GAL1, B4GALT6, FUT6, ST3GAL2, GALNT5), as well as three glycosphingolipid biosynthesis pathways (ganglioseries, lactoseries and globoseries) as significantly enriched (p=4.0E−4-2.1E−6) in subjects with schizophrenia (Figure 2). In separate microarray studies investigating genome-wide expression profiles in schizophrenia using Affymetrix Human Genome U133 Plus 2.0 arrays, we found several categories related to glycobiology among our gene lists using IPA and DAVID pathways analysis. Significantly enriched Gene Ontology (GO) biological process terms included “glycoprotein biosynthesis” and “biopolymer glycosylation”, while DAVID significant Functional Clustering groups included “glycoprotein” and “glycosylation” (Narayan et al. 2008).
Table 3.
Ingenuity Systems Pathways Analysis “Functions” annotation tool identifies different significant systems associated with schizophrenia.
| Function: | p-value: | #genes: | Genes: |
|---|---|---|---|
| Post-Translational Modification | 1.08E-05 | 18 | NCSTN, ST3GAL2, KITLG, RPN2, LGALS1, TSTA3, MAN2B1, CD74, AGA, CHST4, FGF7, DPM2, FUT6, DPM3, B3GALT4, B4GALT6, GALNT5, PSEN1 |
| Carbohydrate Metabolism | 1.88E-05 | 15 | ST3GAL2, TSTA3, CHST4, FGF7, MGAT3, GNS, GAA, GALT, B4GALT6, B3GAT3, PSEN1, ARSB, GALNT5, HEXA, GALE |
| Cancer | 3.44E-05 | 10 | MGAT3, KITLG, TSTA3, LGALS1, SELP, CD74, ERBB3, FGF7, EEF2, PSEN1 |
| Cell-To-Cell Signaling and Interaction | 3.44E-05 | 14 | CHGA, KITLG, LGALS1, CSF2RB, TSTA3, SELP, ERBB3, TNFRSF9, CHST4, CCR6, FGF7, CNTFR, CLEC11A, PSEN1 |
| Lipid Metabolism | 3.52E-05 | 14 | KITLG, LGALS1, CSF2RB, CD74, FGF7, LAMP1, MGAT3, LTA4H, DPM3, B3GALT4, CLEC11A, UGT8, ST6GALNAC4, HEXA |
| Hematological System Development | 7.81E-05 | 13 | KITLG, CSF2RB, LGALS1, CHRD, ERBB3, SELP, CD74, TNFRSF9, CHST4, CCR6, FGF7, LAMP1, CLEC11A |
| Tissue Morphology | 7.81E-05 | 14 | KITLG, CSF2RB, ERBB3, SELP, TNFRSF9, AGA, CHST4, CCR6, FGF7, MGAT3, CNTFR, GAA, CLEC11A, PSEN1 |
| Cellular Assembly and Organization | 8.51E-05 | 10 | CHGA, LGALS1, GAA, SELP, CD74, B4GALT6, LAMP1, PSEN1, HEXA, ARSB |
| Cellular Growth and Proliferation | 1.78E-04 | 8 | KITLG, LGALS1, CSF2RB, CLEC11A, ERBB3, TNFRSF9, CCR6, FGF7 |
| Protein Synthesis | 2.33E-04 | 10 | NCSTN, RPL39, RPL8, RPL38, RPL7A, EEF1A1, RPS2, RPL3, PSEN1, EEF2 |
P-values were calculated using the right-tailed Fisher Exact Test in order to determine statistically significant enrichment of the indicated genes in each functional category. UniGene gene symbols for each gene are shown.
Figure 2.
The top significantly enriched canonical pathways displayed as a bar chart. Significantly enriched canonical pathways were determined using Ingenuity Systems Pathways Analyses and are displayed along the x-axis. The left y-axis displays the negative log of the p-value (gray bars), which was calculated using the right-tailed Fisher Exact Test. The right y-axis displays the Ratio (white bars), which was calculated by the # of genes in a given pathway that meet cutoff criteria, divided by the total # of genes that make up that particular pathway. The threshold for significance is indicated.
