Abstract
The sodium-dependent multivitamin transporter (SMVT) is essential for mediating and regulating biotin entry into mammalian cells. In cells, holocarboxylase synthetase (HCS) mediates covalent binding of biotin to histones; biotinylation of lysine-12 in histone H4 (K12BioH4) causes gene repression. Here we propose a novel role for HCS in sensing and regulating levels of biotin in eukaryotic cells. We hypothesize that nuclear translocation of HCS increases in response to biotin supplementation; HCS then biotinylates histone H4 at SMVT promoters, silencing biotin transporter genes. We show that nuclear translocation of HCS is a biotin-dependent process that might involve tyrosine kinases, histone deacetylases, and histone methyltransferases in human lymphoid (Jurkat) cells. The nuclear translocation of HCS correlated with biotin concentrations in cell culture media; the relative enrichment of both HCS and K12BioH4 at SMVT promoter 1 (but not promoter 2) increased by 91% in cells cultured in medium containing 10 nmol/L biotin compared with 0.25 nmol/L biotin. This increase of K12BioH4 at the SMVT promoter was inversely linked to SMVT expression. Biotin homeostasis by HCS-dependent chromatin remodeling at the SMVT promoter 1 locus was disrupted in HCS knockdown cells, as evidenced by abnormal chromatin structure (K12BioH4 abundance) and increased SMVT expression. The findings from this study are consistent with the theory that HCS senses biotin, and that biotin regulates its own cellular uptake by participating in HCS-dependent chromatin remodeling events at the SMVT promoter 1 locus in Jurkat cells.
Gene regulation at the chromatin level by histone biotinylation
Biotin is attached covalently to at least 2 classes of proteins, carboxylases and histones. Acetyl-CoA carboxylases 1 and 2, propionyl-CoA carboxylase, 3-methylcrotonyl-CoA carboxylase, and pyruvate carboxylase mediate the covalent binding of bicarbonate to organic acids and play essential roles in the metabolism of fatty acids, leucine, and glucose (1). The attachment of biotin to the ɛ-amino group of a specific lysine residue in carboxylases is catalyzed by holocarboxylase synthetase (HCS).4
In mammals and most other eukaryotes, 5 major classes of histones (H1, H2A, H2B, H3, and H4) play a predominant role in the folding of DNA into chromatin (2). Histones consist of a globular domain and a more flexible amino terminus (histone “tail”). DNA and histones form repetitive nucleoprotein units, or nucleosomes (2). Each nucleosome (“nucleosomal core particle”) consists of 146 basepairs of DNA wrapped around an octamer of core histones (1 H3-H3-H4-H4 tetramer and 2 H2A-H2B dimers). The DNA located between nucleosomal core particles binds histone H1.
The amino terminal tail of histones protrudes from the nucleosomal surface; covalent modifications of this tail affect the structure of chromatin and form the basis for gene regulation (3,4). Histone tails are modified by covalent acetylation, methylation, phosphorylation, ubiquitination, and poly(ADP-ribosylation) of ɛ-amino groups (lysine = “K”), guanidino groups (arginine), carboxyl groups (glutamate), and hydroxyl groups (serine) (2–5). The various modifications of histones have distinct functions. For example, trimethylation of K4 in histone H3 (K4Me3H3) is associated with transcriptional activation of surrounding DNA, whereas dimethylation of K9 (K9Me2H3) is associated with transcriptional silencing (3,4). Covalent modifications of histones are reversible (4).
Recently, our laboratory and other laboratories identified a novel modification of histones in humans: binding of biotin to lysine residues via an amide bond (6–9). In these studies we identified 11 distinct biotinylation sites: K9, K13, K125, K127, and K129 in histone H2A (10); K4, K9, K18, and perhaps K23 in histone H3 (9,11); and K8 and K12 in histone H4 (8). Importantly, we demonstrated that biotinylation of K12 (and perhaps K8) in histone H4 (K12BioH4) participates in heterochromatin formation and gene repression in human cells (12), and that biotinylation of histones depends on biotin supply (13,14).
An early study (6) suggested that biotinylation of histones is mediated by biotinidase. However, subsequently it was shown that HCS (15) and its microbial ortholog BirA (11) may also biotinylate histones, and that HCS is more important for histone biotinylation than biotinidase (16). HCS is abundant in the cell nucleus where biotinylation of histones takes place (10,15). A study in Drosophila melanogaster (16) suggested that HCS is a chromosome-associated protein, consistent with a role in chromatin structure and remodeling. Nuclear localization and chromosome binding of HCS are difficult to explain, given that HCS lacks both a classical nuclear localization sequence and a DNA-binding domain. Ongoing studies in our laboratory suggest that both nuclear localization and binding to DNA are mediated by interactions of HCS with other proteins (see below).