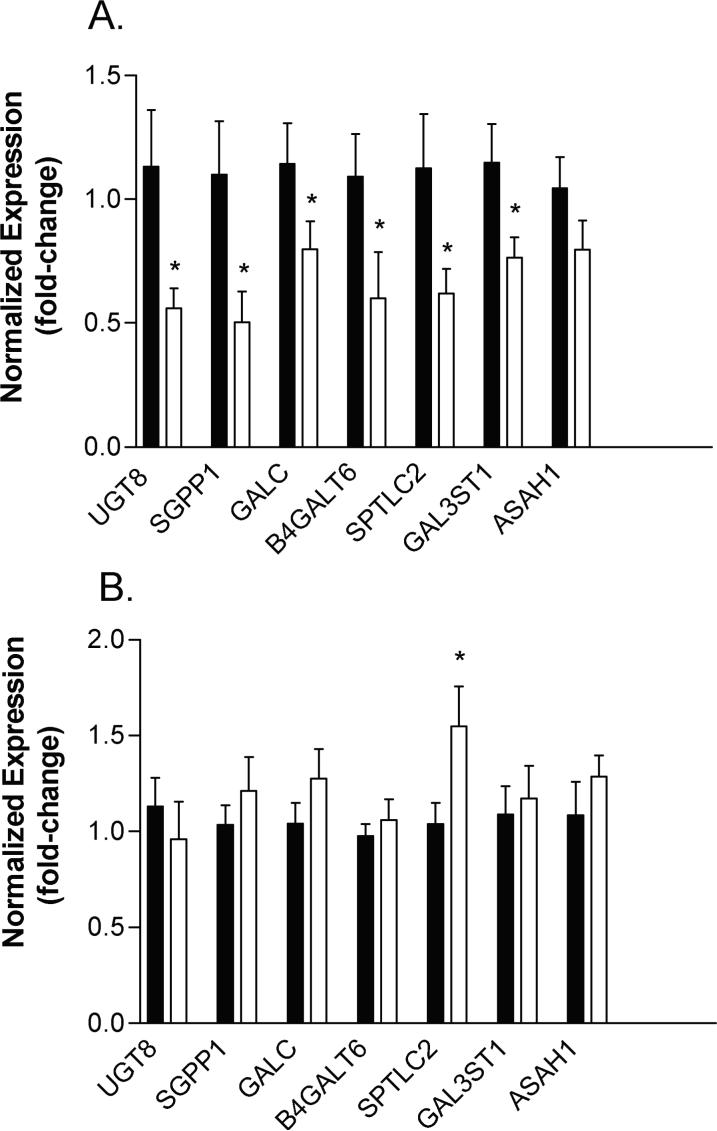
From the data output of both GLYCOv2 and whole genome Affymetrix arrays, 18 genes encoding proteins involved in the sphingolipid pathway were found to be altered in schizophrenic subjects early in illness. The top seven genes showing the greatest expression differences from the array analysis were validated by real-time PCR in subjects with short-term DOI. Our results revealed significant decreases in expression for UDP glycosyltransferase 8 (UGT8), sphingosine-1-phosphate phosphatase 1 (SGPP1), galactosylceramidase (GALC), UDPGal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 (B4GALT6), serine palmitoyltransferase, long chain base subunit 2 (SPTLC2), and galactose-3-O-sulfotransferase 1 (GAL3ST1) (Figure 3). The expression of N-acylsphingosine amidohydrolase (acid ceramidase) 1 (ASAH1) was lower in schizophrenic subjects, but this decrease did not reach statistical significance (p=0.09) (Figure 3).
Figure 3.
Genes involved in sphingolipid metabolism are altered in schizophrenic subjects. Real-time PCR expression levels in control subjects (black bars) and schizophrenic subjects (white bars) are shown (fold-change) for the indicated genes: UDP glycosyltransferase 8 (UGT8), sphingosine-1-phosphate phosphatase 1 (SGPP1), galactosylceramidase (GALC), UDPGal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 (B4GALT6), Serine palmitoyltransferase, long chain base subunit 2 (SPTLC2), Galactose-3-O-sulfotransferase 1 (GAL3ST1) and N-acylsphingosine amidohydrolase (acid ceramidase) 1 (ASAH1). The relative abundance of each gene expression was normalized by beta-2 microglobulin (B2M) in subjects with short-term DOI (panel A) and by TUBB in subjects with long-term DO1 (panel B). Asterisks denote significant decreases in expression using Student's t test, one-tailed, p<0.05.
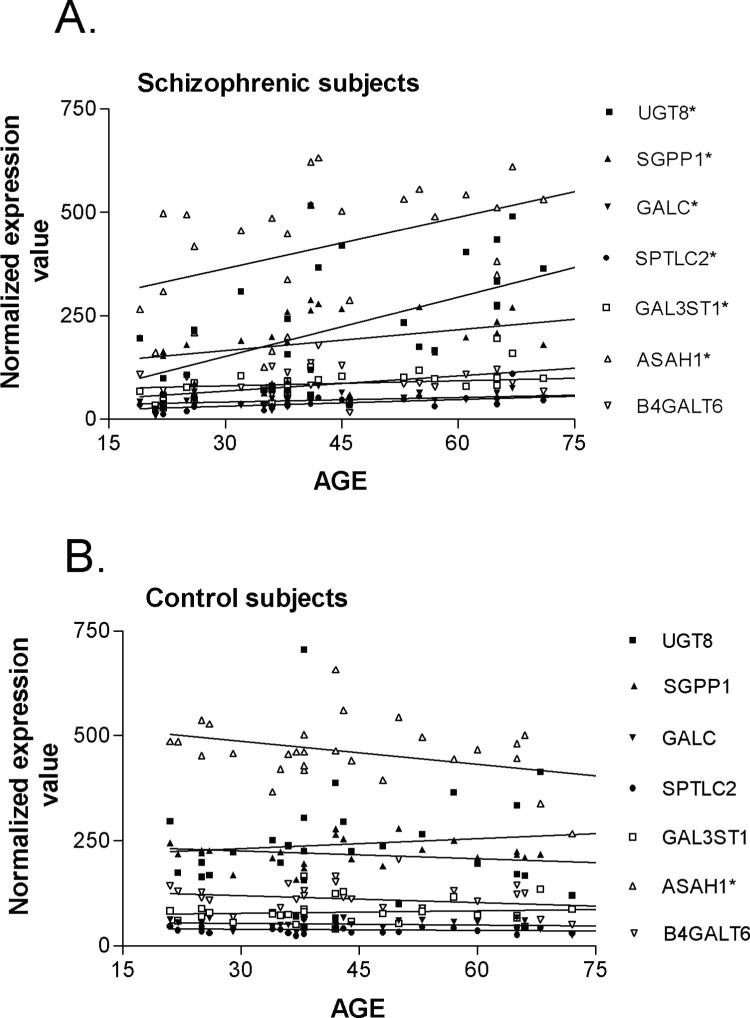
Covariate analyses on the expression values for these genes found no significant correlations to pH, PMI, or drug dose (as chlorpromazine equivalents) and no significant differences between males and females (Table 4). However, when assessing for age, statistically significant linear relationships between gene expression and age were detected for three genes when all subjects were tested. To further tease out this effect, we re-analyzed the age correlations in the control and schizophrenic subjects separately. Pearson's analysis revealed statistically significant positive correlations between gene expression and age in schizophrenic subjects (Pearson's r values = 0.388−0.585; p<0.001−0.05) (Figure 4A), but not in the control subjects (Pearson's r values = −0.260−0.106) (Figure 4B), with the exception of N-acylsphingosine amidohydrolase (acid ceramidase) 1 (ASAH1), which was negatively correlated with age in control subjects (r = −0.396; p<0.05). Some of the r-values less than 0.50 (r2<0.25), although significant, would suggest only a moderate correlation between gene expression and age, hence, qPCR analysis was performed on subjects with long duration illness to determine how the expression of these genes changes in chronic illness. SPTLC2 showed a significant increase in expression (1.03 ± 0.11 vs. 1.54 ± 0.20; p=0.03) in subjects with long-term DOI, while no significant differences, or only trends towards increased expression, were observed for the remaining genes, similar to the microarray results (Figure 3).
Table 4.
Pearson's Product Moment correlation values (r values) of gene expression to demographic data parameters.