Biotin transport and its relationship to biotin supply
Biotin transport across plasma membranes is a carrier-mediated process (17). Both Ganapathy and Said (18–21) have independently shown that uptake of biotin is mediated by the sodium-dependent multivitamin transporter, SMVT. Although SMVT is undoubtedly the primary biotin transporter in mammals, evidence has been provided that the monocarboxylate transporter 1 (MCT1) may also contribute to biotin transport in some tissues (22,23).
SMVT and MCT1 are crucial checkpoints for regulating the cellular uptake of biotin, decreasing the risk for deficiency and overdose. For example, we have demonstrated that the expression of biotin transporters in human cells correlates inversely with biotin concentrations in culture media (24–26). Moreover, biotin transport rates increase in response to cell proliferation (27) to meet increased biotin requirements for biotinylation of carboxylases (28) and histones (7). Increased biotin uptake into proliferating cells is mediated by increased abundance of biotin transporters on the cell surface rather than by increased affinity of transporters for biotin (27). A previous study (26) has linked the cellular uptake of biotin to the abundance of mRNA encoding biotin transporters.
Hypothesis
In Escherichia coli and other enteric bacteria, a complex of BirA and the intermediate biotinyl-AMP binds to promoter regions in the biotin operon, a cluster of genes mediating biotin biosynthesis; binding of biotinyl-AMP/BirA represses the transcription of these genes (29). This feedback loop prevents excessive biosynthesis of biotin. Human cells cannot synthesize biotin and depend on regulating biotin uptake to maintain homeostasis. For our studies, we proposed and tested the following model to explain how mammalian cells maintain biotin homeostasis in response to changes in biotin supply (Fig. 1).
1) HCS serves as biotin sensor in the cytoplasm.
2) Cellular influx of biotin triggers HCS-mediated biotinylation of HCS-binding proteins, phosphorylation of HCS, or both, thereby causing nuclear translocation of HCS; translocation is mediated by chaperones.
3) Nuclear proteins recruit HCS to specific regions in chromatin, including biotin transporter (SMVT, MCT1) loci.
4) HCS catalyzes biotinylation of histones (K12BioH4), mediating gene silencing.
5) The increased abundance of K12BioH4 at biotin transporter loci in response to biotin supplementation is associated with chromatin remodeling events that decrease the transcription of biotin transporter genes (checkpoints for biotin entry into cells).
6) Vice versa, biotinylation of histones at biotin transporter loci decreases in biotin-deficient cells, increasing transcriptional activity of biotin transporter genes.
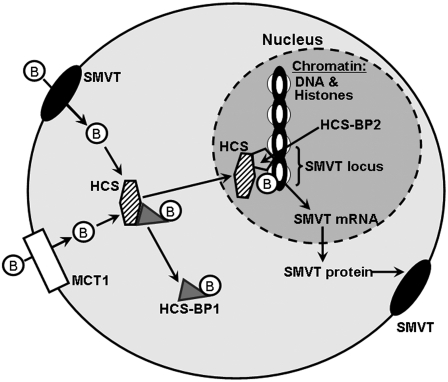
FIGURE 1 .
Regulation of biotin transporter genes at the chromatin level by histone biotinylation in eukaryotic cells. B, biotin; HCS-BP1, (putative) HCS-binding protein 1; HCS-BP2, (putative) HCS-binding protein 2.
Collectively, this model presents an intriguing mechanism by which intracellular biotin directly controls the expression of biotin transporters, mediated by HCS-dependent chromatin remodeling.
Biotin homeostasis
We tested the above hypothesis by using human lymphoid (Jurkat) cells, which were cultured in the following biotin-defined media for at least 5 wk prior to sample collection (13): 0.025 nmol/L of biotin (deficient), 0.25 nmol/L of biotin (physiological), and 10 nmol/L of biotin (pharmacological). These concentrations represent plasma from biotin-deficient individuals, normal subjects, and users of biotin supplements (30,31). HCS-deficient Jurkat cells were generated by using siRNA and served as controls (13). Biotin deficiency and HCS knockdown were confirmed using the abundance of holocarboxylases (streptavidin blot) and the expression of HCS (Western blot and real-time PCR) as markers.