| Pearson's r-values: | p-value: | |||||
|---|---|---|---|---|---|---|
| Gene Symbol: | pH: | PMI: | AGE-control: | AGE-schiz: | Drug Dose: | SEX |
| UGT8 | 0.0097 | −0.0238 | 0.1033 | 0.5552* | 0.2091 | 0.5060 |
| SGPP1 | −0.1904 | −0.2765 | −0.2692 | 0.3880* | 0.1957 | 0.0556 |
| GALC | 0.0909 | −0.2271 | −0.1796 | 0.4094* | 0.1790 | 0.4317 |
| SPTLC2 | −0.0427 | 0.1644 | −0.1452 | 0.5267* | −0.0724 | 0.3261 |
| GAL3ST1 | 0.1711 | −0.0101 | 0.1069 | 0.5854* | 0.0491 | 0.4529 |
| ASAH1 | −0.0168 | −0.2721 | −0.3965* | 0.4750* | 0.0727 | 0.6824 |
| B4GALT6 | −0.2076 | −0.0690 | −0.2441 | 0.1927 | 0.1891 | 0.0972 |
Pearson Product Moment correlations were performed to assess the effects of pH, PMI, and drug dose (in chlorpromazine equivalents) on the microarray gene expression values. Asterisks denote a significant effect on the indicated parameter. Student's t test (two-tailed, unpaired) was used to determine sex differences. The effects of age are shown in Figure 4.
Figure 4.
Correlations between the expression of sphingolipid genes and age in schizophrenic subjects (A) and controls (B) for the indicated genes. Each data point represents a normalized expression value for an individual subject. Pearson Product Moment correlations were performed as described in Methods. Asterisks indicate a significant effect with age.
Discussion
In these studies, we have used a focused microarray, the GLYCOv2 chip, to identify changes in the expression of genes particularly associated with various aspects of glycobiology, an area that has not been widely studies in psychiatric disorders. Our results suggest that abnormalities in sphingolipid metabolism and glycosylation are present in schizophrenic subjects early in illness, hence may represent important pathogenic mechanisms of schizophrenia.
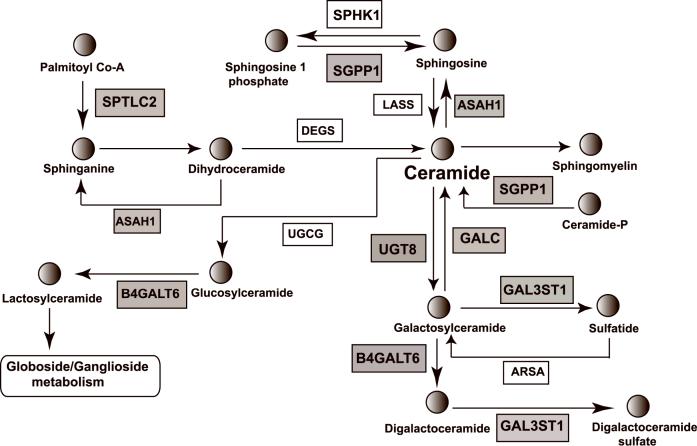
The brain is a lipid-rich organ containing mostly complex phospholipids, sphingolipids, and cholesterol. Glycosphingolipids, including cerebrosides, gangliosides and globosides, are a subtype of sphingolipids, which are derived from a ceramide backbone. We validated decreases in the expression for seven sphingolipid-related genes by quantitative PCR analysis in subjects with short-term illness. Fig. 5 shows a schematic drawing of the sphingolipid/glycosphingolipid pathway, with shaded boxes depicting products of those genes showing altered expression in schizophrenia. Due to the complexity of this pathway, it is not explicitly clear how decreases in the expression of these genes would translate into altered patterns of glycolipid intermediates or end products in this pathway, which were not measured in this study. However, the observed decreases in the expression of UGT8 and GAL3ST1 could result in overall decreases in the levels of galactocerebrosides and sulfatides, which are major lipid components of the myelin sheath (Norton 1977). Consistent with this postulate, previous biochemical studies on postmortem tissue have demonstrated decreases in the levels of galactocerebrosides 1 and 2, as well as sphingomyelin, in the thalamus of schizophrenic subjects compared to normal controls (Schmitt et al., 2004). Another report, dated back to 1969, also found decrease levels of total cerebrosides, as well as sulfatides in the brain of a single schizophrenic subject (Cherayil 1969). Decreased levels of galactocerebrosides and sulfatides would be indicative of decreased myelination and oligodendrocyte dysfunction. In particular, recent studies have indicated that these two glycolipids are essential for glycosynapses between myelin membranes, which are important for myelin-axon communication (Boggs et al., 2008). Hence, abnormalities in the metabolism of these glycolipids could be associated with white matter deficits previously reported in schizophrenia, which include decreased oligodendrocyte number, reduced white matter volumes and decreases in myelin-associated gene expression, including UGT8 (reviewed in (Davis et al., 2003; Kubicki et al., 2005)).
Figure 5.
Schematic depiction of sphingolipid/glycosphingolipid metabolism. The pathway shown was adapted and simplified from the KEGG pathway database (http://www.genome.ad.jp/kegg/pathway.html). The products of those genes whose expression levels are decreased in schizophrenia are indicated by shaded boxes. Note: only a trend towards a decrease was detected for ASAH1. UniGene gene symbols for all genes are shown. Circles depict the indicated glycolipids. Gangliosides and globosides are synthesized from glucosylceramide, and cerebrosides and sulfatides from galactosylceramide.
Furthermore, the decreased expression of UGT8 might initially result in increased accumulation of ceramide, a mediator of apoptotic events in the cell (Hannun 1994; Ruvolo 2003). However, the decreased expression of GALC, SGPP1 and ASAH1, whose enzyme products promote metabolism to ceramide from other glycolipids, would suggest an attempt to balance this deficit. We also observed a decrease in the expression of B4GALT6, whose protein product drives the biosynthesis of gangliosides, suggesting subsequent lowering of this class of glycolipids in the brain. Also included in the 1969 Cherayil study were reportedly lower levels of gangliosides in schizophrenia (Cherayil 1969). With the availability of glycan profiling (see http://www.functionalglycomics.org; Core C), further analysis of glycoconjugate patterns in the cortices schizophrenic subjects should illuminate these possibilities.
Sphingolipids are involved in the structure and function of cell membranes in the brain (Svennerholm 1964; van Heyningen 1974) and are associated with a plethora of biological functions, including cellular recognition and adhesion, signal transduction, growth regulation and differentiation (Yu et al., 2004). A dysfunction in the synthesis and/or breakdown of sphingolipids would be expected to have a major impact on neuronal function. Moreover, such deficits could encompass the diverse pathological effects already described in schizophrenia, namely those related to neurotransmitter receptors, synaptic transmission and cellular signaling, in addition to the myelination/oligodendrocyte deficits mentioned above. The fact that these expression decreases occurred only in subjects within 5 years from initial diagnosis, suggests that they may represent early pathogenic defects. Accordingly, imbalances in this pathway would likely trigger a wide range of secondary effects contributing to and/or attempting to combat pathological corollaries throughout disease. This hypothesis is especially appealing in light of the observations that these expression differences were not detected in subjects with chronic illness, suggesting that compensatory mechanisms, either natural homeostatic or drug-induced, occur with disease progression.
Considering the potential for drug-induced effects, we acknowledge that all of the schizophrenic subjects used in this study were exposed to antipsychotic drugs prior to death, with our chronic subjects having long histories of treatments with antipsychotic drugs. Therefore, we cannot exclude that possibility that differences between cohorts of different disease duration are due to differences in lifetime exposure to such drugs. The fact that the expression decreases of the sphingolipid-related genes, as a whole, were not found in subjects with chronic illness and that six genes were positively correlated with age in schizophrenic subjects, but not controls, suggests that antipsychotic drugs may be acting to reverse these expression decreases. However, as a preliminary argument against our findings being simply due to antipsychotic drug treatment, we did not find significant correlations between the expression levels of any validated gene and antipsychotic drug dose, reported in chlorpromazine equivalents, which takes into account lifetime exposure (Table 4). Rather, we propose the idea that the lack of expression differences in chronic schizophrenic subjects represents an adaptive response to pathology that occurs with disease progression. However, this concept will need further validation in independent datasets
We also found decreased expression of genes encoding glycan transferase enzymes in subjects with schizophrenia, suggesting that a dysfunction in protein glycosylation is important to disease pathology. Glycosylation is the most common form of posttranslational modification of proteins and variations in protein glycosylation patterns often lead to changes in protein function. For example, N-linked glycosylation at residue 109 of the 5HT3 receptor is necessary for receptor assembly, whereas N-linked glycosylation of residues 174 and 190 are important for plasma membrane targeting and ligand binding, respectively (Quirk et al., 2004). Dopamine D2 receptor short and long forms are differentially glycosylated and, as a consequence, are subject to different patterns of intracellular trafficking (Fishburn et al., 1995). Altered serotonergic and dopaminergic neurotransmission have long been associated with schizophrenia. While many receptor subtypes for these neurotransmitters exist, no changes in the expression of the dopamine D2 receptor in the caudate or prefrontal cortex (Harrington et al., 1995; Meador-Woodruff et al., 1997), or the 5HT3 receptor in the amygdala (Abi-Dargham et al., 1993) have been reported in the brains of subjects with schizophrenia (Harrington et al. 1995; Meador-Woodruff et al. 1997). Nonetheless, altered signaling via these receptor subtypes may be attributed to different glycosylation patterns of their receptors.