The nuclear translocation of HCS depended on biotin: biotin-supplemented > biotin normal > biotin-deficient (13). Ongoing studies in our laboratory suggest that nuclear translocation of HCS depends on HCS phosphorylation by a tyrosine kinase, and that targeting of HCS to specific gene loci (such as SMVT) might involve chromatin-remodeling enzymes, such as histone deacetylases and a histone H3 K9-methyltransferase.
K12BioH4 increased at SMVT promoter loci in response to biotin supplementation in wild-type cells but not in HCS knockdown cells (13). For example, if wild-type cells were supplemented with 10 nmol/L biotin, the relative enrichment of SMVT promoter 1 sequences by chromatin immunoprecipitation assay with anti-K12Bio H4 increased by 91 ± 6.3% compared with physiological controls. Note that all values in the text are means ± SD, and that reported differences were statistically different (P < 0.05) unless stated otherwise. Typically, statistical analyses were conducted by 1-way ANOVA and post hoc testing, or by paired t test (see the original references for details). In HCS knockdown cells, the abundance of K12BioH4 at SMVT promoter 1 was <41% of that in wild-type cells at 10 nmol/L biotin. The relative magnitudes of effects of HCS knockdown on K12BioH4 were similar if cells were cultured in media containing 0.025 and 0.25 nmol/L biotin (data not shown) compared with the effects described for 10 nmol/L biotin. The relative enrichment of SMVT promoter 1 sequences by chromatin immunoprecipitation with anti-HCS was similar to that observed for anti-K12BioH4. For example, biotin supplementation (10 nmol/L) was associated with a 146 ± 21% increase of HCS at the SMVT promoter 1 locus compared with physiological controls; this increase was abolished in HCS knockdown cells.
The increase of K12BioH4 and HCS at the SMVT promoter 1 locus in response to biotin supplementation coincided with an increase of the heterochromatin marker K9Me2H3 and a decrease of the euchromatin marker K4Me3H3 (13). HCS knockdown did not affect K9Me2H3 and K4Me3H3 compared with biotin-matched wild-type controls, e.g., the relative abundance of K9Me2H3 and K4Me3H3 at SMVT promoter 1 in HCS knockdown cells was within 3% of that in wild-type cells at 10 nmol/L biotin.
The transcription of SMVT is regulated by 2 promoters, P1 and P2 (21). The relative enrichment of K12BioH4 at SMVT promoter 2 sequences exhibited a relatively weak dependence on biotin supply and HCS expression, suggesting that histone modifications other than K12BioH4 might participate in the regulation of SMVT promoter 2 in Jurkat cells (13). An alternative explanation for this observation would be that P2 is less important than P1 in the transcriptional regulation of SMVT.
Supplementation of wild-type cells with pharmacological doses of biotin was associated with decreased expression of SMVT (13). The abundance of SMVT mRNA in wild-type cells decreased by ∼85% in response to biotin supplementation: 1.0 ± 0.05 arbitrary units (0.25 nmol/L biotin) vs. 0.15 ± 0.09 arbitrary units (10 nmol/L biotin). This feedback control mechanism was impaired in HCS knockdown cells, where the abundance of SMVT mRNA decreased by only 56% in response to biotin supplementation: 1.13 ± 0.06 arbitrary units (0.25 nmol/L biotin) vs. 0.49 ± 0.02 arbitrary units (10 nmol/L biotin). Analysis of biotin transport rates corroborated a role for HCS in the regulation of SMVT. Rates of biotin uptake decreased from 2.6 ± 0.4 fmol biotin/(mg protein × min) in wild-type cells in physiological medium to 1.9 ± 0.1 fmol biotin/(mg protein × min) in wild-type cells in pharmacological medium. In contrast, rates of biotin uptake did not decrease in HCS knockdown cells cultured in pharmacological medium [2.7 ± 0.1 fmol biotin/(mg protein × min)] compared with physiological medium [2.9 ± 0.1 fmol biotin/(mg protein × min)].
We conclude that the nuclear translocation of HCS depends on biotin in human cells, and that increased nuclear translocation of HCS in biotin-supplemented cells increases the enrichment of K12BioH4 at the smvt P1 locus, thereby repressing SMVT expression. HCS-interacting proteins such as kinases, histone deacetylases, and K9H3-methyltransferases might play important roles in this regulatory loop. This mechanism of biotin homeostasis is analogous to the regulation of biotin biosynthesis in microbes by BirA.