In summary, we find that various aspects of glycobiology are perturbed in schizophrenia. Notably, disruption in sphingolipid metabolism early in illness may represent a pathogenic mechanism in schizophrenia that triggers diverse downstream consequences. Dysfunction in this pathway may represent an important link in uniting diverse theories of schizophrenia pathology. In particular, these findings support the hypothesis that decreased myelination and disruption of connectivity in schizophrenia might be related to decreases in lipid membrane components. These findings also suggest that new treatment approaches might consider glycosyltransferase and glycolipid targets.
Acknowledgements
We thank Lana Schaffer for help with statistical analyses. BD is an NHMRC Senior Research Fellow (400016).
Grant Information: This study was funded by grants from the National Institutes of Health (NS44169 and MH069696 to E.A.T.). Microarray analysis was performed by the Microarray Core of the Consortium for Functional Glycomics funded by the National Institutes of Health (GM62116).
References
- Abi-Dargham A, Laruelle M, Lipska B, Jaskiw GE, Wong DT, Robertson DW, Weinberger DR, Kleinman JE. Serotonin 5-HT3 receptors in schizophrenia: a postmortem study of the amygdala. Brain Res. 1993;616:53–57. doi: 10.1016/0006-8993(93)90191-o. [DOI] [PubMed] [Google Scholar]
- Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry. 2005;10:309–322. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- Axford J. The impact of glycobiology on medicine. Trends Immunol. 2001;22:237–239. doi: 10.1016/s1471-4906(01)01890-7. [DOI] [PubMed] [Google Scholar]
- Benes FM, Matzilevich D, Burke RE, Walsh J. The expression of proapoptosis genes is increased in bipolar disorder, but not in schizophrenia. Mol Psychiatry. 2006;11:241–251. doi: 10.1038/sj.mp.4001758. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Bezchlibnyk-Butler K, Jeffries Je. Clinical Handbook of Psychotropic Drugs. Hogrefe & Huber Publishers; Toronto: 1999. [Google Scholar]
- Boggs JM, Gao W, Hirahara Y. Myelin glycosphingolipids, galactosylceramide and sulfatide, participate in carbohydrate-carbohydrate interactions between apposed membranes and may form glycosynapses between oligodendrocyte and/or myelin membranes. Biochim Biophys Acta. 2008;1780:445–455. doi: 10.1016/j.bbagen.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Brunngraber EG, Davis LG, Rieser C, Reddy MV. Urinary glycoconjugates in schizophrenic patients. Biol Psychiatry. 1981;16:741–751. [PubMed] [Google Scholar]
- Cherayil GD. Estimation of glycolipids in four selected lobes of human brain in neurological diseases. J Neurochem. 1969;16:913–920. doi: 10.1111/j.1471-4159.1969.tb08980.x. [DOI] [PubMed] [Google Scholar]
- Davis KL, Stewart DG, Friedman JI, Buchsbaum M, Harvey PD, Hof PR, Buxbaum J, Haroutunian V. White matter changes in schizophrenia: evidence for myelin-related dysfunction. Arch Gen Psychiatry. 2003;60:443–456. doi: 10.1001/archpsyc.60.5.443. [DOI] [PubMed] [Google Scholar]
- Dean B, Keriakous D, Scarr E, Thomas EA. Gene expression profiling in Brodmann's area 46 from subjects with schizophrenia. Australian and New Zealand Journal of Psychiatry. 2007;41:308–320. doi: 10.1080/00048670701213245. [DOI] [PubMed] [Google Scholar]
- Desplats PA, Denny CA, Kass KE, Gilmartin T, Head SR, Sutcliffe JG, Seyfried TT, E.A. Glycolipid and Ganglioside Metabolism Imbalances In Huntington's Disease Neurobiol Dis. 2007 doi: 10.1016/j.nbd.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats PA, Kass KE, Gilmartin T, Stanwood GD, Woodward EL, Head SR, Sutcliffe JG, Thomas EA. Selective deficits in the expression of striatal-enriched mRNAs in Huntington's disease. J Neurochem. 2006;96:743–757. doi: 10.1111/j.1471-4159.2005.03588.x. [DOI] [PubMed] [Google Scholar]
- Fishburn CS, Elazar Z, Fuchs S. Differential glycosylation and intracellular trafficking for the long and short isoforms of the D2 dopamine receptor. J Biol Chem. 1995;270:29819–29824. doi: 10.1074/jbc.270.50.29819. [DOI] [PubMed] [Google Scholar]
- Grimm MO, Grimm HS, Patzold AJ, Zinser EG, Halonen R, Duering M, Tschape JA, De Strooper B, Muller U, Shen J, Hartmann T. Regulation of cholesterol and sphingomyelin metabolism by amyloid-beta and presenilin. Nat Cell Biol. 2005;7:1118–1123. doi: 10.1038/ncb1313. [DOI] [PubMed] [Google Scholar]
- Hannun YA. The sphingomyelin cycle and the second messenger function of ceramide. J Biol Chem. 1994;269:3125–3128. [PubMed] [Google Scholar]
- Harrington KA, Augood SJ, Faull RL, McKenna PJ, Emson PC. Dopamine D1 receptor, D2 receptor, proenkephalin A and substance P gene expression in the caudate nucleus of control and schizophrenic tissue: a quantitative cellular in situ hybridisation study. Brain Res Mol Brain Res. 1995;33:333–342. doi: 10.1016/0169-328x(95)00169-s. [DOI] [PubMed] [Google Scholar]
- Harrison PJ. The neuropathology of schizophrenia. A critical review of the data and their interpretation. Brain. 1999;122:593–624. doi: 10.1093/brain/122.4.593. [DOI] [PubMed] [Google Scholar]
- Haselhorst U, Schenk H, Beyer I, Uebelhack R, Franke E, Kielstein V. Abnormality of gangliosides in erythrocyte membranes of schizophrenic patients. Clin Physiol Biochem. 1988;6:281–284. [PubMed] [Google Scholar]
- Hill C, Keks NA, Roberts S, Opeskin K, Dean B, Copolov DL. Postmortem brain studies in schizophrenia: The problems of diagnosis. Am J Psychiatry. 1996;153:533–537. doi: 10.1176/ajp.153.4.533. [DOI] [PubMed] [Google Scholar]
- Katsel P, Li C, Haroutunian V. Gene expression alterations in the sphingolipid metabolism pathways during progression of dementia and Alzheimer's disease: a shift toward ceramide accumulation at the earliest recognizable stages of Alzheimer's disease? Neurochem Res. 2007;32:845–856. doi: 10.1007/s11064-007-9297-x. [DOI] [PubMed] [Google Scholar]
- Kingsbury AE, Foster OJ, Nisbet AP, Cairns N, Bray L, Eve DJ, Lees AJ, Marsden CD. Tissue pH as an indicator of mRNA preservation in human post-mortem brain. Mol Brain Res. 1995;28:311–318. doi: 10.1016/0169-328x(94)00219-5. [DOI] [PubMed] [Google Scholar]
- Kolter T, Sandhoff K. Sphingolipid metabolism diseases. Biochim Biophys Acta. 2006;1758:2057–2079. doi: 10.1016/j.bbamem.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Kubicki M, McCarley RW, Shenton ME. Evidence for white matter abnormalities in schizophrenia. Curr Opin Psychiatry. 2005;18:121–134. doi: 10.1097/00001504-200503000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meador-Woodruff JH, Haroutunian V, Powchik P, Davidson M, Davis KL, Watson SJ. Dopamine receptor transcript expression in striatum and prefrontal and occipital cortex. Focal abnormalities in orbitofrontal cortex in schizophrenia. Arch Gen Psychiatry. 1997;54:1089–1095. doi: 10.1001/archpsyc.1997.01830240045007. [DOI] [PubMed] [Google Scholar]
- Narayan S, Tang B, Head SR, Gilmartin TJ, Dean B, Sutcliffe JG, Thomas EA. Gene Expression Differences in Schizophrenia at Early, Intermediate and Late Stages of Illness. 2008 Under revision. [Google Scholar]
- Navon R. Molecular and clinical heterogeneity of adult GM2 gangliosidosis. Dev Neurosci. 1991;13:295–298. doi: 10.1159/000112200. [DOI] [PubMed] [Google Scholar]
- Nawa H, Takei N. Recent progress in animal modeling of immune inflammatory processes in schizophrenia: implication of specific cytokines. Neurosci Res. 2006;56:2–13. doi: 10.1016/j.neures.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Norton WT. In: Isolation and characterization of myelin. Morell P, editor. Plenum Press; New York: 1977. pp. 161–199. [Google Scholar]