Other articles in this supplement include references (32–35).
Presented as part of the symposium “Advances in Understanding of the Biological Role of Biotin at the Clinical, Biochemical, and Molecular Level” given at the 2008 Experimental Biology meeting on April 7, 2008, in San Diego, CA. The symposium was sponsored by the American Society for Nutrition and supported by an educational grant from Mead Johnson Nutritionals.
Author disclosures: J. Zempleni, M. Gralla, G. Camporeale, and Y. I. Hassan, no conflicts of interest.
Supported by a contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act. Additional support was provided by NIH grants DK 063945 and ES 015206, USDA grant 2006-35200-17138, and National Science Foundation Experimental Program to Stimulate Competitive Research grant EPS-0701892 (to J.Z.).
Abbreviations used: HCS, holocarboxylase synthetase; K, lysine; K4Me3H3, K4-trimethylated histone H3; K9Me2H3, K9-dimethylated histone H3; K12BioH4, K12-biotinylated histone H4; MCT1, monocarboxylate transporter 1; SMVT, sodium-dependent multivitamin transporter.
References
- 1.Camporeale G, Zempleni J. Biotin. In: Bowman BA, Russell RM, editors. Present Knowledge in Nutrition. 9th ed. Washington, D.C.: International Life Sciences Institute; 2006. p. 314–26.
- 2.Wolffe A. Chromatin. 3th ed. San Diego, CA: Academic Press; 1998.
- 3.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–83. [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. [DOI] [PubMed] [Google Scholar]
- 5.Boulikas T, Bastin B, Boulikas P, Dupuis G. Increase in histone poly(ADP-ribosylation) in mitogen-activated lymphoid cells. Exp Cell Res. 1990;187:77–84. [DOI] [PubMed] [Google Scholar]
- 6.Hymes J, Fleischhauer K, Wolf B. Biotinylation of histones by human serum biotinidase: assessment of biotinyl-transferase activity in sera from normal individuals and children with biotinidase deficiency. Biochem Mol Med. 1995;56:76–83. [DOI] [PubMed] [Google Scholar]
- 7.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur J Biochem. 2001;268:5424–9. [DOI] [PubMed] [Google Scholar]
- 8.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur J Biochem. 2004;271:2257–63. [DOI] [PubMed] [Google Scholar]
- 9.Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, Zempleni J. K4, K9, and K18 in human histone H3 are targets for biotinylation by biotinidase. FEBS J. 2005;272:4249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chew YC, Camporeale G, Kothapalli N, Sarath G, Zempleni J. Lysine residues in N- and C-terminal regions of human histone H2A are targets for biotinylation by biotinidase. J Nutr Biochem. 2006;17:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobza K, Sarath G, Zempleni J. Prokaryotic BirA ligase biotinylates K4, K9, K18 and K23 in histone H3. BMB Rep. 2008;41:310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camporeale G, Oommen AM, Griffin JB, Sarath G, Zempleni J. K12-biotinylated histone H4 marks heterochromatin in human lymphoblastoma cells. J Nutr Biochem. 2007;18:760–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gralla M, Camporeale G, Zempleni J. Holocarboxylase synthetase regulates expression of biotin transporters by chromatin remodeling events at the SMVT locus. J Nutr Biochem. 2008;19:400–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith EM, Hoi JT, Eissenberg JC, Shoemaker JD, Neckameyer WS, Ilvarsonn AM, Harshman LG, Schlegel VL, Zempleni J. Feeding Drosophila a biotin-deficient diet for multiple generations increases stress resistance and lifespan and alters gene expression and histone biotinylation patterns. J Nutr. 2007;137:2006–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narang MA, Dumas R, Ayer LM, Gravel RA. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum Mol Genet. 2004;13:15–23. [DOI] [PubMed] [Google Scholar]
- 16.Camporeale G, Giordano E, Rendina R, Zempleni J, Eissenberg JC. Drosophila holocarboxylase synthetase is a chromosomal protein required for normal histone biotinylation, gene transcription patterns, lifespan and heat tolerance. J Nutr. 2006;136:2735–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowers-Komro DM, McCormick DB. Biotin uptake by isolated rat liver hepatocytes. In: Dakshinamurti K, Bhagavan HN, editors. Biotin. New York: New York Academy of Sciences; 1985. p. 350–8. [DOI] [PubMed]