- Quirk PL, Rao S, Roth BL, Siegel RE. Three putative N-glycosylation sites within the murine 5-HT3A receptor sequence affect plasma membrane targeting, ligand binding, and calcium influx in heterologous mammalian cells. J Neurosci Res. 2004;77:498–506. doi: 10.1002/jnr.20185. [DOI] [PubMed] [Google Scholar]
- Radmacher MD, McShane LM, Simon R. A paradigm for class prediction using gene expression profiles. J Comput Biol. 2002;9:505–511. doi: 10.1089/106652702760138592. [DOI] [PubMed] [Google Scholar]
- Rapoport JL, Addington AM, Frangou S, Psych MR. The neurodevelopmental model of schizophrenia: update 2005. Mol Psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- Rosebush PI, MacQueen GM, Clarke JT, Callahan JW, Strasberg PM, Mazurek MF. Late-onset Tay-Sachs disease presenting as catatonic schizophrenia: diagnostic and treatment issues. J Clin Psychiatry. 1995;56:347–353. [PubMed] [Google Scholar]
- Rutishauser U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat Rev Neurosci. 2008;9:26–35. doi: 10.1038/nrn2285. [DOI] [PubMed] [Google Scholar]
- Ruvolo PP. Intracellular signal transduction pathways activated by ceramide and its metabolites. Pharmacol Res. 2003;47:383–392. doi: 10.1016/s1043-6618(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Saetre P, Emilsson L, Axelsson E, Kreuger J, Lindholm E, Jazin E. Inflammation-related genes up-regulated in schizophrenia brains. BMC Psychiatry. 2007;7:46. doi: 10.1186/1471-244X-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A, Wilczek K, Blennow K, Maras A, Jatzko A, Petroianu G, Braus DF, Gattaz WF. Altered thalamic membrane phospholipids in schizophrenia: a postmortem study. Biol Psychiatry. 2004;56:41–45. doi: 10.1016/j.biopsych.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Turecki G. Genome wide gene expression studies in mood disorders. Omics. 2006;10:444–454. doi: 10.1089/omi.2006.10.444. [DOI] [PubMed] [Google Scholar]
- Svennerholm L. The Gangliosides. J Lipid Res. 1964;5:145–155. [PubMed] [Google Scholar]
- Tamboli IY, Prager K, Barth E, Heneka M, Sandhoff K, Walter J. Inhibition of glycosphingolipid biosynthesis reduces secretion of the beta-amyloid precursor protein and amyloid beta-peptide. J Biol Chem. 2005;280:28110–28117. doi: 10.1074/jbc.M414525200. [DOI] [PubMed] [Google Scholar]
- Thompson JL, Pogue-Geile MF, Grace AA. Developmental pathology, dopamine, and stress: a model for the age of onset of schizophrenia symptoms. Schizophr Bull. 2004;30:875–900. doi: 10.1093/oxfordjournals.schbul.a007139. [DOI] [PubMed] [Google Scholar]
- van Heyningen W. Gangliosides as membrane receptors for tetanus toxin, cholera toxin and serotonin. Nature. 1974;249:415–417. [Google Scholar]
- Woo TU, Crowell AL. Targeting synapses and myelin in the prevention of schizophrenia. Schizophr Res. 2005;73:193–207. doi: 10.1016/j.schres.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y. Glycobiology of the synapse: the role of glycans in the formation, maturation, and modulation of synapses. Biochim Biophys Acta. 2002;1573:369–376. doi: 10.1016/s0304-4165(02)00405-1. [DOI] [PubMed] [Google Scholar]
- Yu RK, Bieberich E, Xia T, Zeng G. Regulation of ganglioside biosynthesis in the nervous system. J Lipid Res. 2004;45:783–793. doi: 10.1194/jlr.R300020-JLR200. [DOI] [PubMed] [Google Scholar]
- Zelnik N, Khazanov V, Sheinkman A, Karpati AM, Peleg L. Clinical manifestations of psychiatric patients who are carriers of tay-sachs disease. Possible role of psychotropic drugs. Neuropsychobiology. 2000;41:127–131. doi: 10.1159/000026644. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ding L, Sandford AJ. Selection of reference genes for gene expression studies in human neutrophils by real-time PCR. BMC Mol Biol. 2005;6:4. doi: 10.1186/1471-2199-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]