- 18.Prasad PD, Ramamoorthy S, Leibach FH, Ganapathy V. Characterization of a sodium-dependent vitamin transporter mediating the uptake of pantothenate, biotin and lipoate in human placental choriocarcinoma cells. Placenta. 1997;18:527–33. [DOI] [PubMed] [Google Scholar]
- 19.Prasad PD, Wang H, Kekuda R, Fujita T, Fei Y-J, Devoe LD, Leibach FH, Ganapathy V. Cloning and functional expression of a cDNA encoding a mammalian sodium-dependent vitamin transporter mediating the uptake of pantothenate, biotin, and lipoate. J Biol Chem. 1998;273:7501–6. [DOI] [PubMed] [Google Scholar]
- 20.Prasad PD, Wang H, Huang W, Fei Y-J, Leibach FH, Devoe LD, Ganapathy V. Molecular and functional characterization of the intestinal Na+-dependent multivitamin transporter. Arch Biochem Biophys. 1999;366:95–106. [DOI] [PubMed] [Google Scholar]
- 21.Said HM. Recent advances in carrier-mediated intestinal absorption of water-soluble vitamins. Annu Rev Physiol. 2004;66:419–46. [DOI] [PubMed] [Google Scholar]
- 22.Daberkow RL, White BR, Cederberg RA, Griffin JB, Zempleni J. Monocarboxylate transporter 1 mediates biotin uptake in human peripheral blood mononuclear cells. J Nutr. 2003;133:2703–6. [DOI] [PubMed] [Google Scholar]
- 23.Grafe F, Wohlrab W, Neubert RH, Brandsch M. Transport of biotin in human keratinocytes. J Invest Dermatol. 2003;120:428–33. [DOI] [PubMed] [Google Scholar]
- 24.Manthey KC, Griffin JB, Zempleni J. Biotin supply affects expression of biotin transporters, biotinylation of carboxylases, and metabolism of interleukin-2 in Jurkat cells. J Nutr. 2002;132:887–92. [DOI] [PubMed] [Google Scholar]
- 25.Scheerger SB, Zempleni J. Expression of oncogenes depends on biotin in human small cell lung cancer cells NCI-H69. Int J Vitam Nutr Res. 2003;73:461–7. [DOI] [PubMed] [Google Scholar]
- 26.Crisp SERH, Camporeale G, White BR, Toombs CF, Griffin JB, Said HM, Zempleni J. Biotin supply affects rates of cell proliferation, biotinylation of carboxylases and histones, and expression of the gene encoding the sodium-dependent multivitamin transporter in JAr choriocarcinoma cells. Eur J Nutr. 2004;43:23–31. [DOI] [PubMed] [Google Scholar]
- 27.Zempleni J, Mock DM. Mitogen-induced proliferation increases biotin uptake into human peripheral blood mononuclear cells. Am J Physiol. 1999;276:C1079–84. [DOI] [PubMed] [Google Scholar]
- 28.Stanley JS, Griffin JB, Mock DM, Zempleni J. Biotin uptake into human peripheral blood mononuclear cells increases early in the cell cycle, increasing carboxylase activities. J Nutr. 2002;132:1854–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cronan JE, Jr. The E. coli bio operon: transcriptional repression by an essential protein modification enzyme. Cell. 1989;58:427–9. [DOI] [PubMed] [Google Scholar]
- 30.Mock DM, Lankford GL, Mock NI. Biotin accounts for only half of the total avidin-binding substances in human serum. J Nutr. 1995;125:941–6. [DOI] [PubMed] [Google Scholar]
- 31.Zempleni J, Helm RM, Mock DM. In vivo biotin supplementation at a pharmacologic dose decreases proliferation rates of human peripheral blood mononuclear cells and cytokine release. J Nutr. 2001;131:1479–84. [DOI] [PubMed] [Google Scholar]
- 32.Mock DM, Said HM. Introduction to Advances in Understanding of the Biological Role of Biotin at the Clinical, Biochemical, and Molecular Level. J Nutr. 2008;138:152–3. [DOI] [PubMed] [Google Scholar]
- 33.Mock DM. Marginal biotin deficiency is common in normal human pregnancy and is highly teratogenic in mice. J Nutr. 2008;138:154–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Said HM. Cell and molecular aspects of the human intestinal biotin absorption process. J Nutr. 2008;138:158–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beckett D. Biotin sensing at the molecular level. J Nutr. 2008;138:167–70. [DOI] [PMC free article] [PubMed] [Google Scholar]